Abstract
The single celled eukaryote Trypanosoma cruzi, a parasite transmitted by numerous species of triatomine bug in the Americas, causes Chagas disease in humans. T. cruzi generally reproduces asexually and appears to have a clonal population structure. However, two of the six major circulating genetic lineages, TcV and TcVI, are TcII-TcIII inter-lineage hybrids that are frequently isolated from humans in regions where chronic Chagas disease is particularly severe. Nevertheless, a prevalent view is that hybridisation events in T. cruzi were evolutionarily ancient and that active recombination is of little epidemiological importance. We analysed genotypes of hybrid and non-hybrid T. cruzi strains for markers representing three distinct evolutionary rates: nuclear GPI sequences (n = 88), mitochondrial COII-ND1 sequences (n = 107) and 28 polymorphic microsatellite loci (n = 35). Using Maximum Likelihood and Bayesian phylogenetic approaches we dated key evolutionary events in the T. cruzi clade including the emergence of hybrid lineages TcV and TcVI, which we estimated to have occurred within the last 60,000 years. We also found evidence for recent genetic exchange between TcIII and TcIV and between TcI and TcIV. These findings show that evolution of novel recombinants remains a potential epidemiological risk. The clearly distinguishable microsatellite genotypes of TcV and TcVI were highly heterozygous and displayed minimal intra-lineage diversity indicative of even earlier origins than sequence-based estimates. Natural hybrid genotypes resembled typical meiotic F1 progeny, however, evidence for mitochondrial introgression, absence of haploid forms and previous experimental crosses indicate that sexual reproduction in T. cruzi may involve alternatives to canonical meiosis. Overall, the data support two independent hybridisation events between TcII and TcIII and a recent, rapid spread of the hybrid progeny in domestic transmission cycles concomitant with, or as a result of, disruption of natural transmission cycles by human activities.
Author Summary
Trypanosoma cruzi is a parasite that causes Chagas disease in humans, an often fatal condition affecting at least 8 million people. The clinical outcome of Chagas disease is variable, which is probably partially attributable to genetic differences between T. cruzi strains. Differences between T. cruzi strains can arise by gradual accumulation of mutations in independent lineages (clonal reproduction) or by recombination between strains, which generates new combinations of existing alleles (sexual reproduction). Although sex has been observed experimentally, and epidemiologically important T. cruzi subgroups (TcV TcVI) originated via hybridisation events, active recombination is considered to be extremely rare. Our aim was to determine when recombination events have occurred during the evolution of T. cruzi. We analysed differences within and between T. cruzi subgroups in DNA sequences that evolve at different speeds. We found multiple recombination events, several of which were very recent in evolutionary terms. We show that TcV and TcVI most likely originated as a result of human activities that promoted mixing between subgroups, suggesting a continuing risk for emergence and spread of new genotypes generated by sexual reproduction. We also found evidence that sexual reproduction in T. cruzi could involve mechanisms different from those seen in multi-cellular eukaryotes.
Introduction
Trypanosoma cruzi is a single celled eukaryotic parasite, which is transmitted to vertebrate hosts via the faeces of blood-sucking triatomine bugs. It is the aetiological agent of Chagas disease in humans, which results in the death of ∼13,000 people and the loss of 649,000 disability-adjusted life years (DALYs) per year [1]. Transmission can also occur congenitally or through contaminated blood products and organs. T. cruzi is currently split into six genetic lineages or discrete typing units (DTUs), previously named TcI, TcIIa, IIb, IIc, IId and IIe [2], [3] but recently revised by broad consensus to TcI, TcIV, TcII, TcIII, TcV and TcVI respectively [4]. Analysis of multiple molecular markers shows that a lack of inter-lineage recombination generally preserves the individuality of each DTU [2], [5]–[7]. Recent studies of large samples of TcI and TcIII using highly variable microsatellite markers have shown that clonal population structure, may persist at an intra-lineage level [8]. However, recombination at the scale of active transmission cycles is known to occur [9] and the failure to detect it more frequently is potentially a result of high rates of inbreeding [10], gene conversion [8] or insufficient sampling of infra-populations [11].
Many pathogenic eukaryotic microorganisms have essentially clonal population structures while also having non-obligate sexual cycles, which may enable them to adapt to environmental changes [12]. Although recombination may be rare in such pathogens, it can lead to the evolution and spread of epidemiologically important traits, including those relating to virulence, transmission dynamics and drug resistance [13]. Indeed, recombination events have shaped the evolution of at least some T. cruzi lineages. Comparison of nucleotide sequences showed that two lineages, TcV and TcVI (which includes the genome strain CL Brener), have arisen through hybridisation between genetically distinct parents [14]–[17]. The identification of these lineages as hybrid was unequivocal because they possess fully intact alleles from two other DTUs (TcII and TcIII) that are so distinct that they could not have arisen independently. It is not clear whether TcV and TcVI are the products of independent hybridisation events [15] or a single hybridisation event followed by clonal divergence [17]. Furthermore, the resolution afforded by the genetic markers used so far has been insufficient to resolve whether these hybridisation(s) were evolutionarily ancient [14], [18] or recent events [16], [17]. The ecological circumstances of the origin of TcV and TcVI are not known. While 5S rDNA sequences suggested the TcII parent may have originated in south-western South America [17], too few TcIII isolates have been characterised sufficiently to permit similar investigation of the other putative parent.
Generation of intra-lineage TcI hybrids in vitro proved that the capacity for genetic exchange has been retained [19], however, there are important differences between these experimental hybrids and natural TcV and TcVI isolates [20]. Genetic characterisation of the experimental hybrids indicated they were formed by diploid-diploid fusion, forming a tetraploid intermediate, followed by genome erosion resulting in progeny with DNA contents approximately 70% higher than the parental strains [19], [20]. TcV and TcVI strains, by contrast, have DNA contents equivalent to those found in TcII and TcIII. The experimental TcI hybrids inherited >2 alleles at multiple genetic loci [19], whereas TcV and TcVI typically have only one or two alleles per locus [20]. Finally, uniparental inheritance of kDNA maxicircles occurred in both natural and experimental hybrids [16], [19]. The cytological mechanism of hybridisation in natural T. cruzi populations therefore remains undefined and is yet to be reconciled with that documented for in vitro hybrids. However, the discovery of conserved meiotic gene homologues in several basally diverging protists, including kinetoplastids, Giardia and Trichomonas implies that the common ancestor of all extant eukaryotes was capable of meiotic recombination [21], [22].
In this study molecular markers from three sequence classes (nuclear coding sequence, mitochondrial (kinetoplast maxicircle) coding sequence, and multiple microsatellite loci), each with inherently different mutation rates, were used to analyse the origins and evolution of T. cruzi at several overlapping levels of resolution. Recent advances in the capacity for large scale multilocus microsatellite typing [8] permitted high resolution analysis of both parental and hybrid lineages leading to novel insights into the timing, ecological circumstances and cytological mechanism of hybridisation in T. cruzi.
Materials and Methods
Samples
DNA samples for a core panel of 35 T. cruzi clones representing hybrid lineages TcV and TcVI and parental lineages TcII and TcIII were used for all analyses; up to 53 additional strains representing all six lineages were used in selected analyses as indicated (Tables 1 and S1). DTU-level genotypes were determined using a previously described triple-marker assay [23]. The samples were derived from diverse hosts and vectors from geographical locations representative of the distribution of each lineage.
Table 1. Genotypes for core samples analysed in the study.
| GPI | COII-ND1 | MLMT (19 loci) | MLMT (28 loci) | |||||
| Haplotype code(s) | ||||||||
| Strain | DTU | Location | Host/Vector | Allele 1 | Allele 2 | Haplotype code | MLG code | MLG code |
| Tu18 cl2 | TcII | Tupiza, BO | Triatoma infestans | Hap nG-21 | Hap nG-22 | Hap kCN-26 | 19/B01 | 28/B01 |
| Rita cl5 | TcII | Bahia, BR | Homo sapiens | Hap nG-21 | Hap kCN-27 | 19/B02 | 28/B02 | |
| CBB cl2 | TcII | Region IV, CL | Homo sapiens | Hap nG-21 | Hap nG-22 | Hap kCN-27 | 19/B03 | 28/B03 |
| Pot7a cl1 | TcII | Boqueron, PY | Triatoma infestans | Hap nG-21 | Hap kCN-27 | 19/B04 | 28/B04 | |
| Pot7b cl5 | TcII | Boqueron, PY | Triatoma infestans | Hap nG-21 | Hap kCN-27 | 19/B04 | 28/B04 | |
| IVV cl4 | TcII | Cuncumen, CL | Homo sapiens | Hap nG-21 | Hap nG-22 | Hap kCN-26 | 19/B05 | 28/B05 |
| Chaco23 col4 | TcII | Pr.Hayes, PY | Triatoma infestans | Hap nG-21 | Hap nG-22 | Hap kCN-25 | 19/B06 | 28/B06 |
| Esm cl3 | TcII | Bahia, BR | Homo sapiens | Hap nG-21 | Hap kCN-27 | 19/B04 | 28/B04 | |
| T665 cl1 | TcII | Pr.Hayes, PY | Triatoma infestans | Hap nG-22 | Hap kCN-28 | 19/B07 | 28/B07 | |
| ARMA13 cl1 | TcIII | Boqueron, PY | Dasypus novemcinctus | Hap nG-14 | Hap kCN-15 | 19/C01 | 28/C01 | |
| JA2 cl2 | TcIII | Amazonas, BR | Monodelphis sp. | Hap nG-15 | Hap kCN-14 | 19/C02 | 28/C02 | |
| SABP19 cl1 | TcIII | Vitor, PE | Triatoma infestans | Hap nG-16 | Hap nG-17 | Hap kCN-22 | 19/C03 | 28/C03 |
