Abstract
Transposable elements (TEs) have a unique ability to mobilize to new genomic locations and the major advance of next-generation DNA sequencing has provided insights into the dynamic relationship between TEs and their hosts. It now is clear that TEs have adopted diverse strategies – such as specific integration sites or patterns of activity - to thrive in host environments that are replete with mechanisms – such as small RNAs or epigenetic marks - to combat their amplification. Emerging evidence suggests that TE mobilization might sometimes benefit host genomes by enhancing genetic diversity, but TEs are also implicated in diseases such as cancer. Here, we discuss recent findings about how, where, and when TEs insert in diverse organisms.
Subject categories: Evolutionary Biology, Genomics, Epigenetics, genomic instability, small RNAs
Introduction
Through her pioneering work in maize, Barbara McClintock was the first to realize that eukaryotic genomes are not static entities and contain transposable elements (TEs) that have the ability to move from one chromosomal location to another1. It now is clear that virtually all organisms harbor TEs that have amplified in copy number over evolutionary time via DNA or RNA intermediates. On occasion, TEs sporadically have been co-opted by the host to perform critical cellular functions(e.g., 2–5). However, most TEs likely are finely tuned genomic parasites that mobilize to ensure their own survival6–9. The genomic revolution, coupled with new DNA sequencing technologies, now provides an unprecedented wealth of data documenting TE content and mobility in a broad array of organisms.
In multi-cellular eukaryotes, TEs must mobilize within gametes or during early development to be transmitted to future generations. In humans, there are at least 65 documented cases of diseases resulting from de novo TE insertions; these events account for approximately 1/1000 spontaneous cases of disease in humans5, 10. Indeed, new genomic technologies combined with cell culture based experiments have demonstrated that active TEs are more prevalent in the human population than previously appreciated11–18. A growing body of evidence further suggests that mammalian TE integration occurs during early development19–21. In addition, studies of neurogenesis and some forms of cancer have raised the intriguing possibility that TE activity may impact the biology of certain somatic cells12, 22–24. It is likely that we only have observed the tip of the iceberg and still are underestimating the contribution of TE-mediated events to inter-and intra-individual structural variation in mammalian genomes.
TE mobility poses a serious challenge to host fitness. Paradoxically, TE insertions that are harmful to the host jeopardize TE survival. Thus, many TEs have evolved highly specific targeting mechanisms that direct their integration to genomic “safe havens,” thereby minimizing their damage to the host(e.g., 25–29, and references mentioned below). Nevertheless, host genomes have evolved potent restriction mechanisms, such as the methylation of TE DNA sequences and the expression of small RNAs or cytidine deaminases, to restrict TE activity in the germline and perhaps somatic cells(e.g., 30–33, and references mentioned below).
Interestingly, a growing number of examples suggest that TEs may become activated under certain environmental conditions, such as stress. Stress has been shown to induce TE transcription or integration, or redirect TE integration to alternative target sites34–38. These findings are consistent with Barbara McClintock’s hypothesis that environmental challenges may induce transposition, and that transposition, in turn, may create genetic diversity to overcome threats to host survival39.
We begin this review with a brief description of the types of TEs and their modes of mobility. We then describe the latest understanding of TE integration mechanisms and how the host defends against these attacks. Finally, we discuss exciting new research that suggests TE mobility may impact the biology of somatic cells. From the growing understanding of target site selection to the discovery of new active TE copies in human populations, it is clear that the field of transposon biology continues to yield new insights about genome biology.
The diversity and abundance of transposons
Mobilization mechanisms
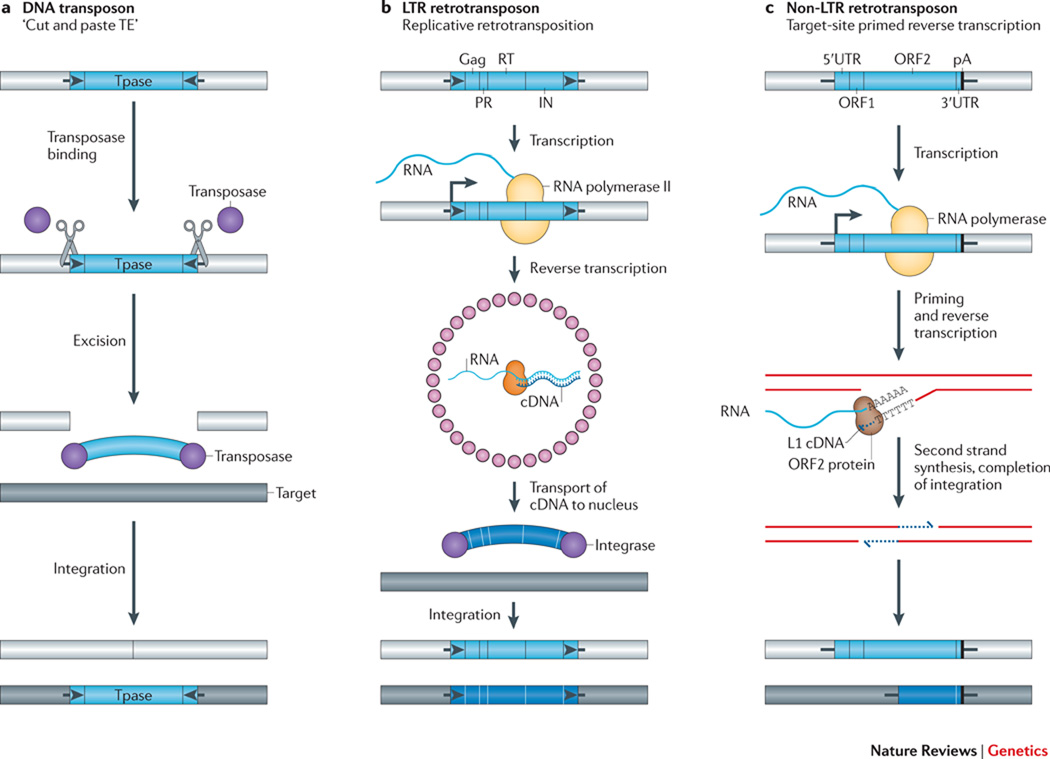
TEs mobilize by remarkably diverse replication strategies (Table 1 and Figure 1)40. Many DNA transposons mobilize by a non-replicative “cut and paste” mechanism, whereby an element-encoded enzyme, the transposase, recognizes sequences at or near TE inverted terminal repeats to “cut” the TE from its existing genomic location and then acts to “paste” the excised DNA into a new genomic location (Fig. 1a)41.
Table 1.
Classes of transposable elements and their mobility mechanisms
| Class of TE | Structural features | Replication mechanism |
Variant forms | Active examples |
|---|---|---|---|---|
| DNA transposons |
|
Transposase-mediated excision of donor dsDNA followed by insertion into the target site |
|
|
| LTR retrotransposons |
|
Within virus-like particles, reverse transcriptase copies the mRNA of the TE into a full-length cDNA; integrase inserts the cDNA into target sites |
|
|
| Non-LTR retrotransposons |
|
An element-encoded endonuclease mediates TPRT. The endonuclease nicks the DNA at the target site and uses the 3′ nicked end for the primer as it reverse transcribes TE mRNA |
|
|
Figure 1. The diverse mechanisms of transposon mobilization.
A. DNA transposons: Many DNA transposons are flanked by terminal inverted repeats (TIRs, black arrows), encode a transposase (TPase, purple circles), and mobilize by a “cut and paste” mechanism (scissors). Transposase binds at or near the TIRs, excises the transposon from its existing genomic location (light gray bar), and pastes it into a new genomic location (dark gray bar). The cleavages of the two strands at the target site are staggered, resulting in a target site duplication (TSD), typically 4 to 8 bp as specified by the TPase (Short black lines flanking the TE). B & C. Retrotransposons: Retrotransposons mobilize by a replicative mechanism that requires the reverse transcription of an RNA intermediate. B. LTR-retrotransposons contain two long terminal repeats (LTRs, black arrows) and encode Gag, protease (PR), reverse transcriptase (RT), and integrase (IN) activities critical for retrotransposition. The 5’ LTR contains a promoter that is recognized by the host RNA polymerase II and produces the mRNA of the TE (start of transcription indicated by a black vertical line attached to a right facing arrow). In step #1 of the reaction, Gag (small pink circles) assembles into virus like particles containing TE mRNA, RT, and IN. The RT copies the TE mRNA into a full-length double stranded DNA (wide blue arc). In step #2 of the reaction, IN (purple circles) inserts the DNA into the new target site. Similar to the TPases of DNA transposons, INs create staggered cuts at the target sites that result in TSDs. C. Non-LTR retrotransposons (right) lack LTRs and encode either one or two open reading frames. The transcription of non-LTR retrotransposons (indicate arrow as in panel b) also leads to the production of a full-length mRNA (blue wavy line). However, these elements mobilize by target-site primed reverse transcription (TPRT). An element-encoded endonuclease generates a single-strand “nick” in genomic DNA, liberating a 3’OH that is used to prime reverse transcription of the RNA. The proteins encoded by autonomous non-LTR retrotransposons also can mobilize non-autonomous retrotransposon RNAs, as well as other cellular RNAs (see text). The TPRT mechanism of an L1 element is depicted in the figure; the new L1 retrotransposition event is 5’ truncated and is retrotransposition-defective (bottom dark gray rectangle with blue line). Some non-LTR retrotransposons lack a poly A tails at their 3’ ends. The integration of non-LTR retrotransposons can lead to target-site duplications (TSDs) and small deletions at the target site in genomic DNA. For example, L1s are generally flanked by 7–20 bp TSDs.
Retrotransposons mobilize via the reverse transcription of an RNA intermediate; however, different types of retrotransposons carry out this process by distinct mechanisms (see Fig. 1b and c). Long terminal repeat (LTR) retrotransposons42–44 (Fig. 1b) and non-LTR retrotransposons45 (Fig. 1c) use element-encoded enzymes to mediate their mobility. In addition, the endonuclease and reverse transcriptase activities of non-LTR retrotransposons also play a central role in mobilizing non-autonomous Short INterspersed Elements (SINEs)46–48, certain classes of non-coding RNAs49–52, and messenger RNAs, which can result in the formation of processed pseudogenes53, 54.
Transposon activity across species
Examples of DNA TEs include Tn5 and Tn7 of E. coli55, 56, P elements of Drosophila57, and Tc1 elements of C. elegans58. Though they thrive in prokaryotes and simpler eukaryotes, DNA TE activity appears to be extinct in most mammals, which fuelled speculation that DNA TEs play a limited role in the ongoing evolution of mammalian genomes59. However, recent studies suggest that DNA TEs, namely non-autonomous hobo/Activator/TAM (nhAT transposons and helitrons), are active in certain bat species60–62. Thus, these studies highlight how new DNA sequencing technologies can facilitate fundamental discoveries about the impact of different TE families on genome evolution and serve as a cautionary note against deriving general conclusions regarding TE activity from relatively few “reference” sequences.
