Abstract
We have recently shown that MHC class II-dependent thymocyte–thymocyte (T-T) interaction successfully generates CD4+ T cells (T-T CD4+ T cells), and that T-T CD4+ T cells expressing promyelocytic leukemia zinc finger protein (PLZF) show an innate property both in mice and humans. In this article, we report that the thymic T-T interaction is essential for the conversion of CD8+ T cells into innate phenotype in the physiological condition. CD8+ T cells developed in the presence of PLZF+ CD4+ T cells showed marked upregulation of eomesodermin (Eomes), activation/memory phenotype, and rapid production of IFN-γ on ex vivo stimulation. Their development was highly dependent on the PLZF expression in T-T CD4+ T cells and the IL-4 secreted by PLZF+ T-T CD4+ T cells. The same events may take place in humans, as a substantial number of Eomes expressing innate CD8+ T cells were found in human fetal thymi and spleens. It suggests that PLZF+ T-T CD4+ T cells in combination with Eomes+ CD8+ T cells might actively participate in the innate immune response against various pathogens, particularly in human perinatal period.
Innate or nonconventional T cells represent a distinct lineage of T cells restricted by nonclassical MHC class Ib molecules expressed mainly on hematopoietic cells (1–5). During thymic ontogeny, these cells acquire memory markers (CD44hi CD122hiNK1.1int) as a result of a maturation process in the absence of antigenic encounters. In general, innate T cells have several features in common, such as a rapid response on antigenic encounter and dependence on IL-15, the B7–CD28 interaction, and the signaling lymphocytic activation molecule-associated protein (SLAM–SAP) signaling pathway (3, 4). To date, invariant NKT (iNKT) cells, γδT cells, mucosa-associated invariant T (MAIT) cells, H2-M3–restricted CD8+ T cells, and CD8αα+ intraepithelial lymphocytes have been described (2–8). Recently, CD8αβ+ T cells in Itk−/− or Itk−/−Rlk−/− mice (4, 9, 10) and CD4+ T cells that resulted from the MHC class II-dependent thymocyte–thymocyte (T-T) interaction (T-T CD4+, also referred to as T-CD4+) were added to the category of innate T cells (11–14).
Two transcriptional factors are known to regulate development of innate T cells: promyelocytic leukemia zinc finger protein (PLZF) and eomesodermin (Eomes). PLZF, which was first identified being expressed in iNKT cells and subsequently in T-T CD4+ T cells, modulates the cellular maturation and the cytokine production of both Th1 and Th2 types (15–17). In the absence of PLZF, iNKT cells remain immature and lose their innate function, whereas transgenic overexpression of PLZF converts naive T cells into the memory phenotype. In CD8 lineage cells, the T-box transcription factor Eomes, together with T-bet, is sufficient to invoke the attributes of the effector program including Th1 cytokine secretion, IL-2Rβ (CD122) upregulation, and IL-15 dependency (18–20). CD8αβ+ innate T cells in Itk−/− or Itk−/−Rlk−/− mice showed enhanced expression of Eomes during the thymic maturation process, indicating that Eomes is a marker of innate CD8+ T cells in the thymus (10).
By developing a mouse model in which MHC class II molecules are present only on T cells (CIITAtgpIV−/− mice), we and others have demonstrated the development of a distinct CD4+ T cell repertoire (T-T CD4+ T cells) via the MHC class II-dependent homotypic interaction between thymocytes (12, 13). In recent subsequent studies, some proportion of T-T CD4+ T cells was found to express PLZF, which mainly exerts innate function (14, 21, 22). Most importantly, PLZF+ CD4+ T cells existed in humans, having a characteristic developmental kinetics in the early fetal stage. This strongly suggested that PLZF+ T-T CD4+ T cells might work as potent innate immune cells in the perinatal period, that is, before the establishment of sufficient memory pools elicited by conventional T cells.
In this study, we found that the novel CD8+ T cell population was generated in CIITAtgpIV−/− mice, which acquired innate characteristics with the upregulation of Eomes via IL-4, which is mainly secreted by PLZF+ T-T CD4+ T cells. Interestingly, the CD8+ T cell population of the same phenotype was found in the fetal human thymus and spleen, as was in the case of PLZF+ T-T CD4+ T cells. These findings provide compelling evidence that the T-T interaction generates functionally competent innate CD8+ and CD4+ T cell populations, which compose a part of the complex immune defense against multiple pathogens, especially viruses in the perinatal and early childhood period.
Materials and Methods
Mice
The plck-CIITAtg mice were previously generated in our laboratory (12). CD45.1 congenic B6, PLZF−/−, TCRCα−/−, and IL-4−/− mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice carrying a deletion of promoter IV of the Mhc2ta gene (pIV−/−) were provided by Hans Acha-Orbea (University of Lausanne, Lausanne, Switzerland). B6. CD1d−/− mice were provided by Hua Gu (Columbia University, New York, NY). The plck-CIITAtg mice were backcrossed to pIV−/−, CD1d−/−, PLZFLu/Lu, TCRCα−/−, and IL-4−/− mice to generate CIITAtgpIV−/−, CIITAtgCD1d−/−, CIITAtgPLZFLu/+, CIITAtgPLZFLu/Lu, CIITAtgTCRCα−/−, and CIITAtgIL-4−/− mice, respectively. PLZFtg mice driven by CD4 promoter were a kind gift from Dr. Albert Bendelac (Chicago, IL). All of the mice were maintained under specific pathogen-free conditions in the animal facility at the Center for Animal Resource Development, Seoul National University College of Medicine (Seoul, Korea). Experiments were performed after receiving approval from the Institutional Animal Care and Use Committee of the Institute of Laboratory Animal Resources, Seoul National University.
