Abstract
We dissected the complete genome sequence of the O1 serotype strain Vibrio anguillarum 775(pJM1) and determined the draft genomic sequences of plasmidless strains of serotype O1 (strain 96F) and O2β (strain RV22) and V. ordalii. All strains harbor two chromosomes, but 775 also harbors the virulence plasmid pJM1, which carries the anguibactin-producing and cognate transport genes, one of the main virulence factors of V. anguillarum. Genomic analysis identified eight genomic islands in chromosome 1 of V. anguillarum 775(pJM1) and two in chromosome 2. Some of them carried potential virulence genes for the biosynthesis of O antigens, hemolysins, and exonucleases as well as others for sugar transport and metabolism. The majority of genes for essential cell functions and pathogenicity are located on chromosome 1. In contrast, chromosome 2 contains a larger fraction (59%) of hypothetical genes than does chromosome 1 (42%). Chromosome 2 also harbors a superintegron, as well as host “addiction” genes that are typically found on plasmids. Unique distinctive properties include homologues of type III secretion system genes in 96F, homologues of V. cholerae zot and ace toxin genes in RV22, and the biofilm formation syp genes in V. ordalii. Mobile genetic elements, some of them possibly originated in the pJM1 plasmid, were very abundant in 775, resulting in the silencing of specific genes, with only few insertions in the 96F and RV22 chromosomes.
INTRODUCTION
Vibrio anguillarum is a marine pathogen that causes vibriosis in close to 50 species of fish, including cultured and wild fish, mollusks, and crustaceans, in marine, brackish, and fresh water (1). Vibriosis is a hemorrhagic septicemia with dire consequences for fish rearing, especially in countries that depend heavily on fish for their food consumption. Despite the fact that V. anguillarum is a dramatic cause of vibriosis in fish, little is known about the genomic composition of this important pathogen. Although 23 serotypes have been reported in V. anguillarum, the O1 and O2 serotypes are the major causative agent of fish vibriosis (32, 66, 75). Many O1 serotype strains harbor 65-kb pJM1-type plasmids, which carry the siderophore anguibactin biosynthesis and transport genes, a main virulence factor of V. anguillarum, while one of the O1 serotype strains and other serotypes, such as all of the O2 strains, are plasmidless (1, 13, 33, 76). Many virulence factors have been characterized, but we are still far from getting the whole picture of the virulence mechanisms of this pathogen (1, 50). The fact that the pJM1 plasmid is an important component of virulence for the 775 strain but that the other three strains examined do not harbor this plasmid and are still virulent indicates that they must have different mechanisms to cause disease. Moreover, O1 serotype strains cause disease in salmonid fish, whereas O2β strains are usually isolated from cod and other nonsalmonids (1, 32, 43). We have previously performed random genome sequencing of V. anguillarum 775 genomic DNA and identified potential virulence factors (61). Some of the genes identified in that study, such as hemolysins and the Rtx toxin, were later confirmed as virulence factors of this bacterium (34, 60).
In this study, we have determined the whole genome sequence of V. anguillarum 775(pJM1) and have done comparative genomics with virulent plasmidless strains of serotype O1 and O2β and with V. ordalii. The last is highly related to V. anguillarum and was previously recognized as V. anguillarum biotype 2 (64). Schiewe et al. (63) proposed the name V. ordalii based on cultural and biochemical characteristics, as well as DNA homology with biotype 1. These experiments demonstrated that V. anguillarum and V. ordalii are 58 to 69% related based on DNA-DNA hybridization (64). Histopathological studies have shown that vibrioses caused by V. anguillarum and V. ordalii are dramatically different (59). In early stages of infection, V. anguillarum strains cause histopathological changes in blood, loose connective tissue, kidney, spleen, gills, and posterior gastrointestinal tract, and these bacteria are most abundant in the blood, although they appear uniformly dispersed throughout the affected tissues. Conversely, V. ordalii develops the bacteremia only at late stages of disease, and the concentration of bacterial cells in blood is normally less than in the V. anguillarum infection. Tissues displaying histopathological changes after infection with V. ordalii are skeletal and cardiac muscles, anterior and posterior gastrointestinal tract, and gills, and the bacteria are not evenly dispersed but are present within tissues as colonies or aggregates of cells. In this work, we identified common and strain-specific virulence genes from V. anguillarum and V. ordalii that could explain the virulence mechanisms of these bacteria and that would be useful for future development of vaccines.
MATERIALS AND METHODS
Genome sequence data.
We sequenced the genomes of Vibrio anguillarum 775 (ATCC 68554), 96F, and RV22 and V. ordalii ATCC 33509. The V. anguillarum 775 strain was isolated from Coho salmon (Oncorhynchus kisutch) in the U.S. Pacific Ocean coast (14), strain 96F was isolated from striped bass (Morone saxatilis) in the United States (76), and strain RV22 was isolated from turbot (Scophthalmus maximus) in Spain (33). In addition, we sequenced a strain of V. ordalii ATCC 33509 isolated from Coho salmon (Oncorhynchus kisutch) kidney in Puget Sound, WA (63). There is some confusion in the literature as to the genus name for Vibrio anguillarum, because it was suggested that it should be renamed Listonella (37). However, workers in the vibrio field have been using the classical Vibrio name for the genus. Recently, we demonstrated that Vibrio anguillarum is a bona fide Vibrio species, and we proposed the use of V. anguillarum instead of the more recent heterotype Listonella for this bacterium (69).
Library, sequencing, gap closure, and assembly.
To complete the genome, V. anguillarum 775 was sequenced using Roche 454 technology. Whole genomic DNA was subjected to 454 Titanium sequencing with 32-fold coverage. For gap closure, we generated amplicons using long-range PCR and sequence assembly by primer walking and prepared cosmid and fosmid libraries for fosmid (cosmid) end sequencing, scaffold assembly on gapped genome, and primer walking sequence assembly. For homopolymers, we searched for regions with >5 identical bases in a row and then cloned the region and confirmed by Sanger capillary sequencing using an AB 3130XL instrument in the MMI Core Facility.
The reads were assembled using Newbler software version 2.3 (Roche/454 Life Sciences, Branford, CT) combined with the generation of cosmid and fosmid libraries using SuperCos1 (Stratagene, La Jolla, CA) and a CopyControl fosmid library production kit (Epicentre, Madison WI) using total DNA from V. anguillarum 775. The cosmid and fosmid DNA was purified and subjected to end sequencing by the Sanger method. For gap closure of the initial 71 contigs, comparative genomics with closely related Vibrio genomes was used to design amplicons using primers from the contig ends. The amplicons were amplified using PCR methods. Draft genome sequencing of V. anguillarum 96F and RV22 and V. ordalii ATCC 33509 was also performed using the Roche 454 technology, and the reads were also assembled using Newbler software version 2.3 (Roche/454 Life Sciences) of the 454 Titanium pyrosequencing approach, resulting in the following contigs: 220 for 96F, 362 for RV22, and 281 for V. ordalii. The average coverage for all four genomes was 32-fold.
Annotation and comparative genomics.
The complete genome sequence of V. anguillarum 775 and the draft genomic sequences of 96F, RV22, and V. ordalii ATCC 33509 were annotated by the Rapid Annotation System Technology (RAST) server (4). The annotation is based on subsystems, i.e., a set of functional roles that an annotator has decided should be thought of as related. Frequently, subsystems represent the collection of functional roles that make up a metabolic pathway, a complex (e.g., the ribosome), or a class of proteins (e.g., two-component signal transduction proteins). The steps in annotation by RAST are as follows. First, the call of tRNAs and rRNAS, then an initial call of open reading frames (ORFs), is performed using GLIMMER2 (17). After an initial prediction, a set of representative sequences universal or nearly universal in prokaryotes, for example, the tRNA synthetases, is used to search protein-encoding genes from the new genome. The outcome can be used to estimate the closest phylogenetic neighbors of the newly sequenced genome and to confirm the putative gene prediction by GLIMMER2. The next step is an initial metabolic reconstruction, reflecting basic divisions of function, producing detailed estimates of the genome contents that were successfully connected to the subsystems. To perform the comparative analysis, we use the RAST Compare Metabolic Reconstruction tool. This tool utilizes the BLAST algorithm to perform the analysis. In addition, we used the GeneWiz browser 0.94 server to visualize all of the genomes (24).