| ARMA18 cl3 | TcIII | Boqueron, PY | Dasypus novemcinctus | Hap nG-14 | Hap nG-18 | Hap kCN-13 | 19/C04 | 28/C04 |
| CM25 cl2 | TcIII | Carimaga, CO | Dasyprocta fugilinosa | Hap nG-15 | Hap nG-19 | Hap kCN-13 | 19/C05 | 28/C05 |
| M5631 cl5 | TcIII | Para, BR | Dasypus novemcinctus | Hap nG-15 | Hap nG-20 | Hap kCN-18 | 19/C06 | 28/C06 |
| M6241 cl6 | TcIII | Para, BR | Homo sapiens | Hap nG-15 | Hap nG-20 | Hap kCN-17 | 19/C07 | 28/C07 |
| 85/847 cl2 | TcIII | Alto Beni, BO | Dasypus novemcinctus | Hap nG-15 | Hap kCN-11 | 19/C08 | 28/C08 | |
| X9/3 | TcIII | Pr. Hayes, PY | Canis familiaris | Hap nG-14 | Hap kCN-22 | 19/C09 | 28/C09 | |
| X109/2 | TcIII | Pr.Hayes, PY | Canis familiaris | Hap nG-16 | Hap kCN-22 | 19/C10 | 28/C10 | |
| Sc43 cl1 | TcV | Santa Cruz, BO | Triatoma infestans | Hap nG-22 | Hap nG-23 | Hap kCN-19 | 19/D01 | 28/D01 |
| Para6 cl4 | TcV | Paraguari, PY | Triatoma infestans | Hap nG-22 | Hap nG-23 | Hap kCN-20 | 19/D01 | 28/D01 |
| 92-80 cl2 | TcV | Santa Cruz, BO | Homo sapiens | Hap nG-22 | Hap nG-23 | Hap kCN-21 | 19/D01 | 28/D01 |
| Para4 cl3 | TcV | Paraguari, PY | Triatoma infestans | Hap nG-22 | Hap nG-23 | Hap kCN-20 | 19/D01 | 28/D01 |
| Chaco2 cl3 | TcV | Boqueron, PY | Triatoma infestans | Hap nG-22 | Hap nG-23 | Hap kCN-20 | 19/D01 | 28/D01 |
| PAH179 cl5 | TcV | Chaco, AR | Homo sapiens | Hap nG-22 | Hap nG-23 | Hap kCN-20 | 19/D02 | 28/D02 |
| Vinch101 cl1 | TcV | Limari, CL | Triatoma infestans | Hap nG-22 | Hap nG-23 | Hap kCN-20 | 19/D01 | 28/D01 |
| Bug2148 cl1 | TcV | Rio Gr. do Sul, BR | Triatoma infestans | Hap nG-22 | Hap nG-23 | Hap kCN-20 | 19/D01 | 28/D01 |
| CL Brener | TcVI | Rio Gr. do Sul, BR | Triatoma infestans | Hap nG-22 | Hap nG-14 | Hap kCN-24 | 19/E01 | 28/E01 |
| VFRA1 cl1 | TcVI | Francia, CL | Triatoma infestans | Hap nG-22 | Hap nG-14 | Hap kCN-24 | 19/E01 | 28/E01 |
| Chaco17 col1 | TcVI | Pr. Hayes, PY | Triatoma infestans | Hap nG-22 | Hap nG-14 | Hap kCN-24 | 19/E01 | 28/E01 |
| Tula cl2 | TcVI | Tulahuen, CL | Homo sapiens | Hap nG-22 | Hap nG-14 | Hap kCN-24 | 19/E02 | 28/E02 |
| P251 cl7 | TcVI | Cochabamba, BO | Homo sapiens | Hap nG-22 | Hap nG-14 | Hap kCN-24 | 19/E03 | 28/E03 |
| LHVA cl4 | TcVI | Chaco, AR | Triatoma infestans | Hap nG-22 | Hap nG-14 | Hap kCN-24 | 19/E01 | 28/E01 |
| EPV20-1 cl1 | TcVI | Chaco, AR | Triatoma infestans | Hap nG-22 | Hap nG-14 | Hap kCN-24 | 19/E02 | 28/E02 |
| Chaco9 col15 | TcVI | Pr. Hayes, PY | Triatoma infestans | Hap nG-22 | Hap nG-14 | Hap kCN-23 | 19/E01 | 28/E01 |
DTU, discrete typing unit; MLMT, multi-locus microsatellite typing; MLG, multi-locus genotype; Hap, Haplotype; BO, Bolivia; BR, Brazil; CL, Chile; PY, Paraguay; PE, Peru; CO, Colombia; AR, Argentina.
Nucleotide sequences
Nucleotide sequences were determined for a fragment of glucose-6-phosphate isomerase (GPI), a single copy nuclear gene, and for a region of the kDNA maxicircle, spanning regions of two adjacent genes, cytochrome oxidase subunit II (COII) and NADH dehydrogenase subunit 1 (ND1). PCR amplification of GPI was performed as described previously [23]. PCR amplification reactions for COII-ND1 contained 1× quantity NH4 buffer, 0.2 mM of dNTPs, 3.5 mM MgCl2, 1 pmol/µl of each primer, 1 Unit of Taq DNA polymerase (all Bioline, UK) and 10–100 ng genomic DNA using primers ND1.3A and COII.2A [16]. Amplification conditions were 94°C for 3 mins, 30 cycles of 94°C for 30 s, 58°C for 1 min, 72°C for 2 mins, followed by a final elongation step at 72°C for 10 mins.
Sequencing reactions were performed using the ABI Prism BigDye3.1 terminator cycle sequencing kit (Applied Biosystems, UK) using PCR primers and additional internal sequencing primers in some cases: for GPI, primer GPI.1 [19]; for COII-ND1, primers COII.A400 [16], COII-INTL1 (5′-CAAAAGATAATAACACTATAACAGAATC-3′) and COII-INTL2 (5′-CAAAAGATAATAACACTATAACAG-3′). For resolution of GPI haplotypes, PCR products were cloned using the pGEM-T Easy vector kit (Promega, UK) and inserts from up to 12 clones were sequenced. Sequences for outgroups and 43 additional T. cruzi strains were obtained from GenBank; accession numbers are given in Table S1.
Phylogenetic analysis of GPI and COII-ND1
Sequences were aligned using CLUSTAL_X [24] and sequence diversity statistics calculated using DnaSP v.5 [25]. Unique haplotypes were identified and aligned with outgroup sequences to produce final alignments. Each alignment was used to reconstruct multiple alternative tree topologies in MEGA4 [26] using (a) Neighbour Joining (NJ) using Kimura's 2-parameter (K2P) [27], Tamura-Nei (TN93) [28], and maximum composite likelihood [29] substitution models and (b) Maximum Parsimony (MP) using the close neighbour interchange procedure, which generated 143 and 98 trees of equal length for GPI and COII-ND1 respectively. Bootstrap support for clades was estimated using 1000 pseudo-replicate datasets. Maximum likelihood (ML) analysis was conducted for each topology using PAML v.4 [30] implementing Shimodaira-Hasegawa (SH) tests [31] to test for statistically significant differences in likelihoods between topologies. The program FINDMODEL (http://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html) was used to select the most appropriate nucleotide substitution model for each data set: GTR (REV)+gamma for GPI and TN93+gamma for COII-ND1. In each ML analysis a discrete gamma distribution model with four rate categories was applied to account for rate heterogeneity among sites; the gamma distribution shape parameter, α, was estimated from the data. To test for rate constancy (i.e. a molecular clock) ML analysis was performed using topologies rooted using either Trypanosoma brucei sequences (GPI) or T. cruzi marinkellei sequences (COII-ND1), with all branches constrained to a single rate of evolution. The resulting likelihood was compared to the unconstrained model using a likelihood ratio test (LRT).
Estimation of divergence times
BEAST v1.5.3 [32] was used to estimate divergence times. This program implements a Bayesian Markov chain Monte Carlo (MCMC) procedure to sample the posterior probability of trees generated under a specified prior evolutionary model. One advantage of this approach is that by sampling from a large number of trees, the statistical credibility (the 95% highest posterior density (HPD) interval) associated with parameter estimates (e.g. mutation rates, divergence times) can be obtained, thereby more accurately reflecting the uncertainties of phylogenetic inference than analyses generating a single tree. For each analysis the same nucleotide substitution model as implemented in the ML analysis was used. For GPI we applied a strict molecular clock and for COII-ND1 a relaxed clock with uncorrelated log-normally distributed rates [33]. A constant population coalescent prior was used as the demographic model. Random tree topologies were used as initial trees. To calibrate the rate estimation for GPI we applied a normally distributed prior on the divergence time between T. brucei sequences and T. cruzi clade sequences, i.e. the age of the root of the tree, with a mean of 100 million years ago (MYA) and SD of 2.0, according to the well supported evidence for the divergence of these groups concomitant with the separation of the African and South American land masses [34]. For COII-ND1, calibration was achieved in the same way but with a time applied to the T. cruzi – T. cruzi marinkellei ancestral node (the root) with a mean of 6.511 MYA and SD of 1.17, as inferred from the GPI results. For each data set two MCMC chains of 1×107 iterations were run, with parameters logged every 1000 iterations and removal of the first 10% of states as burn-in. Log-files were checked for sufficient effective sampling sizes (ESSs) and convergence of chains on similar posterior distributions using TRACER v1.5 [35].
Microsatellite analysis
Genotypes were obtained for the core 35 samples at 28 microsatellite loci and for an additional 46 strains at a subset of 19 microsatellite loci using previously described protocols [8], [20]; physical positions and primer details are given in Table S2. Microsatellite genotypes for some strains at some loci were from previously published datasets as indicated [8], [20], [36]. The genotypes were used to infer a measure of genetic distance between all possible pairs of strains (pairwise distance [D AS]) under the assumptions of the infinite-alleles model (IAM) [37] as previously [8]. Multi-allelic genotypes (≥3 alleles per locus), observed for 7 of 1826 genotypes, were treated as missing data. For each DTU, heterozygosity indices were calculated in ARLEQUIN 3.0 [38]. Allelic richness (Ar) was used as a sample size-corrected measure of diversity and was calculated in FSTAT [39]. In hybrid groups TcV and TcVI, genotypes at individual loci were classed as hybrid (TcII/TcIII) or non-hybrid (TcII/TcII or TcIII/TcIII) based on the presence or absence of alleles in the TcII and TcIII parental groups. In order to minimize the possibility of null alleles affecting classification of TcV/VI genotypes, all instances of homozygosity in these samples were confirmed across three replicates. For inference of geographical relationships between TcV/VI hybrid and TcII/III parent DTUs, the genetic identity between each hybrid multilocus genotype (MLG) and all parent MLGs was calculated using 1-D AS. Then for TcV and TcVI separately, the mean identities with parental MLGs from countries with ≥2 measurements (Brazil, Bolivia, Chile and Paraguay) were calculated and compared using t-tests.
Results
GPI nucleotide sequence
Sequences for GPI were obtained for 86 T. cruzi strains allowing a gap-free alignment of 1038 nucleotides to be assembled. Heterozygous strains were found in all DTUs: TcI (n = 19/29), TcII (n = 4/9), TcIII (n = 8/25), TcIV (n = 1/7), TcV (n = 8/8) and TcVI (n = 8/8). A total of 34 distinct haplotypes and 58 polymorphic sites (5.59%) were identified (Tables 1 and S1). Comparing cloned and uncloned sequence reads we classified 39/247 (15.8%) as mosaics, most likely chimaeric products of PCR-mediated recombination [40], [41]. Once these were discarded two haplotypes per strain with mutually exclusive single nucleotide (SNP) patterns and compatible with the uncloned sequences were consistently identifiable.
