Abstract
Thioredoxins are 12-kDa proteins functional in the regulation of cellular processes throughout the animal, plant, and microbial kingdoms. Growing evidence with seeds suggests that an h-type of thioredoxin, reduced by NADPH via NADP-thioredoxin reductase, reduces disulfide bonds of target proteins and thereby acts as a wakeup call in germination. A better understanding of the role of thioredoxin in seeds as well as other systems could be achieved if more were known about the target proteins. To this end, we have devised a strategy for the comprehensive identification of proteins targeted by thioredoxin. Tissue extracts incubated with reduced thioredoxin are treated with a fluorescent probe (monobromobimane) to label sulfhydryl groups. The newly labeled proteins are isolated by conventional two-dimensional electrophoresis: (i) nonreducing/reducing or (ii) isoelectric focusing/reducing SDS/PAGE. The isolated proteins are identified by amino acid sequencing. Each electrophoresis system offers an advantage: the first method reveals the specificity of thioredoxin in the reduction of intramolecular vs. intermolecular disulfide bonds, whereas the second method improves the separation of the labeled proteins. By application of both methods to peanut seed extracts, we isolated at least 20 thioredoxin targets and identified 5—three allergens (Ara h2, Ara h3, and Ara h6) and two proteins not known to occur in peanut (desiccation-related and seed maturation protein). These findings open the door to the identification of proteins targeted by thioredoxin in a wide range of systems, thereby enhancing our understanding of its function and extending its technological and medical applications.
Keywords: peanut allergens, desiccation-related protein, seed maturation protein, Ara h allergens, proglycinin
There is a growing body of evidence that, as in photosynthesis, the regulatory protein thioredoxin (1–4) plays a role in heterotropic processes in plants. In this capacity, the disulfide group of a thioredoxin of the h-type is reduced by NADPH via the flavin enzyme, NADP-thioredoxin reductase (NTR) (1, 5, 6) (Eq. 1).
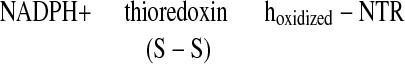 |
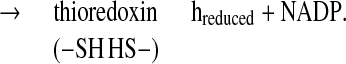 |
1 |
Biochemical studies initiated a decade ago with wheat have provided evidence for a function of thioredoxin h in germination and seedling development. The results suggest that thioredoxin h, reduced via NTR with metabolically generated NADPH, acts early in the imbibed seed to initiate the mobilization of nitrogen and carbon in the endosperm, the major repository of storage protein and carbohydrate in cereals (7, 8). The NADPH needed for this reduction can be generated enzymatically from carbohydrate stored in the endosperm via glucose 6-phophate and 6-phosphogluconate dehydrogenases (8).
Through the reduction of intramolecular disulfide bonds (Eq. 2), thioredoxin h was shown to promote the degradation of major storage proteins, the inactivation of small proteins that inhibit amylolytic enzymes, and the activation of a novel calcium-dependent substrate-specific protease (1, 7, 9, 10). The results provide evidence that thioredoxin h acts as a wakeup call in germination and seedling development.
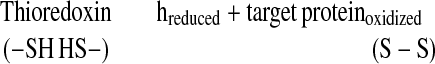 |
 |
2 |
It has become clear that more complete information on target proteins will facilitate our understanding of the role of thioredoxin. We previously have addressed the question in seeds using transgenic barley overexpressing thioredoxin h and have confirmed and extended our original biochemical observations. The results show that thioredoxin h overexpressed in the endosperm increases the activity of starch debranching enzyme (pullulanase) (11).
Although overexpression is a valid approach, we have sought a more comprehensive strategy to serve as a guide in the transgenic work. To this end, we have applied a combination of well-known two-dimensional gel-based separation methods (12, 13) with a sulfhydryl probe and amino acid sequencing to peanut extracts. In this article, we describe this strategy and demonstrate its application in the isolation of at least 20, and the identification of five, proteins targeted by thioredoxin h, among them three allergens. Although this article addresses seeds, the strategy potentially has broad application and could be used to identify thioredoxin target proteins not only in other plant systems, but in animals and microorganisms as well.
Materials and Methods
Materials.
Peanut (Arachis hypogaea L.) seeds were obtained from a local market. Chemicals and biochemicals were purchased from commercial sources and were of the highest quality available.
Preparation of Extracts.
Protein was extracted according to a modification of the protocol of Shokraii and Esen (14) as follows. The meal was prepared by grinding 12 randomly selected deskinned peanut seeds to a fine powder in a prechilled mortar with a pestle. The meal then was defatted three times with ethyl ether using a solvent-to-meal ratio of 1:10 (wt/vol). The defatted meal was extracted for 2 h by shaking in 0.1 M Tris⋅HCl buffer (pH 7.5) containing 1.5 M NaCl, 2 mM PMSF, and 0.2% NaN3. The meal-to-buffer ratio was 1:20 (wt/vol). The slurry was clarified by centrifugation (14,000 g, 5 min) and the supernatant fraction (protein extract) was saved.