Prediction of replication origins.
To predict the origins of replication of the chromosomes, we used the DoriC tool, which utilizes z-curve analysis for nucleotide distribution asymmetry, DnaA box distribution, genes adjacent to candidate oriCs, and phylogenetic relationships (89).
GIs and determination of dinucleotide relative abundance values.
To predict the genomic islands (GIs), we used the IslandViewer tool (31). The tool utilizes SIGI-HMM and IslandPath-DIMOB methods (26, 79). The SIGI-HMM method uses a hidden Markov model (HMM) and measures codon usage to identify possible genomic islands. The IslandPath method identifies genomic islands by visualizing several common characteristics of GIs, such as abnormal sequence composition or the presence of genes that are functionally related to mobile elements. We determined the dinucleotide relative abundance value for each island predicted by IslandViewer. Mononucleotide and dinucleotide frequencies were calculated using COMPSEQ (EMBOSS). Dinucleotide relative abundances (ρ*XY) were calculated using the equation ρ*XY = fXY/fXfY where fXY denotes the frequency of dinucleotide XY, and fX and fY denote the frequencies of X and Y, respectively (28). Statistical theory and data from previous studies indicate that the normal range of ρ*XY is between 0.78 and 1.23 (28). The difference in genome signature between two sequences is expressed by the genomic dissimilarity (δ*), which is the average of the relative abundance in the dinucleotide difference between two sequences. The dissimilarities in relative abundance of dinucleotides between both sequences were calculated using the equation described by Karlin et al. (28): δ*(f,g) = 1/16Σ∣ρ*XY(f) − ρ*XY(g)∣ (multiplied by 1,000 for convenience), where g indicates the island genome and the sum extends over all dinucleotides.
Nucleotide sequence accession numbers.
The complete genome of V. anguillarum 775 has been deposited at DDBJ/EMBL/GenBank under the accession numbers CP002284 to CP002285. The sequences of the genomes of V. anguillarum 96F and RV22 and V. ordalii ATCC 33509 have been deposited at DDBJ/EMBL/GenBank under the accession numbers AEZA00000000, AEZB00000000, and AEZC00000000, respectively.
RESULTS
Vibrio anguillarum 775 genome.
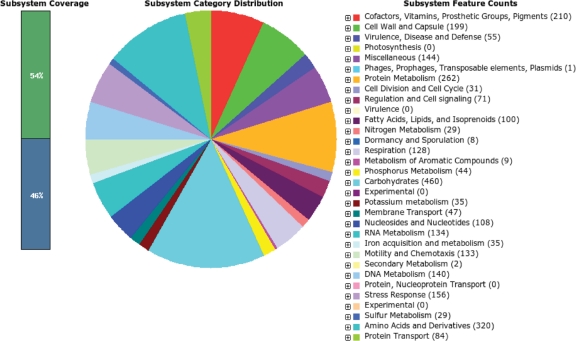
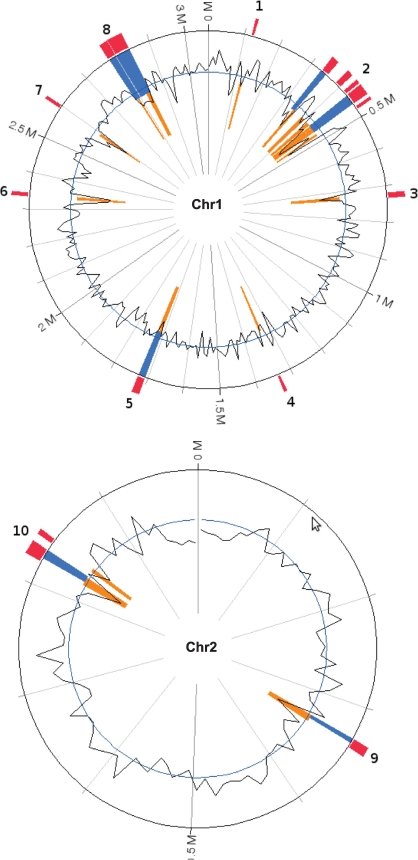
Table 1 and Fig. 1 show that the complete genome sequence of V. anguillarum 775 consists of two circular chromosomes, chromosome 1 (Chr1) of 3,063,912 bp and chromosome 2 (Chr2) of 988,135 bp, in addition to the 65,009-bp pJM1 plasmid, which carries anguibactin biosynthesis and ferric anguibactin transport genes (19). V. anguillarum 775 contains 2,093 (54%) products (protein) identified in subsystems and 1,783 (46%) not included in subsystems, indicating a large portion of proteins with unknown function according to the RAST method. The GC contents of Chr1 and Chr2 are 45% and 44%, respectively, and that of the pJM1 plasmid is 43%. The total number of subsystems found in the 775 genome is 396. In V. cholerae El Tor N16691, the total number of subsystems is 423. The lower number in V. anguillarum could be due to the fact that this is the first complete sequencing of the V. anguillarum genome and that, therefore, knowledge about the subsystems needs to be enhanced. This total genome was predicted to have 3,876 ORFs in the two chromosomes and 65 ORFs on the pJM1 plasmid. Approximately 1,007 (26%) of the total number of chromosomal and plasmid genes represent hypothetical proteins. Figure 2 shows that the majority of gene products were classified into categories such as production and/or utilization of carbohydrates, amino acids, and derivatives, as well as protein metabolism. Table S1 in the supplemental material shows the comparison of the molecular parameters of the genomes of V. anguillarum 775 (4.1 Mb), 96F (4.0 Mb), and RV22 (4.0 Mb) and V. ordalii (3.4 Mb).
Table 1.
General features of the V. anguillarum 775 genome
| Feature | Result for: |
||
|---|---|---|---|
| Chromosome 1 | Chromosome 2 | pJM1 | |
| Size (bp) | 3,063,912 | 988,135 | 65,009 |
| No. of CDSb | 2,864 | 951 | 65 |
| GC content (%) | 44.65 | 44.09 | 42.57 |
| No. of tRNAs | 82 | 4 | 0 |
| No. of rRNAs | 22 | 0 | 0 |
| oriC localization (nt)c | 219251 . . . 219628 | 535047 . . . 535332 | Ori1 |
| oriC GC content (%) | 39.95 | 30.42 | |
| Length of oriC (bp) | 377 | 285 | |
| No. of dnaA boxes | 5 | 4 | |
| Genbank accession no. for: | |||
| ParA | VAA_00834 | VAA_00096 | AY312585.1a |
| ParB | VAA_00833 | VAA_00097 | AY312585.1a |
| Soj-like protein | VAA_03384 | ||
| MukBEF | VAA_02164-VAA_02166 | ||
| StbDE toxin-antitoxin | VAA_00010-VAA_00011 | ||
Accession number of plasmid pJM1.
CDS, coding sequences.
nt, nucleotides.
Fig. 1.
Atlas visualization of chromosomes 1 and 2 of strain 775. The GeneWiz browser 0.94 server was used to visualize all of the genomes (24). The reference genome is V. anguillarum 775, and the four outer circles represent the genomes from the other Vibrio strains. The Roman numerals represent genes that are absent from the 96F and RV22 strains and V. ordalii. The numbers in the table at left correspond to the intensity of the colors.
Fig. 2.
Distribution of subsystems found in the V. anguillarum 775 genome. Subsystem coverage is indicated by green and blue bars. The green bar corresponds to the percentage of the proteins included in the subsystems, and the blue bar corresponds to the percentage of the proteins that are not included in the subsystems. The complete genome sequence of V. anguillarum 775 and the draft genomic sequences of 96F, RV22, and V. ordalii ATCC 33509 were annotated using the Rapid Annotation System Technology (RAST) server (4).
Distribution of origins of chromosomes, plasmids, and partition genes in the 775 strain.