LTR-retrotransposons are particularly abundant in eukaryotes. For example, Drosophila contains approximately 20 distinct families of LTR-retrotransposons that comprise ~1% of the genome63, while maize contains ~400 families of LTR-retrotransposons that comprise ~ 75% of the genome64, 65. In addition, the mouse genome contains multiple active LTR-retrotransposon families. Indeed, the ongoing retrotransposition of both autonomous LTR-retrotransposons and their non-autonomous derivatives is estimated to account for approximately ~10–12 percent of sporadic mutations in mouse66. By comparison, there appears to be little LTR-retrotransposon activity in human genomes59; however, a small number of human endogenous retroviruses are polymorphic with respect to presence/absence at a given genomic location, suggesting that they have retrotransposed relatively recently in human evolution67.
Non-LTR retrotransposons are widespread among eukaryotes, but have been especially prolific in mammalian genomes. For example, L1 elements and the non-autonomous SINEs that they mobilize (e.g., Alu and SINE-R/VNTR/Alu (SVA) sequences)47, 48, comprise approximately 30% of human genomic DNA sequence59. Furthermore, recent research using a combination of transposon display68, 69, second-generation DNA sequencing12, 15–17 and analyses of genomic DNA sequences from the Human Structural Variation project13, 70–72, the 1000 genomes project18, 73, 74, and clinical cohorts14, have revealed that L1 presence/absence dimorphisms, as well as non-allelic recombination between L1 and Alu elements, account for an appreciable proportion of the inter-individual structural variation observed among humans and continue to have a profound effect on the human genome (see ref. 5 for a detailed review).
The diverse patterns of integration sites
Transposons exhibit a remarkable diversity of integration behaviors. Some TEs preferentially integrate into gene-dense regions of the genome, others target regions such as heterochromatin, telomeres, or ribosomal DNA arrays, and some appear to insert throughout the genome. Below, we describe several examples of TE integration and what is known about how TEs target specific sites in genomic DNA.
Integration into gene rich regions
Many TEs integrate into gene rich regions although they use mechanisms that prevent the disruption of open reading frames (ORFs). An extreme example is the E. coli Tn7 DNA TE. Tn7 encodes a sequence specific DNA binding protein, TnsD, which mediates integration into a specific position in the host chromosome, termed attnTn7, and thereby avoids damaging the host genome75, 76. A second targeting protein, TnsE, can alter Tn7 target preference by directing integration to plasmid DNAs that are transferred between E. coli by conjugation77 or to double strand breaks and DNA structures formed during DNA replication78. By comparison, the Drosophila P element avoids disrupting ORFs by integrating within the 500 bp upstream of transcription start sites of genes79. However, the mechanism by which P-elements target these sites requires elucidation.
Certain non-LTR retrotransposons encode endonucleases that target specific sites in genomic DNA. For example, the R1 and R2 elements of insects encode sequence-specific endonucleases that cleave at specific positions within the 28S rDNA locus to initiate target-primed reverse transcription (TPRT)28, 45. However, these endonucleases operate by distinct mechanisms. R1 encodes an endonuclease that shares sequence similarity to an apurinic/apyrimidinic (APE) DNA repair endonuclease80, 81, whereas R2 encodes a type II-S restriction endonuclease82. Thus, these elements apparently have evolved convergent mechanisms to integrate into ribosomal DNA arrays.
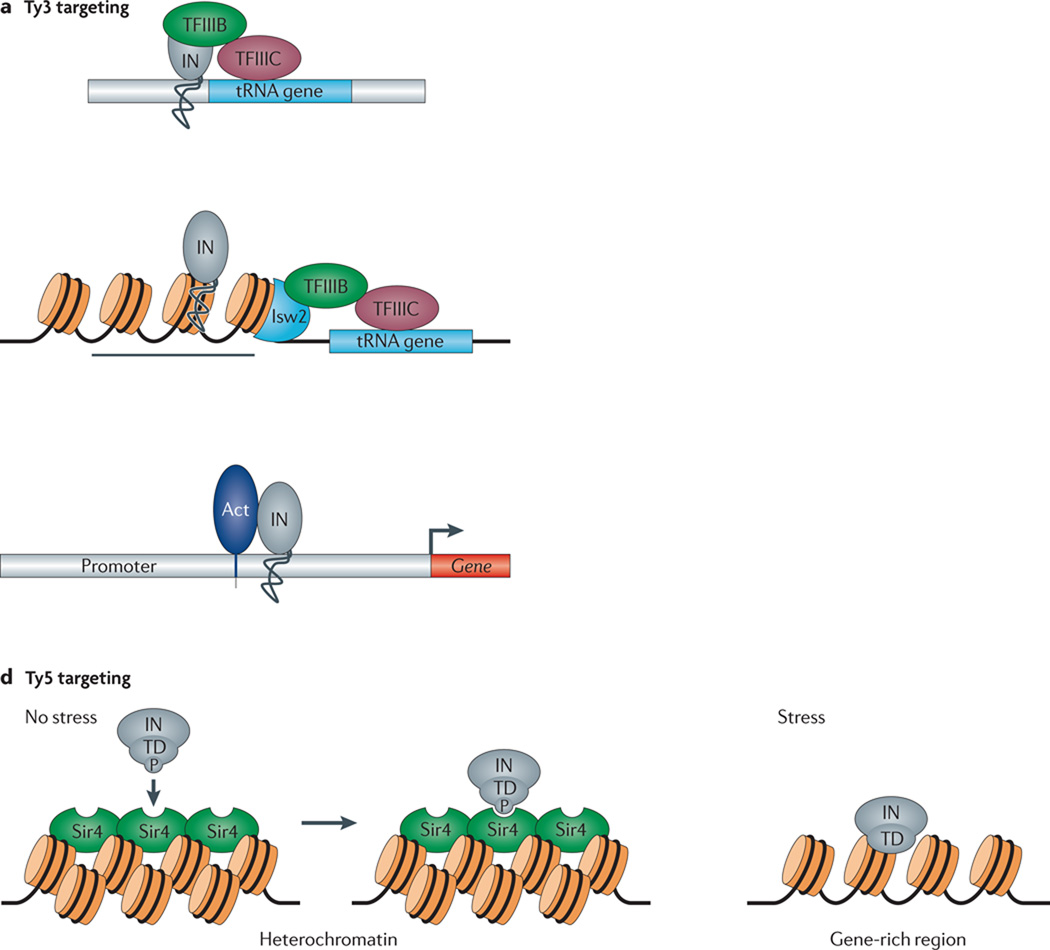
LTR-retrotransposons also have evolved strategies to integrate into gene rich regions, while ensuring minimal damage to their hosts. For example, the Ty1 and Ty3 retrotransposons of S. cerevisiae specifically target gene-free windows located immediately upstream of RNA polymerase III transcribed genes, such as tRNAs25, 27, 83, 84; Ty3 is directed to integration sites 2 or 3 bp upstream of such genes by transcription factors TFIIIB and TFIIIC (Fig. 2a)85, 86. However, in the case of the SNR6 gene, which does not depend on TFIIIC for its expression, the TFIIIB factors Brf1 and TBP are sufficient to direct Ty3 integration87, 88.
Figure 2. Mechanisms that position integration.
A. The Ty3 targeting mechanism: Integration of Ty3 occurs one or two bp upstream of tRNA genes. This pattern requires Brf1 and TBP, components of TFIIIB that recruit integrase (IN, gray oval) to the target site. B. The Ty1 targeting mechanism: The factor Bdp1 is a component of TFIIIB (green circles) that is required to recruit the chromatin remodeling complex Isw2 (light blue semi-circle). Integration of Ty1 occurs with an 80 bp periodicity in a 700 bp window upstream of tRNA genes. The periodicity requires both Bdp1 and Isw2. C. The mechanism of Tf1 targeting: Tf1 integrates into promoters transcribed by RNA polymerase II. Transcription factors (Act), such as Atf1p, bind to the promoter and recruit IN to the insertion sites. D. The mechanism of Ty5 integration: In the absence of a stress condition, the phosphorylation of the IN targeting domain (blue knob on TD) directs integration to heterochromatin by binding a structural component of the heterochromatin, Sir4 (green ovals). Orange cylinders indicated condensed nucleosomes; black lines represent DNA. When cells are exposed to environmental stress the IN is dephosphorylated and integration occurs in gene rich regions.
Ty1 integrates into a ~700 bp window upstream of tRNA genes with a periodicity of 80 bp (Fig. 2b)27, 89. Although the factors that direct Ty1 to tRNA genes remain unknown, the unusual periodicity of integration depends on the amino-terminal domain of Bdp1, another TFIIIB factor90. The ability to integrate upstream of RNA polymerase III transcribed genes also can regulate host and TE gene expression. For example, Ty1 and Ty3 insertions can stimulate the transcription of downstream RNA polymerase III transcribed genes and transcription of the RNA polymerase III target genes can reciprocate by repressing Ty1 transcription91, 92. Clearly, determining how Ty1 target integration sites and exploring how integration alters gene regulation remain areas for future study.
The ability to target RNA polymerase III transcribed genes is not peculiar to LTR-retrotransposons. For example, the Dictyostelium discoideum non-LTR retrotransposon DRE (also known as TRE-5A) preferentially inserts ~48bp upstream of tRNA genes, whereas the retrotransposon Tdd3 (also known as TRE-3A) inserts downstream of tRNA genes29, 93. Indeed, experimental evidence suggests that the TRE-5A ORF1-encoded protein directly interacts with subunits of TFIIIB to direct its integration to tRNA genes94.
The Schizosaccharomyces pombe retrotransposon, Tf1, preferentially integrates into the promoters of RNA polymerase II transcribed genes and provides another example of how TEs target gene-rich regions95–97 (Fig. 2c). Tf1 integration has been studied by examining integration into promoters contained within extrachromosomal replicating plasmids26. For example, the fbp1 promoter is induced when the activating transcription factor Atf1p binds to an eight base pair upstream activating sequence (UAS1)98. Tf1 integration generally occurs 30 bp and 40 bp downstream of UAS1; however, mutating six nucleotides of UAS1 or deleting the Atf1p gene26 disrupts Tf1 integration specificity, causing integration to occur throughout the plasmid. Although cells lacking Atf1p show little reduction in the overall Tf1 retrotransposition frequency99, the above data, as well as the finding that Atf1p forms a complex with Tf1 IN, indicate that specific transcription factors such as Atf1p can play a critical role in directing Tf1 integration to a specific target site. Notably, experiments conducted with a synthetic promoter revealed that RNA polymerase II transcription is not sufficient to target Tf1 integration99.