Protein Assay.
Protein concentration was determined according to Bradford with bovine gamma globulin as standard (15).
In Vitro Protein Reduction.
Reduction of the disulfide bonds of proteins was determined with: (i) the NADP/thioredoxin system, consisting of 0.125 μmol NADPH, 2.4 μg Chlamydomonas reinhardtii thioredoxin h, and 2.1 μg Arabidopsis thaliana NTR (both proteins kind gifts of J.-P. Jacquot, Université de Nancy I, Vandoeuvre, France) or (ii) the NADP/glutathione system, composed of 0.125 μmol NADPH, 0.3 μmol reduced glutathione, and 1.5 μg yeast glutathione reductase (Sigma). As indicated, the plant NTR and thioredoxin h were replaced with their counterparts from Escherichia coli. The reaction was carried out for 3 or 5 h at 37°C in 50 mM Tris⋅HCl buffer, pH 7.9, in a final volume of 100 μl, using 20 μl (50 μg) peanut extract. For complete reduction, samples were boiled with DTT for 5 min.
Monobromobimane (mBBr) Labeling of Proteins.
Reduction of the disulfide bonds of proteins was performed as described (16). Sulfhydryl groups were visualized as their fluorescent mBBr derivatives. After incubation, mBBr, 0.2 μmol in 10 μl acetonitrile, was added to each sample, which then was incubated for 20 min at room temperature.
Nonreducing/Reducing Two-Dimensional SDS/Gel Electrophoresis.
Thioredoxin-reduced mBBr-labeled protein samples were dissolved in SDS sample buffer free of reducing agents (17). Gels (10–20% acrylamide gradient, 1.0-mm thickness) were prepared according to Laemmli (17) and subjected to electrophoresis in the first dimension for 16 h at constant current (7 mA). After electrophoresis, the narrow gel lane containing the separated proteins was excised from the gel and immersed in SDS sample buffer containing 5% mercaptoethanol for 20 min at room temperature. The gel strip then was applied horizontally to another gel (10–20% acrylamide gradient, 1.5-mm thickness), and electrophoresis was carried out in the second dimension (16 h at a constant current of 7 mA). The gel was immersed in 20% methanol that contained 5% acetic acid and examined under 365-nm UV light (Spectroline, Spectronic, Westbury, NY) to detect mBBr-labeled proteins. The gel then was incubated overnight at room temperature in 20% methanol containing 5% acetic acid and 0.025% Coomassie brilliant blue R-250, and finally was destained with a solution of 20% methanol and 5% acetic acid until the protein bands were visible.
Isoelectric Focusing/Reducing SDS/PAGE.
Isoelectric focusing and the subsequent SDS/PAGE were performed by using the Protean IEF Cell and Criterion Precast System (Bio-Rad) according to the instruction manual provided by the manufacturer. IPG (immobilized pH gradient gel) strips (pH 3–10) were swollen in rehydration buffer composed of 0.5% CHAPS (3-[3-cholamidopropyl)dimethylammonio]-1-propane-sulfonate), 8 M urea, 10 mM DTT, 0.1% Bio-Lytes, and 0.001% bromophenol blue. Thioredoxin-reduced mBBr-labeled protein samples were dissolved in 30 μl of rehydration buffer and applied to the rehydrated gel strip. Isoelectric focusing was performed in Protean IEF Cell (Bio-Rad) with 35,000 total voltage-hour and an upper voltage limit of 8,000 V. After that, the IPG strip was dipped in SDS-sample buffer containing 6 M urea and 130 mM DTT, and electrophoresis in the second dimension was performed on a Criterion Precast System.
In-Gel Digestion and Fractionation of Peptides.
Reduction/alkylation and trypsin in-gel digestion of mBBr-labeled proteins were carried out essentially by the procedure described by Shevchenko et al. (18). Extracted trpysin-digested peptides from gels were separated by microbore C18 reverse-phase column (1 mm × 25 cm; Vydac, Hesperia, CA) on ABI 172 HPLC system (Applied Biosystems). After injection of the sample, the column was washed with 95% solvent A (0.1% trifluoroacetic acid in water), 5% solvent B (0.075% trifluoroacetic acid in 70% acetonitrile) for 5 min for column equilibration and was eluted first with a gradient from 5% to 10% solvent B for 10 min, second with a linear gradient from 10% to 70% solvent B for 70 min that increased to 90% solvent B over 15 min.
Amino Acid Sequence Analysis of Peptides.