Abs and flow cytometric analysis
The following fluorochrome- or biotin-labeled mAbs were purchased from BD Pharmingen (San Diego, CA), eBioscience (San Diego, CA), or DiNonA (Seoul, Korea): anti-mouse CD4 (RM4.5), CD8α (53–6.7), CD8β (341), CD24 (M1/69), CD25 (3C7), I-Ab (AF6-120.1), CD62L (MEL-14), CD44 (IM7), Ki-67 (B56), NK1.1 (PK136), TCRβ (H57-597), TCRγδ (GL3), CD69 (H1.2F3), CD122 (TM-β1), CD127 (SB/199), IFN-γ (XMG1.2), IL-4 (11B11), and Eomes (WD1928) and anti-human CD4 (RPA-T4 or OKT-4), CD8α (RPA-T8 or OKT-8), CD8β (2ST8.5H7), CD25 (M-A251), CD69 (FN50), CD45RA (HI100), CD45RO (UCHL1), CD122 (Mik-β3), CD161 (DX12), IFN-γ (45.15), and Eomes (Dan11-mag). The anti-mouse Vβ2 (B20.6), Vβ3 (KJ25), Vβ4 (KT4), Vβ5.1&5.2 (MR9-4), Vβ6 (RR4-7), Vβ7 (TR310), Vβ8 (F23.1), Vβ10 (B21.5), Vβ11 (RR3-15), and Vβ13 (MR12-3) Abs were purchased from BD Pharmingen. FITC-conjugated anti-human Vβ1 (BL37.3), Vβ2 (MPB2D5), Vβ5.1 (IMMU157), Vβ8 (56C5.2), Vβ11 (C21), Vβ13.1 (IMMU222), Vβ17 (E17.5F3.15.13), Vβ21.3 (IG125), and Vβ22 (IMMU546) Abs were purchased from Beckman Coulter. Allophycocyanin-conjugated anti-mouse α-galactosylceramide-loaded or unloaded CD1d tetramers were purchased from ProImmune (Bradenton, FL). Human samples were stained with mouse CD1d tetramers, as they are cross-reactive (23, 24). Fresh cell suspensions of thymocytes, splenocytes, or lymph node cells were resuspended in flow cytometry buffer, which consisted of PBS with 0.1% BSA and 0.1% sodium azide. After staining with fluorophore-conjugated Abs for 30 min at 4°C, the cells were analyzed using a FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA) and CellQuest Pro software (Becton Dickinson).
Adoptive T cell transfer
Splenic CD8+ T cells from 8 wk-old wild-type (WT; 2.6 × 107) and CIITAtgpIV−/− mice (2.17 × 107) were purified using MACS according to manufacturer’s guide and injected into congenic mice (1 × 107). One week later, recipient mice were sacrificed, and we analyzed the content of donor T cells in thymi, spleens, and lymph nodes.
Intracellular staining
For intracellular flow cytometry of PLZF and Eomes, cells were fixed with the fixation and permeabilization buffers from the Foxp3 staining buffer set (eBioscience). Intracellular PLZF was detected using mouse mAb D-9 (Santa Cruz, CA), and in some experiments, a biotin-conjugated D-9 Ab (DiNonA) was used. For cytokine staining, cells were stimulated with 50 ng/ml PMA and 1.5 μM ionomycin (Sigma, St. Louis, MO) for 5 h; 10 μg/ml brefeldin A (Sigma) was added during the last 3 h of stimulation. The stimulated cells were surface stained, fixed, and stained with anti–IL-4 (11B11), anti–IFN-γ (XMG1.2 or 45.15), anti-PLZF (D-9), or anti-Eomes (WD1928 or Dan11mag), followed by fluorophore-conjugated goat anti-mouse IgG1 (A85-1; BD Pharmingen) or streptavidin (BD Pharmingen) using the Foxp3 staining buffer set (eBioscience).
Quantitative real-time and RT-PCR
Total RNA was extracted from MACS-sorted thymocytes using the RNeasy mini kit (Qiagen, CA) according to the manufacturer’s instructions, and cDNA was synthesized using oligo(dT) primers. Sphingosine-1-phosphate 1 (S1P1) real-time RT-PCR was performed using premade primers from Applied Bioscience. The primers used for Eomes RT-PCR were: 5′-ACGGCCGCAGGTAACTAAACTGAA-3′ (forward) and 5′-AGAGCCAGCCCTACAACAAATGGT-3′ (reverse); and for β-actin, 5′-GGAAATCGTGCGTGACATTAAGG-3′ (forward) and 5′-GGCTTTTAGGATGGCAAGGGAC-3′ (reverse).
Affymetrix gene array
The total RNA was prepared for the microarray and hybridized according to standard Affymetrix protocols (Affymetrix GeneChip Mouse Gene 1.0 ST Array: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE26090, accession number GSE26090). Arrays were scanned on the Affymetrix GeneChip scanner. Microarray images were visually inspected for quality, and the probes with low signal intensity and excessively noisy background were removed before further analysis using the Affymetrix Microarray Analysis Suite v5.0. The signal values were determined using the GeneChip Operating System 1.2 (Affymetrix).
BrdU incorporation
BrdU staining was performed with the “Allophycocyanin Mouse Anti-BrdU set” according to the manufacturer’s protocol (BD Pharmingen).
Fetal thymic organ culture
On embryonic day 15.5 (E15.5), fetal thymuses from C57BL/6 mice were removed and cultured on polycarbonate filters (pore size, 0.8 μm; Millipore, Medford, MA) in RPMI 1640 medium supplemented with 10% fetal bovine calf serum (HyClone, Logan, UT), 1% penicillin and streptomycin (HyClone), and 50 nM 2-ME (Sigma) in the presence or absence of murine rIL-4 (1–10 ng/ml; PeproTech). After 7 d, the thymuses were harvested and single-cell suspensions were prepared and analyzed for the expression of CD4, CD8, PLZF, and Eomes by flow cytometry.
Bone marrow chimeras
Recipient CIITAtgpIV−/− mice were exposed to 900 rad total body irradiation from a [137Cs] source administered in two doses given 4 h apart. The mice were rested for 4–24 h before receiving bone marrow (BM) cells. Total BM cells were prepared from the femurs and tibiae of donor mice, and mature T cells were depleted using a mixture of CD4 and CD8 microbeads and magnetic sorting (Miltenyi Biotech, Auburn, CA). Each recipient mouse received 3 × 106 T cell-depleted BM cells in a volume of 300 μl PBS via lateral tail vein injection. The mice were analyzed 5–12 wk later.
Human tissues and samples
Postnatal thymuses were obtained during cardiac surgery at Seoul National University Hospital, and fetal spleen and thymus samples were obtained from aborted fetuses (16–26 wk of gestation) from Seoul National University Hospital or Jang’s Women’s Hospital (Seoul, Korea). Human umbilical cord blood cells were collected during normal full-term deliveries from Jang’s Women’s Hospital. All samples were obtained with written informed consent in accordance with the guidelines set forth by the Institutional Review Board of the Clinical Research Institute, Seoul National University Hospital. The gestational age of the fetus was calculated from the last menstrual period. Thymic tissues were teased into single-cell suspensions, and splenic mononuclear cells were separated by density gradient centrifugation over lympho M solution (Cedarlane, Burlington, ON, Canada). Cord blood mononuclear cells were isolated from whole blood using Ficoll-Hypaque density gradient centrifugation (GE Health-care).
Statistical analysis
All data were analyzed using GraphPad Prism software (GraphPad Software). Bar graphs denoting the percentage of each cell or concentration of each cytokine represent the mean value ± SEM, and the data were compared using an unpaired t test, one-way ANOVA, and Tukey’s multiple-comparison test.