The development of second-generation sequencing technology recently has allowed the in vivo examination of Tf1 integration sites en masse. Characterization of 73,125 Tf1 integration events from four independent experiments revealed a highly reproducible pattern — approximately 95% of integration events are clustered upstream of ORFs96. Interestingly, the most frequently targeted promoters are associated with genes that are induced by environmental stressors. The targeting of genes that respond to stress, coupled with the ability of Tf1 to induce the expression of adjacent genes26, suggests that Tf1 integration has the potential to improve survival of specific cells that are exposed to environmental stress. Likewise, the transcription of Tf2, another LTR-retrotransposon in S. pombe, is induced by oxidative and osmotic stress or by growth in low oxygen34, 100. Clearly, understanding the consequences of stress-induced retrotransposition will yield insights about how TE mobility can lead to genetic diversity, which may affect the ability of an organism to cope with stress.
Finally, certain retroviruses, which are descended from LTR-retrotransposons101, also exhibit preferential integration in gene rich regions. For example, human immunodeficiency virus-1 (HIV-1) preferentially integrates into RNA polymerase II transcribed genes, whereas murine leukemia virus shows a strong integration preference near transcriptional start sites102–104. Structural and biochemical data demonstrate that HIV-1 IN interacts with the cellular lens epithelium-derived growth factor (LEDGF/p75) host factor, and there is evidence that this interaction plays an important role in proviral DNA integration105, 106.
Integration into heterochromatin
Some TEs target heterochromatic sequences that contain relatively few genes. For example, chromoviruses, which are related to Ty3/Gypsy LTR-retrotransposons, reside in heterochromatin of eukaryotes from fungi to vertebrates107. They contain a chromodomain near the carboxyl-terminus of IN that is related to HP1, a heterochromatin protein that binds to histone H3 methylated at lysine 9107. Furthermore, chromovirus chromodomains fused to green fluorescent protein (GFP) co-localize with heterochromatin108, suggesting that the chromodomain plays a principal role in directing integration. Indeed, fusion of one such chromodomain to the carboxyl-terminus of Tf1 IN directs integration of this TE to heterochromatin108.
The Ty5 LTR-retrotransposon also targets gene poor regions in S. cerevisiae. Approximately 90% of Ty5 integration events occur within the silent mating type loci or near silent heterochromatin at telomeres (Fig. 2d)109–111. Genetic and biochemical experiments indicate that a nine amino acid targeting domain (TD) in the Ty5 integrase (IN) carboxyl-terminus directly binds to a structural component of heterochromatin, Sir4p, to target integration35, 112, 113. Moreover, fusing Sir4p to the DNA binding domain of the LexA repressor protein causes Ty5 integration to be redirected to Lex A binding sites114. Thus, the Ty5 TD, by interacting with Sir4p, directs integration to heterochromatin. Interestingly, genetic and biochemical evidence indicate that the Ty5 TD evolved its interaction with Sir4p by mimicking residues in a host factor, Esc1p, that binds to the same amino acids of Sir4p115.
Although the ability of Ty5 IN to target heterochromatin suggests that the TE dictates target site integration, there also are indications that TE-host interactions can alter Ty5 target-site preference. Mass spectroscopy revealed that phosphorylation of the integrase TD at S1095 is critical for binding Sir4p35 and mutating S1095 redirects integration to expressed regions of the genome. Although the host-encoded kinase has not been identified, studies using a phospho-specific antibody indicate that stressors, such as nitrogen deprivation, can down-regulate S1095 phosphorylation35. Thus, stress conditions may alter the phosphorylation state of Ty5 IN, thereby redirecting Ty5 integration specificity. This elegant example provides a plausible mechanism for how stress can alter transposon mobilization in a manner that might provide an advantage for the host. It remains to be determined whether retargeting Ty5 to gene rich regions benefits Ty5 by allowing newly retrotransposed copies to reside in permissive expression contexts or benefits the host by generating genetic diversity, offering the potential to adapt to stress.
Integration into Telomeres
Some TEs exclusively integrate at or near telomeric ends. For example, the Het-A, TART, and TAHRE non-LTR retrotransposons comprise the ends of Drosophila chromosomes and likely substitute for the function of telomerase in maintaining chromosome end integrity2, 116, 117. The SART1 and TRAS1 non-LTR retrotransposons may have a similar role in Bombyx mori118. The proteins encoded by TART, TAHRE, SART1, and TRAS1 have a Apurinic/apyrimidinic (AP)-like endonuclease domain118, 119 and it is likely that the SART1 and TRAS1 endonuclease proteins direct their integration into telomeric repeats118; however, the functional role of the putative endonuclease domain in TART and TAHRE remains unknown.
Excitingly, recent studies have revealed that certain retrotransposons can target telomeric sequences for integration. For example, by an alternative endonuclease-independent retrotransposition mechanism, human L1 retrotransposons containing missense mutations in the L1 EN active site can integrate at endogenous DNA lesions and dysfunctional telomeres in Chinese Hamster Ovary cell lines that are deficient for factors important in the non-homologous end-joining pathway of DNA repair as well as p53 function120, 121. Similarly, members of the Penelope clade of retrotransposons, which encode an RT that lacks an obvious endonuclease domain, reside at telomeres in organisms from four eukaryotic kingdoms122. The RNAs encoded by these terminal Penelope elements also contain sequences that are complementary to telomeric DNA sequences, suggesting that base pairing between the TE RNA and single stranded telomeric DNA is critical for integration. Interestingly, both of the above cases can be considered as a type of RNA-mediated DNA repair that appears curiously similar to the mechanism used by telomerase120, 122. Future studies should elucidate whether host factors are critical for the localization of these retrotransposons to DNA lesions and/or chromosomal termini.
Dispersed patterns of integration
In contrast to elegant mechanisms that target integration of some TEs into specific regions of the genome, other TEs appear to lack target site specificity. For example, L1s and the non-autonomous elements they mobilize are interspersed throughout the genome59. Indeed, ~30% of engineered human L1 retrotransposition events in cultured cells, and a similar proportion of recently discovered full-length, dimorphic human-specific L1s, are near or within the introns of genes13, 50, 123, 124. Since protein-coding genes constitute ~40% of the human genome125, 126, these findings suggest a lack of robust mechanisms employed by L1s or the host to prevent L1 retrotransposition into genes.
The interspersed nature of L1 and Alu sequences probably reflects the fact that the L1 endonuclease has relatively weak target-site specificity, preferentially cleaving the sequence 5’-TTTT/A-3’ (and variants of that sequence), to initiate TPRT80, 121, 127, 128. Interestingly, while “young” Alu and L1 insertions exhibit similar interspersed integration patterns, cytogenetic studies and examination of the human genome reference sequence revealed that evolutionarily “older” L1s and Alus show distinct genomic distributions59, 129. Older L1s preferentially reside in gene-poor AT-rich sequences, whereas older Alus are preferentially reside in gene-rich GC-rich regions of the genome59.
The distinct distributions of older L1s and Alus likely result from post-integration selective processes that have operated on the genome for millions of years59. However, how these skewed distributions arose remains a mystery. Some researchers have suggested that Alus may possibly play an advantageous, albeit undefined, role in gene-rich regions of the genome59. Others have suggested that L1 retrotransposition events into genic regions may exert a greater fitness cost to the host than Alu insertions130. If so, negative selection would lead to the removal of detrimental L1 alleles from the population. Consistent with this hypothesis, data suggest that evolutionary recent human full-length L1s insertions are detrimental to the host131, 132, whereas in vitro studies have revealed that L1s contain cis-acting sequences that can reduce gene expression133, 134. Clearly, further studies are needed to explain how the distributions of L1 and Alu have diverged over evolutionary time.
Despite their interspersed distribution, a small body of evidence suggests that there may be preferred, albeit rare, L1 integration sites. For example, independent L1-mediated retrotransposon insertions at the same nucleotide position in the BTK gene (i.e., an SVA and an Alu element) have resulted in two sporadic cases of X-linked agammaglobulinemia135. Similarly, independent L1 and Alu insertions associated with colorectal and desmoid tumors, respectively, have occurred at the same nucleotide position in the APC gene22, 135, 136, whereas two independent Alu insertions at the same nucleotide position in the Factor IX gene have caused hemophilia B135, 137. Thus, it would not be surprising to find that chromatin structure and accessibility impact L1-mediated retrotransposon target preference138.
How the host defends against transposons
Although many TEs have evolved mechanisms to limit genome damage, TE integration still poses a potential threat to the host. Thus, it is not surprising that host organisms have evolved a diverse array of mechanisms to combat TE activity. However, the host must be able to discriminate TE sequences from host genes to accomplish this feat. Below, we discuss mechanistic strategies employed by the host to restrict TE mobilization.
DNA methylation
Cytosine methylation (5-methylcytosine) is an important DNA modification in eukaryotes with genomes larger than 5×108 bp, which includes vertebrates, flowering plants, and some fungi. The majority of cytosine methylation in plants and mammals, and almost all cytosine methylation in Neurospora crassa, occurs within repetitive elements and is correlated with the transcriptional repression of retrotransposons in somatic and germline cells139, 140.
Experiments in mammals and plants demonstrate that global demethylation of genomic DNA strongly reactivates TE transcription141–144. For example, deletion of DNA cytosine-5-methyltransferase 3-like gene (Dnmt3L) in mice leads to loss of de novo cytosine methylation of both LTR and non-LTR retrotransposons, reactivation of transposable element expression in spermocytes and spermatogonia, and meiotic catastrophe in male germ cells145. Determining whether TE mobilization directly is responsible for the meiotic defects requires further study. Moreover, recent data demonstrates that inactivation of cytosine methylation in Arabidopsis thaliana causes a burst of retrotransposon and DNA TE activity and results in substantial increases in TE copy number144. Thus, epigenetic mechanisms act to control the expression, and perhaps mobility of various TEs.
Multiple lines of evidence indicate that DNA methylation inhibits TE transcription. Patterns of DNA methylation are established during gametogenesis and are mediated by Dnmt3a and the non-catalytic paralog, Dnmt3L, in mammals, but how TEs are recognized as methylation substrates requires further study146. By comparison, during plant development, small 24 nt RNAs target paralogous DNA sequences that share high levels of homology (such as TEs) for cytosine methylation. The mechanism of RNA-directed DNA methylation (RdDM) is not fully understood, but it appears to require the canonical RNA interference (RNAi) machinery (see below and Fig. 3), the DNA methyltransferase DRM2, and two plant specific RNA polymerases, Pol IV and Pol V146.