Sequence analysis of C18-purified peptides was performed by automated Edman degradation on an ABI model 494 Procise sequencer (Applied Biosystems). Nontarget proteins, including peanut Gly1, also were analyzed by nano-electrospray ionization tandem mass spectrometry (nano ESI/MS/MS) using a hybrid mass spectrometer QSTAR (Perkin–Elmer). Nano-spray capillaries were obtained from Protana (Odense, Denmark). For nano-ESI/MS/MS, in-gel digested peptide mixture was analyzed directly without any C18 column fractionation.
Results
Reduction of Proteins by Thioredoxin.
As seen previously with proteins from other seeds, thioredoxin h, reduced with NADPH via NTR, was effective in the reduction of peanut proteins. Based on results with one-dimensional electrophoresis gels, thioredoxin h appeared to show a preference for intramolecular disulfide bonds (data not shown).
To isolate and characterize the proteins targeted by thioredoxin and confirm the specificity of their disulfide bonds, we subjected the preparation to more complete two-dimensional separation procedures. We first applied nonreducing/reducing SDS/PAGE to identify the thioredoxin-linked proteins and determine the nature of their disulfide bonds. Then we analyzed the samples by isoelectric focusing/reducing SDS/gel electrophoresis for better resolution and a more complete analysis of the target proteins. In accord with longstanding findings from this laboratory, reduced glutathione was consistently found without effect (data not shown) (1, 7, 19).
Isolation of Thioredoxin Target Proteins: Nonreducing/Reducing Two-Dimensional SDS/Gel Electrophoresis.
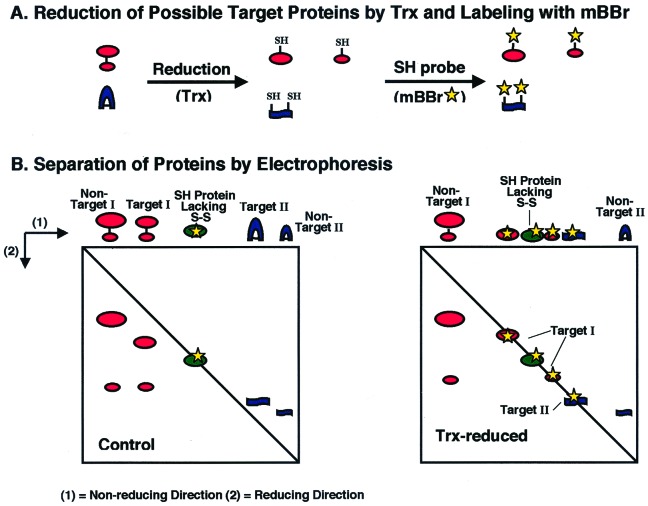
As shown in Fig. 1, the nonreducing/reducing SDS/gel electrophoresis system provides a direct means both to identify disulfide proteins and to determine the nature of their disulfide bonds. Proteins with either intermolecular or intramolecular disulfide bonds (potential targets I and II, respectively) that have undergone reduction before application to the gel (Fig. 1A) resemble proteins without disulfide bonds and are recovered on the diagonal line (line of protein monomers) (Fig. 1B Right). By contrast, if the proteins are not reduced before electrophoresis in the first dimension, those having intermolecular disulfide bonds are recovered below the diagonal line, owing to dissociation and the attendant decrease in molecular mass after reduction in the second dimension (Fig. 1B Left). Counterparts with intramolecular disulfide bonds, on the other hand, are recovered above the diagonal line as a result of the change in the apparent migration in reducing SDS/gel electrophoresis after reduction. An analysis of protein-stained gels thus can serve to identify not only proteins with disulfide bonds, but also the nature of these bonds. The nonreducing/reducing two-dimensional gel system has been widely used in the past for this purpose (20).
Figure 1.
Distinction between proteins with intramolecular vs. intermolecular disulfide bonds after labeling with mBBr (A) and nonreducing/reducing two-dimensional SDS/gel electrophoresis (B). Trx = thioredoxin.
In the present study, the primary objective was somewhat different. The gel system was used to identify not all disulfide proteins, but only those targeted by a specific reductant, thioredoxin h, using mBBr as a probe. The bond that had been reduced in these target proteins could then be shown to be either intermolecular or intramolecular (Fig. 1B Right).
Specificity of Proteins for Thioredoxin.