Results
The MHC class II-dependent T-T interaction facilitates the development of innate-like CD8+ T cells
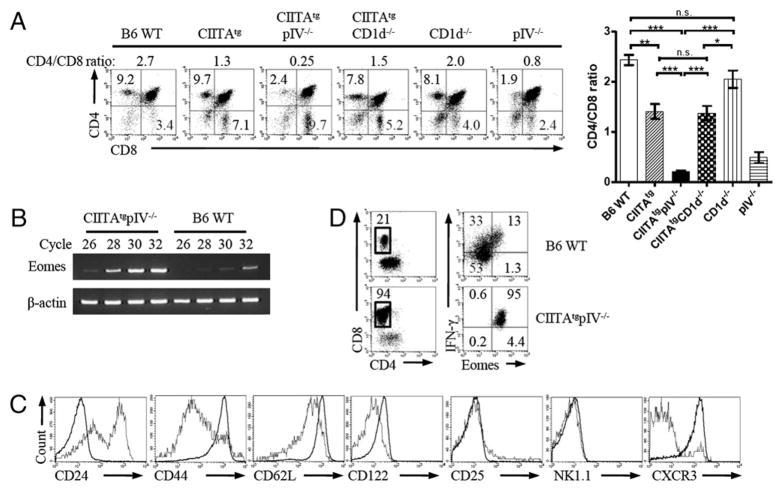
In a mouse model system, where MHC class II molecules are present only on T cells to envisage MHC class II-dependent homotypic T-T interaction (CIITAtgpIV−/− mice) (12, 15), we unexpectedly found that the proportions and total numbers of CD8 single-positive (SP) thymocytes increased (CD4/CD8 ratio ≈ 1:2–1:3) compared with the WT control (CD4/CD8 ratio ≈ 2:1–3:1) (12, 13) (Fig. 1A, 1B). This phenomenon was not observed in CD1d−/− mice, in which iNKT cells are nearly absent (Fig. 1A). The Affymetrix gene expression profile of these CD8+ SP thymocytes revealed a dramatic increase of Eomes transcripts (a 14.4-fold increase), which is normally expressed in NK, γδ T, and memory CD8+ T cells (19, 20, 25). Most CD8 SP thymocytes in CIITAtgpIV−/− mice showed an activated/memory phenotype, showing increased levels of CD44, CD62L, CD122, and CXCR3 (Fig. 1C). These cells were distinct from intraepithelial CD8αα+ T cells, because this CD8+ T cell population expresses the CD8αβ heterodimer (data not shown). Splenic CD8+ T cells of CIITAtg and CIITAtgpIV−/− mice also showed a similar phenotype (Supplemental Fig. 1). Moreover, when these CD24loCD8 SP thymocytes were stimulated with PMA/ionomycin, Eomes+ CD8 SP thymocytes from both WT and CIITAtgpIV−/− mice immediately produced IFN-γ (Fig. 1D). It is notable that >90% of CD8+ cells were IFN-γ+ Eomes+ in thymi of CIITAtgpIV−/− mice, suggesting that these CD8 SP thymocytes belong to a category of innate CD8+ T cells.
FIGURE 1.
Innate CD8+ T cell development in CIITAtgpIV−/− mice. A, Thymic T-T interaction facilitates the development of CD8 SP thymocytes. Single-cell suspensions of total thymocytes from the indicated mice were analyzed to assess CD4/CD8 ratios (n = 6, 8–12 wk old, respectively). Representative data from three independent experiments (left panel) and cumulative data (right panel) are shown; bars indicate the mean ± SEM. B, RT-PCR was performed to confirm the Eomes expression in CD8 SP thymocytes. Eomes expression was compared with that of β-actin. C, Eomes+ CD8 SP thymocytes from CIITAtgpIV−/− mice have the memory phenotype. Eomes+ and Eomes− CD8 SP thymocytes from CIITAtgpIV−/− mice were analyzed for their expression of CD24, CD44, CD62L, CD122, CD25, NK1.1, and CXCR3. The histograms show Eomes+ (thick lines) or Eomes− (thin lines) CD8 SP populations. Data are representative of one of two experiments. D, Eomes+ CD8 SP thymocytes produce IFN-γ on ex vivo stimulation. MACS purified CD24lo thymocytes from the WT and CIITAtgpIV−/− mice were stimulated with PMA and ionomycin for 5 h followed by intracellular IFN-γ staining. Data are representative of one of two experiments. Numbers in regions or quadrants indicate the percentage of cells in each. *p < 0.05, **p < 0.01, ***p < 0.001. n.s., not significant.
Eomes+ CD8+ T cells of CIITAtgpIV−/− mice are preferentially committed and developed intrathymically
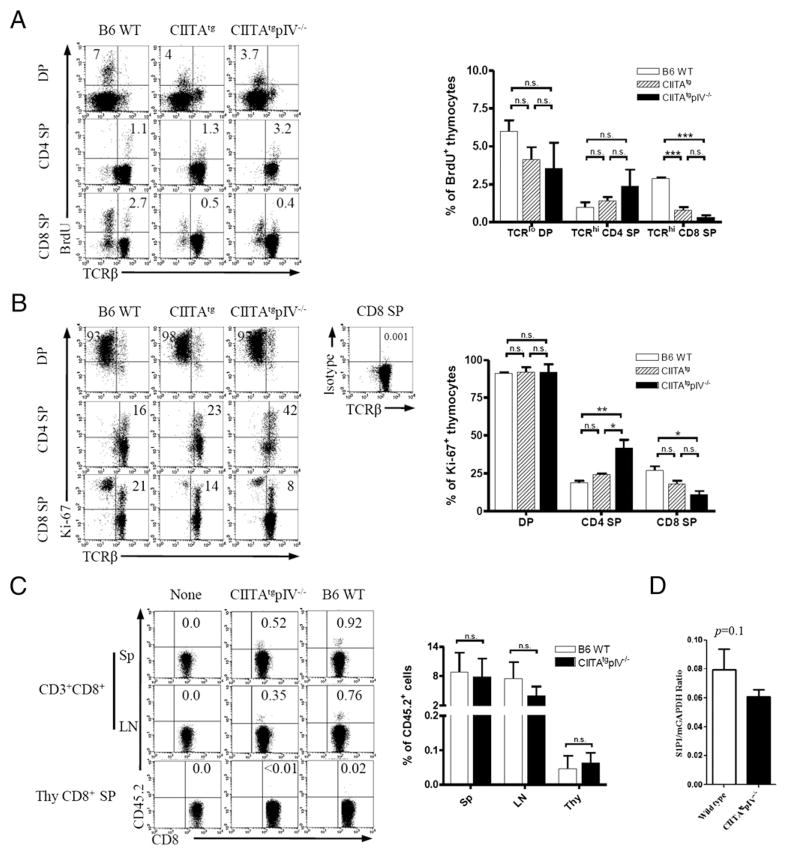
Next, we investigated whether the increased proportion of CD8 SP thymocytes is due to their enhanced proliferation after the completion of CD8 lineage commitment. The divisional capacity of the CD8 SP thymocytes from CIITAtg mice and CIITAtgpIV−/− mice, however, was even decreased compared with WT control mice (Fig. 2A, 2B), suggesting that the increase in CD8 SP thymocytes is not likely caused by the active expansion after the lineage commitment into CD8+ T cells. To further investigate the possibility that the peripheral memory CD8+ T cells in CIITAtgpIV−/− mice preferentially migrate into the thymus, we isolated splenic CD8+ T cells (CD45.2+) from WT and CIITAtgpIV−/− mice, and 1 × 107 cells from each strain were adoptively transferred into CD45.1+ congenic host mice (Fig. 2C, Supplemental Fig. 2). However, few transferred cells were found within the host thymi, in contrast with the relative abundance in the periphery such as lymph nodes and spleens. Next, to investigate whether CD8 SP thymocytes in CIITAtgpIV−/− mice were accumulated in thymus, we evaluated S1P1 expression level, which is known to regulate T cell egress (26). The S1P1 level was not significantly different between CD8+ SP thymocytes from CIITAtgpIV−/− mice and those from WT mice (Fig. 2D), which supported the assumption that the CD8+ SP thymocytes of CIITAtgpIV−/− mice were not retained in thymus because of the block of the peripheral migration.