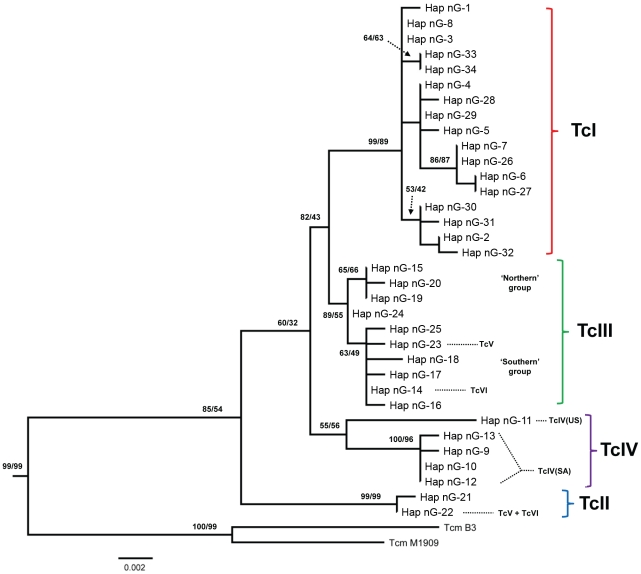
Sequence analysis (Figure 1) identified four major clades of T. cruzi sequences, equating to the T. cruzi lineages TcI, TcII, TcIII and TcIV. Each TcV and TcVI strain had one TcII-derived and one TcIII-derived haplotype. Three of these clades (TcI, II and III) had robust bootstrap support. Sequences from North American (NA) and South American (SA) TcIV strains were clearly divergent and monophyly of these taxa was only moderately supported. Examination of mean intra-lineage pair-wise genetic distances showed that in our sample of strains the predominantly domestic lineages TcII, TcV and TcVI were much more homogeneous than strains from the predominantly sylvatic lineages TcI, TcIII and TcIV. TcIII and TcIV sequences contained fixed SNPs that were otherwise restricted to TcI or TcII, as well as multiple lineage specific polymorphisms.
Figure 1. Maximum likelihood tree of GPI sequences (unique haplotypes).
Four major clades are indicated according to corresponding T. cruzi lineage; TcV and TcVI haplotypes are within the parental TcII and TcIII clades from which they are derived. Where clear geographic associations were evident these are indicated: US (samples from USA); SA (samples from South America); ‘Northern’ and ‘Southern’ groups within TcIII are as defined previously [36]. Branches separating T. cruzi from T. rangeli and T. brucei (outgroup) are not shown. Bootstrap support values for branches (if >50%) are shown and were calculated for the equivalent nodes of the bootstrap consensus NJ/MP trees (1000 replicates). Scale units are substitutions/site.
All TcV and TcVI strains shared the same intact TcII haplotype (haplotype nG-22), which was also present in 5/9 TcII isolates (Tu18 cl2, CBB cl2, IVV cl4, T665 cl1 and Chaco23 col4) (Table 1). The low diversity in the TcII clade made it difficult to infer geographical associations between the present isolates and the TcII parent of TcV and TcVI but it is noteworthy that haplotype nG-22 was not found in either of the two isolates from Northern Brazil (Esm and Rita). The TcIII clade haplotype found in all TcVI strains (haplotype nG-14) was also found in 13/25 of the TcIII isolates (Table 1). A different TcIII haplotype (haplotype nG-23) was found in all TcV isolates but not in any other DTU. However, haplotype nG-23 and haplotype nG-14 were closely related, with only one distinguishing SNP (834C/G). There was evidence for some substructure within the TcIII clade, with separation between a ‘Southern’ group [36] containing all strains from Paraguay (11/11), Peru (1/1) and a subset of those from Bolivia (4/7) and a ‘Northern’ group [36], containing strains from Brazil (3/3), Colombia (2/2), Venezuela (1/1), and the remainder from Bolivia (3/7). The haplotypes from the hybrids TcV and TcVI were both unambiguously in the ‘Southern’ group (Figure 1).
COII-ND1 nucleotide sequence
Mitochondrial COII-ND1 sequences were obtained for 105 T. cruzi strains allowing an alignment of 1118 nucleotides to be assembled. Thirty-nine unique haplotypes were identified with 198 polymorphic sites (17.7%) (Tables 1 and S1). Numerous small indels (1–3 nt) were identified, as well as some larger indels, including a large deletion of 245 nt (positions 671–915) in haplotype kCN-26, previously found in Tu18 cl2 [16], and which we also found in IVV cl4. Intra-lineage nucleotide diversities (Table S3) revealed that hybrid groups TcV and TcVI had very low genetic diversity. All TcV sequences were identical bar private SNPs in Sc43 cl1 (position 818) and 92-80 cl2 (position 1073). Likewise, all TcVI had identical sequences bar a private SNP in Chaco9 col15 (position 1025). The mean genetic distance between TcV and TcVI sequences was low (0.43%), but fixed inter-lineage SNPs were found at three positions (105, 629 and 671).
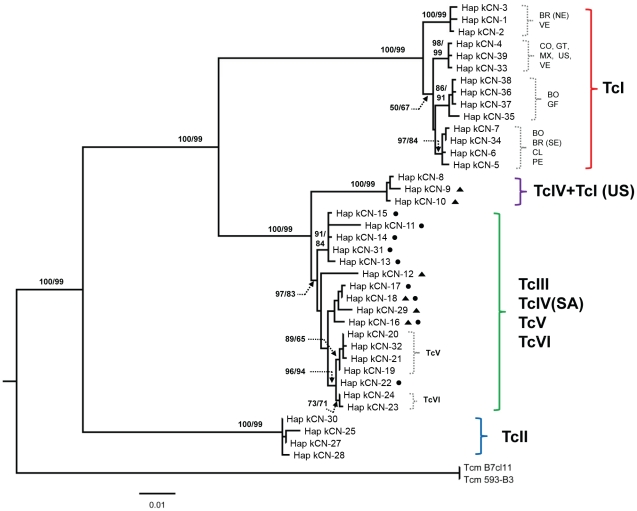
Four major clades were recovered: (i) TcI, (ii) TcIV(NA)+three TcI(NA), (iii) TcIII+TcV+TcVI+TcIV(SA), (iv) TcII (Figure 2). Within TcI four well-supported sub-clades corresponded broadly to different geographical regions: first, all TcI sequences from Central America (5/6), Colombia (2/2) and a subset of those from Venezuela (2/3) and USA (2/5); second, sequences from South of the Amazon (Bolivia (4/7), Chile (2/2), Peru (4/4) and southern Brazil (3/3)); third, sequences from northern Brazil (5/5) and Venezuela (1/3); and fourth, sequences from French Guyana (2/2) and from a subset of isolates from Bolivia (3/7). No clear correlations between different clades and different host species or transmission cycles were apparent.
Figure 2. Maximum likelihood tree of COII-ND1 sequences (unique haplotypes).
Clades and subclades are indicated according to corresponding major T. cruzi lineage (see text). Black circles indicate TcIII haplotypes and black triangles indicate TcIV haplotypes, clearly showing paraphyly of TcIV and TcIII sequences and sharing of identical haplotypes between subsets of TcIII and TcIV samples. Where clear geographic associations were evident these are indicated using standard two letter country codes; TcIV(SA) are TcIV samples from South America. Bootstrap support values for branches (if >50%) are shown and were calculated for the equivalent nodes of the bootstrap consensus NJ/MP trees (1000 replicates). Scale units are substitutions/site.
As for GPI, COII-ND1 sequences showed clear divergence between North and South American TcIV strains. Moreover, TcIV(SA) strains were paraphyletic and interspersed between TcIII haplotypes. There were three separate instances of sharing of identical or nearly identical mitochondrial haplotypes between TcIV strains and strains from other DTUs (TcIII or TcI) for which nuclear GPI sequences were highly divergent (Figures 1 and 2). Such gross incongruence between mitochondrial and nuclear phylogenies is likely indicative of historical genetic exchange events resulting in mitochondrial introgression between DTUs.
The TcV and TcVI COII-ND1 sequences and TcIII COII-ND1 haplotype kCN-22 were more closely related to each other than to any other TcIII sequence, forming a very strongly supported monophyletic group. Haplotype kCN-22 differed from the TcV and TcVI consensus sequences at only three or one nucleotide position(s) respectively. By comparison, the next most similar TcIII isolates had fixed differences at twelve positions compared to TcVI or thirteen positions compared to TcV. Haplotype kCN-22 was found in 6/18 TcIII strains: one human isolate from southern Peru (SABP19 cl1), three were isolated from domestic dogs in the Paraguayan Chaco (X9/3, X110/8, X109/2) [42], and two were isolated from sylvatic mammals, again from the Paraguayan Chaco (Sp4, Sp16) [36], [43].
Estimation of divergence times
For GPI sequences, rooted using T. brucei, the molecular clock model was not significantly worse than the unconstrained model (LRT p = 0.83; Table 2). Thus we used a time-calibrated strict molecular clock in the Bayesian analysis of mutation rate and divergence times for nodes in the GPI tree. For COII-ND1 sequences, rooted using T. cruzi marinkellei, the molecular clock was rejected for the ML tree (LRT p = 0.032; Table 2). However, for 46/98 (46.9%) of the topologies tested the molecular clock was not rejected (p>0.05) and none of these topologies gave likelihood scores significantly worse than the ML tree (SH tests, p>0.05) under the unconstrained model. Thus, instead of a strict molecular clock we chose to apply a relaxed molecular clock model [33] permitting rate variation across the tree to be factored into the Bayesian estimation of divergence times.
