Abstract
It has been less than two decades since the underlying genetic defects in Niemann-Pick disease Type C were first identified. These defects impair function of two proteins with a direct role in lipid trafficking, resulting in deposition of free cholesterol within late endosomal compartments and a multitude of effects on cell function and clinical manifestations. The rapid pace of research in this area has vastly improved our overall understanding of intracellular cholesterol homeostasis. Excessive cholesterol buildup has also been implicated in clinical manifestations associated with a number of genetically unrelated diseases including cystic fibrosis. Applying knowledge about anomalous cell signaling behavior in cystic fibrosis opens prospects for identifying similar previously unrecognized disease pathways in Niemann-Pick disease Type C. Recognition that Niemann-Pick disease Type C and cystic fibrosis both impair cholesterol regulatory pathways also provides a rationale for identifying common therapeutic targets.
Keywords: Cholesterol homeostasis, Cystic fibrosis, Molecular motors, Niemann-Pick disease Type C, Organelle motility, Rab GTPases
Introduction
Endolysosomal processing of cholesterol and other lipids as a contributing factor to the progression of a variety of diseases is a growing area of research. These studies are beginning to encompass conditions apparently unrelated to lysosomal storage diseases (LSD) where these abnormalities were first recognized. This phenotype may arise because of primary defects in genes directly involved in cholesterol trafficking or secondary to mutations impairing other mechanisms. Recently, similarities in cholesterol processing defects have been identified in two genetic diseases at opposite ends of the spectrum, Niemann-Pick disease Type C (NPC)5 characterized by defects in cholesterol trafficking and cystic fibrosis (CF) which affects chloride transport [1, 2]. Cells from NPC and CF patients and animal models exhibit accumulation of free cholesterol in perinuclear membrane compartments despite respective genetic mutations affecting unrelated proteins. The goal of this review is to compare prevailing thought on the mechanisms leading to cholesterol accumulation in NPC and CF and the consequences of impaired cholesterol processing on disease progression. Comparing the molecular lesions and phenotypic outcomes in these disparate diseases could provide insight into new regulatory interactions at various points in cholesterol processing pathways.
Overview of genetic lesions and protein families
LSDs comprise more than 40 human genetic disorders [3]. Although a majority of LSDs involve mutations in lysosomal acid hydrolases, others such as NPC have underlying defects in intracellular trafficking [4]. NPC is an autosomal recessive condition caused primarily by loss of function of NPC1, a polytopic membrane protein localized to late endosomes (LE) and lysosomes (Ly) that facilitates cholesterol movement between different intracellular membrane compartments [5]. NPC1 has a conserved sterol sensing domain (SSD) similar to those found in other integral membrane proteins involved in cholesterol homeostasis [6] (Figure 1). NPC1 is believed to work in tandem with the soluble intralysosomal protein NPC2 which mobilizes cholesterol from LE/Ly internal vesicles. These internal vesicles are enriched for the unconventional phospholipid lysobisphosphatidic acid (LBPA) which controls cholesterol capacity in endosomes [7]. Addition of exogenous LBPA partially reverses the cholesterol storage phenotype suggesting LBPA becomes rate-limiting in NPC cells [7]. NPC2 is thought to bind free cholesterol first and deliver it to NPC1 on LE limiting membrane [8]. The exact mechanism of NPC1-mediated cholesterol egress is unknown, however it could involve vesicular or non-vesicular pathways [9]. NPC1 and NPC2 are both required for the effective movement of cholesterol from LE/Ly and NPC2 mutations account for approximately 5% of NPC patients [4]. Loss of NPC1 or NPC2 function leads to increased cholesterol content in endosomal membranes and the eventual accumulation of free cholesterol in abnormal lysosomal storage organelles (LSOs) [10]. A number of other lipids accumulate in LSOs including various gangliosides, sphingolipids, and LBPA.
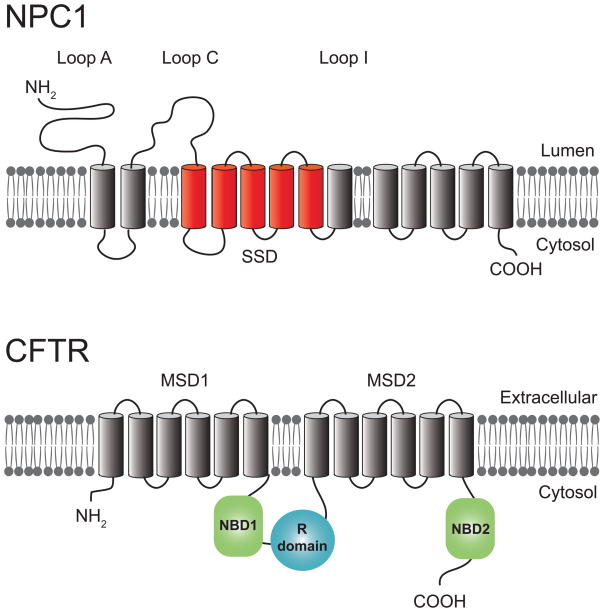
Figure 1.
Diagrammatic views of NPC1 and CFTR membrane topologies. NPC1 is a 1278 amino acid protein with 13 putative transmembrane helices and three prominent luminal glycosylated loops (Loops A, C, and I) that resides in the LE limiting membrane [144]. NPC1 also has a putative sterol sensing domain (SSD) mapped to transmembrane segments 3 to 7 (red). More than 250 NPC1 disease-causing mutations have been identified to date. These include nonsense mutations encoding truncated proteins and point mutations throughout the length of the protein. The highest relative frequency of point mutations reside in cysteine-rich Loop I. CFTR is a 1440 amino acid protein with two membrane-spanning domains (MSD) each composed of six transmembrane helices found primarily on apical membranes in epithelial cells [18]. Two nucleotide-binding domains (NBD1 and NBD2) and a central regulatory (R) domain reside in the cytosol. More than 1,700 disease causing mutations have been identified, with the most common (Δ508, located in NBD1) accounting for 30–80% of mutant alleles depending on ethnic group [145].
In addition to cholesterol accumulation, recent studies indicate that LE/Ly calcium also has a prominent role in the NPC phenotype [11]. Although extracellular calcium can accumulate in endosomes by fluid-phase uptake, early endosomes are depleted of calcium as a consequence of acidification and LE/Ly are subsequently filled with calcium via a proton-dependent mechanism [12]. Intralysosomal calcium then fuels local elevations in cytosolic calcium that drive heterotypic LE/Ly fusion and Ly reformation from LE/Ly hybrid organelles [13]. Cells from NPC patients and animal models have reduced LE/Ly calcium stores and the cholesterol storage phenotype is reversed by treating cells with curcumin, a plasma membrane calcium channel blocker that elevates intracellular calcium levels [14]. In addition to their synergistic roles in cholesterol egress from LE/Ly, NPC1 controls LE/Ly fusion and NPC2 is required for Ly reformation and release of lysosomal cargo-containing membrane vesicles [15]. These findings have prompted some investigators to postulate that NPC defects in cholesterol transport are secondary to abnormalities in calcium homeostasis [14]. Furthermore, the calcium homeostasis model invokes accumulation of sphingosine downstream of NPC1 mutations as the primary cause of impaired LE/Ly calcium levels in NPC cells [16].
CF is an autosomal recessive condition caused by mutations to the gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR) [17]. CFTR is a chloride channel that influences salt and fluid transport localized primarily at the apical membrane of epithelial cells. CFTR is a member of the ATP binding cassette (ABC) protein family consisting of two nucleotide binding (NBD) domains, two membrane spanning domains (MSD), and a regulatory domain that contains multiple consensus protein phosphorylation sites [18] (Figure 1). Regulatory domain phosphorylation by cAMP-dependent protein kinase A is a prerequisite for channel opening. Chloride transport is believed to be gated by ATP binding to NBD1 and NBD2 that promotes NBD dimerization leading to conformational changes in the MSDs [19, 20].
NPC1 and CFTR are both synthesized and acquire N-linked glycans in the endoplasmic reticulum (ER) [21] (Figure 2). Both proteins transit the Golgi where N-glycans are modified and then exit the trans-Golgi network (TGN) in vesicular carriers that deliver them to their final destination. CFTR localization is also regulated by endocytosis, and the cell surface CFTR pool is rapidly internalized and then recycled back to the plasma membrane [22–25]. A number of canonical sorting signals have been identified that regulate different steps in NPC1 and CFTR trafficking. For instance NPC1 has multiple signals that work in concert to target it to LE, including a consensus dileucine-based signal in its cytosolic tail [26]. Both proteins also have sorting signals required for efficient exit from the ER via COPII vesicles [26, 27]. Disease causing mutations impair some of these sorting signals resulting in NPC1 and CFTR mislocalization. Lack of Phe508 (Δ508) in NBD1 of CFTR is the molecular basis for the most common form of CF [18]. The CFTR Δ508 mutant fails to reach the plasma membrane and is retained in the ER due to misfolding [21]. ER buildup of misfolded Δ508 elicits an unfolded protein response (UPR) that has been suggested to be a contributory factor to CF airway inflammatory responses [28]. The Δ508 mutation is believed to block ER exit by impairing conformation of the ER exit code linking newly synthesized CFTR to COPII vesicles [27]. Although disease causing NPC1 mutations are scattered throughout the protein [29], some NPC1 mutations result in protein misfolding and ER retention, suggesting these NPC1 mutations could elicit UPR similar to CFTR Δ508.