FIGURE 2.
Accumulation of Eomes+ CD8 SP thymocytes in the thymus is not due to enhanced proliferation, retention, or recirculation of peripheral memory T cells. A and B, Proliferative capacity of CD8 SP thymocytes from indicated mice. After 40 min of BrdU injection (1 mg i.p.), 8-wk-old-mice (n = 3) of each strain were sacrificed and single-cell suspension of thymocytes was stained with anti-CD4, CD8, BrdU (A), and Ki-67(B). Numbers in quadrants in DP thymocytes indicate the percentage of BrdU+ or Ki-67+ cells per TCRβlo cells, and numbers in quadrants in SP thymocytes indicate the percentage of BrdU+ or Ki-67+ cells/TCRβhigh cells. Representative data from two independent experiments (left panel) and cumulative data (right panel) are shown; bars indicate the mean ± SEM. C, Splenic CD8+ T cells (1 × 107) from WT and CIITAtgpIV−/− mice were adoptively transferred into 8-wk-old CD45.1 congenic hosts (n = 3, respectively). Seven days later, mice were sacrificed and CD45.2+ donor CD8+ T cells were analyzed. Numbers in quadrants indicate the percentage of cells in each. Representative data from two independent experiments (left panel) and cumulative data (right panel) are shown; bars indicate the mean ± SEM. D, S1P1 expression in CD8 SP thymocytes from the WT and CIITAtgpIV−/− mice. Quantitative RT-PCR was performed with purified CD8 SP thymocytes from WT and CIITAtgpIV−/− mice. Results were expressed as ratios relative to the housekeeping gene GAPDH. Data shown are representative of two independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001. n.s., not significant; Sp, spleen; Thy, thymus.
The development of innate-like CD8+ T cells is dependent on the PLZF expression and IL-4 production of CD4+ T cells in CIITAtgpIV−/− mice
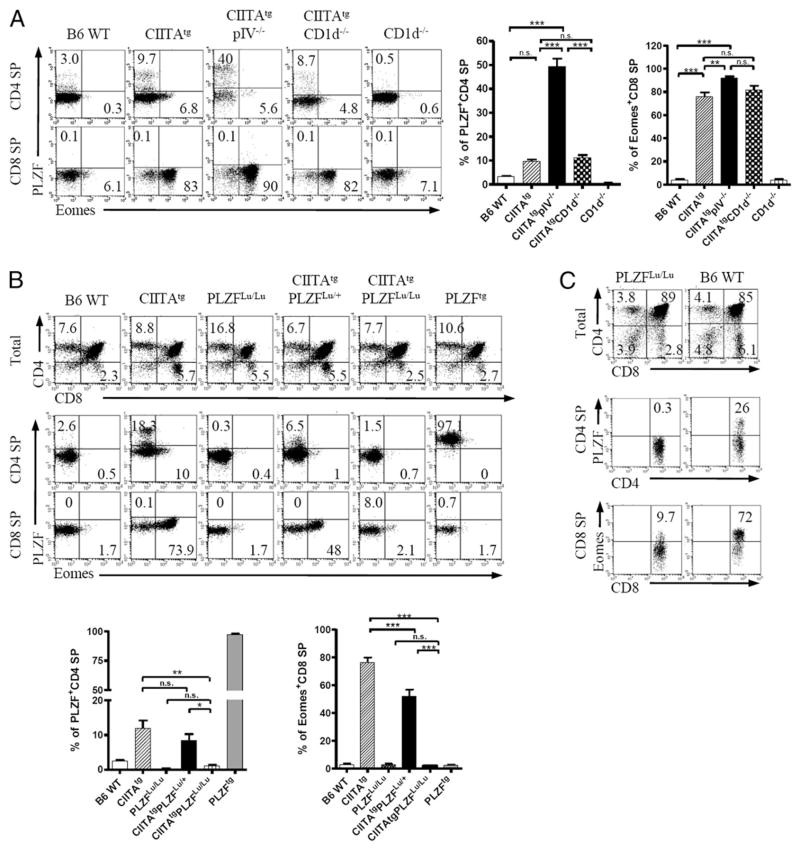
Recently, we showed that a substantial number of T-T CD4+ T cells in CIITAtgpIV−/− mice expressed PLZF, which directs the innate characteristics of these CD4+ T cells as in the case of iNKT cells (15–17). We confirmed that most CD8 SP thymocytes in CIITAtg pIV−/− mice, but not in B6 WT control and CD1d−/− mice, expressed large amounts of Eomes (Fig. 3A, bottom), and we investigated the functional relation between PLZF+ CD4+ T cells and Eomes+ CD8+ T cells. To determine the role of PLZF expression in T-T CD4+ T cells for the development of Eomes expressing innate CD8+ T cells, we generated CIITAtgPLZFLu/+ and CIITAtgPLZFLu/Lu mice in which the PLZF expression was fairly decreased in T-T CD4+ T cells (16). In these mice, the proportion of Eomes+ CD8 SP thymocytes significantly decreased with the concomitant restoration of the CD4/CD8 ratio to the WT level, indicating that PLZF expression is essential for the generation of innate CD8+ T cells (Fig. 3B). However, simple PLZF expression in the absence of T-T interaction was not sufficient to generate innate CD8+ T cells, because Eomes+ CD8+ T cells were not enhanced in PLZFtg mice (Fig. 3B). To further substantiate the role of PLZF in the Eomes+ CD8+ T cell development, PLZFLu/Lu BM cells were mixed with CIITAtgTCRCα−/− BM cells and transferred into lethally irradiated CIITAtgpIV−/− hosts. In these mice, because PLZFLu/Lu thymocytes are to be selected by MHC class II on CIITAtgTCRCα−/− thymocytes, T-T CD4+ T cells would be generated in the absence of PLZF expression. Indeed, CD8 SP thymocytes drastically reduced Eomes expression despite being developed in the presence of T-T CD4+ T cells (Fig. 3C). Therefore, it is evident that PLZF expression in T-T CD4+ T cell is essential for the induction of Eomes in CD8 SP thymocytes.