Table 2. Molecular clock likelihood ratio tests.
| Locus | Haplotypes (n) | Outgroup | Model | Clock? | Log Ln | 2*ΔLn (D) | Δdf | χ2 p value |
| GPI | 39 | Unrooted | GTR+G4 | No | −3236.663433 | |||
| GPI | 39 | T. brucei | GTR+G4 | Yes | −3251.047356 | 28.76785 | 37 | 0.831 |
| COII-ND1 | 40 | Unrooted | TN93+G4 | No | −3151.651149 | |||
| COII-ND1 | 40 | T. cruzi marinkellei | TN93+G4 | Yes | −3179.461322 | 55.62035 | 38 | 0.032 |
The mean estimated mutation rates were 3.55×10−9 nucleotide substitutions/site/year (95% HPD limits 2.56–4.60×10−9) for GPI and 1.94×10−8 (95% HPD limits 1.05–3.04×10−8; rate coefficient of variation = 0.34) for COII-ND1. Divergence time estimates are given in Table 3. The time of the most recent common ancestor (tMRCAs) for T. cruzi sensu stricto (i.e. excluding T. cruzi marinkellei) was estimated at 3.35 MYA (GPI) or 4 MYA (COII-ND1). For this and several other important nodes the two markers yielded similar age estimates with overlapping 95% HPD limits, including tMRCAs for TcI and TcII as well as the divergence of North American and South American TcIV. The divergence of TcI and TcIII COII-ND1 sequences, estimated at 2.44 MYA (4.03–0.986), represents an estimate for the minimum age for a founding hybridisation event between TcI and TcII, assuming uniparental inheritance of the ancestral TcI maxicircle genome [17]. We considered the possibility that if TcIII-VI strains are the products of historical genetic exchange [17] then their inclusion could introduce error into the dating estimates. However, we found this not to be the case since data sets with the TcIII-VI strains included and excluded yielded virtually identical estimates of both substitution rates and divergence times (not shown). GPI sequences did not provide sufficient resolution to date the emergence of the hybrid DTUs TcV and TcVI but the more variable maxicircle sequences supported a very recent origin with tMRCAs estimated at 60,700 (944–126,000) years ago and 33,900 (523–91,300) years ago respectively.
Table 3. Divergence times for selected species and T. cruzi subgroups.
| Alignment | Node/Group | tMRCA (MYA) | 95% HPD Limits (MYA) |
| GPI | T. brucei - T. cruzi (root) | 99.9 | 104 - 96 |
| T. cruzi - T. rangeli | 31 | 39.3 - 22.9 | |
| T. cruzi - T. cruzi marinkellei | 6.51 | 8.91 - 4.33 | |
| T. cruzi | 3.35 | 4.76 - 2.06 | |
| TcI+TcIII–TcIV | 2.24 | 3.17 - 1.38 | |
| TcI–TcIII | 1.78 | 2.59 - 1.04 | |
| TcIV(NA) - TcIV(SA) | 1.65 | 2.48 - 0.888 | |
| TcI | 0.875 | 1.33 - 0.469 | |
| TcIII | 0.827 | 1.32 - 0.353 | |
| TcIV(SA) | 0.378 | 0.732 - 0.0968 | |
| TcII | 0.221 | 0.543 - 0.00363 | |
| COII-ND1 | T. cruzi - T. cruzi marinkellei (root) | 6.04 | 8.47 - 3.69 |
| T. cruzi | 4 | 6.35 - 1.71 | |
| TcI–TcIII+IV+V+VI | 2.44 | 4.03 - 0.986 | |
| TcIV(NA) - TcIII+IV(SA)+V+VI | 0.847 | 1.46 - 0.317 | |
| TcI | 0.517 | 0.911 - 0.184 | |
| TcII | 0.148 | 0.303 - 0.0307 | |
| TcV+VI+Hap22 | 0.138 | 0.26 - 0.0336 | |
| TcV | 0.0607 | 0.126 - 0.00944 | |
| TcVI | 0.0339 | 0.0913 - 0.000523 |
MLMT
We applied MLMT to gain higher resolution of the genetic diversity within and between hybrid and parental lineages. The 35 core TcII, TcIII, TcV and TcVI samples were typed at 28 polymorphic microsatellite loci (28 locus-analysis; Tables 1, S1 and S4), of which eight were described previously for these strains [20]. Genotypes for additional TcI, TcIII and TcIV strains for 19 of these loci were included for comparison in some cases (19-locus analysis; Tables 1, S1 and S4). Unless additional sized peaks were observed, microsatellite profiles were considered to represent homozygous (one peak) or heterozygous (two peaks) diploid genotypes. The majority of samples gave either one or two peak sizes. Triple peak profiles were observed for CBB cl2 (TcII) at 6 loci: 6529(TA)b, 6529(CA)c, 6529(CA)a, 6529(TCC), (all linked on a 17 kb region of chromosome 6), 10101(CA)c, (chromosome 27) 11283(TA)a (chromosome 40); DNA content analysis has shown this strain is probably aneuploid [20]. The only other multi-allelic profile was found for P251 cl7 (TcVI), which had three 10101(TA)/Set0 alleles (chromosome 27).
Pairwise genetic distances (D AS) were used to reconstruct phylogenetic trees using the NJ method (Figure 3). Parental and hybrid DTUs exhibited highly dissimilar patterns of genetic diversity. Most TcII and TcIII strains had unique MLGs, with high pairwise D AS genetic distances (>50%) and numerous private alleles (Table 4; Figure 4). In contrast TcV and TcVI were genetically uniform, with few, highly similar MLGs, low pairwise genetic distances (2.2 and 1% respectively) and far fewer private alleles (Table 4; Figure 4). Allelic richness (Ar) provided a measure of genetic diversity independent of sample size and supported markedly lower diversity in TcV/TcVI compared to TcII/TcIII (Table 4). In the 19-locus analysis TcI, TcII, TcIII and TcIV all showed similar patterns of high genetic differentiation between strains with strong support for four monophyletic clusters (Figure 3).
Figure 3. Unrooted neighbour-joining microsatellite D AS trees showing lack of diversity within hybrid lineages TcV and TcVI.
A) Tree based on 28 loci typed across 35 samples from parental (TcII, TcIII) and hybrid (TcV, TcVI) lineages. B) Tree based on 19 loci typed across 81 samples from all six major lineages TcI-VI. D AS-based bootstrap values were calculated over 1000 trees from pseudo-replicate datasets and those >70% are shown for the relevant nodes.
Table 4. Multilocus microsatellite summary statistics.
| Data set | DTU | N/G | Alleles | Ho | He | MNA | MNA SD | Ar | Mean pairwise genetic distance (D AS) | Mean pairwise genetic distance (D AS) SD |
| 28 loci | TcII | 9/7 | 110 | 0.640 | 0.644 | 3.929 | 1.184 | 3.670 | 0.501 | 0.264 |
| TcIII | 10/10 | 131 | 0.382 | 0.554 | 4.679 | 3.151 | 4.185 | 0.525 | 0.099 | |
| TcV | 8/2 | 54 | 0.817 | 0.450 | 1.929 | 0.466 | 1.919 | 0.022 | 0.039 | |
| TcVI | 8/2 | 52 | 0.786 | 0.424 | 1.857 | 0.525 | 1.821 | 0.010 | 0.009 | |
| 19 loci | TcI | 26/26 | 85 | 0.235 | 0.454 | 4.474 | 2.855 | 3.341 | 0.429 | 0.106 |
| TcIV | 3/3 | 47 | 0.175 | 0.575 | 2.474 | 0.905 | ND | 0.701 | 0.105 | |
| TcII | 9/7 | 71 | 0.601 | 0.600 | 3.737 | 1.327 | 3.548 | 0.472 | 0.251 | |
| TcIII | 25/24 | 107 | 0.381 | 0.493 | 5.632 | 4.044 | 3.766 | 0.443 | 0.106 | |
| TcV | 8/2 | 35 | 0.836 | 0.449 | 1.842 | 0.375 | 1.842 | 0.007 | 0.011 | |
| TcVI | 8/3 | 36 | 0.842 | 0.456 | 1.895 | 0.459 | 1.895 | 0.014 | 0.014 |
DTU, discrete typing unit; N/G, number of samples/number of genotypes; Ho observed heterozygosity; He expected heterozygosity; MNA, mean number of alleles; Ar Allelic richness; ND, not determined.
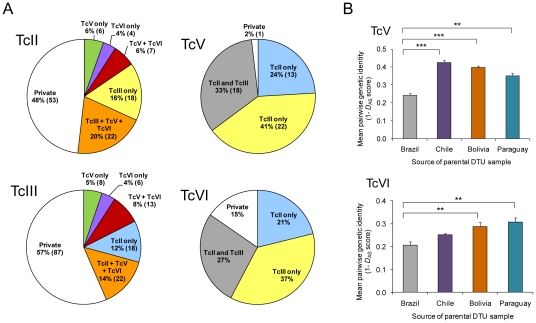
Figure 4. Patterns of microsatellite allelic inheritance in hybrid lineages TcV and TcVI.
Based on analysis of 19 locus and 28 locus data sets combined. A) For each parental (TcII, TcIII) and hybrid (TcV, TcVI) group the charts show the proportion of microsatellite alleles that were also found in each of the other three groups and the proportion of private alleles – those not found in any of the other three groups. B) Mean pairwise genetic identities (1-D AS) between hybrid group (TcV and TcVI) MLGs and parental group (TcII and TcIII) MLGs from different geographical regions; DTU, discrete typing unit; error bars are S.E.M; brackets represent statistically significant 2-tailed t-tests with asterisks indicating: ** p<0.01, *** p<0.001.
Hybrid and non-hybrid DTUs were also easily distinguishable in terms of heterozygosity. TcV and TcVI were both highly heterozygous (H O = 0.82/0.79). TcI and TcIII were far less heterozygous (H O = 0.38/0.25). TcII displayed intermediate H O, and although this was close to overall H E, only 12/28 loci were in Hardy-Weinberg equilibrium (not shown). Identical genotypes were observed from geographically distant locations for domestic isolates from TcII, TcV and TcVI (e.g. Esm cl3 from north-eastern Brazil was identical to Pot7a cl1 and Pot7b cl5 from the Paraguayan Chaco region).
Analysis of hybridisation
For many of the heterozygous loci in TcV and TcVI strains one of the alleles was frequently observed in TcII strains while the other allele was frequently observed in TcIII strains (Figures 4 and S1). Inheritance through hybridisation and without subsequent mutation is the most likely explanation, and was observed at too many independent loci to attribute to homoplasy. Comparison of the MLGs for TcV and TcVI also revealed fixed inter-DTU genotypic differences at 75% (21/28) of microsatellite loci and of the alleles that distinguished between TcV and TcVI 84% (37/44) were present in parental DTU strains. Coupled with the striking lack of intra-DTU diversity in both lineages this strongly suggests the two DTUs are products of separate hybridisation events.
The pairwise D AS distances between each hybrid MLG and each TcII (28 locus analysis) and TcIII (28 and 19 locus analysis) sample were used to measure identity between members of each parental DTU and each hybrid DTU. We then used these distances to infer the approximate geographical location for the origin of TcV and TcVI by grouping hybrid-parent distances according to the country of origin of parental samples. This revealed that TcV and TcVI genotypes were most closely related to parental lineage strains from Chile and Paraguay respectively, and furthermore, TcII/III strains from Brazil were significantly less closely related to TcV/VI than those from Bolivia, Paraguay or Chile (TcV only) (Figure 4B).