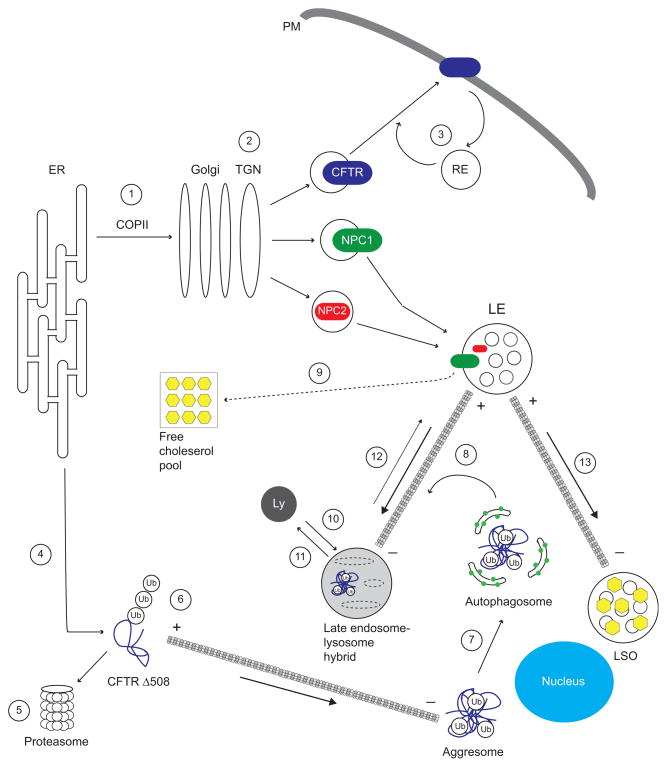
Figure 2.
NPC and CF converge on multiple intracellular trafficking pathways. CFTR, NPC1, and NPC2 exit the ER via COPII vesicles (1), and are delivered to final destinations by transport vesicles derived from TGN (2). Internalized CFTR is sorted back to the plasma membrane from recycling endosomes (RE) (3). The CFTR misfolding mutation Δ508 cannot exit the ER and undergoes retrotranslocation to the cytosol (4) where it is degraded by the ubiquitin-proteasome pathway (5). Polyubiquitin chains link overexpressed CFTR Δ508 to minus-end directed MT dynein-dynactin motors (6) to facilitate formation of pericentriolar aggresomes. Aggresomes are engulfed by LC3-positive (green circles) autophagosomes (7) which then fuse with endosomes en route to Ly where its contents are degraded (8). NPC1 and NPC2 act synergistically to mobilize free cholesterol from LE generating a pool of free cholesterol (yellow hexagons) available for delivery to membranes throughout the cell (9). NPC1 and NPC2 also have independent roles, regulating LE-Ly fusion (10) and Ly reformation (11), respectively. LE motility and positioning is normally controlled by the net balance of vectorial movements toward opposite ends of MTs (12). LEs accumulate at MT minus ends leading to formation of abnormal LSOs in NPC and some forms of CF (13).
In contrast to NPC1 and CFTR polytopic membrane proteins, NPC2 is a soluble protein with a canonical mannose 6-phosphate tag that follows a mannose-6 phosphate receptor (MPR)-dependent TGN-to-Ly sorting route [30, 31]. Experimental disruption of NPC2 trafficking by MPR knockdown triggers LE/Ly accumulation of free cholesterol and LBPA [31, 32]. However, naturally occurring mutations affecting this process have not yet been identified. Although NPC2 is trafficked by both MPR isoforms, the MPR300 isoform appears to be more efficient at transporting NPC2 than MPR46 [31]. Thus NPC1 mutations may adversely affect NPC2 trafficking secondarily since they cause abnormal accumulation of MPR300 in the TGN [33].
Clinical manifestations
Two of the key clinical manifestations of NPC are hepatosplenomegaly due to excessive lipid accumulation in the liver and spleen and progressive cerebellar neurological decline which is what proves to be fatal in this disease [34]. Cerebellar neurodegeneration is initiated by the early loss of Purkinje cells [35, 36]. The accumulation of free cholesterol is a major contributor to the onset of neurological decline as correction of cholesterol processing has yielded impressive improvements in disease progression in both mouse models and human studies. Recent studies demonstrate the ability of 2-hydroxypropyl-β-cyclodextrin (HPCD) to bypass defective NPC1 and NPC2 proteins [37]. HPCD improves lipid mobilization from LE/Ly membranes and has a significant impact on organ lipid load, neurological function, and longevity in an NPC1 mouse model [38–40]. Similar to other neurodegenerative diseases, activation of the innate immune system occurs in the NPC brain resulting in neuro-inflammation, suggesting anti-inflammatory agents may also be beneficial [41].
Classical symptoms of CF are directly related to impaired chloride transport. The loss of CFTR-mediated chloride absorption across sweat duct epithelium results in elevated sweat chloride content that is used as a primary diagnostic marker of CF [42]. Occlusion of pancreatic ducts due to loss of chloride and fluid movement causes necrosis of the pancreas [43, 44]. The resulting pancreatic insufficiency leads to an inability to properly secrete digestive enzymes and the clinical consequence of failure to thrive. In the airways, glandular secretion is impaired resulting in poor mucous clearance. Airways also exhibit increased absorption of sodium that reduces epithelial lining fluid levels, further dehydrating the airways and impairing airway clearance [45].
Poor mucociliary clearance in CF is associated with chronic bacterial infection, chronic and aggressive inflammation, airway tissue destruction, and eventually respiratory failure [46, 47]. However, the development of these airway pathologies is not completely due to reduced salt and fluid transport. Susceptibility to bacterial infection has recently been attributed in part to loss of CFTR function in macrophages leading to impaired bacterial clearance and increased cytokine production. Increased inflammatory signaling and cytokine production in CF epithelial cells has been reported in several studies using multiple model systems [47]. Deficient redox regulation in CF cells has also been postulated to contribute to inflammatory responses characteristic of CF airways [48, 49]. How these inflammatory signaling pathways are specifically caused by CFTR deficiency is unknown and the subject of ongoing investigations.
NPC and CF both affect multiple body systems. Despite some overlap in which organs are involved, pathological manifestations have different origins. Although CF patients do not exhibit overt neurological deficits, growth hormone secretion from the pituitary may be impaired in the absence of CFTR [50]. NPC patients, particularly those with NPC2 mutations, can exhibit severe pulmonary disease [51]. However, the etiology of NPC lung disease is different from pulmonary manifestations in CF. NPC is characterized by macrophage infiltration of the airways and lipid pneumonitis, whereas CF lungs predominantly exhibit neutrophil infiltration and mucous plugging [52, 53]. These differences notwithstanding, NPC and CF cells both accumulate free cholesterol in abnormal endosome compartments associated with perturbed signaling through common pathways.
Cholesterol trafficking and homeostasis
Cellular cholesterol trafficking is an important process to maintain the proper distribution and abundance of cholesterol in appropriate membranes. At the cellular level, cholesterol levels are controlled by the influx of exogenous cholesterol in the form of low density lipoprotein (LDL), de novo cholesterol synthesis, cholesterol catabolism to bile acids and other metabolites, and cholesterol efflux out of cells. The LDL receptor was first identified from the study of familial hypercholesterolemia by the team of Brown and Goldstein who also demonstrated that the receptor-ligand complex is internalized by clathrin-mediated endocytosis [54]. As endosomes acidify, LDL dissociates from the LDL receptor and the receptor recycles back to the plasma membrane to be reutilized for additional rounds of LDL uptake. LDL is delivered to LE/Ly where cholesterol esters are hydrolyzed to form free cholesterol which is subsequently exported out of these compartments via the combined action of NPC1 and NPC2. From there free cholesterol is delivered to various membranes in the cell, with the highest concentration in the plasma membrane and the lowest in the ER.
Cholesterol homeostasis is regulated by sterol-sensing proteins in the ER and is under tight feedback control that is sensitive to small changes in cholesterol concentration. The sterol regulatory element binding protein (SREBP) is a transmembrane protein that is normally resident in the ER but which can be trafficked to the Golgi where it is cleaved to liberate active transcription factors. The cleaved products then translocate to the nucleus where they activate genes required for cholesterol synthesis and LDL uptake [55]. During periods of low sterol load, SREBP cleavage-activating protein (SCAP) binds to SREBP, clustering it in COPII-coated vesicles that escort it to the Golgi allowing SREBP cleavage [56]. As a consequence, target gene transcription increases and cholesterol levels rise. When sterol levels are high, cholesterol binds SCAP and alters its conformation allowing another ER membrane protein called Insig to bind the complex and occlude SCAP from interacting with COPII proteins. Thus SREBP Golgi transport is blocked and cholesterol levels decline due to the down-regulation of gene transcription [57]. Insig is also a sensor for 25-hydroxycholesterol (25-HC), a cholesterol metabolite formed in the ER [58]. 25-HC triggers a conformational change in Insig that allows it to disrupt SCAP-COPII interactions and block SREBP Golgi transport and processing [59]. As described above, NPC1 is believed to work in tandem with NPC2 to transfer cholesterol to intracellular membrane compartments including ER [8, 60]. NPC1 or NPC2 mutations block the export of cholesterol from LE/Ly, and the ER is unable to sense endosomal cholesterol load leading to loss of SREBP feedback control further compounding the NPC cholesterol storage phenotype. Several groups report that unresolved ER stress activates SREBP processing leading to cellular cholesterol accumulation by bypassing cholesterol inhibition of SREBP trafficking to the Golgi. For instance, several ER stressors eliminate SCAP-SREBP ER anchoring by depleting the Insig-1 isoform [61]. These findings point to a critical role for stringent ER quality control in cholesterol homeostasis.