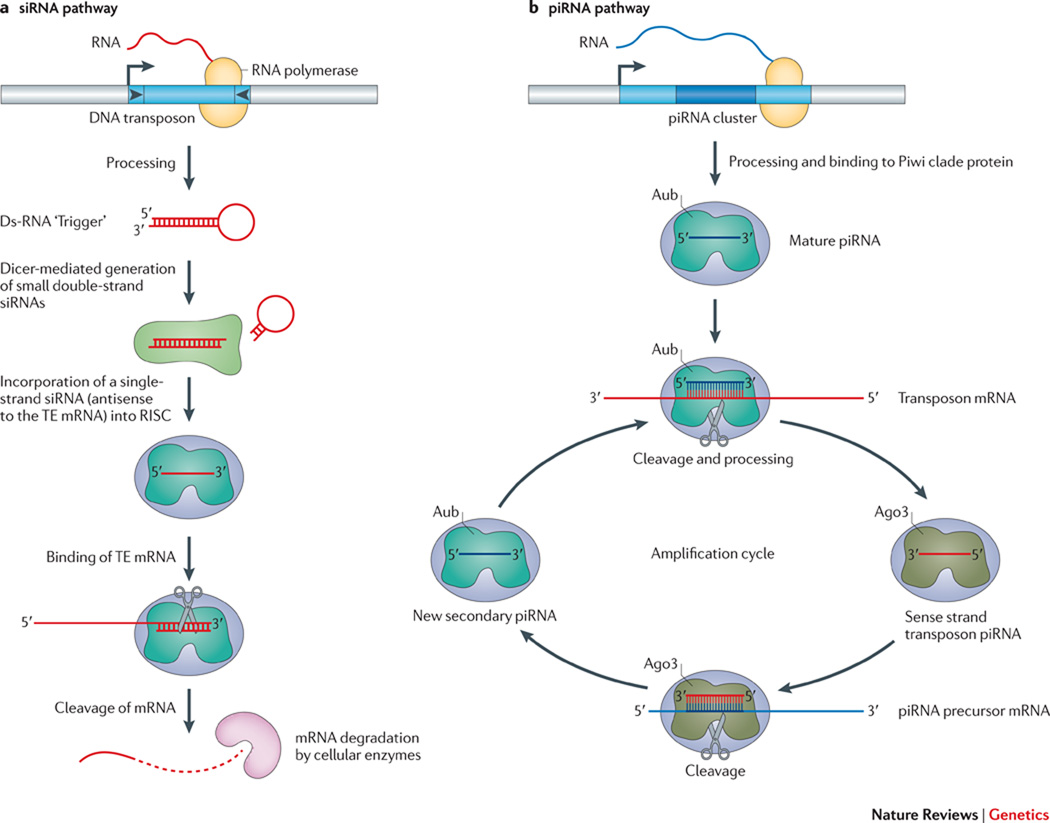
Figure 3. The degradation of transposon mRNA by RNAi.
A. The siRNA pathway: Double-stranded “trigger” RNAs (hairpin), derived from the inverted terminal repeats of a DNA transposon in the case illustrated, are processed and then cleaved into 21 to 24 nt siRNAs by the dicer family of proteins (light green amorphous shape). A single-strand siRNA (short red line) complementary to the transposon mRNA is selectively incorporated into the Argonaute containing RISC complex (blue amorphous shape with short red line). The siRNA directs RISC to complementary sequences in transposon mRNA (long red line), leading to its post-transcriptional destruction. The figure was drawn based on concepts presented in the following reviews147, 148, 152. B. The piRNA pathway: A primary piRNA transcript (wavy blue line) generated from a piRNA cluster (blue rectangle) that contains sequences derived from TEs (darker blue rectangle inset) is processed into mature 24–35 nt piRNAs (small blue line). Binding of the mature piRNA by the Piwi/Aubergine protein (color?…make different from Ago3 and panel A) allows it to be directed to complementary sequences in TE mRNA (red line). Endonucleolytic cleavage (scissor) 10 nt from the 5’ end of the small RNA and 3’ processing liberates a secondary sense strand transposon piRNA (red line), which associates with the Argonaute 3 protein (color?…make different from Aubergine and panel A). The binding of this complex to complementary sequences in the original precursor piRNA, followed by endonucleolytic cleavage and 3’ processing liberates, generates a secondary antisense piRNA that can be directed to TE mRNA. This reiterative cycle (e.g., a “ping-pong” cycle) can lead to the destruction of transposon mRNA in the germ line. The piRNA model was redrawn based on concepts and models presented in the following papers30, 151, 157, and for the example illustrated is from Drosophila; however, a similar pathway likely operates in mammalian cells (see text for details).
Small RNAs inhibit TEs
Small RNA-based mechanisms (including, endogenous small interfering RNAs (endo-siRNAs) and Piwi-interacting RNAs (piRNAs)), also act to defend eukaryotic cells against TEs. The mechanisms by which these small RNAs are generated and how they inhibit TEs remain an active area of investigation in various model organisms. Mechanistic details regarding these processes can be found in many outstanding reviews on this topic(e.g., 30, 147–152); here we briefly summarize common themes that have emerged from the above studies.
Endo-siRNAs have the potential to inhibit TE mobility through the post-transcriptional disruption of transposon mRNA. For example, double-strand “trigger” RNAs (ds-RNAs) can be derived from the complementary inverted terminal repeats in DNA transposons, from structured mRNA transcripts, or from overlapping regions contained within convergent transcription units30, 147, 148, 153. The resultant ds-RNAs then can be processed into ~21–24 nt endo-siRNAs by members of the Dicer family of proteins (Fig. 3a)30, 154. These endo-siRNAs are loaded onto an Argonaute protein, and the “passenger” RNA strand (typically the sense strand of a TE) is degraded. The remaining complex of a single stranded RNA and Argonaute is called the RNA-Induced Silencing Complex (RISC); the RNA directs RISC to complementary sequences in target mRNAs, leading to their post-transcriptional degradation. Importantly, the RNAi machinery has the capability to inhibit any TE that generates a ds-RNA “trigger” that can serve as a substrate for the RNAi machinery.
By a different mechanism, piRNAs can be generated from genomic loci encoding long precursor RNAs that contain the remnants of different families of TEs151. In general, processing of these precursor RNAs leads to the production of a mature ~24–35 nt piRNAs (Fig 3b). A subfamily of Argonaute proteins, known as the Piwi clade of proteins, predominantly binds mature antisense piRNAs and directs them to complementary sequences in TE mRNA. An endonuclease activity associated with the Piwi protein cleaves the TE mRNA to release a sense strand piRNA, which can interact with other Piwi clade proteins. Binding of this complex to the original piRNA precursor RNA then reiterates this amplification cycle by a “ping-pong” mechanism151, 155. In addition to this type of mechanism that restricts TE mobility in the germline, recent studies suggest that specialized piRNA pathways, which do not operate via a ping-pong mechanism, might restrict somatic TE activity32, 155–157.
Examples of TEs that are controlled by small RNA-based mechanisms include Tc1 transposons in C. elegans, and P elements in Drosophila153, 158. Also, 21nt small interfering RNAs (siRNA) derived from the Athila family LTR-retrotransposons in the vegetative nucleus of the pollen grains in Arabidopsis thaliana are delivered to the sperm cells to inhibit expression of transposons that, in principle, could mobilize in the germline159.
Small RNA based mechanisms also may be critical for silencing mammalian L1s. For example, an antisense promoter located within the human L1 5’UTR allows the production of an antisense RNA that, in principle, could base pair with sense strand L1 mRNA to establish a ds-RNA substrate for Dicer160. Furthermore, mouse mutants lacking the murine Piwi family proteins MILI or MIWI2 exhibit a loss of L1 and intracisternal A particle (IAP) LTR-retrotransposon DNA methylation,; this loss correlates with their transcriptional activation in male germ cells161. Similarly, mice lacking a MILI interacting protein, Tudor containing protein-1, exhibit a similar loss of L1 DNA methylation and a reactivation of L1 expression162. Finally, mouse mutants lacking the non-canonical Maelstrom protein, a component of the nuage complex that may be important for small RNA biogenesis, exhibit de-repression of L1 transcription, an increase of L1 ribonucleoprotein particle intermediates in spermatids, and a chromosomal synapsis defect during male meiosis163. Together, the above examples provide compelling data that small RNA-based pathways likely act to control the expression of certain TEs in the mammalian germline.
Finally, it is noteworthy that other antisense RNA-based mechanisms may be involved in TE silencing. For example, antisense transcripts from S. cerevisiae Ty1 elements reduce Ty1 IN and RT protein levels by a post-translational mechanism; this leads to inhibition of Ty1 mobility and thus controls Ty1 copy number164. Since S. cerevisiae lacks RNAi machinery, these results suggest that genomes have evolved other RNA-dependent strategies to tame TEs.
Cytosine deaminases and DNA repair factors restrict TEs
Proteins involved in nucleic acid metabolism and/or DNA repair can also restrict TE mobility. For example, members of the APOBEC3 family of cytidine deaminases can restrict the retrotransposition of a various retroviruses and LTR and non-LTR retrotransposons33. For retroviruses and LTR-retrotransposons, APOBEC3 proteins generally deaminate cytidines during the first strand cDNA synthesis, which leads to either cDNA degradation or the integration of a mutated provirus. The mechanisms by which certain APOBEC3 proteins restrict non-LTR retrotransposons require elucidation. Similarly, over-expression of the 3’-repair exonuclease 1 (Trex1) gene, mutations in which cause Aicardi-Goutieres syndrome, can inhibit L1 and IAP retrotransposition in cultured cell assays165, 166, but the mechanism of Trex1-mediated TE repression requires elucidation.
Other mechanisms are also likely to restrict the mobility of non-LTR retrotransposons. For example, the overwhelming majority of L1 elements in mammalian genomes are 5’ truncated and are essentially “dead on arrival” because they cannot synthesize proteins critical for retrotransposition167. It has been proposed that 5’ truncation may be due to the low processivity of the L1-encoded reverse transcriptase. However, recent work on the reverse transcriptase encoded by the R2 non-LTR retrotransposon of Bombyx mori demonstrated that this enzyme is more processive than the reverse transcriptases encoded by retroviruses168. Alternatively, L1 5’ truncation might result if host factors cause the dissociation of the L1 reverse transcriptase from the nascent cDNA and/or degrade the L1 mRNA during integration. In this scenario, to generate a full-length insertion the L1 RT would need to complete integration before the TPRT intermediate is recognized as DNA damage by the host50, 169. Indeed, proteins involved in the non-homologous end-joining pathway of DNA repair seem to act to restrict the retrotransposition of a zebrafish LINE-2 element in DT40 chicken cells170, whereas members of DNA excision repair pathway (that is, ERCC1/XPF1) might restrict L1 retrotransposition in cultured human cells171.
Finally, in addition to recognizing the L1 integration intermediate as a form of DNA damage, recent data suggests that retrotransposition indicator cassettes delivered by engineered L1s in human embryonic carcinoma cell lines can be epigenetically silenced during or immediately after their integration into genomic DNA172. Given that L1 is an ancient “stowaway” in mammalian genomes, it is likely that the host has evolved multiple mechanisms to combat L1 mobility at discrete steps in the retrotransposition pathway, and that some of these mechanisms operate in a context dependent manner. Clearly, continued studies will reveal new and more diverse host mechanisms to restrict TE mobility.