The genotypes at each individual microsatellite locus in the TcV/VI isolates were examined to predict the parental origin of each allele given its presence in or absence from TcII/III isolates. In this way the predicted ancestry of hybrid alleles could be classified as (i) TcII, (ii) TcIII, (iii) TcII or TcIII, or (iv) unknown (i.e. parent not sampled or de novo mutation) (Figures 4A and S1). For TcV 65%, and for TcVI 58% of alleles were assigned to either TcII or TcIII; either 33% (TcV) or 27% (TcVI) were found in both TcII and TcIII; 2% (TcV) or 15% (TcVI) were found in neither parental DTU. Based on this information there was evidence of non-hybrid genotypes (i.e. both alleles derived from the same parental DTU) at three loci in TcV: 6855(TTA)(GTT) for one strain and 6789(TA) and 11283(TA)a for all strains; and two loci in TcVI 6789(TA) and 11283(TA)a for all strains; in all cases the genotypes comprised only TcIII-restricted alleles.
Discussion
Origin of T. cruzi
The subgenus Schizotrypanum comprises approximately half a dozen described trypanosome species, most of which evolved on the South American continent after the break-up of Gondwanaland, and are referred to as the “T. cruzi clade” [34], [44]. By analysing nuclear coding (GPI), mitochondrial coding (COII-ND1) and nuclear non-coding (microsatellites) genetic markers, we were able to resolve the evolutionary processes within this clade at multiple, over-lapping levels of resolution. GPI sequences had the slowest rate of evolution, estimated to have a nucleotide substitution rate of 3.55×10−9 substitutions/site/year, reflecting its role as a house-keeping gene. The mitochondrial locus COII-ND1 had a faster evolutionary rate, estimated at 1.86×10−8 substitutions/site/year on average, in keeping with the generally higher rate of evolution in mtDNA than in nuclear DNA for eukaryotes [45]. Microsatellites typically have mutation rates at least three orders of magnitude higher than protein coding genes [46]. Accordingly the microsatellite loci used in this study afforded the highest resolution of intra-lineage diversity.
We estimated the dates of key events in the evolution of T. cruzi and related species, including divergence of T. rangeli and T. cruzi ancestral populations approximately 30 MYA, divergence of T. cruzi s.s. and the Chiropteran-restricted subspecies T. cruzi marinkellei at 6.5 – 6 MYA, and the tMRCA for T. cruzi s.s. at 4.0 – 3.35 MYA when the TcI and TcII lineages diverged. This date closely coincides with the formation of the land bridge between South and North America 3.5 – 3.1 MYA [47] suggesting that T. cruzi diversification may have been influenced by the subsequent changes in fauna known as the Great American Interchange [48]. Previous estimates for the age of T. cruzi range from 3–16 MYA, a date also obtained by calibration on the breakup of Gondwanaland, and 10.6 MYA calibrated using a universal rate of mutation of 1% MY−1 for mitochondrial CYTB [14]. TcIII and TcIV GPI sequences contained numerous SNPs that were otherwise only found in TcI or TcII, as well as lineage-specific polymorphisms. This pattern of SNPs is compatible with an ancient hybridisation event between TcI and TcII as previously suggested [17], [49]. Such reticulate evolution explains the large discrepancies between MP (based on individual sites) and NJ (based on overall genetic distance) bootstrap values for relevant nodes of the GPI phylogeny. The finding that the COII-ND1 sequences of TcI and TcIII/IV diverged an estimated 2.44 MYA implies additional complexity because analysis of both COII-ND1 [16] and CYTB [14] shows there is far less mitochondrial diversity both within and between TcIII and TcIV(SA) than would be expected in light of the considerable divergence observed for slower-evolving nuclear genes. This implies a mechanism acting to homogenise maxicircle sequences while nuclear sequences remain free to diverge. A partial explanation may stem from our finding of multiple instances of clearly recent mitochondrial introgression between TcIII and TcIV in South America, indicating additional recombination events involving either substantial backcrossing or kDNA exchange without exchange of nuclear material.
The sequence analysis showed clear divergence between North and South American TcIV populations, estimated at 0.85–1.65 MYA, again most likely a result of faunal migrations between the two continents. Migration of TcI from South to North America may have occurred more recently as suggested by a strong reduction in microsatellite diversity among North American TcI isolates [8]. Additionally, this study builds on evidence [16] supporting recent mitochondrial introgression between a subset of TcI and TcIV strains in North America, which share almost identical mitochondrial COII-ND1 sequences while displaying extensive divergence for nuclear GPI sequences. Typing of further nuclear loci is required to determine whether such events also involved transfer of any nuclear material, but the nuclear GPI and microsatellite data presented here indicate that it did not.
Our estimates of substitution rates and divergence times depend on the prior assumption that T. brucei and T. cruzi diverged due to the break-up of Gondwanaland ∼100MYA. Biogeographic data indicate this is a robust assumption. For example, Stevens et al [34] showed that, with the exception of those associated with bats, T. cruzi clade species are found exclusively in the New World and are most closely related to a kangaroo trypanosome. This is consistent with the connection of South America to Australia via Antarctica during the late Cretaceous [50]. Molecular data show this biogeographic event also led to the evolution of the Triatominae [51]. However, the recent finding of T. conorhini-like and T. vespertilionis-like trypanosomes in two terrestrial mammals in West Africa [52] illustrates the need for caution. In this case, introduction of these trypanosomes from the New World seems a plausible explanation given the capacity for host-switching in trypanosomes and the well documented inter-continental dispersal of T. cruzi clade species by bats (T. vespertilionis, T. dionisii, T. cruzi marinkellei) and rats (T. conorhini) [53].
Origin and clonal expansion of TcV and TcVI
The multilocus genotypes of the T. cruzi hybrid lineages TcV and TcVI fit the expectations of relatively recent hybridisation: they are highly heterozygous, exhibit minimal intra-lineage diversity and have accumulated few, if any, private alleles by de novo mutation. This contrasts with TcI, TcII and TcIII, which are genetically diverse, have abundant private alleles and generally have lower than expected heterozygosity [8], [36]. TcV and TcVI have undergone dramatic clonal expansion: identical MLGs were found in samples originating across a vast geographic area. Any selfing or outcrossing would have produced homozygous genotypes rather than the uniform heterozygosity observed at most loci. The minority of TcV/VI microsatellite genotypes that were homozygous can be explained by several phenomena, including inheritance of identical alleles from both parents, loss of heterozygosity (LOH) due to gene conversion, null alleles, or non-meiotic inheritance (see below).
High resolution microsatellite data allowed us to address the question of whether TcV and TcVI arose by a single event followed by clonal diversification [17] or by separate hybridisation events [15]. Our data support two independent hybridisation events between distinct TcII and TcIII strains as the most plausible scenario because they have fixed inter-lineage genotypic differences at 75% of microsatellite loci and display virtually no intra-lineage diversity. A single event leading to a hybrid population from which TcV and TcVI are the only two surviving lineages is unlikely. Firstly, the majority of microsatellite alleles that distinguish TcV and TcVI were also present in parental strains; independent inheritance is far more parsimonious than repeated homoplasy. Secondly, TcV and TcVI have distinct GPI and COII-ND1 alleles, MLEE profiles [42], [54], [55], LSU rDNA sequences [23], [56] and MLST profiles [57]; if these differences were the result of divergence of TcV and TcVI from the same ancestral hybrid population then a higher frequency of independently derived (private) alleles at rapidly evolving microsatellite loci would be expected. Alternatively, TcV and TcVI could have evolved from distinct progeny from the same hybridisation event between heterozygous TcII and TcIII parents i.e. siblings, with different alleles inherited by the two hybrid lineages.
TcV and TcVI are almost exclusively found in domestic transmission cycles and are the predominant genotypes in some hyper-endemic regions such as the Gran Chaco area where severe forms of Chagas disease are common [42], [55], [58]–[64]. Their clonal reproductive mode and negligible genetic diversity have potentially important biological and medical implications. For instance, innate phenotypic variability for traits such as resistance to drugs used to treat Chagas disease might be lower in TcV and TcVI than in other, more diverse lineages. Equally, under certain conditions the availability of diverse alleles may result in enhanced fitness compared to either parental lineage (heterosis).
Geographic, temporal and ecological aspects of hybridisation events
Fully intact or highly similar parental alleles identifiable in TcV and TcVI permitted phylogenetic analysis to identify TcII and TcIII strains most closely related to their actual parents. Nuclear GPI sequences and microsatellite genotypes both supported a significantly closer genetic relationship between TcII isolates from Chile, Paraguay and Bolivia than with TcII from Brazil. This is congruent with a study of 5S rDNA sequences [65], which linked TcV/VI to TcII from Bolivia and Chile based on the presence of an ‘invader sequence’ not found in TcII from Brazil. For the other parental DTU, TcIII, GPI and microsatellites supported a closer genetic relationship with TcIII from Bolivia, Paraguay and Peru than with TcIII from Brazil, Colombia or Venezuela. A previous analysis of mitochondrial COII-ND1 sequences [16] showed that three TcIII isolates isolated from domestic dogs in the Paraguayan Chaco were most similar to TcV/VI. We identified the same COII-ND1 sequence in a human isolate from southern Peru and in two Paraguayan Chaco sylvatic isolates. Overall, these data support the Gran Chaco and adjoining Andean valleys in south-western South America as the most probable origin of TcV and TcVI, with mitochondrial sequences pointing more specifically to the Paraguayan Chaco. Further analysis using additional strains and nuclear coding loci that afford sufficient resolution will be required to confirm this hypothesis.
This leads to the questions of how and when the hybridisation events occurred. From our mtDNA sequence analysis we conclude that the ages of the TcV and TcVI MRCAs are within approximately the last 60,000 years. At this scale, however, COII-ND1 provided relatively low resolution, reflected by wide 95% HPD limits. The lack of allelic and genotypic diversity across so many fast-evolving microsatellite loci and the finding that the TcII and TcIII-derived genetic material appears unchanged, indicates a date towards the more recent end of the mtDNA-inferred range would be most parsimonious. Plasmodium vivax provides a useful comparison since it displays a similarly low level of microsatellite diversity to that found here for TcV and TcVI and is estimated to have expanded from a small founding population less than 10,000 years ago [66]. When the distribution of TcV and TcVI is considered, the data are indicative of a recent and rapid spread of what are effectively two clonal genotypes. This fits well the hypothesis [14] that the present distribution of TcV and TcVI is the product of changes in the distribution of the vector Triatoma infestans, which is itself thought to have spread from an initial sylvatic focus in Bolivia, across the Southern cone region and into North-eastern Brazil as a result of human-mediated dispersal and adaptation to synanthropic and domestic niches [67]–[69].