Table 1 shows the origins of replication identified as follows. V. anguillarum ori1 (ori1Va) in Chr1 is approximately 39.95% GC, with 5 dnaA boxes, and is close to the dnaA gene. ori2Va in Chr2 is approximately 30.42% GC, with 4 dnaA boxes, and is close to the parA gene. We also identified parA and parB homologues in both chromosomes and in the pJM1 plasmid, although Chr1 encodes exclusively partition proteins, such as Soj-like protein and MukBEF, and Chr2 has StbDE as a toxin-antitoxin system. The origin of pJM1 DNA replication has been located adjacent to ORF50 (9).
Genes of interest in V. anguillarum 775.
We first investigated the location of housekeeping and other essential genes and found the majority located on Chr1, as is the case in other vibrios (data not shown). We also analyzed proven and potential virulence-associated and environmentally associated genes and their distribution in the 775 chromosomes and the plasmid pJM1. Table 2 shows that the majority of the ferric anguibactin biosynthesis and transport genes are present in the pJM1 plasmid, with the exception of vabA, which is essential for anguibactin biosynthesis but resides on Chr1 (3). The TonB1 and TtpC-TonB2 clusters, which are essential for iron transport, are encoded on Chr1 (67, 68). The fur gene is located on Chr1, which also harbors the heme determinants, although we have also identified a heme-related V. cholerae hutR homologue on Chr2 (42, 48, 49, 70). Genes involved in ferric vanchrobactin and/or ferric enterobactin transport are located on Chr1, although vanchrobactin biosynthesis in 775 is abrogated by a mutation in vabF and there are no biosynthesis genes for enterobactin (51). It is of interest that potential ABC transporters FvtB-FvtE for vanchrobactin and enterobactin uptake are found on Chr2. Following the trend of other genes essential for iron transport, Chr1 also harbors the genes feoABC for ferrous iron, fbpABC for ferric iron, and fhuABCDE for ferrichrome transport, which are homologues to those of V. cholerae (62, 85).
Table 2.
V. anguillarum and V. ordalii genes proven to be or potentially associated with virulence or the environment
| Gene category and name | Presence/absence of geneb in V. anguillarum 775: |
GenBank accession no. | Presence/absence of geneb in: |
||||
|---|---|---|---|---|---|---|---|
| Chr1 | Chr2 | pJM1 | 96F | RV22 | V. ordalii | ||
| Anguibactin biosynthesis and iron transport | |||||||
| angA-angE, angGHMNRTU | — | — | x | AY312585.1a | — | — | — |
| vabA-vabE | x | — | — | VAA_02392-VAA_02395, VAA_02076 | x | x | x |
| fatA-fatD | — | — | x | AY312585.1a | — | x | — |
| ttpC, tonB2, exbB2, exbD2 | x | — | — | VAA_01800-VAA_01804 | x | x | — |
| tonB1, exbB, exbD1 | x | — | — | VAA_01389-VAA_01387 | x | x | x |
| Utilization protein for unknown catechol siderophore | — | x | — | VAA_01367 | x | x | x |
| fur | x | — | — | VAA_03429 | x | x | x |
| Heme transport | |||||||
| TonB-dependent heme receptor HutR gene | — | x | — | VAA_02922 | x | x | — |
| huvA-huvD, huvSXZ | x | — | — | VAA_01392, VAA_01385-VAA_01383, VAA_01390-VAA_01391 | x | x | x (except huvXZ) |
| Ferric enterobactin and ferric vanchrobactin transport | |||||||
| fvtA | x | — | — | VAA_02079 | x | x | x |
| fvtB-fvtE | — | x | — | VAA_03000-VAA_03003 | x | x | — |
| fetAB | x | — | — | VAA_00188-VAA_00189 | x | x | — |
| Ferrous and ferric transport | |||||||
| feoABC | x | — | — | VAA_03398-VAA_03400 | x | x | x |
| fbpCBA | x | — | — | VAA_01820-VAA_01823, VAA_00769-VAA_00767 | x | x | x |
| Ferrichrome | |||||||
| fhuACBD | x | — | — | VAA_02423-VAA_02426 | x | x | Only FhuA |
| Hemolysins | |||||||
| Hemolysin genes | x | — | — | VAA_00718 | x | x | x |
| Putative hemolysin genes | x | — | — | VAA_02701 | x (4) | x (4) | x (1) |
| 21-kDa hemolysin precursor gene | x | — | — | VAA_03731 | x | x | x |
| vah1 | — | x | — | VAA_02992 | x | x | x |
| vah2-vah5 | x | — | — | VAA_01090, VAA_03209, VAA_03298, VAA_00294 | x | x (except vah4) | x (except vah4) |
| mltD | x | — | — | VAA_03554 | x | x | x |
| RTX toxins | |||||||
| rtxEDBHCGA | x | — | — | VAA_03091-VAA_03085 | x | x | x |
| RTX toxin and related Ca2+-binding protein genes | — | x | — | VAA_02949 | x | x | x |
| RTX toxin gene | x | — | — | VAA_02070 | x | — | — |
| Metalloproteases | |||||||
| empA | — | x | — | VAA_01306 | x | x | x |
| prtV | — | x | — | VAA_02995 | x | x | x |
| Zinc protease, insulinase family gene | — | x | — | VAA_01003 | x | x | — |
| Secreted enzymes | |||||||
| Phospholipase C gene | — | x | — | VAA_01330 | x | — | — |
| Nonhemolytic enterotoxin lytic component L1 genec | — | x | — | VAA_00241-VAA_00240 | x | x | x |
| Microbial collagenase gene, secreted | — | x | — | VAA_02998 | x | x | — |
| Lysophospholipase L2 gene | x | — | — | VAA_01963 | x | x | x |
| Lipase family protein gene | — | x | — | VAA_01024 | x | x | x |
| Motility and chemotaxis | |||||||
| flaEDBGI | x | — | — | VAA_03469-VAA_03465 | x | x | x |
| fliS, fleSQ | x | — | — | VAA_03463-VAA_03459 | x | x | x |
| fliE-fliR, flhB | x | — | — | VAA_03459-VAA_03443 | x | x | x |
| flgTOPNMA | x | — | — | VAA_03532-VAA_03526 | x | x | x |
| cheVR | x | — | — | VAA_03525-VAA_03524 | x | x | x |
| flgB-flgL | x | — | — | VAA_03522-VAA_03512 | x | x | x |
| flaAC | x | — | — | VAA_03511-VAA_03510 | x | x | x |
| flhA | x | — | — | VAA_03392 | x | x | x |
| fliL | x | — | — | VAA_00455 | x | x | x |
| rpoHN | x | — | — | VAA_00451, VAA_02739 | x | x | x |
| motYBA | x | — | — | VAA_02089, VAA_01161-VAA_01162 | x | x | x |
| Methyl-accepting chemotaxis gene | x (23) | x (19) | — | x (40) | x (45) | x (28) | |
| Type IV pilus | |||||||
| pilABCD | x | — | — | VAA_00732-VAA_00728 | x | x | x |
| pilVWE | x | — | — | VAA_01191, VAA_01193, VAA_01195 | x | x | x |
| Putative type IV pilin gene | x | — | — | VAA_01194 | x | x | x |
| pilUT | x | — | — | VAA_01655-VAA_01656 | x | x | x |
| pilF | x | — | — | VAA_03067 | x | x | x |
| Multimodular transpeptidase-transglycosylase gene | x | — | — | VAA_02627, VAA_00759 | x | x | x |
| pilMNOPQ | x | — | — | VAA_02628-VAA_02632 | x | x | x |
| MSHA type 4 | |||||||
| mshQPODCABF | x | — | — | VAA_01721-VAA_01728 | x | x | x |
| mshGENMLKJIH | x | — | — | VAA_01729-VAA_01738 | x | x | x |
| Type VI secretion system | |||||||
| vtsA-vtsH system I | x | — | — | VAA_03042-VAA_03021 | x | x | — |
| vtsA-vtsH system II | — | x | — | VAA_02833-VAA_02849 | x | x | — |
| Quorum sensing | |||||||
| luxS | x | — | — | VAA_00716 | x | x | x |
| luxR | x | — | — | VAA_01741 | x | x | x |
| vanMN | — | x | — | VAA_00956-VAA_00957 | x | x | x |
| vanUO | x | — | — | VAA_02384-VAA_02385 | x | x | x |
| vanRI | — | x | — | VAA_01245-VAA_01246 | x | x | x |
| vanQP | — | x | — | VAA_01548-VAA_01547 | x | x | x |
| vanT | x | x | — | VAA_00743, VAA_00909 | x | x | x |
| Regulators | |||||||
| virK, slyA | — | x | — | VAA_02801-VAA_02800 | x | x | x |
| DNA-binding protein H-NS gene | x | — | — | VAA_02203 | x | x | x |
| Transcriptional activator HlyU gene | — | x | — | VAA_00407 | x | x | x |
| Transcriptional regulator VpsT gene | — | x | — | VAA_01253 | x | x | x |
| hfq | x | — | — | VAA_00118 | x | x | x |
GenBank accession number of plasmid pJM1.