Developmental triggers of transposition
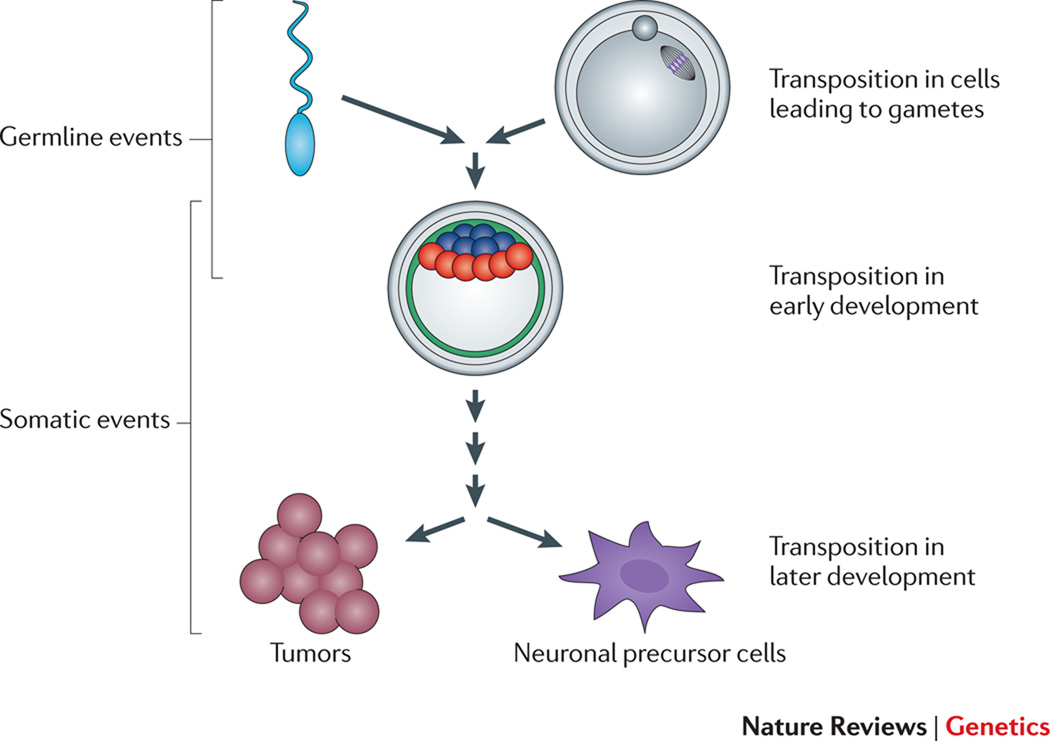
Despite mechanisms to combat TE mobility, TEs continue to thrive in many host genomes. Thus, TEs must have evolved ways to either overwhelm or counteract these host defenses. TEs must mobilize in germ cells or during early development to ensure their survival (Figure 4). However, some TEs can mobilize in somatic cells, providing a potential mechanism to generate intra-individual genetic variation.
Figure 4. Timing of transposition.
TE mobility in cells giving rise to gametes, as well as TE mobility post-fertilization during early development, can lead to germline TE integration events. Embryonic TE mobility in cells that do not contribute to the germline, or at later times in development can, in principle, lead to somatic TE integration events. The overlapping brackets signify that some TE insertions in early development can contribute to the germline, whereas others may not. Endogenous L1 retrotransposition events can occur in certain tumors, and experiments using engineered human L1s suggest that L1 retrotransposition also may occur during mammalian neurogenesis. Examples of the developmental timing of TE integration events are described in the main text.
Transposition in the germ line or during early development
Drosophila P elements provide one of the best-studied cases of cell type specific transposition173. P element transposition occurs when females lacking P elements (the M cytotype) mate with males carrying P elements (the P cytotype); P element mobilization can cause hybrid dysgenesis in the offspring. In the reciprocal cross, eggs from P cytotype females produce a repressor protein and piRNAs that inhibit P element transposition57, 158. The repressor is an alternatively spliced truncated form of the transposase. Importantly, the repressor not only controls which crosses produce germline integration, but also inhibits transposition in the soma.
The Drosophila gypsy element is another example of a TE that exhibits tissue specific control174, 175. Gypsy transcription is induced in somatic follicle cells that surround the oocyte. The TE mRNA assembles into virus-like particles that are thought to traffic to the oocyte to carry out transposition. It remains unclear whether the transfer of gypsy virus-like particles to the oocyte occurs via an enveloped particle (similar to retroviruses) or by a form of endocytosis. However, the Flamenco locus encodes piRNAs that silence gypsy elements in follicle cells, thereby preventing the spread of these TEs to the surrounding germ cells155.
Relatively little is known about the developmental timing of L1 retrotransposition in mammals. The sheer numbers of L1 and Alu retrotransposons that populate mammalian genomes provide prima facie evidence that they mobilize in the germline. Various studies, using endogenous and engineered L1s, provide strong experimental evidence to back this assertion. For example, full-length mouse L1 RNA and the mouse L1 ORF1-encoded protein are co-expressed in leptotene and zygotene spermatocytes during meiotic prophase176. In addition, the mouse ORF1 protein is expressed in the cytoplasm during specific stages of development in oocytes177. Similarly, human oocytes express L1 RNA and support the retrotransposition of an engineered human L1178. Finally, transgenic mouse experiments demonstrated that an engineered human L1 retrotransposon, whose expression is driven from a heterologous pPol II promoter, can retrotranspose in male germ cells179.
Unexpectedly, a growing body of experimental evidence suggests that L1 retrotransposition also might occur frequently during early development (Figure 4) (also reviewed in ref. 5). For example, human embryonic stem cells can express L1 RNA and ORF1 protein, and accommodate the retrotransposition of engineered L1s, albeit at lower levels than in other types of transformed human cells19, 180. In addition, studies of a male patient with X-linked choroideremia revealed that his mother had mosaicism for the mutagenic L1 insertion in both germline and somatic tissues20. Thus, the initial retrotransposition event must have occurred during early embryogenesis in the mother. Finally, recent transgenic experiments conducted in rats and mice led to the conclusion that most L1 retrotransposition occurs during early embryogenesis and that most of the resultant events are not heritable21. Intriguingly, these data suggest that L1 ribonucleoprotein particles can be deposited into zygotes by either the sperm or egg to undergo retrotransposition during early development, thereby providing a possible mechanism to generate somatic mosaicism and intra-individual genetic variation (see below).
Somatic transposition
Classical experiments in maize revealed that DNA TE activity in somatic tissues could lead to variegated corn color phenotypes1, 181. Since that time, somatic TE events also have been reported in other organisms. For example, it is well established that Tc1 transposition in the Bergerac strain of C. elegans preferentially occurs in somatic cells182. Similarly, a recent study has revealed that somatic transposition of a DNA TE (Hatvine1-rrm) into the promoter region of the VvTFL1A gene of the grapevine cultivar Carnigan affects the grapevine branching pattern and size of fruit clusters183. Also, a mutagenic L1 insertion was identified in the adenomatous polyposis coli (APC) gene in tumor tissue, but not in the surrounding tissue, of a patient with colon cancer, suggesting a role for the insertion in cancer development22. Together with the transgenic L1 experiments (discussed above), these findings establish that somatic TE mobility can lead to phenotypic changes in the host.
Intriguingly, several lines of evidence suggest that somatic L1 retrotransposition may also occur in the mammalian nervous system (also reviewed in ref. 5). First, an engineered human L1 can retrotranspose in neurogenic zones of the brain in transgenic mice24 when its expression is driven by a promoter contained within its native 5’ UTR184. Second, engineered human L1s can retrotranspose in cultured rat neuronal progenitor cells (NPCs), human embryonic stem cell-derived NPCs, and at low levels in human fetal derived NPCs23, 24. Third, sensitive multiplex quantitative PCR experiments suggest a modest increase in L1 copy number in post mortem brain tissue, when compared to heart and liver tissue derived from the same individual23. Finally, retrotransposition of an engineered human L1 is elevated in a mouse model of Rett syndrome (a neurodevelopmental disorder), and induced pluripotent stem cells derived from Rett syndrome patients exhibit an increase in L1 DNA copy number when compared to normal controls, suggesting a potential increase in endogenous L1 retrotransposition185.
The above studies strongly suggest that certain neuronal cells may be permissive for L1 retrotransposition. However, additional research is needed to truly understand the impact of L1 retrotransposition in the brain. For example, recent advances in DNA sequencing technology should provide a means to directly test whether L1 DNA copy number changes detected in quantitative PCR experiments represent actual de novo endogenous retrotransposition events or result from other forms of genomic instability reported in neurons186, 187. Similarly, it remains unclear whether endogenous L1 retrotransposition events represent a type of “genomic noise” or whether they have any functional impact on neuronal development. Finally, it remains a mystery why neuronal cells may accommodate L1 retrotransposition at apparently higher levels than other somatic cells. Nonetheless, these studies have unveiled a new area of investigation that surely will be the subject of future work.
Deregulated L1 retrotransposition in cancer cells
A growing body of evidence suggests that L1 retrotransposition may become deregulated in certain cancers. For example, early studies revealed that hypomethylation of the L1 promoter is correlated with increased L1 expression and/or the production of the L1 ORF1-encoded protein in certain tumors188–190. Moreover, engineered human L1s readily retrotranspose in a variety of transformed human and mouse cell lines, but generally show lower levels of retrotransposition activity in “normal” human cells such as fibroblasts(e.g.,11, 191, 192). Consistent with this, recent findings using second-generation DNA sequencing revealed a total of 9 de novo L1 retrotransposition events in 6 of 20 examined non-small cell lung tumors12. Intriguingly, the tumors containing the new L1 insertions also exhibited a specific genome-wide hypomethylation signature, which is consistent with the notion that altering the epigenome can create a permissive environment for L1 expression and/or retrotransposition, and perhaps the retrotransposition of other classes of non-LTR retrotransposons. Clearly, further innovations in DNA sequencing of heterogeneous cell populations will be critical to reveal patterns of TE activity in diverse tumors. The challenge then will be to determine whether all these TE insertions are “passenger” mutations that are a consequence of the altered cellular milieu of cancer cells or whether some act as “drivers” to promote tumorigenesis.
Closing Remarks
It is undeniable that TEs have played important roles in structuring genomes and generating genetic diversity. By understanding how, when, and where TEs integrate, and how the host responds to this ever-present threat, we will unveil the dynamic forces that shape our genomes. Indeed, we are now able to critically evaluate the McClintock doctrine and future experiments should allow valuable insight into whether the increases in TE transcription caused by environmental stress lead to higher levels of TE integration, and whether these insertions impact host phenotypes and/or survival.
It remains a curiosity why sequences without any apparent purpose continue to thrive in genomes. What is clear is that an understanding of TE biology is necessary to understand genome biology. It is intriguing to speculate that some phenotypic differences among organisms and/or between individuals are due to the effects of TEs. These speculations require rigorous experimental tests. However, the coming years should be an exciting time for TE biology.