Two scenarios for TcV and TcVI origins can be considered. Firstly, limited diversity in hybrids now found in domestic cycles could result from a genetic bottleneck associated with invasion of domestic niches from more diverse, unsampled TcV/VI sylvatic populations. Records of non-domestic TcV and TcVI are, however, extremely rare. Multiple foci of wild T. infestans occur in the Gran Chaco, Chile and especially in highland Bolivia (reviewed in [70]), but only TcI genotypes have so far been documented [71]. Alternatively the hybrids are anthropogenic with human activities leading to a set of conditions initially promoting hybridisation between TcII and TcIII and then establishment of hybrid populations in newly available niches. Subsequent bottlenecks may have also contributed to low TcV/VI diversity. Humans had arrived in South America by 14,600 years ago and possibly earlier [72]; transmission of T. cruzi to humans was a frequent occurrence from at least 9,000 years ago as shown by analysis of mummified specimens [73].
Co-infection of a single host or vector would have been prerequisite for hybridisation and current distributions suggest a domestic/synanthropic setting is more likely. Like TcV and TcVI, TcII occurs predominantly in domestic transmission cycles [7], [74]–[76]. TcIII is most frequently associated with Dasypus armadillos but also has domestic foci involving dogs and T. infestans in the Gran Chaco [43], [76], [77]. The mtDNA sequences showed that the most genetically similar TcIII isolates to the hybrids were isolated mainly from domestic sources. Human activities potentially promoting co-infection include provision of habitats favourable for triatomine domiciliation [69] and practices associated with hunting [73].
When the geographic, ecological and palaeontological data are considered in the context of the extreme lack of microsatellite diversity in TcV and TcVI and the possible timescale for their emergence inferred from our data, we propose that anthropogenic hybridisation events are more plausible than invasion from a sylvatic source. TcII and TcIII are ancient lineages that would have been well adapted to sylvatic niches and so any inter-lineage hybrids would likely be out-competed. Therefore, an anthropogenic origin for the hybrids may also explain their establishment and success, which is evident from their widespread current distribution, association with domiciliated T. infestans and human infectivity. TcV in particular predominates in human infections in Bolivia and appears to be well-adapted to congenital transmission [78]–[81].
Mechanism of hybridisation
T. cruzi laboratory hybrids produced to date are products of diploid fusion followed by genome erosion [19]. The mechanism of gene loss is incompletely characterised but does not involve a rapid return to diploidy [20]. It is yet to be determined whether such a mechanism operates in the field. Diploid fusion in T. cruzi is in contrast with T. brucei and Leishmania major for which laboratory crosses normally generate diploid progeny resembling standard meiotic F1 hybrids [82]–[86]. However, no haploid stages have been observed and polyploid and/or aneuploid hybrid progeny occur frequently [82], [87], [88]. There are numerous precedents for non-meiotic tetraploid to diploid transitions, for example, in Saccharomyces cerevisiae, as a result of repeated mitoses [89], [90], or in Candida albicans, in response to stress conditions [91], [92].
An expected consequence of diploid fusion followed by a 4n→2n transition is the presence of non-recombinant (non-hybrid) genotypes in 2n progeny at one third of unlinked loci, assuming that loss of chromosomal homologues from the 4n intermediate is random with respect to parentage, as observed in the C. albicans parasexual cycle [93]. In this context we examined the TcV and TcVI marker data but identified only 3/28 microsatellite loci with evidence for non-hybrid genotypes. Two of these loci were also homozygous and two were physically linked to other loci with hybrid, heterozygous genotypes. Therefore, LOH by gene conversion or the presence of alleles in unsampled TcII or TcIII strains are more likely explanations than uniparental inheritance. Multilocus sequence analysis has now shown evidence for relatively frequent LOH in TcV and TcVI coding sequences [57]. Some alleles may have been wrongly classified as parental due to homoplasy; however, the frequency of such errors is likely to be low. For example, of 196 alleles in TcII and TcIII only 31 were found in both DTUs, showing that independent evolution has tended to create divergent rather than convergent genotypes.
TcV and TcVI have hybrid genotypes for most markers examined in a sufficiently representative sample [16], [17], [94]–[96]. Non-hybrid (TcII/TcII or TcIII/TcIII) genotypes have been identified in TcV/VI for a small proportion of other markers, including SL-RNA, 24Sα rDNA (TcVI only), and HSP70 [3], [17], [23]. Again though, LOH better explains the absence of one parental allele rather than differential loss during a 4n→2n transition, particularly since these sequences all correspond to multicopy genes, which appear more susceptible to gene conversion [97]. The CL Brener (TcVI) genome analysis gives additional support to our conclusions since equal proportions of its genome are derived from TcII as from TcIII with only 3% being classified as homozygous [98].
Thus, overall there is strong evidence that TcV and TcVI strains do not deviate significantly from the 1∶1 ratio of TcII∶TcIII alleles expected for the clonal progeny of standard meiotic F1 hybrids and so the operation of meiosis in natural cycles should not be ruled out. Scenarios involving fusion followed by either random loss of chromosomes (parasexual reduction) or by a meiotic reductive division (i.e. [4n→8n→2(4n)→4(2n)]) with independent assortment of parental chromosomes, would be expected to result in a far higher proportion of non-hybrid genotypes than is observed. Nevertheless, the lack of an observed haploid stage and recombination via diploid fusion in vitro suggest that the programme of sexual reproduction in T. cruzi may differ from canonical meiosis yet still enable production of viable diploid recombinants, for example by meiosis with constraints on chromosomal assortment [99] or one-step meiosis [100].
While the details of the cytological mechanism remain somewhat unclear, the epidemiological consequences of recent hybridisation are not: TcV and TcVI are important agents of Chagas disease that are frequently isolated from domestic transmission cycles in parts of South America where severe manifestations (e.g. megasyndromes) and congenital transmission are prevalent. It will be important to determine why the hybrids and one of their parents are well adapted to domestic transmission while the other parent has remained largely sylvatic. Modern human activities continue to disrupt sylvatic T. cruzi transmission cycles and may promote the emergence of novel, virulent recombinant strains.
Supporting Information
Inferred patterns of allelic inheritance in hybrid multilocus genotypes. Unique multilocus genotypes (MLGs) for TcV (28/D01, 28/D02) and TcVI (28/E01, 28/E02, 28/E03) are shown with loci ordered in synteny based on the position of loci in the CL Brener (TcVI) reference genome sequence. Underlined loci contain fixed differences between TcV and TcVI; alleles in bold italic type indicate intra-DTU genotypic variability. Alleles are shaded according to their presence or absence among parental DTU (TcII and TcIII) samples: yellow, TcIII-restricted; blue, TcII-restricted; green, both TcII and TcIII; white, neither TcII nor TcIII. Boxed genotypes indicate putative non-hybrid genotypes.
(PDF)
Genotypes for all samples.
(PDF)
Microsatellite locus information.
(PDF)
Sequence diversity summary statistics.
(PDF)
Multilocus microsatellite genotypes.
(PDF)
Acknowledgments
We are grateful to Hernán Carrasco, Patricio Diosque, Vera Valente, Sebastião Valente, Michel Tibayrenc and Christian Barnabé for kindly providing T. cruzi samples. We also thank Isabel Mauricio, Sinead Fitzpatrick and Rania Baleela for insightful discussions and Louisa Messenger for assistance with sequencing and suggestions on the manuscript. We thank three anonymous reviewers for their constructive comments.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the BBSRC (http://www.bbsrc.ac.uk/), the Wellcome Trust (http://www.wellcome.ac.uk/) and the European Union Seventh Framework Programme (http://cordis.europa.eu/fp7) grant 223034 (“ChagasEpiNet”). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. The World Health Report, 2002. Geneva: World Health Organisation; 2002. [Google Scholar]
- 2.Brisse S, Barnabé C, Tibayrenc M. Identification of six Trypanosoma cruzi phylogenetic lineages by random amplified polymorphic DNA and multilocus enzyme electrophoresis. Int J Parasitol. 2000;30:35–44. doi: 10.1016/s0020-7519(99)00168-x. [DOI] [PubMed] [Google Scholar]
- 3.Brisse S, Verhoef J, Tibayrenc M. Characterisation of large and small subunit rRNA and mini-exon genes further supports the distinction of six Trypanosoma cruzi lineages. Int J Parasitol. 2001;31:1218–1226. doi: 10.1016/s0020-7519(01)00238-7. [DOI] [PubMed] [Google Scholar]
- 4.Zingales B, Andrade SG, Briones MRS, Campbell DA, Chiari E, et al. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz. 2009;104:1051–1054. doi: 10.1590/s0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- 5.Tibayrenc M, Neubauer K, Barnabé C, Guerrini F, Skarecky D, et al. Genetic characterization of six parasitic protozoa: parity between random-primer DNA typing and multilocus enzyme electrophoresis. Proc Natl Acad Sci USA. 1993;90:1335–1339. doi: 10.1073/pnas.90.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliveira RP, Broude NE, Macedo AM, Cantor CR, Smith CL, et al. Probing the genetic population structure of Trypanosoma cruzi with polymorphic microsatellites. Proc Natl Acad Sci USA. 1998;95:3776–3780. doi: 10.1073/pnas.95.7.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnabé C, Brisse S, Tibayrenc M. Population structure and genetic typing of Trypanosoma cruzi, the agent of Chagas disease: a multilocus enzyme electrophoresis approach. Parasitology. 2000;120:513–526. doi: 10.1017/s0031182099005661. [DOI] [PubMed] [Google Scholar]