“x”, present genes; “—”, absent genes. The numbers in parentheses indicate the number of copies in the genome.
Similar to VCA0884.
The genes related to the repeat-in-toxin gene rtx are located on Chr1. This toxin was also implicated in V. anguillarum virulence (34). In addition, we identified two more rtx toxin genes, one on Chr1 and the other on Chr2. As in V. vulnificus, only one RTX cluster of a total of three contains a complete set of rtx genes (8, 34). The hemolysin genes are located mainly on Chr1, except for vah1, which was located on Chr2 (25, 60, 86). The metalloprotease genes are located on Chr2 (44, 46). Genes for other secreted enzymes are located on Chr2, except for lysophospholipase L2, which is on Chr1. We also identified flagellar and chemotaxis genes previously demonstrated to be associated with virulence in immersion challenges (41, 45). These genes were identified on Chr1, whereas chemotaxis genes were found on both chromosomes. We identified V. cholerae type 4 pilus and mannose-sensitive hemagglutinin (MSHA) pilus homologues (39). These genes were located on Chr1, which also harbored the type VI secretion system locus containing the vstA-vstH genes (81). In addition, we found another novel locus harboring a second type VI secretion system in Chr2. These two systems are highly related; however, the genes in the two systems are organized differently.
Quorum sensing-related genes are distributed in both chromosomes, although Chr2 carries most of the genes described in the literature (16). Chr2 also harbored genes for homologues of regulators, such as slyA and virK, as adjacent genes transcribing in opposite directions. These genes were demonstrated to be important for virulence in different bacteria, such as Salmonella enterica serovar Typhimurium for slyA and Campylobacter jejuni, Salmonella Typhimurium, and Shigella flexneri for virK (18, 21, 52, 54). Regulators hlyU and hns were located on Chr2 and Chr1, respectively. We recently reported that homologues of these genes in V. vulnificus operated in silencing-antisilencing regulation of the rtxA1 gene, which we proved to be an important virulence factor in this bacterium (35, 36). Chr2 also harbors a homologue of the V. cholerae vpsT gene, which regulates matrix production and motility in that bacterium (30). The RNA chaperone Hfq has previously been shown to be necessary for V. anguillarum NB10 virulence (80). This gene was located on Chr1.
GIs.
Ten putative GIs were identified in V. anguillarum 775, with the majority (eight) on chromosome 1 and two on chromosome 2, totaling 199 genes (Fig. 3; also see Table S2 in the supplemental material). To predict the islands, we utilized the IslandViewer tool, which uses a hidden Markov model and measures codon usage. The 10 islands ranged in size from 4 kb to 140 kb, and the largest genomic island (GI2) had 74 genes, whereas the smallest (GI7) had 5 genes (Fig. 3; Table S2). The largest island had a GC content of approximately 39%, indicating a region of easy transcription but difficult translation due to codon usage bias (84). The largest island comprised genes for transposase, dTDP-rhamnose synthesis (lipopolysaccharide [LPS] component), glycosyltransferase, aminotransferases, and acetoin and butanediol metabolism. Three islands, GI3, GI6, and GI8, are inserted adjacent to tRNA-Leu, tRNA-Tyr, and tRNA-Met loci, respectively. Six of the islands contain integrase and transposase genes, which may indicate a possible mechanism of insertion. The genomic signature between the genomes of V. anguillarum 775 and 96F strains is 1.5, whereas the genomic signature for each GI compared with the chromosomes ranged from 37 to 116, showing the high genomic dissimilarity between the islands and the genomes.
Fig. 3.
Genomic islands of strain 775 predicted by IslandViewer. The colors red, blue, and orange represent the three methods for island detection (integrated detection, SIGI-HMM, and IslandPath-DIMOB, respectively). The black line represents the GC content (%).
Vibrio anguillarum 775 superintegron.
The superintegron region of strain 775 was approximately 53 kb, with a GC content of 41%, and contained homologues of the V. cholerae infC gene (translation initiation factor 3), rpmL and rplT (ribosomal proteins L20 and L35), the integrase gene, and the attI and attC recombination sites as found in the V. cholerae superintegron (40). The superintegron has 96 genes, of which 72 (75%) were identified as hypothetical proteins (see Table S3 in the supplemental material). The V. anguillarum strains 96F and RV22 and V. ordalii have the same genes as the 775 superintegron.
Comparative analysis of V. anguillarum strains 775, 96F, and RV22 and V. ordalii.
Table S1 shows the comparison of the molecular parameters of the genomes of V. anguillarum 775 (4.1 Mb), 96F (4.0 Mb), and RV22 (4.0 Mb) and V. ordalii (3.4 Mb). What leaps to the eye is the reduced size of the V. ordalii genome. The difference in genome size is similar to the value we obtained using DNA-DNA hybridization analysis, which demonstrated that the V. ordalii genome is about 70% of the genome size of the O1 and O2 strains (64). In order to determine the similarity between the strains and strain-specific genes, a comparison was performed using BLAST Atlas for both chromosomes (Fig. 1). In Fig. 1, the outermost circle (red) represents the V. ordalii ATCC 33509 genome, which is the most distinct in relation to strain 775, followed by the plasmidless strains of serotype O2β RV22 (green circle) and serotype O1 96F (blue circle). The genomes of V. anguillarum strains 775 and 96F had higher numbers of genes in common. Chr1 shows higher gene conservation among these strains than does Chr2, underscoring the higher plasticity of Chr2. There are 5 specific regions in Chr1 of strain 775 in comparison with the other strains. The Roman numerals in Fig. 1 represent genes that are absent from the 96F and RV22 strains and V. ordalii (also see Table S4 in the supplemental material). In total, these regions corresponded to 173 unique genes for V. anguillarum 775. All five specific regions of strain 775 have integrases and transposases. The regions I, II, III, and V include the islands GI2, GI3, GI5, and GI8, respectively (Fig. 3). The GC content of region II, at 39%, is lower than that for the total chromosome. This region contains several tRNA and rRNA genes. The genes in each of these regions are shown in Table S4. There are 2,681 genes common to all of the strains, using a cutoff of 60% identity. The majority of the 775 chromosome-specific genes are in the five major regions. There are 65 genes carried on the pJM1 plasmid that are also unique for 775, and because we did not close the gaps on 96F, RV22 and V. ordalii, we did not search for smaller clusters or individual chromosomal genes, since they could have been missed in the draft sequence of the latter genomes.
Distribution of virulence-related and environmentally related genes in the chromosomes of 775, 96F, RV22, and V. ordalii.
Table 2 shows the comparison of putative virulence gene distributions in the three V. anguillarum strains and V. ordalii. The majority of the genes were conserved throughout the strains; however, the anguibactin biosynthesis and transport genes were found only on the pJM1 plasmid in the 775 strain, although the RV22 strain had homologues of the fatABCD genes. These genes appeared not to be expressed (5, 51). The tonB1 and tonB2-ttpC clusters were found in all of the V. anguillarum strains, but only the tonB1 system was identified in V. ordalii. The vanchrobactin system was found in all strains, but V. ordalii lacks the ABC transporters fvtB-fvtE and fetAB. HutR homologues were identified only in the V. anguillarum strains. All hemolysin genes were conserved except for vah4, and the third copy of the rtx toxin gene and the phospholipase C gene were identified only in O1 serotype strains. Type VI secretion systems and the gene for microbial collagenase are present in all of the V. anguillarum strains but absent from V. ordalii, while the chemotaxis genes are distributed in the two chromosomes in all strains.