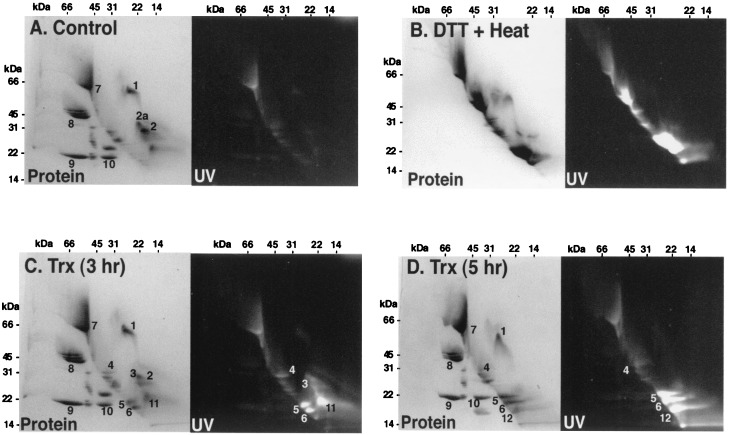
Application of the nonreducing/reducing two-dimensional gel system to peanut extract revealed a preponderance of proteins with intermolecular disulfide bonds (below the line) but a significant number of proteins with intramolecular counterparts (above the line) (Fig. 2A). Relatively few of the proteins had free sulfhydryl groups. When the sample was boiled with DTT, both the intermolecular and intramolecular bonds were fully reduced so that all visible proteins became highly labeled and migrated on the diagonal line (Fig. 2B). By contrast, in the presence of NADPH, NTR, and thioredoxin h, fewer proteins were reduced. Thioredoxin h reduced 3 of the 4 visible proteins with intramolecular disulfide bonds (nos. 4–6) after 3-h incubation (Fig. 2C). However, even with this prolonged incubation time, some proteins appeared to be only partially reduced as they still appeared above the diagonal line (nos. 3 and 11). Extending the incubation time to 5 h gave essentially full reduction as most of the proteins with intramolecular disulfide bonds then fell on the diagonal line (nos. 4–6 and 12) (Fig. 2D). Even after a 5-h incubation, one of the major proteins containing intramolecular disulfide bonds (no. 1) was not reduced by thioredoxin h.
Figure 2.
Analysis of peanut seed proteins after reduction by the NADP/thioredoxin system or DTT and labeling with mBBr using nonreducing/reducing two-dimensional SDS/gel electrophoresis. Fifty micrograms of peanut protein (20 μl extract) was incubated in 50 mM Tris⋅HCl buffer, pH 7.9, in a final volume of 100 μl. (A) Control: no addition. (B) DTT + heat: the sample was heated in boiling water after addition of 5 mM DTT. (C) Thioredoxin (Trx) (3 h): incubation for 3 h at 37°C in the presence of 0.125 μmol NADPH, 2.4 μg C. reinhardtii thioredoxin h and 2.1 μg A. thaliana NTR. (D) Thioredoxin (Trx) (5 h): incubation under the same conditions as C, except for 5 h.
The length of time required for full reduction indicates that some peanut proteins are difficult to reduce relative to other proteins that have been examined (7–9, 16, 21–23). An alternate explanation is that peanut proteins are highly specific for native thioredoxin h. Although possible, this explanation seems unlikely in view of the finding that E. coli thioredoxin gave essentially the same reduction profile as that in Fig. 2 for thioredoxin h from Chlamydomonas (data not shown).
After 3 and 5 h, five proteins with intramolecular disulfide bonds were reduced by thioredoxin (nos. 3–6 and 11, that apparently was converted to 12), whereas only one such protein (no. 1) was resistant to reduction and was recovered above the diagonal line (Fig. 2D). By contrast, it appeared that all visible proteins with intermolecular disulfide bonds resisted reduction by thioredoxin based on the finding that none showed a change in migration so as to travel on the diagonal line (e.g., nos. 8–10, Fig. 2 A and D). This finding confirms earlier results with the castor seed 2S protein in showing that thioredoxin preferentially reduced intramolecular disulfide bonds (21). As expected, all of the disulfide proteins—those with intramolecular or intermolecular disulfide bonds—were reduced when heated in the presence of DTT (Fig. 2B).
Isolation of Thioredoxin Target Proteins: Isoelectric Focusing/Reducing SDS/Gel Two-Dimensional Electrophoresis.