Excess free cholesterol is typically stored as cholesterol esters or removed from cells by cholesterol efflux [62]. Cholesterol esterification is primarily catalyzed by ER-localized acyl-coenzyme A: cholesterol acyltransferase (ACAT) and newly formed cholesterol esters are stored in cytoplasmic lipid droplets. NPC1/NPC2 mutations also impair cholesterol storage by depleting ACAT cholesterol pools. Cholesterol efflux to apolipoprotein A-1 is mediated by ABCA1, a member of the ABC protein family located in plasma membrane and other intracellular compartments [62]. ABCA1 is transcriptionally regulated by oxysterol-dependent activation of the nuclear receptor liver X receptor (LXR) [63]. Defective cholesterol trafficking impairs LXR-dependent gene regulation further exacerbating buildup of free cholesterol in NPC [64].
Cholesterol modulates Rab GTPase function
The Rab family of small GTPases plays a major role in intracellular trafficking [65–67]. Rab GTPases function as molecular switches that are active in their GTP-bound state and inactive in their GDP-bound state. In the active state, Rab GTPases interact with a variety of effector proteins that carry out diverse functions including vesicle formation, motility along microtubules (MT), vesicle tethering, and membrane fusion [68]. Furthermore, organelle identity is thought to be specified by the presence of distinct Rab proteins [69–71]. Rab GTPases are cytosolic proteins that require post-translational lipid modification to mediate membrane binding by way of dual prenylation of carboxyl-terminal cysteine motifs. Once associated with membranes Rab GTPases cycle between GDP- and GTP-bound states through the concerted action of a number of accessory proteins including guanine nucleotide exchange factors (GEFs), GTPase activating proteins (GAPs), Rab escort proteins (REPs), and GDP dissociation inhibitors (GDIs).
A number of Rab proteins become entrapped in cholesterol-rich endosomal membranes leading to their inactivation [72]. Many studies with NPC cells have examined Rab7 which controls LE motility and heterotypic LE-Ly fusion. Cholesterol accretion inhibits GDI-mediated Rab7 extraction from LE limiting membranes causing Rab7 inactivation and inhibition of endosomal motility [73]. Furthermore injection of Rab-GDI into fibroblasts inhibits LE/Ly cholesterol mobilization recapitulating the NPC pheneotype [74]. Rab7 overexpression overcomes sequestration induced by high endosomal cholesterol leading to partial rescue of NPC cholesterol storage defects [75]. The cholesterol storage phenotype associated with mutations in NPC1 is also rescued by an adenovirus membrane protein called RIDα which is a GTP-Rab7 mimic capable of binding certain Rab7 effectors [76, 77]. RIDα appears to facilitate an NPC1-independent cholesterol trafficking pathway that overcomes inhibition by high endosomal cholesterol. These data suggest for the first time that adenovirus modulates cholesterol metabolism presumably to benefit replication in the host. The mechanism of RIDα-mediated NPC1 rescue is an active area of investigation.
Cholesterol accumulation also causes LE/Ly sequestration of Rab9 which is required for transport from endosomes to the trans-Golgi network (TGN). Similar to Rab7, Rab9 resists extraction from NPC cell membranes by Rab-GDI [78]. Rab9 sequestration leads to MPR missorting and failure to deliver mannose-6 phosphate-tagged cargo to LE/Ly in NPC cells [78]. Rab9 overexpression partially compensates for cholesterol storage defects in NPC cells [79], and NPC mice carrying an overexpressed Rab9 transgene exhibit a dramatic improvement in cholesterol mobilization and survival [80]. Overexpression of several other Rab GTPases, including Rab8 and Rab11, which are both involved in retrograde transport from recycling endosomes to the plasma membrane, also ameliorates the NPC phenotype [81, 82]. Overexpressed Rab8 and Rab11 appear to redistribute cholesterol from LE to the plasma membrane and stimulate ABCA1-mediated cholesterol efflux.
Rab GTPases have also been shown to play a role in CF. Rab4 regulates retrograde trafficking events from endosomes to the plasma membrane, and Rab4 overexpression inhibits CFTR trafficking to the plasma membrane apparently by restraining CFTR in endosomes via a direct Rab4-CFTR interaction [83]. CFTR Δ508 can be induced to exit the ER and traffic to plasma membrane when cells are kept at reduced temperature. However its appearance at plasma membrane is short-lived suggesting CFTR Δ508 is also defective in endosome-to-plasma membrane recycling. Rab11 overexpression shifts the pool of endocytosed CFTR Δ508 back to the plasma membrane [84]. Rab9 overexpression also causes increased localization of CFTR Δ508 on the cell surface at low temperature [84]. Furthermore, Rab9 overexpression rescues the cholesterol storage phenotype associated with the Δ508 mutation similar to results obtained in NPC cells [85]. However, it is not currently known if Rab9 overexpression alleviates the CF cholesterol storage phenotype by reconstituting MPR trafficking.
Microtubule-dependent endosome motility
Organelle motility is regulated by the coordinate activity of a number of factors including Rab GTPases, MT modification, and MT motor function [86]. Dynein-dynactin and kinesin MT motors drive retrograde transport towards MT minus ends and anterograde transport to MT plus ends, respectively. Kinesin and dynein-dynactin motors constitute large molecular complexes that also regulate MT attachment, cargo binding and exchange, and interactions with Rab proteins [87]. Another key molecule with a potentially important regulatory role in organelle motility is HDAC6, a MT-specific enzyme with α-tubulin deacetylase activity that also binds dynein-dynactin motors [88]. Recent studies have shown that MT acetylation affects the affinity and processivity of MT motors implicating a role in organelle transport. HDAC6 has been shown to have a critical role in mitochondrial translocation [89] and endocytic trafficking [90] through modulation of tubulin acetylation. Furthermore, tubulin acetylation is essential for starvation-induced autophagy in HeLa cells [91].
Studies examining Rab7 have established that defective organelle motility has a critical role in generating the NPC cholesterol storage phenotype. Rocha et al. delve most specifically into the mechanisms responsible for altered organelle transport in NPC cells by focusing on two Rab7 effector molecules - oxysterol binding related protein 1L (ORP1L) and Rab7 interacting lysosomal protein (RILP) [92]. Previous work from this laboratory had shown that Rab7 activates endosomal dynein-dynactin motors involved in minus end-directed MT transport by facilitating a direct interaction between RILP and the p150Glued dynein-dynactin subunit [93]. The study by Rocha et al. demonstrated that under low cholesterol conditions, the conformation of ORP1L allows ER and LE membrane interactions, bringing the ER vesicle-associated membrane protein VAP into direct contact with Rab7-RILP-ORP1L complexes. This interaction releases p150Glued from RILP and allows retrograde endosomal movement to MT plus ends. In the presence of high cholesterol conditions such as in NPC, p150Glued remains associated with Rab7-RILP causing LE to accumulate at MT minus ends at the MT organizing center (MTOC). Thus endosome motility and positioning that is normally controlled by Rab7 sensing of LE cholesterol is significantly impaired in NPC. Visual confirmation that NPC1-containing vesicles fail to traffic correctly in high cholesterol conditions has also been obtained by tracking fluorescently labeled NPC1 movement [94]. The authors of that study describe three types of NPC1-containing organelle movement - vectorial, Brownian, and slow retraction. They also demonstrate that LE lacking functional NPC1 or with elevated cholesterol due to treatment with U18666A, an amphiphilic drug that induces an NPC-like phenotype [95], lack vectorial movement indicating poor MT motor function [94].
Recent studies indicate NPC is also associated with induction of autophagy [96], a regulated and evolutionarily conserved vesicular trafficking pathway that recycles limited or damaged macromolecules to promote cell survival [97]. However, robust activation of autophagy can also provoke cell stress and programmed cell death [98]. NPC1 mutations activate basal autophagy in part by increasing expression of a regulatory protein for the class III phosphoinositide 3-kinase family called Beclin-1 that has a critical role in autophagosome formation [99, 100]. Autophagosomes fuse with endosomes en route to Ly where their contents are degraded [101, 102]. Thus endocytosis and autophagy share many common elements required for LE/Ly trafficking including Rab7-dependent minus-end directed MT transport [103, 104]. Increased Beclin 1 levels and elevated autophagy are emerging as hallmarks in other lipid storage diseases characterized by disruptions in cholesterol trafficking [105]. Furthermore, elevated endosomal cholesterol is thought to restrict autophagosome-Ly fusion leading to an imbalance between induction and flux, thus contributing to cell stress and culminating in cell death associated with many neuropathies [96, 106].