FIGURE 3.
PLZF is an essential factor for the induction of Eomes+ CD8+ SP thymocytes. A, Single-cell suspension of thymocytes from indicated mice was analyzed to compare Eomes and PLZF expression (n = 6; 8 to 12 wk old). CD4 SP (top left panels) and CD8 SP (bottom left panels) thymocytes were shown. Representative data from two independent experiments (left panel) and cumulative data (right panel) are shown; bars indicate the mean ± SEM. B, PLZFLu/Lu (n = 4), CIITAtgPLZFLu/+ (n = 4), CIITAtgPLZFLu/Lu (n = 3), and PLZFtg mice (n = 4) were assessed for CD4/CD8 ratios (top panels) and the expression of Eomes and PLZF in CD4 (top middle panels) and CD8 (top bottom panels) SP thymocytes. Representative data from two independent experiments (top panel) and cumulative data (bottom panel) are shown; bars indicate the mean ± SEM. C, WT or PLZFLu/Lu BM cells were mixed with CIITAtgTCRCα−/− BM cells in a 1:1 ratio and transferred into lethally irradiated CIITAtgpIV−/− hosts (n = 5). Thymocytes were analyzed to assess the expression of PLZF and Eomes in WT or PLZFLu/Lu thymocytes after 12 wk of the transfer. Data are representative of five mice. Numbers in quadrants indicate the percentage of cells in each. *p < 0.05, **p < 0.01, ***p < 0.001. n.s., not significant.
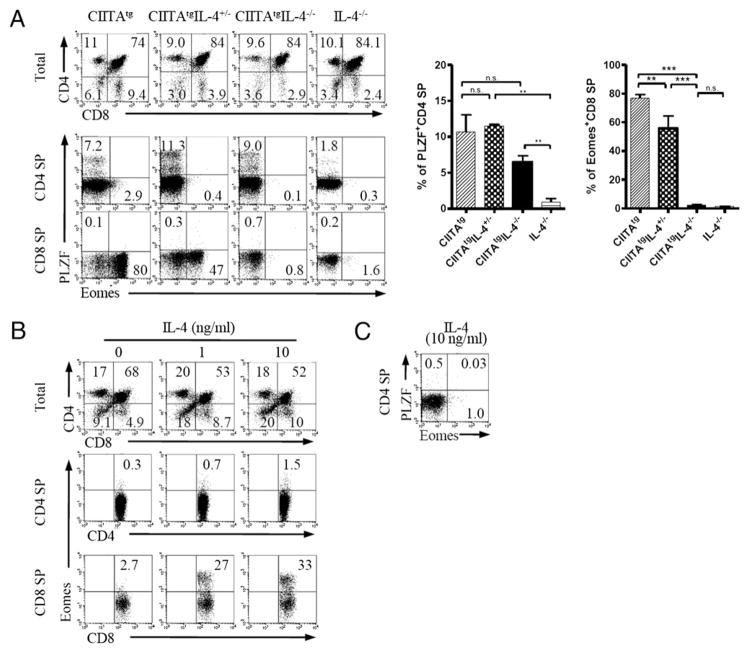
We previously reported that T-T CD4+ T cells produce abundant IL-4 in the thymus (14) and spleen, which was accompanied by PLZF expression (15). Also, it was recently found that IL-4 produced by KLF2-deficient CD4+ T cells regulated Eomes in bystander CD8 SP thymocytes (27). Based on these results, we speculated that IL-4 is a downstream signaling pathway of PLZF for the induction of Eomes. To investigate IL-4 effect on the development of Eomes+ CD8 SP thymocytes, we generated CIITAtg IL-4−/− mice in which the IL-4 expression was abolished in T-T CD4+ T cells. Interestingly, Eomes expression was barely detectable in these mice (Fig. 4A), and CIITAtgIL-4+/− mice showed a 2-fold reduction of Eomes+ CD8+ T cells, suggesting a dose-dependent regulation by IL-4. Moreover, IL-4 alone was sufficient for the Eomes induction in CD8 SP thymocytes even in the absence of PLZF, as shown in WT B6 thymi (E15.5) cultured with variable concentrations of IL-4 for 7 d (Fig. 4B, Supplemental Fig. 3). Eomes was upregulated dramatically in CD8 SP thymocytes in the presence of IL-4 in a dose-dependent manner, although CD4 SP thymocytes did not express PLZF (Fig. 4C). The earlier results indicate that IL-4 is necessary and sufficient for the development of Eomes+ CD8 SP thymocytes independent of PLZF expression.
FIGURE 4.
IL-4 is necessary and sufficient to induce Eomes expression in CD8+ T cells. A, IL-4 is the key mediator for Eomes induction. Single-cell suspension of thymocytes from CIITAtg (n = 3), CIITAtgIL-4+/− (n = 2), CIITAtgIL-4−/− (n = 4), and IL-4−/− (n = 3) mice were analyzed to assess CD4/CD8 ratio (top left panels) and their expression of Eomes and PLZF in CD4 (middle left panels) and CD8 (bottom left panels) SP thymocytes. Representative data from three independent experiments (left panel) and cumulative data (right panel) are shown; bars indicate the mean ± SEM. B and C, IL-4 induces upregulation of Eomes independent of PLZF. Fetal thymuses of E15.5 B6 mice were cultured in the presence or absence of exogenous mouse rIL-4 for 7 d. Three to four fetal thymic lobes were used for each condition, and pooled cells were analyzed. Eomes was markedly upregulated in CD8 SP thymocytes (B), and PLZF was not expressed in CD4 SP thymocytes (C). Representative data from two independent experiments are shown. Numbers in quadrants indicate the percentages of cells in each. *p < 0.05, **p < 0.01, ***p < 0.001. n.s., not significant.