Distribution of transposase and phage proteins in V. anguillarum and V. ordalii.
We compared the chromosome distributions of transposase and phage proteins in the three V. anguillarum strains and V. ordalii (see Table S5a in the supplemental material). V. anguillarum 775 contains more transposase genes (about 53) than 96F (about 23), RV22 (about 42), and V. ordalii (about 18). In pJM1, there are two insertion sequences, ISV-A1 (transposase, ORF2) and ISV-A2 (transposase, ORF11), and many transposases for which insertion sequences have not been identified (73). Table S5b in the supplemental material shows transposases that possibly originated in the pJM1 plasmid in many locations of the chromosomes of V. anguillarum 775. One of these insertions took place in the vabF gene, resulting in the abrogation of vanchrobactin biosynthesis. Two of the pJM1 transposases were not found in either of the two chromosomes. An important and exciting finding was the presence of only one pJM1 transposase on one site of the chromosomes of 96F and RV22, whereas no pJM1 transposase was found in V. ordalii.
Vibrio anguillarum genes specific to 96F compared to 775.
Table S6 in the supplemental material shows that strain 96F had genes not present in strain 775. These included several genes involved in stress response (catalase and peroxidase) and the toxin-antitoxin system (YoeB-YefM), a prophage, RNase H1, Tra system genes, OmpA/MotB, and tRNAs.
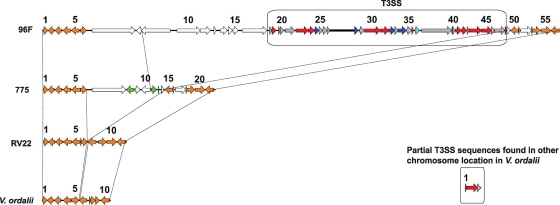
We also identified in V. anguillarum 96F a type III secretion system (T3SS) cluster (Fig. 4; also see Table S7 in the supplemental material) that is highly related to the V. parahaemolyticus type III secretion system 2 (T3SS2), which was also found in V. cholerae and V. mimicus (2, 6, 20, 38, 55). The T3SS has been experimentally shown to be important for virulence in V. parahaemolyticus and V. cholerae (20, 57). The 96F T3SS locus contains genes that are predicted to encode putative T3SS apparatus proteins and putative T3SS regulatory and effector proteins. Those genes are highly conserved with other T3SS2 genes in different Vibrio species. In strain 775, this cluster is missing from both chromosomes, although we find a partial sequence in the same place, indicating that possibly this strain previously carried this locus. Interestingly, this position in 775, corresponding to the 96F T3SS locus, instead contains three sets of transposase sequences, one of which appears to have its origin in pJM1, suggesting that the deletion of the T3SS locus could have been caused by a transposition, deletion, and inversion event. The T3SS cluster was not found in RV22 or in V. ordalii in the 96F location, although V. ordalii has three T3SS homologous genes in a different location.
Fig. 4.
T3SS of V. anguillarum 96F. The box shows the location of the T3SS in 96F: apparatus (red), effectors (blue), and regulator (cyan). Genes conserved among all of the strains are shown in orange. Transposase sequences are shown in green. The numbers correspond to the ORF numbers in Table S7 in the supplemental material, which shows the complete list of genes.
V. anguillarum genes specific to RV22 compared to 775.
The strain RV22 harbors the toxin-antitoxin systems YoeB-YefM, ParE-ParD, and HigB-HigA and has other genes not present in strain 775 (see Table S8 in the supplemental material). These genes comprised a variety of functions, including amino acid and carbohydrate metabolism, 2-keto-3-deoxyoctulosonic acid (KDO)-lipid A biosynthesis (modification and synthesis of lipid A [77]), and cell wall and capsule synthesis (e.g., sialic acid metabolism). In RV22, we also identified a TniA putative transposase and a TniB NTP-binding protein and genes that could be associated with virulence, such as those encoding the accessory V. cholerae enterotoxin (Ace) and the Zonula occludens toxin (Zot). In V. cholerae, Zot decreases ileal tissue resistance by modulating intercellular tight junctions, while Ace increases short-circuit current in Ussing chambers and causes fluid secretion in a ligated rabbit ileal model (22, 78). We have found that zot and ace homologues of Vibrio species exist only in V. anguillarum RV22. Although there is a zot-like gene in the list of 96F-specific genes, our detailed BLAST analysis indicated that it does not encode Zot.
Strain RV22 has genes specifying biosynthetic proteins and transport for a yersiniabactin-like siderophore. This cluster was also found in Vibrio ordalii (see Table S9 in the supplemental material). This cluster is highly conserved in many Vibrio species, and Photobacterium damselae subspecies piscicida carries a highly related cluster (56).
Vibrio ordalii genes specific to ATCC 33509 compared to 775.
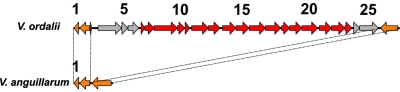
Strain ATCC 33509 has genes not present in strain 775, such as those encoding production of catalase, peroxidase, and superoxide dismutase. In addition, we find genes for a ribosyl nicotinamide transporter and metal transport and toxin-antitoxin systems YefM-YoeB, ParE-ParD, and HigB-HigA. As stated above, the yersiniabactin-like siderophore cluster was also found in V. ordalii (see Table S9 in the supplemental material). We have also identified an entire biofilm formation cluster, the Syp system, in the V. ordalii genome, except for SypE, a negative regulator (Fig. 5; also see Table S10a in the supplemental material). The Syp system has mainly been characterized as an essential factor of V. fischeri to colonize squid organs (47, 87, 88). It can also be found in other pathogenic Vibrio species, such as V. vulnificus and V. parahaemolyticus, and it contributes to biofilm formation in V. vulnificus (29, 88). Although the genes surrounding the Syp system are conserved, the Syp cluster was absent from the 775, 96F, and RV22 genomes (Fig. 5; Table S10a).
Fig. 5.
Syp system in the V. ordalii genome. Shown are syp genes (red), genes encoding hypothetical proteins (gray), and genes found outside the syp genes in V. ordalii and present in V. anguillarum (orange). The numbers correspond to the ORF numbers in Table S10a in the supplemental material, which shows the complete list of syp genes.
Comparison of the O antigen genes in the V. anguillarum strains 775, 96F, and RV22 and V. ordalii.
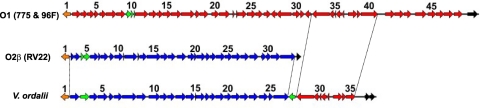
It has been shown that the regions for O-antigen biosynthesis in V. cholerae O1, O22, O31, O37, and O139 are surrounded by gmhD and rjg genes and that this region is also important for the production of capsule of non-O1 V. cholerae O31 and O139 (7, 10, 11, 12). In the V. anguillarum serotype O1 strain, virA (wbhS) and virB (wbhR) are involved in LPS biosynthesis and have been shown to be important for virulence (53). Those genes are located in the cluster adjacent to gmhD (27). Furthermore, LPS genes such as rmlD (wbhC) and rmlC (wbhD) are essential for stability of the outer membrane ferric anguibactin-specific receptor FatA in the 531A strain of V. anguillarum, which carries the pJM1-like plasmid pJHC1. Thus, the LPS genes are necessary for ferric anguibactin transport and virulence (82). Until now, the rjg gene has not been described in V. anguillarum; however, in this work we located the entire locus in Chr1 of V. anguillarum 775, which included the gmhD and rjg genes surrounding the O-antigen cluster genes (Fig. 6; also see Table S10b in the supplemental material). In addition to genes that are responsible for LPS biosynthesis, we found genes such as wza-wzc, wbfB-wbfD, and orf1, shown to be important in exopolysaccharide biosynthesis (15). These genes are also located within this cluster. Very related gmhD and rjg genes are also found in 96F, RV22, and V. ordalii. It is clear from Table S10b that the O-antigen clusters for 775 and 96F are almost identical, as could be expected of these two O1 serotype strains. In RV22, the O2β serotype cluster between gmhD and rjg had genes different from those of 775 and 96F. V. ordalii presents a mosaic situation in which about 25 genes from the gmhD locus are highly homologous to those of the O2β serotype strain RV22. The rest of the genes up to rjg are O1 cluster genes, suggesting that V. ordalii, which can react with O2α serum, is actually a hybrid serotype.