In CF, organelle trafficking defects have been described primarily in the context of misfolded CFTR Δ508 which undergoes retrotranslocation from ER membranes and is degraded by the cytoplasmic ubiquitin-proteasome pathway. Polyubiquitin chains link CFTR Δ508 to dynein-dynactin motors via HDAC6 to facilitate retrograde minus-strand MT transport towards the MTOC where pericentriolar structures called aggresomes are formed [107]. The polyubiquitin chains promote binding to p62 which is also a ligand for LC3, a cytosolic protein that is recruited to nascent autophagosomal membranes after it undergoes lipidation [108]. The p62-LC3 interaction recruits autophagic membranes allowing aggresomes to be engulfed in autophagosomes. These autophagosomes subsequently fuse with the endocytic pathway to facilitate degradation of misfolded proteins by Ly hydrolases. CFTR Δ508 overexpressed in HEK cells redistributes to aggresomes probably because this misfolded protein exceeds proteasome and autophagic degradative capacity [109]. Overexpressed CFTR Δ508 also induces a redistribution of dynein into aggresomes reflecting an increase in net retrograde transport of minus-end directed MT molecular motors. In addition, CFTR Δ508 overexpression leads to LSO-like cholesterol accumulation in CHO cells, in contrast to a mutation that disrupts CFTR function but allows for normal CFTR folding and processing (G551D) that does not [85]. The authors of this study also suggest the CFTR Δ508-aggresome pathway has a secondary impact on endosome cholesterol trafficking although the mechanism by which this occurs is currently unknown. However, cholesterol accumulation and lipid anomalies have been seen in CF cell systems that simply lack CFTR expression [1, 2, 110]. A reduction in the autophagosome marker LC3 and an increase in the polyubiquitin-binding protein p62 have been observed in multiple CF models including primary cells obtained by nasal biopsy [111]. These findings suggest CF cells accumulate aggresomes with a corresponding inhibition of autophagy. Furthermore, CFTR knockdown and CFTR Δ508 overexpression both down-regulate Beclin-1 suggesting a potential mechanistic link between multiple CFTR defects and autophagy inhibition [111].
It is well established that cholesterol accumulation in NPC leads to the redistribution of many membrane proteins that transit LEs en route to other cellular destinations [112]. This appears to also be the case in CF where it has been shown that the Toll-like receptor TLR4 accumulates in endosomes in macrophages isolated from Cftr−/− mice. Similar TLR4 trafficking defects have also been observed in isolated human CF macrophages [113]. As with cholesterol accumulation, TLR4 missorting is not due to CFTR ΔF508 per se but is also seen when CFTR expression is inhibited. These data demonstrate that the influence of CFTR defects on intracellular trafficking is not restricted to epithelial cells, which may help explain changes in inflammatory cell function in CF [48, 49].
Common signaling pathways
A number of changes in cell signaling have been directly attributed to perturbed cholesterol homeostasis in CF cells. These signaling abnormalities include reduced expression of nitric oxide synthase 2 (NOS2), increased expression but reduced function of signal transducer and activator of transcription 1 (STAT1), and increased expression and activation of the small GTPase RhoA involved in cell adhesion and cytoskeletal rearrangements [114]. Inhibition of de novo cholesterol synthesis with statins reverts these signaling profiles to levels more typical of wild-type cells in cellular and mouse CF models [115]. Identification of the cholesterol processing phenotype in CF cells suggests that NPC cells might display similar abnormalities involving these signaling molecules. In support of this hypothesis, NPC fibroblasts also display an inability to induce NOS2 expression as well as increased STAT1 and RhoA protein expression similar to CF cells [1]. The identified role of RhoA signaling and the sensitivity to statins leads to the speculation that increased isoprenoid synthesis and modification of GTPases is the main cause of signaling changes seen in CF cell models. Isoprenoid-dependent regulation of TGF-β1 signaling has also been shown in CF cells providing further support for this mechanism [116]. The role of isoprenoids in these signaling cascades has not been specifically examined in NPC cells. Nevertheless the CF studies were instrumental in identifying previously unrecognized signaling changes in NPC.
Inflammatory signaling is another area of commonality between CF and NPC cells. Although most inflammatory signaling studies in CF involve epithelial responses to bacterial challenge and bacterial products, recent work has focused on the role of macrophage dysfunction in the pathogenesis of CF airway disease. Loss of CFTR function in macrophages has a direct impact on both inflammatory signaling and bactericidal activity [113]. As already mentioned, TLR4 accumulates in LE in CF macrophages thereby avoiding degradation by Ly hydrolases. The failure to degrade TLR4 prolongs LPS-stimulated TLR4 signaling resulting in NF-κB activation and a sustained pro-inflammatory state [113]. A very similar mechanism has also been reported in NPC where LE accumulation of TLR4 in glial cells leads to constitutive secretion of multiple cytokines including IFN-β, IL-6, and IL-8 [117]. Thus TLR4 missorting induced by defects in cholesterol processing may provide a key mechanism for glial cell activation observed in NPC brains. Despite differences in biological outcomes, NPC and CF both provoke sustained TLR4 signaling because of defects in cholesterol processing. These findings may reflect a general paradigm for identifying additional clinically relevant pathways where signaling is normally attenuated by Ly degradation.
Common therapeutic targets
Recognition that these vastly different genetic diseases share the common phenotype of excess free cholesterol storage suggests therapeutic approaches being pursued for the treatment of one disease may improve health of both sets of patients. Here we focus on three potential common mechanisms and therapeutic approaches – LE/Ly function, ER cholesterol homeostasis and quality control, and organelle motility (Figure 3).
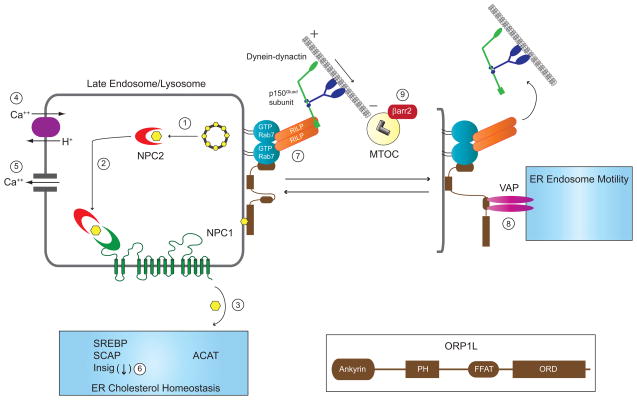
Figure 3.
NPC and CF converge on several mechanisms involved in LE/Ly function, ER cholesterol homeostasis and quality control, and MT-dependent organelle motility. In the prevailing model for NPC protein function, soluble NPC2 mobilizes free cholesterol from LBPA-rich internal membranes in multivesicular LEs (1). NPC2 then delivers free cholesterol to the N-terminal loop of NPC1 (2), and free cholesterol is exported from LE/Ly to other cellular membranes including the ER by an unknown mechanism (3). An alternative model proposes that NPC1 controls proton-dependent LE/Ly calcium filling via an unknown mechanism (4). LE/Ly calcium fuels local elevations in cytosolic calcium required for LE/Ly fusion (5). The ER houses multiple sterol-sensing proteins involved in cholesterol homeostasis. Stringent ER quality control has an important role in cholesterol homeostasis as several ER stressors bypass cholesterol inhibition of SREBP trafficking to the Golgi by depleting Insig-1 (6). LE/Ly motility and positioning is regulated by GTP-Rab7 effectors RILP and ORP1L that reversibly couple these organelles to minus end directed dynein-dynactin MT motors. ORP1L has several protein interaction modules including amino-terminal ankyrin repeats that bind GTP-Rab7, a pleckstrin homology (PH) domain, a FFAT motif capable of binding VAP located on ER membranes, and an oxysterol regulatory domain (ORD) (see inset). When the ORP1L ORD is occupied by LE cholesterol, the Rab7-RILP-ORP1L complex assumes a conformation allowing it to bind minus end directed dynein-dynactin MT motors (7). The ORP1L FFAT motif is exposed under low LE/Ly sterol conditions facilitating interactions with VAP that trigger release of dynein-dynactin MT motor complexes from RILP (8). Unopposed dynein-dynactin MT activity as in NPC impairs LE/Ly motility causing accumulation of cholesterol-filled LSOs at the MTOC. CF conceivably impairs LE/Ly motility triggering a similar response by interfering with normal function of βarr2 at the MTOC (9).
The prevailing model for NPC1/NPC2 function is that they mediate egress of free cholesterol from LE/Ly by working in tandem. HPCD has recently emerged as a promising therapy for NPC in both human and animal studies [39]. HPCD administration is associated with a marked increase in ACAT-mediated cholesterol esterification suggesting HPCD mobilizes LE cholesterol for ER transport [37]. Recent data indicate NPC2 and HPCD rapidly deliver and remove cholesterol from model membranes by similar but not identical mechanisms [118]. Furthermore NPC2 and HPCD each appear to promote membrane-membrane interactions consistent with a hypothesis that they both catalyze cholesterol transfer at tightly apposed sites [118]. However, in contrast to NPC2, LBPA does not enhance HPCD cholesterol membrane transfer efficacy [118]. Although HPCD also interacts with cholesterol at the inner leaflet and catalyzes its diffusion across the LE limiting membrane [119], the mechanism by which this drug bypasses NPC1 remains unclear. Another group of investigators suggests HPCD alleviates the NPC cholesterol storage by acting at the plasma membrane rather than LE/Ly [120]. Their studies indicate HPCD provokes membrane damage by extracting cholesterol from the plasma membrane. This elicits calcium-triggered Ly exocytosis leading to formation of a membrane patch that reseals the membrane lesion and secretion of Ly content [121]. Determining whether HPCD alleviates the cholesterol storage phenotype in CF cells which have wild-type NPC proteins could help distinguish these alternative modes of action.