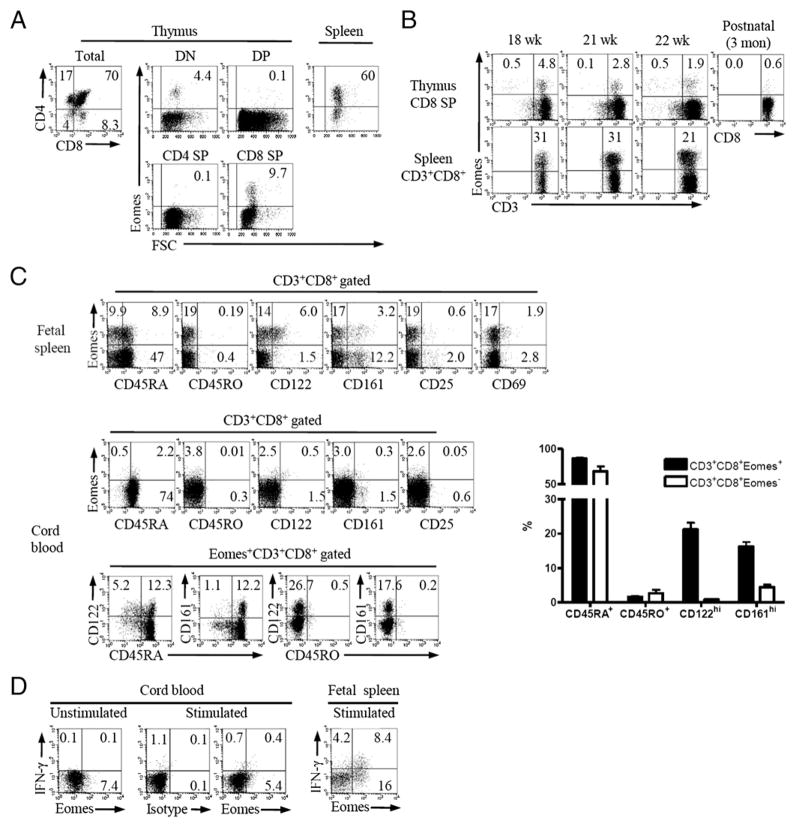
The presence of CD8+ T cells with the innate phenotype in the human fetus
Because PLZF-expressing T-T CD4+ T cells were shown to be present in humans (15), we extended our study to humans to identify whether thymic CD8+ T cells express Eomes and innate phenotype. At 18 wk of gestation, both double-negative and CD8 SP populations clearly expressed Eomes (Fig. 5A), which were not stained with α-galactosylceramide–loaded CD1d tetramers (data not shown). However, the Eomes+ CD8+ T cells were gradually decreased in the course of human fetal thymopoiesis and had almost disappeared 3 mo after birth (Fig. 5B). This pattern correlates well with the expression kinetics of PLZF in CD4 SP thymocytes (15). Fetal splenocytes showed a greater percentages of Eomes+ CD8+ T cells (31, 31, and 21% of CD8+ T cells at 18, 21, and 22 wk of gestational age, respectively) with partial up-regulation of CD122 and CD161 (Fig. 5C, top panel), and a substantial fraction of cord blood CD3+CD8+ T cells (1.7–5.4%, n = 4) still expressed Eomes (Fig. 5C, middle panel). Furthermore, most Eomes-expressing CD3+CD8+ T cells were CD45RAhi CD45ROlow, and the significant proportion of them expressed CD122 and CD161 (Fig. 5C, bottom panel). These Eomes+ T cells immediately produced IFN-γ on ex vivo stimulation (Fig. 5D), showing their functional competence.
FIGURE 5.
Presence of Eomes+ innate CD8+ T cells in the human fetus and spleen. A, Analysis of Eomes expression in the fetal thymocytes and the CD8+ splenocytes of 18-wk human fetus. B, Eomes expression in CD8 SP thymocytes and splenic CD3+CD8+ T cells at the indicated gestation ages and 3 mo after birth. C, Expression of memory markers in Eomes+ CD8+ T cells from human fetal spleen (top left panel, n = 2) and human cord blood (middle and bottom left panels, n = 4). Representative data (left panels) and cumulative data (right panel) are shown; bars indicate the mean ± SEM. D, IFN-γ production from Eomes+ CD8+ T cells. Total fetal splenocytes or Ficoll purified cord blood mononuclear cells were stimulated with PMA and ionomycin for 5 h and stained with IFN-γ. Representative data from two independent experiments (fetal splenocytes) and four independent experiments (cord blood) are shown. Numbers in quadrants indicate the percentages of cells in each.
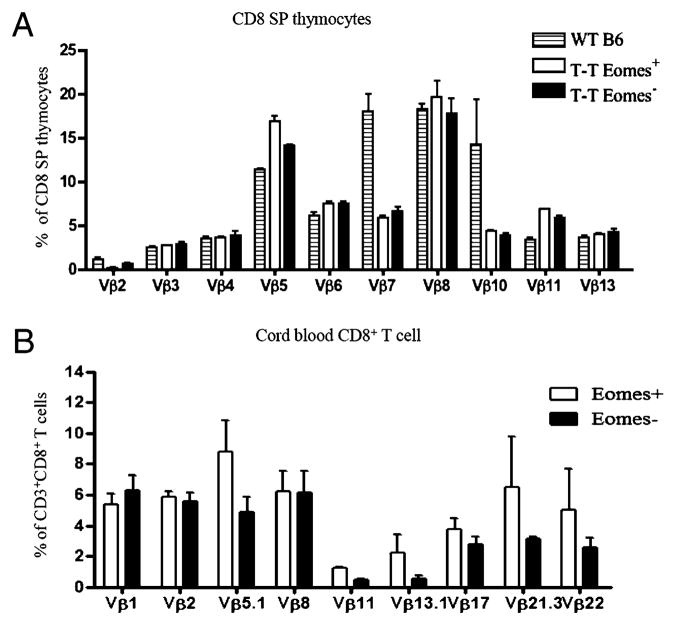
In contrast with other innate T cells including iNKT cells and MAIT cells (8), the TCR repertoire of the Eomes+ CD8+ T cells from both CIITAtgpIV−/− mice and humans was as diverse as that of conventional CD8+ T cells (Fig. 6A, 6B). These results indicate that the functional and the phenotypic nature of this population in humans are developmentally similar to that in CIITAtgpIV−/− mice.
FIGURE 6.