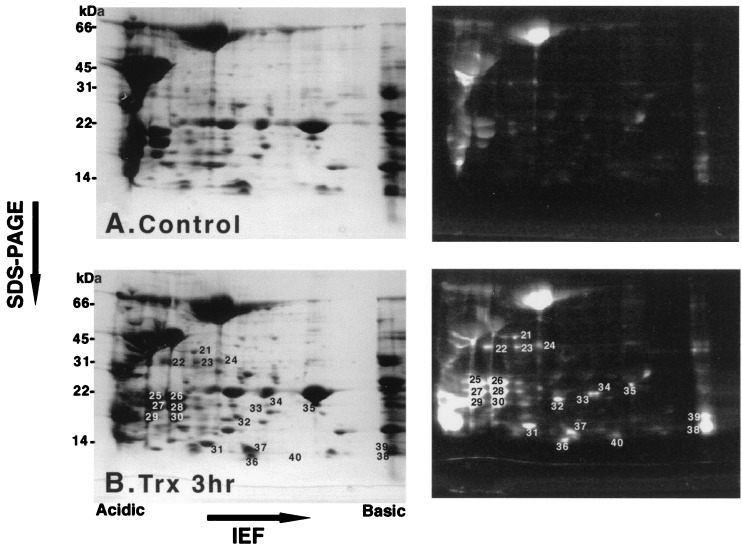
The proteins that had been extracted from peanut seeds and then reduced by thioredoxin also were subjected to isoelectric focusing/reducing SDS/gel two-dimensional electrophoresis after labeling with mBBr. As shown in Fig. 3, at least 20 proteins were reduced by thioredoxin (numbering starts with 21). This system appears, therefore, to give more complete separation than nonreducing/reducing two-dimensional gel electrophoresis (compare Figs. 2 and 3). The latter method, however, offers the advantage of demonstrating reductant specificity for intramolecular vs. intermolecular disulfide bonds.
Figure 3.
Analysis of peanut seed proteins after reduction by the NADP/thioredoxin system and labeling with mBBr using isoelectric focusing/reducing SDS/PAGE/mBBr. One hundred fifty micrograms of peanut protein (60 μl extract) was incubated in 50 mM Tris⋅HCl buffer, pH 7.9, in a final volume of 300 μl. (A) Control: no addition. (B) Thioredoxin (Trx) (3 h): incubation for 3 h in the presence of 0.375 μmol NADPH, 7.2 μg C. reinhardtii thioredoxin h, 6.3 μg A. thaliana NTR. IEF, isoelectric focusing.
Identification of Disulfide Proteins.
The thioredoxin target proteins identified in peanut seed extract after separation by each of the two gel systems are discussed below.
Nonreducing/reducing two-dimensional SDS/gel electrophoresis.
Protein spots 1–10 shown in Fig. 2 were excised from the gels and subjected to N-terminal or internal amino acid sequence analysis (Table 1). The resolved proteins were identified as vicilins, 2S proteins, and glycinins, which, respectively, correspond to allergens Ara h1 (24), h2 (25), h3 (26) plus h4 (27). Of these, thioredoxin reduced intramolecular disulfide bonds of Ara h2 (nos. 2, 3, and 6) such that the nonfluorescent, oxidized (no. 2), partly reduced (no. 3), and fully reduced (no. 6) forms were identified. Thioredoxin also reduced a fragment of an Ara h3 (no. 5) that appeared to be derived from no. 11. Under these conditions, thioredoxin was ineffective in reducing the three forms of Ara h4 (nos. 8–10). Ara h1 (no. 7) also was unaffected.
Table 1.
Internal amino acid sequence analysis of thioredoxin target and nontarget peanut proteins
| No. | Internal sequence | Homologous protein | Amino acid matches |
|---|---|---|---|
| Nonreducing/reducing two-dimensional SDS/PAGE target protein (Fig. 2 no.) | |||
| 2 | QQWELQGDR | Peanut 2S protein (allergen Ara h2, oxidized) | 9/9 |
| 3 | ANLRPCcmEQHLMQ | Peanut 2S protein (allergen Ara h2, partially reduced) | 12/12 |
| NLPQQCcmGLR | 9/9 | ||
| 6 | RQQWEL | Peanut 2S protein (allergen Ara h2, fully reduced) | 6/6 |
| CmBBrCmBBrNELNEFENN | 11/11 | ||
| NLPQQCmBBrGLR | 9/9 | ||
| 5 | SQSENFEYVAFK | Peanut glycinin (allergen Ara h3 basic subunit) | 12/12 |
| 4a | ALGSVLAGDKDSLAYGR | Desiccation-related protein from resurrection plant | 11/17 |
| KLVAGLLGVESGQDAVIR | 15/18 | ||
| NEQVHPYGVSVATFTNR | 9/17 | ||
| ANLDAFTR | 5/8 | ||
| ISILR | 4/5 | ||
| LTXYVGTNPELQNP | 9/13 | ||
| DVILQFAYQEVGHLR | 11/15 | ||
| 4b | IAQFGSDVPMK | Seed maturation protein from soybean | 11/13 |
| AIAADLGYDENCmBBrK | 8/11 | ||
| IAVITGGDSGIGR | 12/13 | ||
| Nontarget proteins (Fig. 2 no.) | |||
| 7 | VQIEAKPNTLVLPK | Peanut vicilin (allergen Ara h1) | 14/14 |
| 8 | GYFGLIFPGCcmPSTYEEP | Peanut glycinin (allergen Ara h4 acidic subunit) | 17/17 |
| 9 | VYDEELQEGHVLVVPQN | Peanut glycinin (allergen Ara h4 basic subunit) | 17/17 |
| 10 | WLGLSAEYGNLYR | Peanut glycinin (allergen Ara h4 basic subunit) | 13/13 |
| 1 | XLSPDRK | Peanut glycinin (Gly1) | 6/6 |
| XFNLAGNHEQEFLR | 13/13 | ||
| XENESEEQGAIV | 12/12 | ||
| SPDIYNPQAGSLK | 13/13 | ||
| TANDLNLLILR | 11/11 | ||
| Isoelectric focusing/SDS/two-dimensional PAGE (Fig. 3 no.) | |||
| 31 | GKEEHKPIVVDF | Thioredoxin h from Chlamydomonas | 12/12 |
| 30 | XMGEQEQYDSYDIR | Peanut 2S protein (allergen Ara h6 isoform) | 10/13 |
The sole protein with an intramolecular disulfide bond that appeared to be resistant to reduction by thioredoxin (no. 1) contained partial amino acid sequences of both acidic and basic subunits of glycinin (Gly1, GenBank accession no. AF125192). Moreover, the molecular mass of this protein (65 kDa) was approximately equivalent to the molecular mass of the sum total of acidic and basic subunits of glycinin (40 and 25 kDa, respectively). These findings suggest that protein no. 1 is a proglycinin (28). It remains to be seen whether the proglycinin is an allergen as are its posttranslational acidic and basic cleavage products represented here by spot nos. 8 and 9 (Ara h3/h4) (26).