- 8.Llewellyn MS, Miles MA, Carrasco HJ, Lewis MD, Yeo M, et al. Genome-scale multilocus microsatellite typing of Trypanosoma cruzi discrete typing unit I reveals phylogeographic structure and specific genotypes linked to human infection. PLoS Pathog. 2009;5:e1000410. doi: 10.1371/journal.ppat.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ocaña-Mayorga S, Llewellyn MS, Costales JA, Miles MA, Grijalva MJ. Sex, Subdivision, and Domestic Dispersal of Trypanosoma cruzi Lineage I in Southern Ecuador. PLoS Negl Trop Dis. 2011;4:e915. doi: 10.1371/journal.pntd.0000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rougeron V, De Meeûs T, Hide M, Waleckx E, Bermudez H, et al. Extreme inbreeding in Leishmania braziliensis. Proc Natl Acad Sci USA. 2009;106:10224–10229. doi: 10.1073/pnas.0904420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prugnolle F, De Meeûs T. Apparent high recombination rates in clonal parasitic organisms due to inappropriate sampling design. Heredity. 2010;104:135–140. doi: 10.1038/hdy.2009.128. [DOI] [PubMed] [Google Scholar]
- 12.Heitman J. Sexual Reproduction and the Evolution of Microbial Pathogens. Current Biology. 2006;16:R711–R725. doi: 10.1016/j.cub.2006.07.064. [DOI] [PubMed] [Google Scholar]
- 13.Awadalla P. The evolutionary genomics of pathogen recombination. Nat Rev Genet. 2003;4:50–60. doi: 10.1038/nrg964. [DOI] [PubMed] [Google Scholar]
- 14.Brisse S, Henriksson J, Barnabé C, Douzery EJP, Berkvens D, et al. Evidence for genetic exchange and hybridization in Trypanosoma cruzi based on nucleotide sequences and molecular karyotype. Infect Genet Evol. 2003;2:173–183. doi: 10.1016/s1567-1348(02)00097-7. [DOI] [PubMed] [Google Scholar]
- 15.de Freitas JM, Augusto-Pinto L, Pimenta JR, Bastos-Rodrigues L, Gonçalves VF, et al. Ancestral Genomes, Sex, and the Population Structure of Trypanosoma cruzi. PLoS Pathog. 2006;2:e24. doi: 10.1371/journal.ppat.0020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machado CA, Ayala FJ. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc Natl Acad Sci USA. 2001;98:7396–7401. doi: 10.1073/pnas.121187198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westenberger SJ, Barnabé C, Campbell DA, Sturm NR. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics. 2005;171:527–543. doi: 10.1534/genetics.104.038745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tibayrenc M, Ayala FJ. The clonal theory of parasitic protozoa: 12 years on. Trends Parasitol. 2002;18:405–410. doi: 10.1016/s1471-4922(02)02357-7. [DOI] [PubMed] [Google Scholar]
- 19.Gaunt MW, Yeo M, Frame IA, Stothard JR, Carrasco HJ, et al. Mechanism of genetic exchange in American trypanosomes. Nature. 2003;421:936–939. doi: 10.1038/nature01438. [DOI] [PubMed] [Google Scholar]
- 20.Lewis MD, Llewellyn MS, Gaunt MW, Yeo M, Carrasco HJ, et al. Flow cytometric analysis and microsatellite genotyping reveal extensive DNA content variation in Trypanosoma cruzi populations and expose contrasts between natural and experimental hybrids. Int J Parasitol. 2009;39:1305–1317. doi: 10.1016/j.ijpara.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik S-B, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM., Jr An Expanded Inventory of Conserved Meiotic Genes Provides Evidence for Sex in Trichomonas vaginalis. PLoS ONE. 2008;3:e2879. doi: 10.1371/journal.pone.0002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramesh MA, Malik SB, Logsdon JM. A phylogenomic inventory of meiotic genes: Evidence for sex in Giardia and an early eukaryotic origin of meiosis. Current Biology. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Lewis MD, Ma J, Yeo M, Carrasco HJ, Llewellyn MS, et al. Genotyping of Trypanosoma cruzi: Systematic Selection of Assays Allowing Rapid and Accurate Discrimination of All Known Lineages. Am J Trop Med Hyg. 2009;81:1041–1049. doi: 10.4269/ajtmh.2009.09-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 27.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 28.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 31.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 32.Drummond A, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drummond AJ, Ho S, Phillips M, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens J, Noyes H, Dover G, Gibson W. The ancient and divergent origins of the human pathogenic trypanosomes, Trypanosoma brucei and T. cruzi. Parasitology. 1999;118:107–116. doi: 10.1017/s0031182098003473. [DOI] [PubMed] [Google Scholar]
- 35.Rambaut A, Drummond A. 2009. Tracer v1.5, Available from http://beast.bio.ed.ac.uk/Tracer.
- 36.Llewellyn MS, Lewis MD, Acosta N, Yeo M, Carrasco HJ, et al. Trypanosoma cruzi IIc: Phylogenetic and Phylogeographic Insights from Sequence and Microsatellite Analysis and Potential Impact on Emergent Chagas Disease. PLoS Negl Trop Dis. 2009;3:e510. doi: 10.1371/journal.pntd.0000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura M, Crow JF. The number of alleles that can be maintained in a finite population. Genetics. 1964;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Excoffier L, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 39.Goudet J. FSTAT (Version 1.2): A Computer Program to Calculate F-Statistics. J Hered. 1995;86:485–486. [Google Scholar]
- 40.Meyerhans A, Vartanian J-P, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanabe K, Sakihama N, Färnert A, Rooth I, Björkman A, et al. In vitro recombination during PCR of Plasmodium falciparum DNA: a potential pitfall in molecular population genetic analysis. Mol Biochem Parasitol. 2002;122:211–216. doi: 10.1016/s0166-6851(02)00095-6. [DOI] [PubMed] [Google Scholar]
- 42.Chapman M, Baggaley R, Godfrey-Fausset P, Malpas T, White G, et al. Trypanosoma cruzi from the Paraguayan Chaco: isoenzyme profiles of strains isolated at Makthlawaiya. J Protozool. 1984;31:482–486. doi: 10.1111/j.1550-7408.1984.tb02999.x. [DOI] [PubMed] [Google Scholar]
- 43.Yeo M, Acosta N, Llewellyn M, Sanchez H, Adamson S, et al. Origins of Chagas disease: Didelphis species are natural hosts of Trypanosoma cruzi I and armadillos hosts of Trypanosoma cruzi II, including hybrids. Int J Parasitol. 2005;35:225–233. doi: 10.1016/j.ijpara.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 44.Simpson AGB, Stevens JR, Lukeš J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 45.Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 46.Ellegren H. Microsatellite mutations in the germline: implications for evolutionary inference. Trends Genet. 2000;16:551–558. doi: 10.1016/s0168-9525(00)02139-9. [DOI] [PubMed] [Google Scholar]
- 47.Coates AG, Obando JA. The geologic evolution of the Central American isthmus. In: Jackson JBC, Budd AF, Coates AG, editors. Evolution and Environment in Tropical America. Chicago: University of Chicago Press; 1996. pp. 21–56. [Google Scholar]
- 48.Pough FH, Janis CM, Heiser JB. Vertebrate Life. Upper Saddle River, New Jersey: Prentice Hall; 1999. [Google Scholar]
- 49.Sturm NR, Campbell DA. Alternative lifestyles: The population structure of Trypanosoma cruzi. Acta Trop. 2010;115:35–43. doi: 10.1016/j.actatropica.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 50.Cox CB, Moore PD. Biogeography. An Ecological and Evolutionary Approach. Oxford, U.K.: Blackwell; 2000. [Google Scholar]
- 51.Patterson JS, Gaunt MW. Phylogenetic multi-locus codon models and molecular clocks reveal the monophyly of haematophagous reduviid bugs and their evolution at the formation of South America. Mol Phylogenet Evol. 2010;56:608–621. doi: 10.1016/j.ympev.2010.04.038. [DOI] [PubMed] [Google Scholar]
- 52.Hamilton PB, Adams ER, Njiokou F, Gibson WC, Cuny G, et al. Phylogenetic analysis reveals the presence of the Trypanosoma cruzi clade in African terrestrial mammals. Infect Genet Evol. 2009;9:81–86. doi: 10.1016/j.meegid.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 53.Hoare C. The Trypanosomes of Mammals. Oxford: Blackwell; 1972. [Google Scholar]
- 54.Tibayrenc M, Ayala F. Isozyme variability of Trypanosoma cruzi, the agent of Chagas' disease: genetical, taxonomical and epidemiological significance. Evolution. 1988;42:277–292. doi: 10.1111/j.1558-5646.1988.tb04132.x. [DOI] [PubMed] [Google Scholar]
- 55.Miles MA, B WA, Widmer G, Povoa MM, Schofield CJ. Isozyme heterogeneity and numerical taxonomy of Trypanosoma cruzi stocks from Chile. T Roy Soc Trop Med H. 1984;78:526–535. doi: 10.1016/0035-9203(84)90076-2. [DOI] [PubMed] [Google Scholar]
- 56.Souto RP, Fernandes O, Macedo AM, Campbell DA, Zingales B. DNA markers define two major phylogenetic lineages of Trypanosoma cruzi. Mol Biochem Parasitol. 1996;83:141–152. doi: 10.1016/s0166-6851(96)02755-7. [DOI] [PubMed] [Google Scholar]
- 57.Yeo M, Mauricio IL, Messenger LA, Lewis MD, Llewellyn MS, et al. Multilocus sequence typing (MLST) for lineage assignment and high resolution diversity studies in Trypanosoma cruzi. PLoS Negl Trop Dis. 2011 doi: 10.1371/journal.pntd.0001049. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miles MA, Feliciangeli MD, de Arias AR. American trypanosomiasis (Chagas disease) and the role of molecular epidemiology in guiding control strategies. Brit Med J. 2003;326:1444–1448. doi: 10.1136/bmj.326.7404.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miles M, Cedillos R, Povoa M, de Souza A, Prata A, et al. Do radically dissimilar Trypanosoma cruzi strains (zymodemes) cause Venezuelan and Brazilian forms of Chagas' disease? Lancet. 1981;317:1338–1340. doi: 10.1016/s0140-6736(81)92518-6. [DOI] [PubMed] [Google Scholar]
- 60.Barnabé C, Neubauer K, Solari A, Tibayrenc M. Trypanosoma cruzi: presence of the two major phylogenetic lineages and of several lesser discrete typing units (DTUs) in Chile and Paraguay. Acta Trop. 2001;78:127–137. doi: 10.1016/s0001-706x(00)00183-2. [DOI] [PubMed] [Google Scholar]
- 61.Bosseno M-F, Barnabé C, Magallon Gastelum E, Lozano Kasten F, Ramsey J, et al. Predominance of Trypanosoma cruzi Lineage I in Mexico. J Clin Microbiol. 2002;40:627–632. doi: 10.1128/JCM.40.2.627-632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brenière SF, Bosseno MF, Noireau F, Yacsik N, Liegeard P, et al. Integrate Study of a Bolivian Population Infected by Trypanosoma cruzi, the Agent of Chagas Disease. Mem Inst Oswaldo Cruz. 2002;97:289–295. doi: 10.1590/s0074-02762002000300002. [DOI] [PubMed] [Google Scholar]