An alternative model for NPC1/NPC2 function is that these proteins are required to maintain LE/Ly calcium homeostasis [11]. Loss-of-function mutations impede LE/Ly heterotypic fusion and Ly reformation, and the cholesterol trafficking defects arise secondarily. Supporting this model, elevating intracellular calcium levels with the plasma membrane calcium channel blocker curcumin ameliorates cholesterol storage in NPC cell and animal models [11]. The effects of calcium mobilization have also been studied in CF. Most of these studies have focused on improving CFTR Δ508 trafficking to the plasma membrane. Several groups have concluded that CFTR Δ508 maturation and trafficking are not improved by either curcumin or the ER calcium pump inhibitor thapsigargin [122–124]. In contrast, other investigators have shown that curcumin leads to impressive improvement of CFTR Δ508 transport to the plasma membrane [125–127]. These conflicting results may reflect the fact that defects in cholesterol trafficking are secondary to CF mutations making it difficult to test a specific molecular mechanism. Furthermore, elevated intracellular calcium may impact various end point assays that monitor the CF phenotype differently. Nevertheless direct comparisons between CF and NPC with respect to Ly calcium mobilization warrant serious analysis given positive results obtained in NPC using curcumin [14].
Many of the steps involved in cholesterol egress from LE/Ly and inter-organellar transport are still debated or ill-defined and remain open areas of active investigation. It is also conceivable the NPC1/NPC2 pathway is not the only mechanism that mediates LE cholesterol egress, a possibility supported by findings that ectopic expression of an adenoviral protein lacking homology to either NPC protein ameliorates LSO formation in NPC1-deficient fibroblasts. NPC2 has been shown to efficiently deliver cholesterol to model membranes by a mechanism that is enhanced by LBPA [128, 129]. Thus NPC2 could mediate LE cholesterol egress independent of NPC1 or in tandem with another membrane protein in cells. Such a complementary cholesterol egress pathway could be defective in CF. NPC1 is also required for sterol transport from LE/Ly to mitochondria where free cholesterol is metabolized to 27-hydroxycholerol involved in LXR-dependent gene regulation [64]. Addition of exogenous non-steroidal LXR agonists up-regulates ABCA1 and lowers cholesterol mass in NPC cells [130]. It has not been determined whether these LXR agonists will also have an impact on the cholesterol storage phenotype in CF.
Another unifying theme in NPC and CF is that improper protein folding is associated with deregulated cholesterol homeostasis. In NPC, cholesterol derived from endocytosis of LDL becomes sequestered in LE/Ly thereby preventing normal ER homeostatic responses. Little is known about NPC1 or NPC2 function in CF cells, therefore defective NPC1/NPC2 mediated cholesterol transport cannot be eliminated as a potential mechanism for the CF lipid storage phenotype. However this possibility is considered to be unlikely as CF cells exhibit increased expression of NPC1 [1]. Interestingly, NPC1 is regulated by SREBP-dependent gene transcription [131] suggesting SREBP feedback control could be impaired in CF. Similar to other ER stress responses, CFTR Δ508 could activate SREBP processing leading to cellular cholesterol accumulation by bypassing cholesterol inhibition of SREBP trafficking to the Golgi. A growing body of evidence suggests CFTR Δ508 trafficking and function can be improved by treating CF cells with pharmacological chaperones that enhance ER folding energetics [132]. Although one recent study showed a lack of UPR in NPC1 deficient mice and in NPC1 knockdown cells, this study did not include any known NPC1 ER retention mutants [133]. A recent report indicates that treating cells expressing the misfolding I1061T mutant form of NPC1 with pharmacological chaperones resulted in improved trafficking of NPC1 to LE/Ly and also ameliorated the cholesterol storage phenotype in cultured cells [134]. Although UPR may not be necessary to initiate NPC or CF cholesterol storage phenotypes, this ER stress response could exacerbate pathologic cellular manifestations. Current efforts to expand the molecular chaperone repertoire by targeting endogenous components to ameliorate ER misfolding diseases are reviewed elsewhere [135]. Recently, studies have shown that CFTR function can be improved with HDAC inhibitor treatment [136]. Interestingly, HDAC inhibitors have recently been shown to rescue the cholesterol storage phenotype in NPC1 mutant cells [137]. These drugs are of great interest since they are already in phase III clinical trials for cancer treatment [138], and could be fast-tracked for treatment of NPC patients.
Another potential point of convergence in NPC and CF involves LE motility which is normally controlled by the net balance of vectorial movements toward opposite ends of MTs. NPC mutations impair LE motility by interfering with normal function of Rab7 effectors causing abnormal LSOs to accumulate at MT minus ends due to unopposed dynein-dynactin motor activity. Loss of LE motility also has a negative impact on the ability of autophagosomes to fuse with Ly causing pathologic buildup of cytosolic protein aggregates in NPC brains [139]. Aggresome accumulation, LE/Ly cholesterol accumulation, and deficient autophagy all suggest organelle motility is also impaired in CF cells [111]. The mechanistic reasons for impaired organelle motility in CF are unclear. Cholesterol accumulation in CHO cells in response to CFTR Δ508 expression could imply a role for the UPR in these CF phenotypes [85]. However, a putative Δ508-induced UPR is not sufficient to account for the cholesterol storage defects associated with CF which are also seen in cells lacking CFTR altogether [1, 2]. Loss of CFTR function may trigger a cellular response that initiates an UPR independently of Δ508 expression suggesting CFTR has a direct role in stringent ER quality control required for cholesterol homeostasis. Alternatively, CFTR may directly impact MT structure or motor function. For instance, Nilsson et al. have shown that CFTR inhibition leads to MT shortening suggesting a distinct relationship between CFTR and MT regulation [140].
One candidate mechanism for CF-related MT regulation is the increased expression of the protein β-arrestin-2 (βarr2) that has been observed in CF cells and tissues [110]. Increased βarr2 expression has been shown to elicit specific signaling responses in CF cells that can be attributed to altered MT function. Increased βarr2 expression leads to activation of cAMP response element binding protein (CREB) by a mechanism that is ERK-dependent [141]. Depletion of βarr2 from CF mice by double knock-out restores normal CREB regulation. This mode of CREB regulation can be mimicked by MT manipulation providing a potential link between βarr2 MT regulation and CF-specific signaling. βarr2 is known to interact directly with MTOCs and influence both centrosomal function and MT elongation [142]. βarr2 null cells exhibit increased centrosomal density and impaired MT elongation. Though speculative, it is conceivable that increased βarr2 expression and MTOC localization in CF influences MT structure leading to net retrograde transport similar to anomalous behavior of Rab7-RILP-ORP1L complexes in NPC. The impact of βarr2 expression on cholesterol phenotypes and MT regulation in CF is an important area for further exploration. Another important regulator of MT function is HDAC6. Although the α-tubulin subunit of MTs remains its best known substrate, the role of HDAC6 in the regulation of MT-based functions remains unknown. Nevertheless there is a growing body of evidence that α-tubulin deacetylation plays a critical role in the cellular response to the accumulation of misfolded and aggregated protein in many age-related neurodegenerative disorders such as Alzheimer’s, Parkinson’s, and Huntington’s diseases [143]. Elucidating mechanisms that restore organelle motility and positioning in NPC and CF will help identify clinically important targets for drug development in a number of human diseases.
Concluding remarks
NPC and CF are clinically divergent diseases that originate from mutations in structurally and functionally unrelated proteins. The shared phenotype of free cholesterol accumulation in LE/Ly offers an opportunity to examine the cellular processes that influence lipid trafficking from different perspectives. Understanding these processes should lead to identifying sites of intervention that can improve the clinical outcome in each disease. Already calcium mobilization via curcumin treatment has been shown to be beneficial in cell and animal models of each disease, although results vary in CF studies. There are three areas of congruence between diseases that could be of interest for future study. The first area is determining the relationship between calcium homeostasis and lipid movement. The role of ER stress in modulating these pathways is suggested in studies for both NPC and CF, but its direct impact is unclear. The relative ease of pharmacologically manipulating this system makes it attractive for potential therapeutic intervention if distinct mechanisms can be further delineated. A second goal is to more fully understand the regulation of vesicular trafficking. Considerable work has been accomplished in this area in NPC research, but more CF-related studies are needed to determine the influence of CFTR on these pathways. In addition, alternative mechanisms that influence these trafficking pathways such as the ability of the adenovirus protein RIDα to circumvent NPC dysfunction merit further examination. Finally, the role of HDACs in regulating these related phenotypes could be a powerful therapeutic target. Studies in both NPC and CF have shown potential benefit in manipulating HDAC function, but these mechanisms need to be further defined to devise new therapeutic strategies. Side-by-side comparisons of NPC and CF have already led to identification of common disease manifestations arising from perturbation in cholesterol trafficking. The next challenge is to understand how NPC and CF genetic lesions converge to cause this phenotype. It will also be exciting to discover how these seemingly unrelated genetic pathways collaborate to regulate cholesterol homeostasis.
Review Highlights.
Niemann-Pick disease Type C and cystic fibrosis display abnormal storage of free cholesterol.
Cholesterol storage affects many cellular functions and signaling pathways.
Finding disease commonalities may lead to new therapeutic targets for both diseases.
Acknowledgments
This work was supported by a Peter G. Pentchev Research Fellowship from the National Niemann-Pick Disease Foundation (NLC), NIH grant RO1 GM081498 (CRC), and NIH grants R21HL104358 and R01EB009481 and grants from the Cystic Fibrosis Foundation (TJK).