Spot no. 4 in Fig. 2C contained two proteins, both reduced by thioredoxin and labeled with mBBr. The two proteins, which were apparently derived from a poorly resolved spot just above no. 2 (no. 2a), were separated by electrophoresis with a 10% acrylamide gel (data not shown). Neither protein has previously been reported to occur in peanut. One of the proteins (no. 4a) showed sequence similarity to pcC 13–62, one of five desiccation-inducible proteins from the desiccation-resistant resurrection plant Craterostigma plantagineum (29, 30) (Table 1). This protein, which was reported to be synthesized de novo when leaves of the resurrection plant were desiccated, has been found to share similarity only with an Arabidopsis cDNA clone (GenBank accession no. AL162651). The finding of a closely related protein in peanut extends its distribution and raises the question of its function in seeds. Moreover, while previously found to have three cyst(e)ine residues, the resurrection protein was not known to have a disulfide bond. The above results show that the desiccation-related protein has an intramolecular disulfide bond that can be reduced by thioredoxin.
The second protein in the original spot no. 4 (no. 4b) shared similarity with seed maturation protein from barley and other seeds (31) (Table 1). The seed maturation protein shows both sequence homology and glucose and ribotol dehydrogenase activity. The present results raise the question whether reduction by thioredoxin h alters activity—i.e., whether thioredoxin acts to regulate the enzyme.
Isoelectric focusing/reducing SDS/gel two-dimensional electrophoresis.
Several proteins also were identified after the thioredoxin-reduced and mBBr-labeled peanut extract was subjected to isoelectric focusing/reducing SDS/gel electrophoresis (Fig. 3). Amino acid sequence analysis showed protein no. 30 to be an Ara h6 (27) isoform, an allergen in peanut, and no. 31 to be the Chlamydomonas thioredoxin that had been added to the preparation. The remaining targets require further investigation.
Discussion
One of the factors limiting our understanding of thioredoxin function is the lack of information on target proteins and enzymes (32). In the original studies on photosynthesis, target enzymes were identified by chance (e.g., chloroplast fructose bisphosphatase) or by showing that an enzyme activated either by light in photosynthetic cells or chloroplasts (e.g., phosphoribulokinase) or by DTT in vitro (e.g., NADP-malate dehydrogenase) could be similarly activated by reduced thioredoxin (33). The application of mBBr to label the sulfhydryl groups newly generated by thioredoxin in either known individual proteins (22, 34) or protein families (7), in combination with one-dimensional gel electrophoresis, led to the identification of a number of targets that are independent of light and primarily serve a degradative, storage, or protective function in seeds (1, 7, 10).
The extension of the application of mBBr to two-dimensional gel systems in combination with amino acid sequencing enables the isolation and identification of unknown thioredoxin target proteins. This strategy, shown here for proteins from peanut, can be applied to all types of cells (plant, animal, microbial) as well as to the soluble and membranous fractions of organelles known to contain thioredoxin—e.g., chloroplasts, mitochondria, nuclei (4). By taking advantage of emerging proteomic technologies, it should be possible in the future to identify target proteins occurring at quite low levels and thereby give insight into how thioredoxin, including different isoforms, acts in processes in which its role remains mysterious, e.g., cell division (35, 36). In uncovering additional thioredoxin target proteins, the present strategy complements gene-based approaches that have been applied successfully to thioredoxin, namely gene inactivation (37), gene overexpression (11), yeast complementation (38), and cassette mutagenesis designed to trap thioredoxin-enzyme intermediates (32, 39). In addition to taking advantage of proteomics, it is possible that the current strategy could help give direction to this emerging field (40) by focusing on redox changes taking place in proteins of various systems, such as those involving abiotic stress or disease.