At a glance summary.
Many TEs employ highly specific mechanisms to direct integration to sites in the host genome that lack coding information. This minimizes damage to the host genome that occurs during integration.
High throughput sequencing allows the identification of a saturated map of targeted integration sites in S. pombe.
Host organisms have evolved a diverse array of mechanisms to combat TE activity. Examples include DNA methylation of TEs, siRNA based degradation of TE mRNA, and APOBEC mediated cytosine deamination of TE sequences.
Studies of diverse human populations revealed significantly higher numbers of active L1 elements than exist in the human genome reference sequence.
Recent experiments unexpectedly discovered TE integration in somatic cells. These include insertions of L1 in non-small cell lung tumors. Several lines of evidence suggest that somatic L1 retrotransposition may also occur in the mammalian nervous system.
Acknowledgements
We thank Dr. John Kim, Dr. José Garcia-Perez, and members of the Moran lab for critical reading of the manuscript. H.L. was supported in part by the Intramural Research Program of the NIH from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. H.L. received additional support from the Intramural AIDS Targeted Antiviral Program. J.V.M. was supported in part by grants from the National Institutes of Health (GM060518 and GM082970). J.V.M. also is an Investigator of the Howard Hughes Medical Institute.
Glossary
- LTR
A terminal repeated sequence present at the ends of LTR-retrotransposons. The LTR contains cis-acting sequences that allow the transcription and polyadenylation of retrotransposon mRNA. The LTRs also play critical roles in the reverse transcription of LTR-retrotransposon mRNA.
- Virus-like particles (VLPs)
A cytoplasmic particle that comprises LTR-retrotransposon mRNA, the LTR-retrotransposon-encoded proteins, and host factors that are required for reverse transcription of LTR-retrotransposon mRNA. LTR-retrotransposon mRNA is reverse transcribed into a double stranded cDNA within VLPs.
- Long Interspersed Element (LINE)
A family of autonomous non-LTR retrotransposons that mobilize by retrotransposition.
- Short Interspersed Elements
A family of non-autonomous retrotransposons that require functional protein(s) encoded by LINE elements to mediate their retrotransposition.
- LINE-1 or L1
An abundant family of autonomous non-LTR retrotransposons in mammalian genomes. In humans, L1 elements comprise roughly 17% of genomic DNA. The vast majority of L1s are inactive; however, it is estimated that an average human genome contains ~80–100 active elements (reviewed in ref. 5).
- Alu elements (Alus)
An abundant class of SINEs that comprise ~10% of human genomic DNA. Alu elements require the endonuclease and reverse transcriptase activities contained within the L1 ORF2-encoded protein to mediate their mobility. Some Alu elements remain active in the human genome.
- SINE-R—VNTR—Alu (SVA) elements
A composite non-autonomous retrotransposon that also requires the L1 encoded proteins to mediate its mobility. SVA elements are less abundant than Alu elements, although certain families of SVA elements remain active in the human genome.
- Target-site primed reverse transcription (TPRT)
The mechanism of mobility generally employed by LINEs and SINEs. An endonuclease encoded by the LINE nicks genomic DNA to expose a 3’ hydroxyl residue at the target site that can be used as a primer to initiate the reverse transcription of the retrotransposon RNA by a LINE-encoded reverse transcriptase.
- RNA-directed DNA methylation
A pathway in which 24 nt small RNAs interact with a de novo methyltransferase to mediate the methylation and transcriptional silencing of homologous genomic loci in plants.
- Dicer
A family of RNAse III proteins that possess an endonuclease activity that can process double strand “trigger” RNAs into small-interfering RNAs (siRNAs) and microRNAs (miRNAs).
- Small interfering RNAs (siRNAs)
A family of small (~21–24 nt) RNAs that are generated from double-strand “trigger” RNAs by Dicer-dependent and Dicer-independent mechanisms. These small RNAs bind Argonaute proteins and guide the resultant complex to complementary mRNAs in a cell to mediate their post-transcriptional destruction.
- Argonaute proteins
The proteins that mediate the small RNA-induced silencing processes. Argonaute proteins bind small RNAs and are the defining component of the RNA Induced Silencing Complex (RISC). The small RNAs guide Argonaute proteins to target mRNAs to mediate post-transcriptional degradation and/or translational silencing. Argonaute proteins possess a single strand RNA binding domain (PAZ) and a ribonuclease domain (Piwi).
- Piwi proteins
A specialized class of Argonaute proteins that interact with piRNAs to mediate transposable element silencing. Members include: Piwi, Aubergine, and Argonaute 3 in Drosophila; MIWI1, MIWI2 and MILI in mice, and HIWI1, HIW12, HIWI3, and HILI in humans.
- Piwi-interacting RNAs (piRNA) cluster
A genomic DNA locus that encodes piRNA precursor RNAs. Many piRNA clusters contain sense and anti-sense sequences derived from mobile genetic elements. An example of a piRNA cluster is the Flamenco locus of Drosophila.
- Piwi-interacting RNAs (piRNAs)
A family of small (~24–35 nt) RNAs that are processed from piRNA precursor mRNAs. The mature piRNAs interact with specialized Argonaute proteins (from the Piwi clade), to mediate RNA silencing.
- Hybrid dysgenesis
A syndrome, which includes sterility, induced by the mobilization of P elements in crosses between females lacking P elements (the M cytotype) and males containing P elements (the P cytotype).
- X-linked choroideremia
A recessive degenerative retinal disease.
- Desmoid tumor
A soft tissue tumor that can arise in the abdomen as well as other parts of the body. Desmoid tumors generally are benign and grow slowly.
- Aicardi-Goutieres Syndrome
A rare, autosomal recessive genetic disorder that leads to brain dysfunction as well as other symptoms. The early onset form of the disease can be caused by mutations in the Trex1 gene and is usually fatal.
Biographies
Henry L. Levin heads the Section on Eukaryotic Transposable Elements in the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH. He received his Ph.D. in molecular biology at the University of California, Berkeley with Dr. Howard Schachman and completed postdoctoral research at Johns Hopkins University School of Medicine with Dr. Jef Boeke. Over the past 18 years Levin’s studies of LTR retrotransposons in fission yeast have identified mechanistic details of particle formation, reverse transcription, and integration. Recently, work in the Levin lab has focused on the mechanisms that control the position of integration.
John V. Moran currently is the Gilbert S. Omenn Collegiate Professor of Human Genetics at the University of Michigan Medical School in Ann Arbor, Michigan, USA. He also is an Investigator of the Howard Hughes Medical Institute. He received his Ph.D. in biochemistry from the University of Texas Southwestern Medical Center in Dallas, Texas, USA, and conducted his postdoctoral studies in the laboratory of Dr. Haig H. Kazazian Jr. at Johns Hopkins University School of Medicine in Baltimore, MD, USA and at the University of Pennsylvania School of Medicine in Philadelphia, PA, USA. His laboratory uses genetic, molecular biological, biochemical, and genomic approaches to understand the biology of human LINE-1 retrotransposons.
References
- 1.McClintock B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci U S A. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 4.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 Elements in Structural Variation and Disease. Annu Rev Genomics Hum Genet. 2011;12 doi: 10.1146/annurev-genom-082509-141802. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orgel LE, Crick FH, Sapienza C. Selfish DNA. Nature. 1980;288:645–646. doi: 10.1038/288645a0. [DOI] [PubMed] [Google Scholar]
- 7.Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 8.Bestor TH. Sex brings transposons and genomes into conflict. Genetica. 1999;107:289–295. [PubMed] [Google Scholar]
- 9.Hickey DA. Selfish DNA: a sexually-transmitted nuclear parasite. Genetics. 1982;101:519–531. doi: 10.1093/genetics/101.3-4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 11.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 12. Iskow RC, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. -- This study, along with references 13–18 and 70–74, used second generation DNA sequencing and/or modern genomic approaches to demonstrate that TEs continue to have an ongoing impact on human genome structural variation.
- 13.Beck CR, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang CR, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witherspoon DJ, et al. Mobile element scanning (ME-Scan) by targeted high-throughput sequencing. BMC Genomics. 2010;11:410. doi: 10.1186/1471-2164-11-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hormozdiari F, et al. Alu repeat discovery and characterization within human genomes. Genome Res. 2011 doi: 10.1101/gr.115956.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewing AD, Kazazian HH., Jr High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20:1262–1270. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ewing AD, Kazazian HH., Jr Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 2011 doi: 10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garcia-Perez JL, et al. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. -- This study (as well as references 20 and 21) provide evidence that L1s can retrotranspose during early embryonic development.
- 20.van den Hurk JA, et al. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16:1587–1592. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- 21.Kano H, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miki Y, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- 23. Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. -- This paper (as well as references 24 and 185) suggest that engineered human L1s, and perhaps endogenous L1s, can retrotranspose in somatic cells of the mammalian nervous system.
- 24.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 25.Sandmeyer S. Integration by design. Proc Natl Acad Sci U S A. 2003;100:5586–5588. doi: 10.1073/pnas.1031802100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leem YE, et al. Retrotransposon Tf1 is targeted to pol II promoters by transcription activators. Molecular Cell. 2008;30:98–107. doi: 10.1016/j.molcel.2008.02.016. -- This paper demonstrates a mechanism by which the S. pombe Tf1 retrotransposon can target RNA polymerase II transcribed genes.
- 27.Devine SE, Boeke JD. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 28.Eickbush TH. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, D.C.: ASM Press; 2002. pp. 813–835. [Google Scholar]
- 29.Winckler T, Dingermann T, Glockner G. Dictyostelium mobile elements: strategies to amplify in a compact genome. Cell Mol Life Sci. 2002;59:2097–2111. doi: 10.1007/s000180200010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bestor TH, Bourc'his D. Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb Symp Quant Biol. 2004;69:381–387. doi: 10.1101/sqb.2004.69.381. [DOI] [PubMed] [Google Scholar]
- 32.Golden DE, Gerbasi VR, Sontheimer EJ. An inside job for siRNAs. Mol Cell. 2008;31:309–312. doi: 10.1016/j.molcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 34.Sehgal A, Lee CY, Espenshade PJ. SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS Genet. 2007;3:e131. doi: 10.1371/journal.pgen.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dai J, Xie W, Brady TL, Gao J, Voytas DF. Phosphorylation Regulates Integration of the Yeast Ty5 Retrotransposon into Heterochromatin. Molecular Cell. 2007;27:289–299. doi: 10.1016/j.molcel.2007.06.010. -- This paper demonstrates that TEs can respond to stress by changing their targeting sites. When cells lack access to nitrogen, Ty5 no longer integrates into heterochromatin, but instead targets coding sequences in the genome.