Footnotes
Abbreviations: ABC, ATP binding cassette; ACAT, acyl-coenzyme A:cholesterol acyltransferase; βarr2, β-arrestin-2; CF, cystic fibrosis; CFTR, cystic fibrosis transmembrane conductance regulator; CREB, cAMP response element binding protein; ER, endoplasmic reticulum; GAP, GTPase activating protein; GDI, GDP dissociation inhibitor; GEF, guanine nucleotide exchange factor; HDAC6, histone deacetylase 6; 25-HC, 25-hydroxycholesterol; HPCD, hydroxypropyl-β-cyclodextrin; LBPA, lysobisphospatidic acid; LDL, low-density lipoprotein; LE, late endosome; LSD, lysosomal storage disease; LSO, lysosomal storage organelle; LXR, liver X receptor; Ly, lysosome; MPR, mannose 6-phosphate receptor; MSD, membrane spanning domain; MT, microtubule; MTOC, MT organizing center; NBD, nucleotide binding domain; NOS2, nitric oxide synthase 2; NPC, Niemann-Pick disease Type C; ORP1L, oxysterol binding related protein 1L; REP, Rab escort protein; RILP, Rab7 interacting lysosomal protein; SCAP, SREBP cleavage-activating protein; SREBP, sterol regulatory element binding protein; SSD, sterol sensing domain; STAT1, signal transducer and activator of transcription 1; TGN, trans-Golgi network; UPR, unfolded protein response.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.White NM, Corey DA, Kelley TJ. Am J Respir Cell Mol Biol. 2004;31:538–543. doi: 10.1165/rcmb.2004-0117OC. [DOI] [PubMed] [Google Scholar]
- 2.White NM, Jiang D, Burgess JD, Bederman IR, Previs SF, Kelley TJ. Am J Physiol Lung Cell Mol Physiol. 2007;292:L476–L486. doi: 10.1152/ajplung.00262.2006. [DOI] [PubMed] [Google Scholar]
- 3.Neufeld EF. Annu Rev Biochem. 1991;60:257–280. doi: 10.1146/annurev.bi.60.070191.001353. [DOI] [PubMed] [Google Scholar]
- 4.Patterson MC, Vanier MT, Suzuki K, Morris JA, Carstea E, Neufeld EB, Blanchette-Mackie JE, Pentchev PG. Niemann Pick Disease Type C: A Lipid Trafficking Disorder. McGraw-Hill; New York: 2001. [Google Scholar]
- 5.Garver WS, Heidenreich RA. Curr Mol Med. 2002;2:485–505. doi: 10.2174/1566524023362375. [DOI] [PubMed] [Google Scholar]
- 6.Beachy PA, Cooper MK, Young KE, von Kessler DP, Park WJ, Hall TM, Leahy DJ, Porter JA. Cold Spring Harb Symp Quant Biol. 1997;62:191–204. [PubMed] [Google Scholar]
- 7.Chevallier J, Chamoun Z, Jiang G, Prestwich G, Sakai N, Matile S, Parton RG, Gruenberg J. J Biol Chem. 2008;283:27871–27880. doi: 10.1074/jbc.M801463200. [DOI] [PubMed] [Google Scholar]
- 8.Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, Goldstein JL. Proc Natl Acad Sci USA. 2008;105:15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukherjee S, Maxfield FR. Biochem Biophys Acta. 2004;1685:28–37. doi: 10.1016/j.bbalip.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Sturley SL, Patterson MC, Pentchev P. Proc Natl Acad Sci USA. 2009;106:2093–2094. doi: 10.1073/pnas.0812934106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lloyd-Evans E, Waller-Evans H, Peterneva K, Platt FM. Biochem Soc Trans. 2010;38:1458–1464. doi: 10.1042/BST0381458. [DOI] [PubMed] [Google Scholar]
- 12.Galione A, Churchill GC. Cell Calcium. 2002;32:343–354. doi: 10.1016/s0143416002001902. [DOI] [PubMed] [Google Scholar]
- 13.Pryor PR, Mullock BM, Bright NA, Gray SR, Luzio JP. J Cell Biol. 2000;149:1053–1062. doi: 10.1083/jcb.149.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loyd-Evans EL, Morgan AJ, He S, Smith DA, Elliot-Smith E, Sillence DJ, Churchill GC, Schuchman EH, Galione A, Platt FM. Nat Med. 2008;14:1247–1255. doi: 10.1038/nm.1876. [DOI] [PubMed] [Google Scholar]
- 15.Goldman SDB, Krise JP. J Biol Chem. 2010;285:4983–4994. doi: 10.1074/jbc.M109.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lloyd-Evans E, Platt FM. Traffic. 2010;11:419–428. doi: 10.1111/j.1600-0854.2010.01032.x. [DOI] [PubMed] [Google Scholar]
- 17.Rommens J, Iannuzzi M, Kerem B, Drumm M, Melmer G, Dean M, Rozmahel R, Cole J, Kennedy D, Hidaka N, et al. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 18.Riordan J, Rommens J, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J, et al. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 19.Anderson MP, Berger HA, Rich DP, Gregory RJ, Smith AE, Welsh MJ. Cell. 1991;67:775–784. doi: 10.1016/0092-8674(91)90072-7. [DOI] [PubMed] [Google Scholar]
- 20.Berger HA, Travis SM, Welsh MJ. J Biol Chem. 1993;268:2037–2047. [PubMed] [Google Scholar]
- 21.Kopito RR. Physiol Rev. 1999;79:S167–173. doi: 10.1152/physrev.1999.79.1.S167. [DOI] [PubMed] [Google Scholar]
- 22.Lukacs GL, Chang XB, Kartner N, Rotstein OD, Riordan JR, Grinstein S. J Biol Chem. 1992;267:14568–14572. [PubMed] [Google Scholar]
- 23.Swiatecka-Urban A, Duhaime M, Coutermarsh B, Karlson KH, Collawn J, Milewski M, Cutting GR, Guggino WB, Langford G, Stanton BA. J Biol Chem. 2002;277:40099–40105. doi: 10.1074/jbc.M206964200. [DOI] [PubMed] [Google Scholar]
- 24.Weixel K, Bradbury NA. Methods Mol Med. 2002;70:323–340. doi: 10.1385/1-59259-187-6:323. [DOI] [PubMed] [Google Scholar]
- 25.Silvis MR, Bertrand CA, Ameen N, Golin-Bisello F, Butterworth MB, Frizzell RA, Bradbury NA. Mol Biol Cell. 2009;20:2337–2350. doi: 10.1091/mbc.E08-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott C, Higgins ME, Davies JP, Ioannou YA. J Biol Chem. 2004;279:48214–48223. doi: 10.1074/jbc.M406090200. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Matteson J, An Y, Moyer B, Yoo J-S, Bannykh S, Wilson IA, Riordan JR, Balch WE. J Cell Biol. 2004;167:65–74. doi: 10.1083/jcb.200401035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribeiro CMP, Boucher RC. Proc Am Thorac Soc. 2010;7:387–394. doi: 10.1513/pats.201001-017AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott C, Ioannou YA. Biochim Biophys Acta. 2004;1685:8–13. doi: 10.1016/j.bbalip.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Naureckiene S, Sleat DE, Lackland H, Fensom A, Vanier MT, Wattiaux R, Jadot M, Lobel P. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- 31.Willenborg M, Schmidt CK, Braun P, Landgrebe J, von Figura K, Saftig P, Eskelinen E-L. J Lipid Res. 2005;46:2559–2569. doi: 10.1194/jlr.M500131-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Reaves BJ, Row PE, Bright NA, Luzio JP, Davidson HW. J Cell Sci. 2000;113:4099–4108. doi: 10.1242/jcs.113.22.4099. [DOI] [PubMed] [Google Scholar]
- 33.Umeda A, Fujita H, Kuronita T, Hirosako K, Himeno M, Tanaka Y. J Lipid Res. 2003;44:1821–1832. doi: 10.1194/jlr.M300153-JLR200. [DOI] [PubMed] [Google Scholar]
- 34.Kelly DA, Portmann B, Mowat AP, Sherlock S, Lake BD. J Pediatr. 1993;123:242–247. doi: 10.1016/s0022-3476(05)81695-6. [DOI] [PubMed] [Google Scholar]
- 35.Higashi Y, Murayama S, Pentchev PG, Suzuki K. Acta Neuropathol. 1993;85:175–184. doi: 10.1007/BF00227765. [DOI] [PubMed] [Google Scholar]
- 36.Ko DC, Milenkovic L, Beier SM, Manuel H, Buchanan J, Scott MP. PLoS Genet. 2005;1:e7. doi: 10.1371/journal.pgen.0010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abi-Mosleh L, Infante RE, Radhakrishnan A, Goldstein JL, Brown MS. Proc Natl Acad Sci USA. 2009;106:19316–19321. doi: 10.1073/pnas.0910916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camargo F, Erickson RP, Garver WS, Hossain GS, Carbone PN, Heidenreich RA, Blanchard J. Life Sci. 2001;70:131–142. doi: 10.1016/s0024-3205(01)01384-4. [DOI] [PubMed] [Google Scholar]
- 39.Liu B, Turley SD, Burns DK, Miller AM, Repa JJ, Dietschy JM. Proc Natl Acad Sci USA. 2009;106:2377–2382. doi: 10.1073/pnas.0810895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davidson CD, Ali NF, Micsenyi MC, Stephney G, Renault S, Dobrenis K, Ory DS, Vanier MT, Walkley SU. PLoS ONE. 2009;4:e6951. doi: 10.1371/journal.pone.0006951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baudry M, Yao Y, Simmons D, Liu J, Bi X. Exp Neurol. 2003;184:887–903. doi: 10.1016/S0014-4886(03)00345-5. [DOI] [PubMed] [Google Scholar]