- 63.Virreira M, Serrano G, Maldonado L, Svoboda M. Trypanosoma cruzi: typing of genotype (sub)lineages in megacolon samples from bolivian patients. Acta Trop. 2006;100:252–255. doi: 10.1016/j.actatropica.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 64.Cardinal MV, Lauricella MA, Ceballos LA, Lanati L, Marcet PL, et al. Molecular epidemiology of domestic and sylvatic Trypanosoma cruzi infection in rural northwestern Argentina. Int J Parasitol. 2008;38:1533–1543. doi: 10.1016/j.ijpara.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Westenberger SJ, Sturm NR, Campbell DA. Trypanosoma cruzi 5S rRNA arrays define five groups and indicate the geographic origins of an ancestor of the heterozygous hybrids. Int J Parasitol. 2006;36:337–346. doi: 10.1016/j.ijpara.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 66.Leclerc MC, Durand P, Gauthier C, Patot S, Billotte N, et al. Meager genetic variability of the human malaria agent Plasmodium vivax. Proc Natl Acad Sci USA. 2004;101:14455–14460. doi: 10.1073/pnas.0405186101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bargues MD, Klisiowicz DR, Panzera F, Noireau F, Marcilla A, et al. Origin and phylogeography of the Chagas disease main vector Triatoma infestans based on nuclear rDNA sequences and genome size. Infect Genet Evol. 2006;6:46–62. doi: 10.1016/j.meegid.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 68.Dujardin JC, Schofield CJ, Tibayrenc M. Population structure of Andean Triatoma infestans: allozyme frequencies and their epidemiological relevance. Med Vet Entomol. 1998;12:20–29. doi: 10.1046/j.1365-2915.1998.00076.x. [DOI] [PubMed] [Google Scholar]
- 69.Cortez MR, Monteiro FA, Noireau F. New insights on the spread of Triatoma infestans from Bolivia - Implications for Chagas disease emergence in the Southern Cone. Infect Genet Evol. 2010;10:350–353. doi: 10.1016/j.meegid.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 70.Noireau F. Wild Triatoma infestans, a potential threat that needs to be monitored. Mem Inst Oswaldo Cruz. 2009;104:60–64. doi: 10.1590/s0074-02762009000900010. [DOI] [PubMed] [Google Scholar]
- 71.Cortez MR, Pinho AP, Cuervo P, Alfaro F, Solano M, et al. Trypanosoma cruzi (Kinetoplastida Trypanosomatidae): Ecology of the transmission cycle in the wild environment of the Andean valley of Cochabamba, Bolivia. Exp Parasitol. 2006;114:305–313. doi: 10.1016/j.exppara.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 72.Goebel T, Waters MR, O'Rourke DH. The Late Pleistocene Dispersal of Modern Humans in the Americas. Science. 2008;319:1497–1502. doi: 10.1126/science.1153569. [DOI] [PubMed] [Google Scholar]
- 73.Aufderheide AC, Salo W, Madden M, Streitz J, Buikstra J, et al. A 9,000-year record of Chagas disease. Proc Natl Acad Sci USA. 2004;101:2034–2039. doi: 10.1073/pnas.0307312101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fernandes O, Souto R, Castro J, Pereira J, Fernandes N, et al. Brazilian isolates of Trypanosoma cruzi from humans and triatomines classified into two lineages using mini-exon and ribosomal RNA sequences. Am J Trop Med Hyg. 1998;58:807–811. doi: 10.4269/ajtmh.1998.58.807. [DOI] [PubMed] [Google Scholar]
- 75.Luquetti AO, Miles MA, Rassi A, De Rezende JM, De Souza AA, et al. Trypanosoma cruzi: zymodemes associated with acute and chronic Chagas disease in central Brazil. T Roy Soc Trop Med H. 1986;80:462–470. doi: 10.1016/0035-9203(86)90347-0. [DOI] [PubMed] [Google Scholar]
- 76.Miles MA, Llewellyn MS, Lewis MD, Yeo M, Baleela R, et al. The molecular epidemiology and phylogeography of Trypanosoma cruzi and parallel research on Leishmania: looking back and to the future. Parasitology. 2009;136:1509–1528. doi: 10.1017/S0031182009990977. [DOI] [PubMed] [Google Scholar]
- 77.Póvoa M, de Souza A, Naiff R, Arias J, Naiff M, et al. Chagas disease in the Amazon basin IV. Host records of Trypanosoma cruzi zymodemes in the states of Amazonas and Rondonia, Brazil. Ann Trop Med Parasit. 1984;78:479–487. [PubMed] [Google Scholar]
- 78.Burgos JM, Altcheh J, Bisio M, Duffy T, Valadares HMS, et al. Direct molecular profiling of minicircle signatures and lineages of Trypanosoma cruzi bloodstream populations causing congenital Chagas disease. Int J Parasitol. 2007;37:1319–1327. doi: 10.1016/j.ijpara.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 79.Corrales R, Mora M, Negrette O, Diosque P, Lacunza D, et al. Congenital Chagas disease involves Trypanosoma cruzi sub-lineage IId in the northwestern province of Salta, Argentina. Infect Genet Evol. 2009;9:278–282. doi: 10.1016/j.meegid.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 80.Diez C, Lorenz V, Ortiz S, Gonzalez V, Racca A, et al. Genotyping of Trypanosoma cruzi Sublineage in Human Samples from a North-East Argentina Area by Hybridization with DNA Probes and Specific Polymerase Chain Reaction (PCR). Am J Trop Med Hyg. 2010;82:67–73. doi: 10.4269/ajtmh.2010.09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Virreira M, Alonso-Vega C, Solano M, Jijena J, Brutus L, et al. Congenital Chagas disease in Bolivia is not associated with DNA polymorphism of Trypanosoma cruzi. Am J Trop Med Hyg. 2006;75:871–879. [PubMed] [Google Scholar]
- 82.Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, et al. Demonstration of Genetic Exchange During Cyclical Development of Leishmania in the Sand Fly Vector. Science. 2009;324:265–268. doi: 10.1126/science.1169464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gibson WC. Analysis of a genetic cross between Trypanosoma brucei rhodesiense and T. b. brucei. Parasitology. 1989;99:391–402. doi: 10.1017/s0031182000059114. [DOI] [PubMed] [Google Scholar]
- 84.MacLeod A, Tweedie A, McLellan S, Taylor S, Cooper A, et al. Allelic segregation and independent assortment in Trypanosoma brucei crosses: Proof that the genetic system is Mendelian and involves meiosis. Mol Biochem Parasitol. 2005;143:12–19. doi: 10.1016/j.molbiopara.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 85.Sternberg J, Turner CMR, Wells JM, Ranford-Cartwright LC, Le Page RWF, et al. Gene exchange in African trypanosomes: frequency and allelic segregation. Mol Biochem Parasitol. 1989;34:269–279. doi: 10.1016/0166-6851(89)90056-x. [DOI] [PubMed] [Google Scholar]
- 86.Turner CMR, Sternberg J, Buchanan N, Smith E, Hide G, et al. Evidence that the mechanism of gene exchange in Trypanosoma brucei involves meiosis and syngamy. Parasitology. 1990;101:377–386. doi: 10.1017/s0031182000060571. [DOI] [PubMed] [Google Scholar]
- 87.Gibson W, Peacock L, Ferris V, Williams K, Bailey M. The use of yellow fluorescent hybrids to indicate mating in Trypanosoma brucei. Parasite Vector. 2008;1:4. doi: 10.1186/1756-3305-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hope M, MacLeod A, Leech V, Melville S, Sasse J, et al. Analysis of ploidy (in megabase chromosomes) in Trypanosoma brucei after genetic exchange. Mol Biochem Parasitol. 1999;104:1–9. doi: 10.1016/s0166-6851(99)00103-6. [DOI] [PubMed] [Google Scholar]
- 89.Gerstein AC, Chun H-JE, Grant A, Otto SP. Genomic Convergence toward Diploidy in Saccharomyces cerevisiae. PLoS Genet. 2006;2:e145. doi: 10.1371/journal.pgen.0020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gerstein AC, McBride RM, Otto SP. Ploidy reduction in Saccharomyces cerevisiae. Biol Letters. 2008;4:91–94. doi: 10.1098/rsbl.2007.0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003;22:2505–2515. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hilton C, Markie D, Corner B, Rikkerink E, Poulter R. Heat shock induces chromosome loss in the yeast Candida albicans. Mol Gen Genet. 1985;200:162–168. doi: 10.1007/BF00383330. [DOI] [PubMed] [Google Scholar]
- 93.Forche A, Alby K, Schaefer D, Johnson AD, Berman J, et al. The Parasexual Cycle in Candida albicans Provides an Alternative Pathway to Meiosis for the Formation of Recombinant Strains. PLoS Biol. 2008;6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Robello C, Gamarro F, Castanys S, Alvarez-Valin F. Evolutionary relationships in Trypanosoma cruzi: molecular phylogenetics supports the existence of a new major lineage of strains. Gene. 2000;246:331–338. doi: 10.1016/s0378-1119(00)00074-3. [DOI] [PubMed] [Google Scholar]
- 95.Augusto-Pinto L, Teixeira SMR, Pena SDJ, Machado CR. Single-Nucleotide Polymorphisms of the Trypanosoma cruzi MSH2 Gene Support the Existence of Three Phylogenetic Lineages Presenting Differences in Mismatch-Repair Efficiency. Genetics. 2003;164:117–126. doi: 10.1093/genetics/164.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tran A-N, Andersson B, Pettersson U, Åslund L. Trypanothione synthetase locus in Trypanosoma cruzi CL Brener strain shows an extensive allelic divergence. Acta Trop. 2003;87:269–278. doi: 10.1016/s0001-706x(03)00067-6. [DOI] [PubMed] [Google Scholar]
- 97.Cerqueira GC, Bartholomeu DC, DaRocha WD, Hou L, Freitas-Silva DM, et al. Sequence diversity and evolution of multigene families in Trypanosoma cruzi. Mol Biochem Parasitol. 2008;157:65–72. doi: 10.1016/j.molbiopara.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 98.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 99.Lutes AA, Neaves WB, Baumann DP, Wiegraebe W, Baumann P. Sister chromosome pairing maintains heterozygosity in parthenogenetic lizards. Nature. 2010;464:283–286. doi: 10.1038/nature08818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kondrashov AS. Evolutionary Genetics of Life Cycles. Annu Rev Ecol Syst. 1997;28:391–435. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Inferred patterns of allelic inheritance in hybrid multilocus genotypes. Unique multilocus genotypes (MLGs) for TcV (28/D01, 28/D02) and TcVI (28/E01, 28/E02, 28/E03) are shown with loci ordered in synteny based on the position of loci in the CL Brener (TcVI) reference genome sequence. Underlined loci contain fixed differences between TcV and TcVI; alleles in bold italic type indicate intra-DTU genotypic variability. Alleles are shaded according to their presence or absence among parental DTU (TcII and TcIII) samples: yellow, TcIII-restricted; blue, TcII-restricted; green, both TcII and TcIII; white, neither TcII nor TcIII. Boxed genotypes indicate putative non-hybrid genotypes.
(PDF)
Genotypes for all samples.
(PDF)
Microsatellite locus information.
(PDF)
Sequence diversity summary statistics.
(PDF)
Multilocus microsatellite genotypes.
(PDF)