Fig. 6.
Comparison of the O-antigen genes in the V. anguillarum strains 775, 96F, and RV22 and V. ordalii, which are flanked by gmhD (orange) and rjg (black) genes. The figure shows the genes for antigens O1 (red) and O2β (blue). Transposase sequences are shown in green. The V. ordalii O-antigen cluster is a hybrid of V. anguillarum O1 and O2β antigen genes. ORF2 to ORF26 of the V. ordalii cluster are highly related to ORF2 to ORF32 of the O2β cluster, and ORF28 to ORF35 of the V. ordalii cluster are highly related to ORF33 to ORF40 of the O1 cluster. The numbers correspond to the ORF numbers in Table S10b in the supplemental material, which shows the complete list of genes.
DISCUSSION
The main serotypes of V. anguillarum associated with virulence are O1 and O2. O1 serotype strains have been reported to be the most important in causing disease in salmonid fish (32). Serotype O2 can be divided into O2α and O2β, and O2α strains have been isolated from both salmonid and marine fish, while O2β strains are usually isolated from cod and other nonsalmonids (1, 32, 43).
The majority of serotype O1 strains of V. anguillarum harbor pJM1-type plasmids carrying the siderophore anguibactin biosynthesis and transport genes and are responsible for epizootics in many parts of the world (13, 58, 65). These virulent strains are classified into two groups based on the geographic locations where they were isolated and special features of the pJM1-like plasmid. Strains from the Pacific coast of the United States and Japan harbor a pJM1 plasmid very similar to pJM1 in 775 and produce similar levels of anguibactin, whereas strains originated in the U.S. Atlantic coast and in Spain, Scotland, and the Scandinavian countries carry pJM1-like plasmids that possess several extra insertion elements and a mutation in the angR gene (71, 74). This mutation results in higher anguibactin production levels than those in Pacific coast strains, although this difference was not translated into an enhanced virulence phenotype and no major differences were found by DNA hybridization and biochemical studies (72, 82). These findings suggest that all O1 strains carrying a pJM1-like plasmid, independent of plasmid type and geographical isolation, are very similar. It is then clear that 775 isolated from Coho salmon in the Pacific Northwest coast of the United States can be used as the paradigm for the plasmid-carrying strains.
Therefore, to understand the genomics of virulence in V. anguillarum, we completely sequenced the genome of the paradigm 775 harboring the virulence plasmid pJM1 and obtained the draft genome sequence of two plasmidless strains, one of serotype O1, 96F isolated from striped bass, and another an O2β serotype, RV22, isolated from turbot in Spain. We also sequenced a V. anguillarum strain of biotype 2 ATCC 3350, now named V. ordalii.
We determined that each of the three V. anguillarum strains and the V. ordalii strain possess two chromosomes, whereas the 775 strain also harbors the plasmid pJM1, carrying the genes to produce anguibactin and cognate transport genes to internalize ferric anguibactin, which requires the Chr1 TonB2-TtpC system. Ferric anguibactin production and transport are the most important virulence factors once the bacterium invades the host fish. Although it does not produce these virulence factors, the 775 strain can transport vanchrobactin, enterobactin, ferrichrome, and heme as well. In the O2β serotype strain RV22 and in V. ordalii, we also identified biosynthesis and transport genes for a yersiniabactin-like siderophore, which potentially could play a role in virulence.
Genomic analysis using Karlin's genomic signature dissimilarity identified the presence of eight genomic islands (GIs) in Chr1 of V. anguillarum 775(pJM1) and two GIs in Chr2 where novel DNA was present. The 10 islands ranged in size from 4 kb to 140 kb. The genomic signature between V. anguillarum 775 and 96F strains is 1.5, whereas the genomic dissimilarity for each island ranged from 37 to 116, showing the high genomic dissimilarity between the islands and the genomes. Three islands, GI3, GI6, and GI8, with the highest coefficient of dissimilarity were inserted adjacent to tRNA-Leu, tRNA-Tyr, and tRNA-Met loci, respectively. The islands contain integrase and transposase genes, which may indicate a possible mechanism of insertion. Another DNA sequence that can operate in the acquisition of foreign DNA, superintegron, in the strain 775 contains integrase genes and attI and attC recombination site. The evolutionary genomic plasticity conferred by these large chromosomal regions unique to V. anguillarum suggests that they may play an essential role in the dynamic adaptation of this pathogen to specific ecological niches. The 775 strain carrying the pJM1 plasmid also shows a genomic variability due to the insertion of phages and transposases, many of them with a likely origin in the pJM1 plasmid.
The 775 strain harbors, in addition to many other potential virulence factors, flagellar and chemotaxis genes that have been demonstrated to be important in virulence for fish by immersion challenge and two extra copies of the rtx gene that has previously been shown to be part of the V. anguillarum virulence repertoire (34, 41, 45). Only the O1 serotype 96F strain possesses an entire complement of type III secretion system genes, whereas only in the O2β serotype RV22 strain are the genes for the V. cholerae Ace and the V. cholerae Zot toxins found. From these results, it appears that in addition to the carriage of the pJM1 virulence plasmid, different serotypes have evolved different potential virulence mechanisms.
V. anguillarum causes acute hemorrhagic septicemia, but V. ordalii causes a very slow infection whose outcome is uneven dispersion within tissues and possibly the formation of biofilms. This result is in accordance with our finding that this bacterium carries the syp genes involved in biofilm formation. V. ordalii has suffered a large deletion of its chromosome (about 600 kb) compared to 775 and is attenuated in virulence in the trout model (59). The process of genome reduction that appears to have occurred in the transition of V. anguillarum to V. ordalii can be encountered in other bacteria, such as Mycobacterium leprae and Mycobacterium tuberculosis, the first being reduced from the second (23). Another example is that of the endosymbiont Wolbachia (83). It is possible that V. ordalii is going through a reductive evolution and could be on its way to becoming an endosymbiont, since it has the smallest genome of the vibrios. Comparisons of known virulence genes and of surface-exposed genes and colonization determinants across all sequenced V. anguillarum and V. ordalii genomes identified not only common genes but also genomic variability, because some genes are restricted to specific serotypes. Our studies will result in an increase of our understanding of the evolution of the V. anguillarum genome and will contribute new insights into virulence factors and conserved proteins, which could lead to new avenues in the exploration of targets for vaccine development.
Supplementary Material
ACKNOWLEDGMENTS
J.H.C. was supported by USDA grant 2007-35600-185-27 and NIH grant RO1-19018, whereas F.L.T., C.C.T., and G.M.D. were supported by grants from CNPq, CAPES, and FAPERJ.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 16 May 2011.