- 42.Di Sant’agnese P, Darling RC, Perara GA, Shea E. AMA Am J Dis Child. 1953;86:618–619. [PubMed] [Google Scholar]
- 43.Durie PR. Acta Paediatr Scand Suppl. 1989;363:41–44. doi: 10.1111/apa.1989.78.s363.41. [DOI] [PubMed] [Google Scholar]
- 44.Tucker JA, Spock A, Spicer SS, Shelburne JD, Bradford W. Ultrastruct Pathol. 2003;27:323–335. [PubMed] [Google Scholar]
- 45.Knowles M, Gatzy J, Boucher R. N Engl J Med. 1981;305:1489–1495. doi: 10.1056/NEJM198112173052502. [DOI] [PubMed] [Google Scholar]
- 46.Chmiel J, Davis P. Respir Res. 2003;4:8. doi: 10.1186/1465-9921-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nichols D, Chmiel J, Berger M. Clin Rev Allergy Immun. 2008;34:146–162. doi: 10.1007/s12016-007-8039-9. [DOI] [PubMed] [Google Scholar]
- 48.Thomas GR, Costelloe EA, Lunn DP, Stacey KJ, Delaney SJ, Passey R, McGlinn EC, McMorran BJ, Ahadizadeh A, Geczy CL, Wainwright BJ, Hume DA. The J Immunol. 2000;164:3870–3877. doi: 10.4049/jimmunol.164.7.3870. [DOI] [PubMed] [Google Scholar]
- 49.Bruscia EM, Zhang P-X, Ferreira E, Caputo C, Emerson JW, Tuck D, Krause DS, Egan ME. Am J Respir Cell Mol Biol. 2009;40:295–304. doi: 10.1165/rcmb.2008-0170OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogan MP, Reznikov LR, Pezzulo AA, Gansemer ND, Samuel M, Prather RS, Zabner J, Fredericks DC, McCray PB, Welsh MJ, Stoltz DA. Proc Natl Acad Sci USA. 2010;107:20571–20575. doi: 10.1073/pnas.1015281107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alymlahi E, Dafiri R. J Postgrad Med. 2004;50:289–290. [PubMed] [Google Scholar]
- 52.Nicholson AG, Florio R, Hansell DM, du Bois RM, Wells AU, Hughes P, Ramadan HK, Mackinlay CI, Brambilla E, Ferretti GR, Erichsen A, Malone M, Lantuejoul S. Histopathology. 2006;48:596–603. doi: 10.1111/j.1365-2559.2006.02355.x. [DOI] [PubMed] [Google Scholar]
- 53.Griese M, Brasch F, Aldana VR, Cabrera MM, Goelnitz U, Ikonen E, Karam BJ, Liebisch G, Linder MD, Lohse P, Meyer W, Schmitz G, Pamir A, Ripper J, Rolfs A, Schams A, Lezana FJ. Clin Genet. 2010;77:119–130. doi: 10.1111/j.1399-0004.2009.01325.x. [DOI] [PubMed] [Google Scholar]
- 54.Brown MS, Goldstein JL. Science. 1974;185:61–63. doi: 10.1126/science.185.4145.61. [DOI] [PubMed] [Google Scholar]
- 55.Brown MS, Goldstein JL. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 56.Brown MS, Goldstein JL. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun LP, Li L, Goldstein JL, Brown MS. J Biol Chem. 2005;280:26483–26490. doi: 10.1074/jbc.M504041200. [DOI] [PubMed] [Google Scholar]
- 58.Javitt NB. Biochem Biophys Res Commun. 2002;292:1147–1153. doi: 10.1006/bbrc.2001.2013. [DOI] [PubMed] [Google Scholar]
- 59.Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Proc Natl Acad Sci USA. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramanian K, Balch WE. Proc Natl Acad Sci. 2008;105:15223–15224. doi: 10.1073/pnas.0808256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colgan SM, Al-Hashimi AA, Austin RC. Expert Rev Mol Med. 2011;13:e4. doi: 10.1017/S1462399410001742. [DOI] [PubMed] [Google Scholar]
- 62.Chang T-Y, Chang CCY, Ohgami N, Yamauchi Y. Annu Rev Cell Dev Biol. 2006;22:129–157. doi: 10.1146/annurev.cellbio.22.010305.104656. [DOI] [PubMed] [Google Scholar]
- 63.Ory DS. Circ Res. 2004;95:660–670. doi: 10.1161/01.RES.0000143422.83209.be. [DOI] [PubMed] [Google Scholar]
- 64.Frolov A, Zielinski SE, Crowley JR, Dudley-Rucker N, Schaffer JE, Ory DS. J Biol Chem. 2003;278:25517–25525. doi: 10.1074/jbc.M302588200. [DOI] [PubMed] [Google Scholar]
- 65.Philips JA, Porto MC, Wang H, Rubin EJ, Perrimon N. Proc Natl Acad Sci USA. 2008;105:3070–3075. doi: 10.1073/pnas.0707206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Somsel Rodman J, Wandinger-Ness A. J Cell Sci. 2000;113:183–192. doi: 10.1242/jcs.113.2.183. [DOI] [PubMed] [Google Scholar]
- 67.Stenmark H, Olkkone VM. Genome Biol. 2001;2:reviews3007.1–reviews3007.7. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 69.Zerial M, McBride H. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 70.Pfeffer SR. Trends Cell Biol. 2001;11:487–491. doi: 10.1016/s0962-8924(01)02147-x. [DOI] [PubMed] [Google Scholar]
- 71.Seabra MC, Wasmeier C. Curr Opin Cell Biol. 2004;16:451–457. doi: 10.1016/j.ceb.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 72.Walter M, Chen FW, Tamari F, Wang R, Ioannou YA. Biol Cell. 2009;101:141–152. doi: 10.1042/BC20070171. [DOI] [PubMed] [Google Scholar]
- 73.Lebrand C, Corti M, Goodson H, Cosson P, Cavalli V, Mayran N, Faure J, Gruenberg J. EMBO J. 2002;21:1289–1300. doi: 10.1093/emboj/21.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holtta-Vuori M, Maatta J, Ullrich O, Kuismanen E, Ikonen E. Curr Biol. 2000;10:95–98. [PubMed] [Google Scholar]
- 75.Choudhury A, Dominguez M, Puri V, Sharma DK, Narita K, Wheatley CL, Marks DL, Pagano RE. J Clin Invest. 2002;109:1541–1550. doi: 10.1172/JCI15420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shah AH, Cianciola NL, Mills JL, Sonnichsen FD, Carlin C. J Cell Biol. 2007;179:965–980. doi: 10.1083/jcb.200702187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cianciola NL, Carlin CR. J Cell Biol. 2009;187:537–552. doi: 10.1083/jcb.200903039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ganley IG, Pfeffer SR. J Biol Chem. 2006;281:17890–17899. doi: 10.1074/jbc.M601679200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walter M, Davies JP, Ioannou YA. J Lipid Res. 2003;44:243–253. doi: 10.1194/jlr.M200230-JLR200. [DOI] [PubMed] [Google Scholar]
- 80.Kaptzan T, West SA, Holicky EL, Wheatley CL, Marks DL, Wang T, Peake KB, Vance J, Walkley SU, Pagano RE. Am J Pathol. 2009;174:14–20. doi: 10.2353/ajpath.2009.080660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Linder MD, Uronen R-L, Holtta-Vuori M, van der Sluijs P, Peranen J, Ikonen E. Mol Biol Cell. 2007;18:47–56. doi: 10.1091/mbc.E06-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Holtta-Vuori M, Tanhuanpaa K, Mobius W, Somerharju P, Ikonen E. Mol Biol Cell. 2002;13:3107–3122. doi: 10.1091/mbc.E02-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saxena SK, Kaur S, George C. Biochem Biophys Res Commun. 2006;341:184–191. doi: 10.1016/j.bbrc.2005.12.170. [DOI] [PubMed] [Google Scholar]
- 84.Gentzsch M, Chang X-B, Cui L, Wu Y, Ozols VV, Choudhury A, Pagano RE, Riordan JR. Mol Biol Cell. 2004;15:2684–2696. doi: 10.1091/mbc.E04-03-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gentzsch M, Choudhury A, Chang X-B, Pagano RE, Riordan JR. J Cell Sci. 2007;120:447–455. doi: 10.1242/jcs.03350. [DOI] [PubMed] [Google Scholar]
- 86.Hehnly H, Stamnes M. FEBS Lett. 2007;581:2112–2118. doi: 10.1016/j.febslet.2007.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ross JL, Ali MY, Warshaw DM. Curr Opin Cell Biol. 2008;20:41–47. doi: 10.1016/j.ceb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dompierre JP, Godin JD, Charrin BC, Cordelières FP, King SJ, Humbert S, Saudou F. J Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen S, Owens GC, Makarenkova H, Edelman DB. PLoS ONE. 2010;5:e10848. doi: 10.1371/journal.pone.0010848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gao Y-S, Hubbert CC, Yao T-P. J Biol Chem. 2010;285:11219–11226. doi: 10.1074/jbc.M109.042754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Geeraert C, Ratier A, Pfisterer SG, Perdiz D, Cantaloube I, Rouault A, Pattingre S, Proikas-Cezanne T, Codogno P, Poüs C. J Biol Chem. 2010;285:24184–24194. doi: 10.1074/jbc.M109.091553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, Neefjes J. J Cell Biol. 2009;185:1209–1225. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuiji C, Olkkonen VM, Neefjes J. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ko DC, Gordon MD, Jin JY, Scott MP. Mol Biol Cell. 2001;12:601–614. doi: 10.1091/mbc.12.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liscum L. Biochim Biophys Acta. 1990;1045:40–48. doi: 10.1016/0005-2760(90)90201-8. [DOI] [PubMed] [Google Scholar]