One of the gel systems used above (two-dimensional nonreducing/reducing SDS/PAGE) permits not only the identification of a new thioredoxin target, but also the type of disulfide bond reduced, intramolecular vs. intermolecular. In peanut, thioredoxin was found to act preferentially on intramolecular disulfide bonds, in keeping with earlier experience (1, 7, 21). Further application of the two-dimensional gel/mBBr/amino acid sequencing strategy should provide corresponding information on thioredoxin-linked proteins in other systems.
The present results warrant comment with respect to peanut proteins per se. Peanut was selected for the current study because of our interest in seeds and allergens, and because, unlike cereals, for example, its proteins are primarily water soluble. The targets identified include three allergens (Ara h2, h3, h6) and two proteins previously not known to occur in peanut (desiccation-related protein, seed maturation protein). Two of the allergens (Ara h3, Ara h6) and the two proteins newly described for peanut were previously not known to be linked to thioredoxin. The finding of such a link to these proteins raises new questions relating to the role of thioredoxin in seeds (1) and to its ability to inactivate allergens reductively (16, 23), an effect of potential technological significance.
Concluding Remarks.
In uncovering additional proteins targeted by thioredoxin h in peanut, the strategy described above opens a door to understanding thioredoxin function in other systems. It should now be possible to gain additional information on the role of thioredoxin in organs and organelles about which little is known, e.g., brain (41) and nuclei (42), as well as in those for which knowledge is more advanced (e.g., seeds and chloroplasts) (1–3). Such information is pivotal both to understanding the role of thioreodxin in biology and to extending its application in technology and medicine (43).
Acknowledgments
We thank Dr. Jean-Pierre Jacquot, Université de Nancy I, Vandoeuvre, France for the gift of Chlamydomonas thioredoxin h and Arabidopsis NTR. This work was supported by funds from Syngenta, Inc. and a fellowship to H.Y. from the Japan Science and Technology Corporation.
Abbreviations
- mBBr
monobromobimane
- NTR
NADP-thioredoxin reductase
References
- 1.Besse I, Buchanan B B. Bot Bull Acad Sin (Taipei) 1997;38:1–11. [Google Scholar]
- 2.Ruelland E, Miginiac-Maslow M. Trends Plant Sci. 1999;4:136–141. doi: 10.1016/s1360-1385(99)01391-6. [DOI] [PubMed] [Google Scholar]
- 3.Schürmann P, Jacquot J P. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:371–400. doi: 10.1146/annurev.arplant.51.1.371. [DOI] [PubMed] [Google Scholar]
- 4.Arner E S J, Holmgren A. Eur J Biochem. 2000;267:6102–6209. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 5.Suske G, Wagner W, Follmann H. Z Naturforsch. 1979;34:214–221. [Google Scholar]
- 6.Florencio F J, Yee B C, Johnson T C, Buchanan B B. Arch Biochem Biophys. 1988;266:496–507. doi: 10.1016/0003-9861(88)90282-2. [DOI] [PubMed] [Google Scholar]
- 7.Kobrehel K, Wong J H, Balogh A, Kiss F, Yee B C, Buchanan B B. Plant Physiol. 1992;99:919–924. doi: 10.1104/pp.99.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lozano R M, Wong J H, Yee B C, Peters A, Kobrehel K, Buchanan B B. Planta. 1996;200:100–106. [Google Scholar]
- 9.Jiao J A, Yee B C, Wong J H, Kobrehel K, Buchanan B B. Plant Phys Biochem. 1993;31:799–804. [Google Scholar]
- 10.Besse I, Wong J H, Kobrehel K, Buchanan B B. Proc Natl Acad Sci USA. 1996;93:3169–3175. doi: 10.1073/pnas.93.8.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho M-J, Wong J H, Marx C, Jiang W, Lemaux P G, Buchanan B B. Proc Natl Acad Sci USA. 1999;96:14641–14646. doi: 10.1073/pnas.96.25.14641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allore R J, Barber B H. Anal Biochem. 1984;137:523–527. doi: 10.1016/0003-2697(84)90121-0. [DOI] [PubMed] [Google Scholar]