Diversity of Eomes+ CD8+ T cells in mice and humans. A, Eomes+ CD8 SP thymocytes use diverse TCR Vβ-chains. Single-cell suspension of total thymocytes from WT B6 and CIITAtgpIV−/− mice were stained with Abs recognizing CD4, CD8, CD8, Eomes, and different TCR Vβ-chains to compare the frequencies of cells expressing each TCR Vβ-chain (n = 3 in each group). B, Diverse TCR Vβ usage of Eomes+ CD8+ T cells in human cord blood samples. Ficoll purified cord blood mononuclear cells were stained with Abs recognizing CD3, CD8, Eomes, and TCR Vβ, and analyzed for frequencies of cells using each TCR Vβ-chain (n = 3 in each group).
Discussion
This study showed that Eomes+ innate CD8+ T cells are intrathymically developed and preferentially committed into innate CD8+ T cells in the presence of MHC class II-dependent T-T interaction. In this process, IL-4 produced by PLZF+ T-T CD4+ T cells and the PLZF expression in T-T CD4+ T cells are essential for controlling the development of Eomes+ CD8+ T cells. Overall, these findings indicate that the MHC class II-dependent T-T interaction has a key role in generating PLZF+ CD4+ T cells and subsequently Eomes+ CD8+ T cells, both of which take on an innate phenotype.
In CIITAtg and CIITAtgpIV−/− mice, a strikingly large population of CD8 SP thymocytes expresses Eomes (85 and 93%, respectively). This proportion is not reduced in CIITAtgCD1d−/− mice, indicating that the iNKT cells are not involved in the development of this CD8+ T cell population. It strongly suggests that the PLZF+ T-T CD4+ T cell population generated in mice with CIITAtg background (CIITAtg, CIITAtgpIV−/−, and CIITAtg CD1d−/−) is a key resource of IL-4 in thymus, which directly controls the generation of Eomes+ CD8+ T cells as shown in the fetal thymic organ culture (FTOC) with IL-4 (Fig. 4B). The adoptive transfer experiment supports the finding that Eomes+ CD8+ T cells are bona fide intrathymic T cells, based on the results that mature CD8+ T cells have little chance of returning to the thymus and the subsequent expansion. The S1P1 expression level was not altered in CD8 SP thymocytes from CIITAtgpIV−/− mice compared with those from WT mice, suggesting that the increased number of CD8+ SP thymocytes from CIITAtgpIV−/− mice was not due to the defective migration to the periphery. However, the higher expression of CXCR3 on CD8+ SP thymocytes from CIITAtgpIV−/− mice (Fig. 1C) seemed to be against this, because CXCR3 in mice may be associated with accumulation of thymocytes, as in the case of mature iNKT thymocytes (28). Nevertheless, it is conceivable that the Eomes+ innate CD8+ T cells are preferentially committed during the late stage of CD8+ T cell development.
Similar to CD8+ T cells generated in the Itk−/− or Itk−/−Rlk−/− mice, Eomes+ CD8+ T cells in CIITAtg and CIITAtgpIV−/− mice showed innate-like characteristics, including the surface expression of CD44hiCD122hi and the rapid production of cytokine. These features are also shared by thymic CD8+ T cells from the CREB binding protein (CBP)-deficient mice (29), and it was proposed that Itk, Rlk, and CBP are likely to regulate TCR signaling pathway to develop innate CD8+ T cells. However, one article that was published recently demonstrated that the development of innate CD8+ T cells in Itk−/− or Itk−/−Rlk−/− mice and CBP-deficient mice was attributable to the IL-4–dependent mechanism in common (30). In that report, KLF2-deficient mice developed an innate-like CD8+ T cell population via IL-4 pathway, which was regulated by PLZF+ T cells in mice. However, it is not clear whether and how KLF deficiency causes the expansion of the PLZF+ population in this model (27, 30).
These recent evidences bear a striking similarity to our current findings on the role of IL-4 for the innate CD8+ T cell development; however, the important difference between the earlier models and the CIITAtg mice is that the CIITAtg condition can mimic human thymic ontogeny. Unlike mouse thymocytes, human thymocytes normally express MHC class II molecules on their surface, and it facilitates the cell interactions between thymocytes via MHC class II to produce PLZF+ CD4+ T cells. It strongly suggests that PLZF+ CD4+ T cells selected by MHC class II-dependent T-T interaction would be one of the main PLZF+ populations for IL-4 production in human thymus, which drives the development of innate CD8+ T cells as shown by our current findings. This possibility is supported by the developmental kinetics of both Eomes+ CD8+ T cells and PLZF+ CD4+ T cells, and by the presence of a considerable proportion of PLZF+ but CD1d tetramer− CD4+ T cells (15) and Eomes+ CD8+ T cells in human fetuses. Based on these results, we believe that the innate CD8+ T cells develop in an early human developmental stage, which is regulated by IL-4 secreted from PLZF+ CD4+ T cells. This whole process seems to be intrinsically controlled by MHC class II-dependent T-T interaction, which occurs only in the human system.
In addition, the diverse TCRβ usage of these Eomes CD8+ T cells, as well as PLZF+ CD4+ T cells (15), in both the human and the mouse models repeatedly suggests the difference between these populations from the previously established innate T cells, including iNKT and MAIT cells. This result proposes that Eomes CD8+ T cells are likely to be selected by MHC class Ia molecules, and that this population actively participates in the innate immune response against various pathogens, such as viruses.
Eomes+ CD8+ T cells, though small in number, are also present in the thymus of WT B6 mice. It has been shown that MHC class Ib-restricted CD8 SP thymocytes are present in KbDb knockout mice (5), and these cells were known to have memory markers in thymus, seemingly innate in their phenotype. Based on this finding, it is assumed that small number of Eomes+ CD8+ T cells in thymi of WT mice might be identical cells in nature with CD8+ T cells in KbDb knockout mice. As exogenous IL-4 dramatically enhanced the generation of Eomes+ CD8+ SP thymocytes in FTOC, it seems that IL-4–producing PLZF+ populations modulate the development of innate CD8+ T cells through Eomes expression. This possibility was verified by the recent report that IL-4 produced by iNKT cells regulates the generation of innate-like Eomes+ CD8+ T cells in BALB/c mice (30).
Taken together, although the physiological role of Eomes+ CD8+ T cells is less clear, one can easily envision that these cells may function as a first line of defense on an infection during the perinatal period of human. Regarding PLZF+ T-T CD4+ T cells, they might function in at least two distinct aspects. First, they serve as effector CD4+ T cells in the periphery by producing Th1 and Th2 cytokines immediately upon an immune response. The other function that was discovered in this study is to facilitate the generation of Eomes+ CD8+ SP thymocytes in the thymus by producing IL-4. In this sense, these two populations developed via MHC class II-dependent T-T interaction might offer a comprehensive coverage of the innate immune response before the full-scale activation of adaptive immunity, which is more critical in the early stage of human development.