- 96.Pacheco CD, Lieberman AP. Expert Rev Mol Med. 2008;10:e26. doi: 10.1017/S146239940800080X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang Z, Klionsky DJ. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kang R, Zeh HJ, Lotze MT, Tang D. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pacheco CD, Kunkel R, Lieberman AP. Hum Mol Genet. 2007;16:1495–1503. doi: 10.1093/hmg/ddm100. [DOI] [PubMed] [Google Scholar]
- 100.Simonsen A, Tooze SA. J Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fader CM, Sanchez D, Furlan M, Colombo MI. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 102.Razi M, Chan EYW, Tooze SA. J Cell Biol. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gutierrez MG, Munafo DB, Beron W, Colombo MI. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 104.Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 105.Liu J-P, Tang Y, Zhou S, Toh BH, McLean C, Li H. Mol Cell Neurosci. 2010;43:33–42. doi: 10.1016/j.mcn.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 106.Wong ASL, Lee RHK, Cheung AY, Yeung PK, Chung SK, Cheung ZH, Ip NY. Nat Cell Biol. 2011;13:568–579. doi: 10.1038/ncb2217. [DOI] [PubMed] [Google Scholar]
- 107.Kopito RR. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 108.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 109.Johnston JA, Ward CL, Kopito RR. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Manson ME, Corey DA, White NM, Kelley TJ. Am J Physiol Lung Cell Mol Physiol. 2008;295:L809–L819. doi: 10.1152/ajplung.90402.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina D, Settembre C, Gavina M, Pulze L, Giardino I, Pettoello-Mantovani M, D’Apolito M, Guido S, Masliah E, Spencer B, Quaratino S, Raia V, Ballabio A, Maiuri L. Nat Cell Biol. 2010;12:863–875. doi: 10.1038/ncb2090. [DOI] [PubMed] [Google Scholar]
- 112.Maxfield FR, Tabas I. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- 113.Bruscia EM, Zhang P-X, Satoh A, Caputo C, Medzhitov R, Shenoy A, Egan ME, Krause DS. J Immunol. 2011;186:6990–6998. doi: 10.4049/jimmunol.1100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kraynack NC, Corey DA, Elmer HL, Kelley TJ. Am J Physiol Lung Cell Mol Physiol. 2002;283:L604–L611. doi: 10.1152/ajplung.00459.2001. [DOI] [PubMed] [Google Scholar]
- 115.Kreiselmeier NE, Kraynack NC, Corey DA, Kelley TJ. Am J Physiol Lung Cell Mol Physiol. 2003;285:L1286–L1295. doi: 10.1152/ajplung.00127.2003. [DOI] [PubMed] [Google Scholar]
- 116.Lee JY, Elmer HL, Ross KR, Kelley TJ. Am J Respir Cell Mol Biol. 2004;31:234–240. doi: 10.1165/rcmb.2003-0447OC. [DOI] [PubMed] [Google Scholar]
- 117.Suzuki M, Sugimoto Y, Ohsaki Y, Ueno M, Kato S, Kitamura Y, Hosokawa H, Davies JP, Ioannou YA, Vanier MT, Ohno K, Ninomiya H. J Neurosci. 2007;27:1879–1891. doi: 10.1523/JNEUROSCI.5282-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McCauliff LA, Xu Z, Storch J. Biochemistry. 2011 doi: 10.1021/bi200574f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ramirez CM, Liu B, Aqul A, Taylor AM, Repa JJ, Turley SD, Dietschy JM. J Lipid Res. 2011;52:688–698. doi: 10.1194/jlr.M013789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen FW, Li C, Ioannou YA. PLoS ONE. 2010;5:e15054. doi: 10.1371/journal.pone.0015054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bi GQ, Alderton JM, Steinhardt RA. J Cell Biol. 1995;131:1747–1758. doi: 10.1083/jcb.131.6.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Grubb BR, Gabriel SE, Mengos A, Gentzsch M, Randell SH, Van Heeckeren AM, Knowles MR, Drumm ML, Riordan JR, Boucher RC. Am J Respir Cell Mol Biol. 2006;34:355–363. doi: 10.1165/rcmb.2005-0286OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dragomir A, Björstad J, Hjelte L, Roomans GM. Biochem Biophys Res Commun. 2004;322:447–451. doi: 10.1016/j.bbrc.2004.07.146. [DOI] [PubMed] [Google Scholar]
- 124.Loo TW, Bartlett MC, Clarke DM. Biochem Biophys Res Commun. 2004;325:580–585. doi: 10.1016/j.bbrc.2004.10.070. [DOI] [PubMed] [Google Scholar]
- 125.Egan ME, Pearson M, Weiner SA, Rajendran V, Rubin D, Glöckner-Pagel J, Canny S, Du K, Lukacs GL, Caplan MJ. Science. 2004;304:600–602. doi: 10.1126/science.1093941. [DOI] [PubMed] [Google Scholar]
- 126.Berger AL, Randak CO, Ostedgaard LS, Karp PH, Vermeer DW, Welsh MJ. J Biol Chem. 2005;280:5221–5226. doi: 10.1074/jbc.M412972200. [DOI] [PubMed] [Google Scholar]
- 127.Wang W, Bernard K, Li G, Kirk KL. J Biol Chem. 2007;282:4533–4544. doi: 10.1074/jbc.M609942200. [DOI] [PubMed] [Google Scholar]
- 128.Cheruku SR, Xu Z, Dutia R, Lobel P, Storch J. J Biol Chem. 2006;281:31594–31604. doi: 10.1074/jbc.M602765200. [DOI] [PubMed] [Google Scholar]
- 129.Xu Z, Farver W, Kodukula S, Storch J. Biochemistry. 2008;47:11134–11143. doi: 10.1021/bi801328u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Boadu E, Choi HY, Lee DWK, Waddington EI, Chan T, Asztalos B, Vance JE, Chan A, Castro G, Francis GA. J Biol Chem. 2006;281:37081–37090. doi: 10.1074/jbc.M606890200. [DOI] [PubMed] [Google Scholar]
- 131.Garver WS, Jelinek D, Francis GA, Murphy BD. J Lipid Res. 2008;49:1090–1102. doi: 10.1194/jlr.M700555-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Loo TW, Clarke DM. Methods Mol Biol. 2011;741:23–37. doi: 10.1007/978-1-61779-117-8_3. [DOI] [PubMed] [Google Scholar]
- 133.Klein A, Mosqueira M, Martinez G, Robledo F, Gonzalez M, Caballero B, Cancino GI, Alvarez AR, Hetz C, Zanlungo S. eurodegener Dis. 2011;8:124–128. doi: 10.1159/000316540. [DOI] [PubMed] [Google Scholar]
- 134.Gelsthorpe ME, Baumann N, Millard E, Gale SE, Langmade SJ, Schaffer JE, Ory DS. J Biol Chem. 2008;283:8229–8236. doi: 10.1074/jbc.M708735200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 136.Hutt DM, Herman D, Rodrigues APC, Noel S, Pilewski JM, Matteson J, Hoch B, Kellner W, Kelly JW, Schmidt A, Thomas PJ, Matsumura Y, Skach WR, Gentzsch M, Riordan JR, Sorscher EJ, Okiyoneda T, Yates JR, Lukacs GL, Frizzell RA, Manning G, Gottesfeld JM, Balch WE. Nat Chem Biol. 2010;6:25–33. doi: 10.1038/nchembio.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pipalia NH, Cosner CC, Huang A, Chatterjee A, Bourbon P, Farley N, Helquist P, Wiest O, Maxfield FR. Proc Natl Acad Sci USA. 2011;108:5620–5625. doi: 10.1073/pnas.1014890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tan J, Cang S, Ma Y, Petrillo R, Liu D. J Hematol Oncol. 2010;3:5. doi: 10.1186/1756-8722-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bi X, Liao G. Autophagy. 2007;3:646–648. doi: 10.4161/auto.5074. [DOI] [PubMed] [Google Scholar]
- 140.Nilsson HE, Dragomir A, Lazorova L, Johannesson M, Roomans GM. Exp Mol Pathol. 2010;88:118–127. doi: 10.1016/j.yexmp.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 141.Manson ME, Corey DA, Rymut SM, Kelley TJ. Biochemistry. 2011;50:6022–6029. doi: 10.1021/bi200015h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shankar H, Michal A, Kern RC, Kang DS, Gurevich VV, Benovic JL. J Biol Chem. 2010;285:8316–8329. doi: 10.1074/jbc.M109.062521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li G, Jiang H, Chang M, Xie H, Hu L. J Neurol Sci. 2011;304:1–8. doi: 10.1016/j.jns.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 144.Davies JP, Ioannou YA. J Biol Chem. 2000;275:24367–24374. doi: 10.1074/jbc.M002184200. [DOI] [PubMed] [Google Scholar]
- 145.Bobadilla JL, Macek M, Fine JP, Farrell PM. Hum Mutat. 2002;19:575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]