- 13.O'Farrel P H. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 14.Shokraii E H, Esen A. J Agric Food Chem. 1992;40:1491–1495. [Google Scholar]
- 15.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 16.Buchanan B B, Adamidi C, Lozano R M, Yee B C, Momma M, Kobrehel K, Ermel R, Frick O L. Proc Natl Acad Sci USA. 1997;94:5372–5377. doi: 10.1073/pnas.94.10.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 19.Buchanan B B, Schürmann P, Kalberer P P. J Biol Chem. 1971;246:5952–5959. [PubMed] [Google Scholar]
- 20.Endler A T, Tracy R P. Cancer Invest. 1987;5:127–149. doi: 10.3109/07357908709018467. [DOI] [PubMed] [Google Scholar]
- 21.Shin S H, Wong J H, Kobrehel K, Buchanan B B. Planta. 1993;189:557–560. [Google Scholar]
- 22.Kobrehel K, Yee B C, Buchanan B B. J Biol Chem. 1991;266:16135–16140. [PubMed] [Google Scholar]
- 23.del Val G, Yee B C, Lozano R M, Buchanan B B, Ermel R W, Lee Y M, Frick O L. J Allergy Clin Immunol. 1999;103:690–697. doi: 10.1016/s0091-6749(99)70244-7. [DOI] [PubMed] [Google Scholar]
- 24.Burks A W, Cockrell G, Stanley J S, Helm R M, Bannon G A. J Clin Invest. 1995;96:1715–1721. doi: 10.1172/JCI118216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanley J S, King N, Burks A W, Huang S K, Sampson H, Cockrell G, Helm R M, West C M, Bannon G A. Arch Biochem Biophys. 1997;342:244–253. doi: 10.1006/abbi.1997.9998. [DOI] [PubMed] [Google Scholar]
- 26.Rabjohn P, Helm E M, Stanley J S, West C M, Sampson H A, Burks A W, Bannon G A. J Clin Invest. 1999;103:535–542. doi: 10.1172/JCI5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleber-Janke T, Crameri R, Appenzeller U, Schlaak M, Becker W M. Int Arch Allergy Immunol. 1999;119:265–274. doi: 10.1159/000024203. [DOI] [PubMed] [Google Scholar]
- 28.Hinz G, Menze A, Hohl I, Vaux D. J Exp Bot. 1997;48:139–149. [Google Scholar]
- 29.Bernacchia G, Salamini F, Bartels D. Plant Physiol. 1996;111:1043–1050. doi: 10.1104/pp.111.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piatkowski D, Schneider K, Salamini F, Bartels D. Plant Physiol. 1990;94:1682–1688. doi: 10.1104/pp.94.4.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexander R, Alamillo J M, Salamini F, Bartels D. Planta. 1994;192:519–525. doi: 10.1007/BF00203590. [DOI] [PubMed] [Google Scholar]
- 32.Meyer Y, Verdoucq L, Vignols F. Trends Plant Sci. 1999;4:388–394. doi: 10.1016/s1360-1385(99)01475-2. [DOI] [PubMed] [Google Scholar]
- 33.Buchanan B B. Annu Rev Plant Physiol. 1980;31:341–374. [Google Scholar]
- 34.Jiao J A, Yee B C, Kobrehel K, Buchanan B B. J Agric Food Chem. 1992;40:2333–2336. [Google Scholar]
- 35.Muller E G D. Arch Biochem Biophys. 1995;318:356–361. doi: 10.1006/abbi.1995.1240. [DOI] [PubMed] [Google Scholar]
- 36.Hartman H, Wu M, Buchanan B B, Gerhart J C. Proc Natl Acad Sci USA. 1993;90:2271–2275. doi: 10.1073/pnas.90.6.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller E G D, Buchanan B B. J Biol Chem. 1989;264:4008–4014. [PubMed] [Google Scholar]
- 38.Mouaheb N, Thomas D, Verdoucq L, Monfort P, Meyer Y. Proc Natl Acad Sci USA. 1998;95:3312–3317. doi: 10.1073/pnas.95.6.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verdoucq L, Vignols F, Jacquot J P, Chartier Y, Meyer Y. J Biol Chem. 1999;274:19714–19722. doi: 10.1074/jbc.274.28.19714. [DOI] [PubMed] [Google Scholar]
- 40.Oliver S. Nature (London) 2000;403:601–603. doi: 10.1038/35001165. [DOI] [PubMed] [Google Scholar]
- 41.Takagi Y, Mitsui A, Nishiyama A, Nozaki K, Sono H, Gon Y, Hashimoto N, Yodoi J. Proc Natl Acad Sci USA. 1999;96:4131–4136. doi: 10.1073/pnas.96.7.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rozell B, Holmgren A, Hansson H A. Eur J Cell Biol. 1988;46:470–477. [PubMed] [Google Scholar]
- 43.Buchanan B B, Schürmann P, Decottignies P, Lozano R M. Arch Biochem Biophys. 1994;314:257–260. doi: 10.1006/abbi.1994.1439. [DOI] [PubMed] [Google Scholar]