- 36.Grandbastien MA, et al. Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae. Cytogenet Genome Res. 2005;110:229–241. doi: 10.1159/000084957. [DOI] [PubMed] [Google Scholar]
- 37.Hirochika H. Activation of tobacco retrotransposons during tissue culture. EMBO J. 1993;12:2521–2528. doi: 10.1002/j.1460-2075.1993.tb05907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Courtial B, et al. Tnt1 transposition events are induced by in vitro transformation of Arabidopsis thaliana, and transposed copies integrate into genes. Mol Genet Genomics. 2001;265:32–42. doi: 10.1007/s004380000387. [DOI] [PubMed] [Google Scholar]
- 39.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 40.Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington, D.C.: ASM Press; 2002. [Google Scholar]
- 41.Kleckner N. Regulation of transposition in bacteria. Annu Rev Cell Biol. 1990;6:297–327. doi: 10.1146/annurev.cb.06.110190.001501. [DOI] [PubMed] [Google Scholar]
- 42.Garfinkel DJ, Boeke JD, Fink GR. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985;42:507–517. doi: 10.1016/0092-8674(85)90108-4. [DOI] [PubMed] [Google Scholar]
- 43.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 44.Eichinger DJ, Boeke JD. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell. 1988;54:955–966. doi: 10.1016/0092-8674(88)90110-9. [DOI] [PubMed] [Google Scholar]
- 45.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 46.Kajikawa M, Okada N. LINEs mobilize SINEs in the eel through a shared 3' sequence. Cell. 2002;111:433–444. doi: 10.1016/s0092-8674(02)01041-3. [DOI] [PubMed] [Google Scholar]
- 47.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 48.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH., Jr Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 2007;17:602–611. doi: 10.1101/gr.5870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buzdin A, et al. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3' terminus of l1. Genomics. 2002;80:402–406. doi: 10.1006/geno.2002.6843. [DOI] [PubMed] [Google Scholar]
- 52.Weber MJ. Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genet. 2006;2:e205. doi: 10.1371/journal.pgen.0020205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei W, et al. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 55.Reznikoff WS. Tn5 transposition: a molecular tool for studying protein structure-function. Biochem Soc Trans. 2006;34:320–323. doi: 10.1042/BST20060320. [DOI] [PubMed] [Google Scholar]
- 56.Peters JE, Craig NL. Tn7: smarter than we thought. Nat Rev Mol Cell Biol. 2001;2:806–814. doi: 10.1038/35099006. [DOI] [PubMed] [Google Scholar]
- 57.Rio DC. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, D.C.: ASM Press; 2002. pp. 484–518. [Google Scholar]
- 58.Plasterk RH. The Tc1/mariner transposon family. Curr Top Microbiol Immunol. 1996;204:125–143. doi: 10.1007/978-3-642-79795-8_6. [DOI] [PubMed] [Google Scholar]
- 59.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 60. Ray DA, Pagan HJ, Thompson ML, Stevens RD. Bats with hATs: evidence for recent DNA transposon activity in genus Myotis. Mol Biol Evol. 2007;24:632–639. doi: 10.1093/molbev/msl192. -- This paper (as well as references 61 and 62) reveal that certain DNA transposons are active in the Myotis bat genus.
- 61.Pritham EJ, Feschotte C. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc Natl Acad Sci U S A. 2007;104:1895–1900. doi: 10.1073/pnas.0609601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ray DA, et al. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res. 2008;18:717–728. doi: 10.1101/gr.071886.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bowen NJ, McDonald JF. Drosophila euchromatic LTR retrotransposons are much younger than the host species in which they reside. Genome Res. 2001;11:1527–1540. doi: 10.1101/gr.164201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schnable PS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 65.SanMiguel P, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [see comments] [DOI] [PubMed] [Google Scholar]
- 66.Maksakova IA, et al. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moyes D, Griffiths DJ, Venables PJ. Insertional polymorphisms: a new lease of life for endogenous retroviruses in human disease. Trends Genet. 2007;23:326–333. doi: 10.1016/j.tig.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 68.Badge RM, Alisch RS, Moran JV. ATLAS: a system to selectively identify human-specific L1 insertions. Am J Hum Genet. 2003;72:823–838. doi: 10.1086/373939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sheen FM, et al. Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome Res. 2000;10:1496–1508. doi: 10.1101/gr.149400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kidd JM, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mills RE, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuduvalli PN, Rao JE, Craig NL. Target DNA structure plays a critical role in Tn7 transposition. EMBO J. 2001;20:924–932. doi: 10.1093/emboj/20.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Craig NL. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, D.C.: ASM Press; 2002. pp. 423–456. [Google Scholar]
- 77.Wolkow CA, DeBoy RT, Craig NL. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 1996;10:2145–2157. doi: 10.1101/gad.10.17.2145. [DOI] [PubMed] [Google Scholar]
- 78.Peters JE, Craig NL. Tn7 transposes proximal to DNA double-strand breaks and into regions where chromosomal DNA replication terminates. Mol Cell. 2000;6:573–582. doi: 10.1016/s1097-2765(00)00056-3. [DOI] [PubMed] [Google Scholar]
- 79.Bellen HJ, et al. The Drosophila Gene Disruption Project: Progress Using Transposons With Distinctive Site-Specificities. Genetics. 2011 doi: 10.1534/genetics.111.126995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feng Q, Moran J, Kazazian H, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 81.Feng Q, Schumann G, Boeke JD. Retrotransposon R1Bm endonuclease cleaves the target sequence. Proc Natl Acad Sci U S A. 1998;95:2083–2088. doi: 10.1073/pnas.95.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang J, Malik HS, Eickbush TH. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc Natl Acad Sci U S A. 1999;96:7847–7852. doi: 10.1073/pnas.96.14.7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bushman FD. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell. 2003;115:135–138. doi: 10.1016/s0092-8674(03)00760-8. [DOI] [PubMed] [Google Scholar]
- 84.Lesage P, Todeschini AL. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet Genome Res. 2005;110:70–90. doi: 10.1159/000084940. [DOI] [PubMed] [Google Scholar]
- 85.Kirchner J, Connolly CM, Sandmeyer SB. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element [see comments] Science. 1995;267:1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- 86.Chalker DL, Sandmeyer SB. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 1992;6:117–128. doi: 10.1101/gad.6.1.117. [DOI] [PubMed] [Google Scholar]
- 87.Yieh L, Hatzis H, Kassavetis G, Sandmeyer SB. Mutational analysis of the transcription factor IIIB-DNA target of Ty3 retroelement integration. Journal of Biological Chemistry. 2002;277:25920–25928. doi: 10.1074/jbc.M202729200. [DOI] [PubMed] [Google Scholar]
- 88.Yieh L, Kassavetis G, Geiduschek EP, Sandmeyer SB. The Brf and TATA-binding protein subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element, Ty3. Journal of Biological Chemistry. 2000;275:29800–29807. doi: 10.1074/jbc.M003149200. [DOI] [PubMed] [Google Scholar]
- 89.Bachman N, Eby Y, Boeke JD. Local definition of Ty1 target preference by long terminal repeats and clustered tRNA genes. Genome Res. 2004;14:1232–1247. doi: 10.1101/gr.2052904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bachman N, Gelbart ME, Tsukiyama T, Boeke JD. TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev. 2005;19:955. doi: 10.1101/gad.1299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bolton EC, Boeke JD. Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: a genomic point of view. Genome Res. 2003;13:254–263. doi: 10.1101/gr.612203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kinsey PT, Sandmeyer SB. Adjacent pol II and pol III promoters: transcription of the yeast retrotransposon Ty3 and a target tRNA gene. Nucleic Acids Res. 1991;19:1317–1324. doi: 10.1093/nar/19.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hofmann J, et al. Transfer RNA genes from Dictyostelium discoideum are frequently associated with repetitive elements and contain consensus boxes in their 5' and 3'-flanking regions. J Mol Biol. 1991;222:537–552. doi: 10.1016/0022-2836(91)90495-r. [DOI] [PubMed] [Google Scholar]
- 94.Chung T, Siol O, Dingermann T, Winckler T. Protein interactions involved in tRNA gene-specific integration of Dictyostelium discoideum non-long terminal repeat retrotransposon TRE5-A. Mol Cell Biol. 2007;27:8492–8501. doi: 10.1128/MCB.01173-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Behrens R, Hayles J, Nurse P. Fission yeast retrotransposon Tf1 integration is targeted to 5' ends of open reading frames. Nucleic Acids Res. 2000;28:4709–4716. doi: 10.1093/nar/28.23.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Guo Y, Levin HL. High-throughput sequencing of retrotransposon integration provides a saturated profile of target activity in Schizosaccharomyces pombe. Genome Res. 2010;20:239–248. doi: 10.1101/gr.099648.109. -- This study describes, for the first time, the sequencing of a saturated set of insertion sites that actively are targeted by a TE.