REFERENCES
- 1. Actis L. A., Tolmasky M. E., Crosa J. H. 2011. Vibriosis, p. 570–605In Woo P. T. K., Bruno D. W. (ed.), Fish diseases and disorders, vol. 3: viral, bacterial, and fungal infections, 2nd ed. CABI International, Oxfordshire, United Kingdom [Google Scholar]
- 2. Alam A., Miller K. A., Chaand M., Butler J. S., Dziejman M. 2011. Identification of Vibrio cholerae type III secretion system effector proteins. Infect. Immun. 79:1728–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alice A. F., Lopez C. S., Crosa J. H. 2005. Plasmid- and chromosome-encoded redundant and specific functions are involved in biosynthesis of the siderophore anguibactin in Vibrio anguillarum 775: a case of chance and necessity? J. Bacteriol. 187:2209–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aziz R. K., et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balado M., Osorio C. R., Lemos M. L. 2009. FvtA is the receptor for the siderophore vanchrobactin in Vibrio anguillarum: utility as a route of entry for vanchrobactin analogues. Appl. Environ. Microbiol. 75:2775–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chatterjee S., et al. 2009. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J. Clin. Microbiol. 47:1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chatterjee S. N., Chaudhuri K. 2004. Lipopolysaccharides of Vibrio cholerae II. Genetics of biosynthesis. Biochim. Biophys. Acta 1690:93–109 [DOI] [PubMed] [Google Scholar]
- 8. Chen C. Y., et al. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Q. 1995. Positive and negative control of the expression of iron uptake genes in Vibrio anguillarum. Ph.D. thesis Oregon Health and Science University, Portland, OR [Google Scholar]
- 10. Chen Y., et al. 2007. The capsule polysaccharide structure and biogenesis for non-O1 Vibrio cholerae NRT36S: genes are embedded in the LPS region. BMC Microbiol. 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Y., Stine O. C., Morris J. G., Johnson J. A. 2007. Genetic variation of capsule/LPS biogenesis in two serogroup O31 Vibrio cholerae isolates. FEMS Microbiol. Lett. 273:133–139 [DOI] [PubMed] [Google Scholar]
- 12. Comstock L. E., Johnson J. A., Michalski J. M., Morris J. G. J., Kaper J. B. 1996. Cloning and sequence of a region encoding a surface polysaccharide of Vibrio cholerae O139 and characterization of the insertion site in the chromosome of Vibrio cholerae O1. Mol. Microbiol. 19:815–826 [DOI] [PubMed] [Google Scholar]
- 13. Crosa J. H. 1980. A plasmid associated with virulence in the marine fish pathogen Vibrio anguillarum specifies an iron-sequestering system. Nature 284:566–568 [DOI] [PubMed] [Google Scholar]
- 14. Crosa J. H., Schiewe M. H., Falkow S. 1977. Evidence for plasmid contribution to the virulence of fish pathogen Vibrio anguillarum. Infect. Immun. 18:509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Croxatto A., Lauritz J., Chen C., Milton D. L. 2007. Vibrio anguillarum colonization of rainbow trout integument requires a DNA locus involved in exopolysaccharide transport and biosynthesis. Environ. Microbiol. 9:370–382 [DOI] [PubMed] [Google Scholar]
- 16. Croxatto A., et al. 2004. A distinctive dual-channel quorum-sensing system operates in Vibrio anguillarum. Mol. Microbiol. 52:1677–1689 [DOI] [PubMed] [Google Scholar]
- 17. Delcher A. L., Bratke K. A., Powers E. C., Salzberg S. L. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Detweiler C. S., Monack D. M., Brodsky I. E., Mathew H., Falkow S. 2003. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 48:385–400 [DOI] [PubMed] [Google Scholar]
- 19. Di Lorenzo M., et al. 2003. Complete sequence of virulence plasmid pJM1 from the marine fish pathogen Vibrio anguillarum strain 775. J. Bacteriol. 185:5822–5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dziejman M., et al. 2005. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc. Natl. Acad. Sci. U. S. A. 102:3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ellison D. W., Miller V. L. 2006. Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 9:153–159 [DOI] [PubMed] [Google Scholar]
- 22. Fasano A., et al. 1991. Vibrio cholerae produces a second enterotoxin, which affects intestinal tight junctions. Proc. Natl. Acad. Sci. U. S. A. 88:5242–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gomez-Valero L., Rocha E. P., Latorre A., Silva F. J. 2007. Reconstructing the ancestor of Mycobacterium leprae: the dynamics of gene loss and genome reduction. Genome Res. 17:1178–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hallin P. F., et al. 2009. GeneWiz browser: an interactive tool for visualizing sequenced chromosomes. Stand. Genomic Sci. 1:204–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hirono I., Masuda T., Aoki T. 1996. Cloning and detection of the hemolysin gene of Vibrio anguillarum. Microb. Pathog. 21:173–182 [DOI] [PubMed] [Google Scholar]
- 26. Hsiao W., Wan I., Jones S. J., Brinkman F. S. 2003. IslandPath: aiding detection of genomic islands in prokaryotes. Bioinformatics 19:418–420 [DOI] [PubMed] [Google Scholar]
- 27. Jedani K. E., Stroeher U. H., Manning P. A. 2000. Distribution of IS1358 and linkage to rfb-related genes in Vibrio anguillarum. Microbiology 146:323–331 [DOI] [PubMed] [Google Scholar]
- 28. Karlin S., Mrazek J., Campbell A. M. 1997. Compositional biases of bacterial genomes and evolutionary implications. J. Bacteriol. 179:3899–3913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim H. S., Park S. J., Lee K. H. 2009. Role of NtrC-regulated exopolysaccharides in the biofilm formation and pathogenic interaction of Vibrio vulnificus. Mol. Microbiol. 74:436–453 [DOI] [PubMed] [Google Scholar]
- 30. Krasteva P. V., et al. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Langille M. G. I., Brinkman F. S. L. 2009. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 25:664–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Larsen J. L., Pedersen K., Dalsgaard I. 1994. Vibrio anguillarum serovars associated with vibriosis in fish. J. Fish Dis. 17:259–267 [Google Scholar]
- 33. Lemos M. L., Salinas P., Toranzo A. E., Barja J. L., Crosa J. H. 1988. Chromosome-mediated iron uptake system in pathogenic strains of Vibrio anguillarum. J. Bacteriol. 170:1920–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li L., Rock J. L., Nelson D. R. 2008. Identification and characterization of a repeat-in-toxin gene cluster in Vibrio anguillarum. Infect. Immun. 76:2620–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu M., Alice A. F., Naka H., Crosa J. H. 2007. The HlyU protein is a positive regulator of rtxA1, a gene responsible for cytotoxicity and virulence in the human pathogen Vibrio vulnificus. Infect. Immun. 75:3282–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu M., Naka H., Crosa J. H. 2009. HlyU acts as an H-NS antirepressor in the regulation of the RTX toxin gene essential for the virulence of the human pathogen Vibrio vulnificus CMCP6. Mol. Microbiol. 72:491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. MacDonell M. T., Colwell R. R. 1985. Phylogeny of the Vibrionaceae, and recommendation for two new genera, Listonella and Shewanella. Syst. Appl. Microbiol. 6:171–182 [Google Scholar]
- 38. Makino K., et al. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743–749 [DOI] [PubMed] [Google Scholar]
- 39. Marsh J. W., Taylor R. K. 1999. Genetic and transcriptional analyses of the Vibrio cholerae mannose-sensitive hemagglutinin type 4 pilus gene locus. J. Bacteriol. 181:1110–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mazel D. 2006. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4:608–620 [DOI] [PubMed] [Google Scholar]
- 41. McGee K., Horstedt P., Milton D. L. 1996. Identification and characterization of additional flagellin genes from Vibrio anguillarum. J. Bacteriol. 178:5188–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mey A. R., Payne S. M. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42:835–849 [DOI] [PubMed] [Google Scholar]
- 43. Mikkelsen H., Lund V., Martinsen L. C., Gravningen K., Schroder M. B. 2007. Variability among Vibrio anguillarum O2 isolates from Atlantic cod (Gadus morhua L.): characterisation and vaccination studies. Aquaculture 266:16–25 [Google Scholar]
- 44. Milton D. L., Norqvist A., Wolf-Watz H. 1992. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J. Bacteriol. 174:7235–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Milton D. L., O'Toole R., Horstedt P., Wolf-Watz H. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mo Z., et al. 2010. Identification and characterization of the Vibrio anguillarum prtV gene encoding a new metalloprotease. Chin. J. Oceanol. Limnol. 28:55–61 [Google Scholar]