Supplementary Material
Acknowledgments
This work was supported by Korean Science and Engineering Foundation Grant R01-2007-000-20165-0 from the Ministry of Knowledge Economy of Korea and National Institutes of Health Grant AI I073677 (to C.-H.C.).
Abbreviations used in this article
- BM
bone marrow
- CBP
CREB binding protein
- E15.5
embryonic day 15.5
- Eomes
eomesodermin
- FTOC
fetal thymic organ culture
- iNKT
invariant NKT
- MAIT
mucosa-associated invariant T
- pIV−/−
deletion of promoter IV of the Mhc2ta gene
- PLZF
promyelocytic leukemia zinc finger protein
- SP
single-positive
- S1P1
sphingosine-1-phosphate 1
- tg
transgenic
- T-T
thymocyte–thymocyte
- WT
wild-type
Footnotes
Disclosures
The authors have no financial conflicts of interest.
The sequence presented in this article has been submitted to Gene Expression Omnibus under accession number GSE26090.
The online version of this article contains supplemental material.
References
- 1.Prince AL, Yin CC, Enos ME, Felices M, Berg LJ. The Tec kinases Itk and Rlk regulate conventional versus innate T-cell development. Immunol Rev. 2009;228:115–131. doi: 10.1111/j.1600-065X.2008.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, Premel V, Devys A, Moura IC, Tilloy F, et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 2009;7:e54. doi: 10.1371/journal.pbio.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veillette A, Dong Z, Latour S. Consequence of the SLAM-SAP signaling pathway in innate-like and conventional lymphocytes. Immunity. 2007;27:698–710. doi: 10.1016/j.immuni.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Berg LJ. Signalling through TEC kinases regulates conventional versus innate CD8(+) T-cell development. Nat Rev Immunol. 2007;7:479–485. doi: 10.1038/nri2091. [DOI] [PubMed] [Google Scholar]
- 5.Urdahl KB, Sun JC, Bevan MJ. Positive selection of MHC class Ib-restricted CD8(+) T cells on hematopoietic cells. Nat Immunol. 2002;3:772–779. doi: 10.1038/ni814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambolez F, Kronenberg M, Cheroutre H. Thymic differentiation of TCR alpha beta(+) CD8 alpha alpha(+) IELs. Immunol Rev. 2007;215:178–188. doi: 10.1111/j.1600-065X.2006.00488.x. [DOI] [PubMed] [Google Scholar]
- 7.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 8.Treiner E, Lantz O. CD1d- and MR1-restricted invariant T cells: of mice and men. Curr Opin Immunol. 2006;18:519–526. doi: 10.1016/j.coi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Broussard C, Fleischacker C, Horai R, Chetana M, Venegas AM, Sharp LL, Hedrick SM, Fowlkes BJ, Schwartzberg PL. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. [Published erratum appears in 2006 Immunity. 25: 849.] Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Atherly LO, Lucas JA, Felices M, Yin CC, Reiner SL, Berg LJ. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25:79–91. doi: 10.1016/j.immuni.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Choi EY, Park WS, Jung KC, Chung DH, Bae YM, Kim TJ, Song HG, Kim SH, Ham DI, Hahn JH, et al. Thymocytes positively select thymocytes in human system. Hum Immunol. 1997;54:15–20. doi: 10.1016/s0198-8859(97)00012-8. [DOI] [PubMed] [Google Scholar]
- 12.Choi EY, Jung KC, Park HJ, Chung DH, Song JS, Yang SD, Simpson E, Park SH. Thymocyte-thymocyte interaction for efficient positive selection and maturation of CD4 T cells. Immunity. 2005;23:387–396. doi: 10.1016/j.immuni.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Li W, Kim MG, Gourley TS, McCarthy BP, Sant’Angelo DB, Chang CH. An alternate pathway for CD4 T cell development: thymocyte-expressed MHC class II selects a distinct T cell population. Immunity. 2005;23:375–386. doi: 10.1016/j.immuni.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Li W, Sofi MH, Yeh N, Sehra S, McCarthy BP, Patel DR, Brutkiewicz RR, Kaplan MH, Chang CH. Thymic selection pathway regulates the effector function of CD4 T cells. J Exp Med. 2007;204:2145–2157. doi: 10.1084/jem.20070321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YJ, Jeon YK, Kang BH, Chung DH, Park CG, Shin HY, Jung KC, Park SH. Generation of PLZF+ CD4+ T cells via MHC class II-dependent thymocyte-thymocyte interaction is a physiological process in humans. J Exp Med. 2010;207:237–246. doi: 10.1084/jem.20091519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T, Reiner SL. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 20.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Sofi MH, Rietdijk S, Wang N, Terhorst C, Chang CH. The SLAM-associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity. 2007;27:763–774. doi: 10.1016/j.immuni.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee YJ, Jung KC, Park SH. MHC class II-dependent T-T interactions create a diverse, functional and immunoregulatory reaction circle. Immunol Cell Biol. 2009;87:65–71. doi: 10.1038/icb.2008.85. [DOI] [PubMed] [Google Scholar]
- 23.Karadimitris A, Gadola S, Altamirano M, Brown D, Woolfson A, Klenerman P, Chen JL, Koezuka Y, Roberts IA, Price DA, et al. Human CD1d-glycolipid tetramers generated by in vitro oxidative refolding chromatography. Proc Natl Acad Sci USA. 2001;98:3294–3298. doi: 10.1073/pnas.051604498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, He W, Kim ST, Tao J, Gao Y, Chi H, Intlekofer AM, Harvey B, Reiner SL, Yin Z, et al. Epigenetic and transcriptional programs lead to default IFN-gamma production by gammadelta T cells. J Immunol. 2007;178:2730–2736. doi: 10.4049/jimmunol.178.5.2730. [DOI] [PubMed] [Google Scholar]
- 26.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 27.Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31:122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drennan MB, Franki AS, Dewint P, Van Beneden K, Seeuws S, van de Pavert SA, Reilly EC, Verbruggen G, Lane TE, Mebius RE, et al. Cutting edge: the chemokine receptor CXCR3 retains invariant NK T cells in the thymus. J Immunol. 2009;183:2213–2216. doi: 10.4049/jimmunol.0901213. [DOI] [PubMed] [Google Scholar]
- 29.Fukuyama T, Kasper LH, Boussouar F, Jeevan T, van Deursen J, Brindle PK. Histone acetyltransferase CBP is vital to demarcate conventional and innate CD8+ T-cell development. Mol Cell Biol. 2009;29:3894–3904. doi: 10.1128/MCB.01598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.