- 97.Singleton TL, Levin HL. A Long Terminal Repeat Retrotransposon of Fission Yeast Has Strong Preferences for Specific Sites of Insertion. Eukaryotic Cell. 2002;1:44–55. doi: 10.1128/EC.01.1.44-55.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Neely LA, Hoffman CS. Protein kinase A and mitogen-activated protein kinase pathways antagonistically regulate fission yeast fbp1 transcription by employing different modes of action at two upstream activation sites. Molecular and Cellular Biology. 2000;20:6426–6434. doi: 10.1128/mcb.20.17.6426-6434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Majumdar A, Chatterjee AG, Ripmaster TL, Levin HL. The determinants that specify the integration pattern of retrotransposon Tf1 in the fbp1 promoter of Schizosaccharomyces pombe. J Virol. 2010;85:519–529. doi: 10.1128/JVI.01719-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen D, et al. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Malik HS, Henikoff S, Eickbush TH. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000;10:1307–1318. doi: 10.1101/gr.145000. [DOI] [PubMed] [Google Scholar]
- 102.Mitchell RS, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ciuffi A. Mechanisms governing lentivirus integration site selection. Curr Gene Ther. 2008;8:419–429. doi: 10.2174/156652308786848021. [DOI] [PubMed] [Google Scholar]
- 104.Ciuffi A, Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006;22:388–395. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 105.Engelman A, Cherepanov P. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 2008;4:e1000046. doi: 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Poeschla EM. Integrase, LEDGF/p75 and HIV replication. Cell Mol Life Sci. 2008;65:1403–1424. doi: 10.1007/s00018-008-7540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Malik HS, Eickbush TH. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J Virol. 1999;73:5186–5190. doi: 10.1128/jvi.73.6.5186-5190.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao X, Hou Y, Ebina H, Levin HL, Voytas DF. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 2008;18:359–369. doi: 10.1101/gr.7146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zou S, Voytas DF. Silent chromatin determines target preference of the Saccharomyces retrotransposon Ty5. Proc Natl Acad Sci U S A. 1997;94:7412–7416. doi: 10.1073/pnas.94.14.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zou S, Wright DA, Voytas DF. The Saccharomyces Ty5 retrotransposon family is associated with origins of DNA replication at the telomeres and the silent mating locus HMR. Proc Natl Acad Sci U S A. 1995;92:920–924. doi: 10.1073/pnas.92.3.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zou S, Ke N, Kim JM, Voytas DF. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 1996;10:634–645. doi: 10.1101/gad.10.5.634. [DOI] [PubMed] [Google Scholar]
- 112.Gai X, Voytas DF. A single amino acid change in the yeast retrotransposon Ty5 abolishes targeting to silent chromatin. Mol Cell. 1998;1:1051–1055. doi: 10.1016/s1097-2765(00)80105-7. [DOI] [PubMed] [Google Scholar]
- 113.Xie W, et al. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol Cell Biol. 2001;21:6606–6614. doi: 10.1128/MCB.21.19.6606-6614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhu Y, Dai J, Fuerst PG, Voytas DF. From the Cover: Controlling integration specificity of a yeast retrotransposon. Proc Natl Acad Sci U S A. 2003;100:5891–5895. doi: 10.1073/pnas.1036705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brady TL, Fuerst PG, Dick RA, Schmidt C, Voytas DF. Retrotransposon target site selection by imitation of a cellular protein. Mol Cell Biol. 2008;28:1230–1239. doi: 10.1128/MCB.01502-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.George JA, Traverse KL, DeBaryshe PG, Kelley KJ, Pardue ML. Evolution of diverse mechanisms for protecting chromosome ends by Drosophila TART telomere retrotransposons. Proc Natl Acad Sci U S A. 2010;107:21052–21057. doi: 10.1073/pnas.1015926107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Biessmann H, et al. HeT-A, a transposable element specifically involved in "healing" broken chromosome ends in Drosophila melanogaster. Mol Cell Biol. 1992;12:3910–3918. doi: 10.1128/mcb.12.9.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fujiwara H, Osanai M, Matsumoto T, Kojima KK. Telomere-specific non-LTR retrotransposons and telomere maintenance in the silkworm, Bombyx mori. Chromosome Res. 2005;13:455–467. doi: 10.1007/s10577-005-0990-9. [DOI] [PubMed] [Google Scholar]
- 119.Gladyshev EA, Arkhipova IR. A subtelomeric non-LTR retrotransposon Hebe in the bdelloid rotifer Adineta vaga is subject to inactivation by deletions but not 5' truncations. Mob DNA. 2010;1:12. doi: 10.1186/1759-8753-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Morrish TA, et al. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–212. doi: 10.1038/nature05560. -- This paper (as well as reference 122) reveal pathways by which retrotransposons can use telomeric sequences as integration substrates.
- 121.Morrish TA, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 122.Gladyshev EA, Arkhipova IR. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci U S A. 2007;104:9352–9357. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 124.Symer DE, et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 125.Harrow J, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;(7 Suppl 1) S4:1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Coffey AJ, et al. The GENCODE exome: sequencing the complete human exome. Eur J Hum Genet. 2011;19:827–831. doi: 10.1038/ejhg.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci U S A. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cost GJ, Boeke JD. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- 129.Korenberg JR, Rykowski MC. Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell. 1988;53:391–400. doi: 10.1016/0092-8674(88)90159-6. [DOI] [PubMed] [Google Scholar]
- 130.Brookfield JF. Selection on Alu sequences? Curr Biol. 2001;11:R900–R901. doi: 10.1016/s0960-9822(01)00547-4. [DOI] [PubMed] [Google Scholar]
- 131.Boissinot S, Entezam A, Furano AV. Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- 132.Boissinot S, Davis J, Entezam A, Petrov D, Furano AV. Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci U S A. 2006;103:9590–9594. doi: 10.1073/pnas.0603334103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 134.Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 135.Conley ME, Partain JD, Norland SM, Shurtleff SA, Kazazian HH., Jr Two independent retrotransposon insertions at the same site within the coding region of BTK. Hum Mutat. 2005;25:324–325. doi: 10.1002/humu.9321. [DOI] [PubMed] [Google Scholar]
- 136.Halling KC, et al. Hereditary desmoid disease in a family with a germline Alu I repeat mutation of the APC gene. Hum Hered. 1999;49:97–102. doi: 10.1159/000022852. [DOI] [PubMed] [Google Scholar]
- 137.Vidaud D, et al. Haemophilia B due to a de novo insertion of a human-specific Alu subfamily member within the coding region of the factor IX gene. Eur J Hum Genet. 1993;1:30–36. doi: 10.1159/000472385. [DOI] [PubMed] [Google Scholar]
- 138.Cost GJ, Golding A, Schlissel MS, Boeke JD. Target DNA chromatinization modulates nicking by L1 endonuclease. Nucleic Acids Res. 2001;29:573–577. doi: 10.1093/nar/29.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Selker EU, et al. The methylated component of the Neurospora crassa genome. Nature. 2003;422:893–897. doi: 10.1038/nature01564. [DOI] [PubMed] [Google Scholar]
- 140.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 141.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 142.Schaefer CB, Ooi SK, Bestor TH, Bourc'his D. Epigenetic decisions in mammalian germ cells. Science. 2007;316:398–399. doi: 10.1126/science.1137544. [DOI] [PubMed] [Google Scholar]
- 143.Maksakova IA, Mager DL, Reiss D. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell Mol Life Sci. 2008;65:3329–3347. doi: 10.1007/s00018-008-8494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Tsukahara S, et al. Bursts of retrotransposition reproduced in Arabidopsis. Nature. 2009;461:423–426. doi: 10.1038/nature08351. -- This paper employed modern genomic approaches to reveal that certain classes of LTR-retrotransposon are mobilized in a DDM1 (decrease in DNA methylation 1) mutant background. It provides an example of how epigenetic changes can lead to TE mobility.
- 145.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 146.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.van Rij RP, Berezikov E. Small RNAs and the control of transposons and viruses in Drosophila. Trends Microbiol. 2009;17:163–171. doi: 10.1016/j.tim.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 148.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 150.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 152.Fischer SE. Small RNA-mediated gene silencing pathways in C. elegans. Int J Biochem Cell Biol. 2010;42:1306–1315. doi: 10.1016/j.biocel.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 153.Sijen T, Plasterk RH. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- 154.Tabara H, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 155. Malone CD, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. -- This paper (as well as references 156, 158, and 159) highlight some pathways by which small RNAs can inhibit TE mobility in either the germline or soma.
- 156.Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010;29:3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zamore PD. Somatic piRNA biogenesis. EMBO J. 2010;29:3219–3221. doi: 10.1038/emboj.2010.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Brennecke J, et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 161. Kuramochi-Miyagawa S, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. -- This paper (as well as references 162 and 163) highlight how the small RNA machinery influences the expression, and perhaps mobility, of certain TEs in mammalian cells.
- 162.Reuter M, et al. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 163.Soper SF, et al. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Matsuda E, Garfinkel DJ. Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc Natl Acad Sci U S A. 2009;106:15657–15662. doi: 10.1073/pnas.0908305106. -- This paper reveals a novel RNA-based mechanism to inhibit Ty1 retrotransposition in S. cerevisiae.
- 165.Crow YJ, et al. Mutations in the gene encoding the 3'-5' DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 166.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Grimaldi G, Singer MF. Members of the KpnI family of long interspersed repeated sequences join and interrupt alpha-satellite in the monkey genome. Nucleic Acids Res. 1983;11:321–338. doi: 10.1093/nar/11.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Babushok DV, Kazazian HH., Jr. Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat. 2007;28:527–539. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- 170.Suzuki J, et al. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 2009;5:e1000461. doi: 10.1371/journal.pgen.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gasior SL, Roy-Engel AM, Deininger PL. ERCC1/XPF limits L1 retrotransposition. DNA Repair (Amst) 2008;7:983–989. doi: 10.1016/j.dnarep.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Garcia-Perez JL, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466:769–773. doi: 10.1038/nature09209. -- This paper describes a host mechanism that is able to epigenetically silence reporter genes delivered into genomic DNA by L1 retrotransposition.
- 173.Rio DC. Regulation of Drosophila P element transposition. Trends Genet. 1991;7:282–287. doi: 10.1016/0168-9525(91)90309-E. [DOI] [PubMed] [Google Scholar]
- 174.Pelisson A, et al. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Prud'homme N, Gans M, Masson M, Terzian C, Bucheton A. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics. 1995;139:697–711. doi: 10.1093/genetics/139.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc Natl Acad Sci U S A. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Georgiou I, et al. Retrotransposon RNA expression and evidence for retrotransposition events in human oocytes. Hum Mol Genet. 2009;18:1221–1228. doi: 10.1093/hmg/ddp022. [DOI] [PubMed] [Google Scholar]
- 179.Ostertag EM, et al. A mouse model of human L1 retrotransposition. Nat Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 180.Macia A, et al. Epigenetic control of retrotransposon expression in human embryonic stem cells. Mol Cell Biol. 2011;31:300–316. doi: 10.1128/MCB.00561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Federoff N. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, D.C.: ASM Press; 2002. pp. 997–1007. [Google Scholar]
- 182.Emmons SW, Yesner L. High-frequency excision of transposable element Tc 1 in the nematode Caenorhabditis elegans is limited to somatic cells. Cell. 1984;36:599–605. doi: 10.1016/0092-8674(84)90339-8. [DOI] [PubMed] [Google Scholar]
- 183.Fernandez L, Torregrosa L, Segura V, Bouquet A, Martinez-Zapater JM. Transposon-induced gene activation as a mechanism generating cluster shape somatic variation in grapevine. Plant J. 2010;61:545–557. doi: 10.1111/j.1365-313X.2009.04090.x. [DOI] [PubMed] [Google Scholar]
- 184.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Muotri AR, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rehen SK, et al. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc Natl Acad Sci U S A. 2001;98:13361–13366. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Westra JW, et al. Neuronal DNA content variation (DCV) with regional and individual differences in the human brain. J Comp Neurol. 2010;518:3981–4000. doi: 10.1002/cne.22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Belancio VP, Roy-Engel AM, Deininger PL. All y'all need to know 'bout retroelements in cancer. Semin Cancer Biol. 2010;20:200–210. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Alves G, Tatro A, Fanning T. Differential methylation of human LINE-1 retrotransposons in malignant cells. Gene. 1996;176:39–44. doi: 10.1016/0378-1119(96)00205-3. [DOI] [PubMed] [Google Scholar]
- 190.Asch HL, et al. Comparative expression of the LINE-1 p40 protein in human breast carcinomas and normal breast tissues. Oncol Res. 1996;8:239–247. [PubMed] [Google Scholar]
- 191.Kubo S, et al. L1 retrotransposition in nondividing and primary human somatic cells. Proc Natl Acad Sci U S A. 2006;103:8036–8041. doi: 10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Shi X, Seluanov A, Gorbunova V. Cell divisions are required for L1 retrotransposition. Mol Cell Biol. 2007;27:1264–1270. doi: 10.1128/MCB.01888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]