- 47. Morris A. R., Visick K. L. 2010. Control of biofilm formation and colonization in Vibrio fischeri: a role for partner switching? Environ. Microbiol. 12:2051–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mourino S., Osorio C. R., Lemos M. L. 2004. Characterization of heme uptake cluster genes in the fish pathogen Vibrio anguillarum. J. Bacteriol. 186:6159–6167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mourino S., Rodriguez-Ares I., Osorio C. R., Lemos M. L. 2005. Genetic variability of the heme uptake system among different strains of the fish pathogen Vibrio anguillarum: identification of a new heme receptor. Appl. Environ. Microbiol. 71:8434–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Naka H., Crosa J. H. 2011. Genetic determinants of virulence in the marine fish pathogen Vibrio anguillarum. Fish Pathol. 46:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Naka H., Lopez C. S., Crosa J. H. 2008. Reactivation of the vanchrobactin siderophore system of Vibrio anguillarum by removal of a chromosomal insertion sequence originated in plasmid pJM1 encoding the anguibactin siderophore system. Environ. Microbiol. 10:265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nakata N., et al. 1992. Identification and characterization of virK, a virulence-associated large plasmid gene essential for intercellular spreading of Shigella flexneri. Mol. Microbiol. 6:2387–2395 [DOI] [PubMed] [Google Scholar]
- 53. Norqvist A., Wolf-Watz H. 1993. Characterization of a novel chromosomal virulence locus involved in expression of a major surface flagellar sheath antigen of the fish pathogen Vibrio anguillarum. Infect. Immun. 61:2434–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Novik V., Hofreuter D., Galan J. E. 2009. Characterization of a Campylobacter jejuni VirK protein homolog as a novel virulence determinant. Infect. Immun. 77:5428–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Okada N., et al. 2010. Presence of genes for type III secretion system 2 in Vibrio mimicus strains. BMC Microbiol. 10:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Osorio C. R., Juiz-Rio S., Lemos M. L. 2006. A siderophore biosynthesis gene cluster from the fish pathogen Photobacterium damselae subsp. piscicida is structurally and functionally related to the Yersinia high-pathogenicity island. Microbiology 152:3327–3341 [DOI] [PubMed] [Google Scholar]
- 57. Park K. S., et al. 2004. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72:6659–6665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pedersen K., Gram L., Austin D. A., Austin B. 1997. Pathogenicity of Vibrio anguillarum serogroup O1 strains compared to plasmids, outer membrane protein profiles and siderophore production. J. Appl. Microbiol. 82:365–371 [DOI] [PubMed] [Google Scholar]
- 59. Ransom D. P., Lannan C. N., Rohovec J. S., Fryer J. L. 1984. Comparison of histopathology caused by Vibrio anguillarum and Vibrio ordalii in three species of Pacific salmon. J. Fish Dis. 7:107–115 [Google Scholar]
- 60. Rodkhum C., Hirono I., Crosa J. H., Aoki T. 2005. Four novel hemolysin genes of Vibrio anguillarum and their virulence to rainbow trout. Microb. Pathog. 39:109–119 [DOI] [PubMed] [Google Scholar]
- 61. Rodkhum C., et al. 2006. Putative virulence-related genes in Vibrio anguillarum identified by random genome sequencing. J. Fish Dis. 29:157–166 [DOI] [PubMed] [Google Scholar]
- 62. Rogers M. B., Sexton J. A., DeCastro G. J., Calderwood S. B. 2000. Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J. Bacteriol. 182:2350–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schiewe M. H., Trust T. J., Crosa J. H. 1981. Vibrio ordalii sp. nov.: a causative agent of vibriosis in fish. Curr. Microbiol. 6:343–348 [Google Scholar]
- 64. Schiewe M. H., Crosa J. H. 1981. Molecular characterization of Vibrio anguillarum biotype 2. Can. J. Microbiol. 27:1011–1018 [Google Scholar]
- 65. Skov M. N., Pedersen K., Larsen J. L. 1995. Comparison of pulsed-field gel electrophoresis, ribotyping, and plasmid profiling for typing of Vibrio anguillarum serovar O1. Appl. Environ. Microbiol. 61:1540–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sorensen U. B., Larsen J. L. 1986. Serotyping of Vibrio anguillarum. Appl. Environ. Microbiol. 51:593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stork M., et al. 2004. Two tonB systems function in iron transport in Vibrio anguillarum, but only one is essential for virulence. Infect. Immun. 72:7326–7329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stork M., Otto B. R., Crosa J. H. 2007. A novel protein, TtpC, is a required component of the TonB2 complex for specific iron transport in the pathogens Vibrio anguillarum and Vibrio cholerae. J. Bacteriol. 189:1803–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thompson F. L., et al. 4 February 2011, posting date The genus Listonella MacDonell and Colwell 1986 is a later heterotypic synonym of the genus Vibrio Pacini 1854 (approved Lists 1980)—a taxonomic opinion. Int. J. Syst. Evol. Microbiol. doi: 10.1099/ijs.0.030015-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tolmasky M. E., Actis L. A., Crosa J. H. 1988. Genetic analysis of the iron uptake region of the Vibrio anguillarum plasmid pJM1: molecular cloning of genetic determinants encoding a novel trans activator of siderophore biosynthesis. J. Bacteriol. 170:1913–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tolmasky M. E., Actis L. A., Crosa J. H. 1993. A single amino acid change in AngR, a protein encoded by pJM1-like virulence plasmids, results in hyperproduction of anguibactin. Infect. Immun. 61:3228–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tolmasky M. E., Actis L. A., Toranzo A. E., Barja J. L., Crosa J. H. 1985. Plasmids mediating iron uptake in Vibrio anguillarum strains isolated from turbot in Spain. J. Gen. Microbiol. 131:1989–1997 [DOI] [PubMed] [Google Scholar]
- 73. Tolmasky M. E., Crosa J. H. 1995. Iron transport genes of the pJM1-mediated iron uptake system of Vibrio anguillarum are included in a transposonlike structure. Plasmid 33:180–190 [DOI] [PubMed] [Google Scholar]
- 74. Tolmasky M. E., Salinas P. C., Actis L. A., Crosa J. H. 1988. Increased production of the siderophore anguibactin mediated by pJM1-like plasmids in Vibrio anguillarum. Infect. Immun. 56:1608–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Toranzo A. E., Barja J. L. 1990. A review of the taxonomy and seroepizootiology of Vibrio anguillarum, with special reference to aquaculture in the northwest Spain. Dis. Aquat. Org. 9:73–82 [Google Scholar]
- 76. Toranzo A. E., et al. 1983. Molecular factors associated with virulence of marine vibrios isolated from striped bass in Chesapeake Bay. Infect. Immun. 39:1220–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Trent M. S. 2004. Biosynthesis, transport, and modification of lipid A. Biochem. Cell Biol. 82:71–86 [DOI] [PubMed] [Google Scholar]
- 78. Trucksis M., Galen J. E., Michalski J., Fasano A., Kaper J. B. 1993. Accessory cholera enterotoxin (Ace), the third toxin of a Vibrio cholerae virulence cassette. Proc. Natl. Acad. Sci. U. S. A. 90:5267–5271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Waack S., et al. 2006. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics 7:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Weber B., Chen C., Milton D. L. 2010. Colonization of fish skin is vital for Vibrio anguillarum to cause disease. Environ. Microbiol. Rep. 2:133–139 [DOI] [PubMed] [Google Scholar]
- 81. Weber B., Hasic M., Chen C., Wai S. N., Milton D. L. 2009. Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ. Microbiol. 11:3018–3028 [DOI] [PubMed] [Google Scholar]
- 82. Welch T. J., Crosa J. H. 2005. Novel role of the lipopolysaccharide O1 side chain in ferric siderophore transport and virulence of Vibrio anguillarum. Infect. Immun. 73:5864–5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Werren J. H., Zhang W., Guo L. R. 1995. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc. Biol. Sci. 261:55–63 [DOI] [PubMed] [Google Scholar]
- 84. Willenbrock H., Ussery D. W. 2007. Prediction of highly expressed genes in microbes based on chromatin accessibility. BMC Mol. Biol. 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wyckoff E. E., Mey A. R., Leimbach A., Fisher C. F., Payne S. M. 2006. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J. Bacteriol. 188:6515–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Xu Z., Wang Y., Han Y., Chen J., Zhang X. H. 2011. Mutation of a novel virulence-related gene mltD in Vibrio anguillarum enhances lethality in zebra fish. Res. Microbiol. 162:144–150 [DOI] [PubMed] [Google Scholar]
- 87. Yip E. S., Geszvain K., DeLoney-Marino C. R., Visick K. L. 2006. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol. 62:1586–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yip E. S., Grublesky B. T., Hussa E. A., Visick K. L. 2005. A novel, conserved cluster of genes promotes symbiotic colonization and sigma-dependent biofilm formation by Vibrio fischeri. Mol. Microbiol. 57:1485–1498 [DOI] [PubMed] [Google Scholar]
- 89. Zhang R., Zhang C. T. 2005. Identification of replication origins in archaeal genomes based on the Z-curve method. Archaea 1:335–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.