SUMMARY
Myxococcus xanthus can vary its phenotype or “phase” to produce colonies that contain predominantly yellow or tan cells that differ greatly in their abilities to swarm, survive, and develop. Yellow variants are proficient at swarming (++) and tend to lyse in liquid during stationary phase. In contrast, tan variants are deficient in swarming (+) and persist beyond stationary phase. The phenotypes and transcriptomes of yellow and tan variants were compared with mutants affected in phase variation. Thirty-seven genes were upregulated specifically in yellow variants including those for production of the yellow pigment, DKxanthene. A mutant in DKxanthene synthesis produced nonpigmented (tan) colonies but still phase varied for swarming suggesting that pigmentation is not the cause of phase variation. Disruption of a gene encoding a HTH-Xre-like regulator, highly expressed in yellow variants, abolished pigment production and blocked the ability of cells to switch from a swarm ++ to a swarm (+) phenotype, showing that HTH-Xre regulates phase variation. Among the four genes whose expression was increased in tan variants was pkn14, which encodes a serine-threonine kinase that regulates programmed cell death in Myxococcus via the MrpC-MazF toxin-antitoxin complex. High levels of phosphorylated Pkn14 may explain why tan cells enjoy enhanced survival.
Keywords: development, phenotypic heterogeneity, swarming, DKxanthene, HTH-Xre, serine-threonine kinase Pkn
INTRODUCTION
Myxococcus xanthus is a Gram negative soil bacterium that displays a complex life cycle. Survival skills have been honed well and include predation of other organisms (Berleman et al., 2008), multiple motility systems that allow cells to adapt to different environmental conditions (Shi & Zusman, 1993), and the ability to develop fruiting bodies filled with heat and desiccation-resistant spores in response to starvation (Kaiser, 2003). Starvation initiates a complicated developmental pathway that involves the (p)ppGpp-dependent stringent response (Harris et al., 1998, Laue & Gill, 1995) and requires elaborate cell-cell signaling (Rolbetzki et al., 2008, Sogaard-Andersen, 2004, Sogaard-Andersen et al., 2003, Shimkets, 1999, Kim & Kaiser, 1990). Fruiting development also can be triggered by the presence of particular nutrients when cells are growing on prey organisms such as E. coli. Kirby and co-workers (Berleman & Kirby, 2007) have coined the term predataxis to describe this unusual response when seeking out prey.
Phase variation is an expression of phenotypic plasticity that allows a single bacterial species to persist in alternate forms by expressing different sets of genes. In some cases, organisms are hardwired to alter the expression of various cellular components at a given frequency by mechanism that involve perturbations in the DNA (van der Woude, 2006). Typically, each of the alternative genetic programs results in production of different combinations of proteins, lipids, or carbohydrates on the cell surface that contribute to phase-specific changes in colony texture, color, or morphology (van der Woude & Baumler, 2004). Pathogenic strains that undergo phase variation produce cell types that typically are not recognized by the immune system, thus affording these pathogens enhanced survival. Alternatively, the mechanism of phase variation might involve an epigenetic switch due to the unimodal noise in the expression levels of a master regulator which results in bistability (Chai et al., 2008, Dubnau & Losick, 2006).
The role of phase variation in non-pathogens has been studied less yet evidence suggests that phase variation contributes to environmental fitness of bacteria. A reversible frame-shift mutation in swrA affects both swimming motility and swarming differentiation in the soil bacterium Bacillus subtilis by regulating the number of flagella available to participate in these processes (Kearns et al., 2004, Calvio et al., 2008, Calvio et al., 2005). Multiple studies have documented the phenomenon of phase variation in M. xanthus and its roles in swarming and development (Laue & Gill, 1995, Burchard & Dworkin, 1966, Burchard et al., 1977). The predominant morphotype, or variant, in M. xanthus produces a matte, richly textured yellow colony. Colonies with predominantly yellow variants are proficient at swarming on agar surfaces (Swr++). Colonies of the other morphotype are tan and have a smooth surface. Tan variants exhibit reduced swarming (Swr(+). Yellow and tan colonies do not represent pure populations of either variant type, but rather are mixtures. Yellow colonies contain cells that yield 75–99% yellow and 1– 25% tan colonies; these ratios are reversed in the colonies of tan variants (Laue & Gill, 1995). The rate of switching from yellow to tan has been reported to be 10−2 to 10−3 per cell per generation; switching from tan to yellow is higher, so colonies of the yellow variant outnumber those of the tan variant (Laue & Gill, 1994).
Phase variation in M. xanthus may provide a mechanism to generate two cell types, both of which are needed for development of spore-filled fruiting bodies when nutrients are exhausted. Cultures that are predominantly yellow form dark fruiting bodies and heat-resistant spores when starved for nutrients. In contrast, the tan variant rarely forms dark fruiting bodies and usually arrests at the soft mound stage of development. Tan variants produce fewer heat-resistant spores, particularly at low cell densities (Meiser et al., 2006). Previous work suggested that tan and yellow variants both participate in the formation of heat-resistant spores. Although the tan variants produce very few spores when developed on their own, Laue and Gill designed an experiment to determine if both cell types are needed during development. They allowed differentially labeled tan and yellow variants to develop as mixtures, then found that tan variants contribute disproportionately to the population of heat-resistant spores from the mixture (Laue & Gill, 1995, Laue & Gill, 1994). Hence tan variants were capable of forming spores and from this experiment they concluded that yellow cells provide some factor that the tan cells need to produce viable spores. One factor needed by tan variants is DKxanthene (DKX), a secondary metabolite that confers the characteristic yellow color of M. xanthus yellow variants (Meiser et al., 2006). A detailed genetic and chemical characterization of DKX synthesis has been carried out in the Müller lab (Meiser et al., 2008, Meiser & Muller, 2008, Meiser et al., 2006). Strains with mutations in the dkx biosynthetic operon are tan in color and are severely deficient in their ability to produce heat-resistant spores. Addition of purified DKX partially rescued the sporulation defect of the dkx mutants (Meiser et al., 2006).
The complex phenotype of phase variants, combined with the frequency of switching, has complicated the analysis of phase variation in the past. Mutational analysis of phase variation has failed to provide a wholesale picture of the phase regulatory process because many types of mutations produce swarming or development phenotypes that resemble one aspect of phase variation. In this study, we used microarray and RT-PCR to identify genes differentially regulated in yellow and tan variant populations that likely contribute to the myriad phenotypes seen in these phase variants. Statistically significant data were collected for genes whose expression was increased in yellow cultures relative to tan cultures and genes whose expression was increased in tan cultures relative to yellow cultures. qRT-PCR data derived using RNA isolated from independent batches of tan or yellow cells confirmed the microarray findings. A dkx mutant was studied to understand the role of DKX pigment on swarming and development. This tan mutant was able to switch between swarm proficient and swarm deficient phenotypes, which demonstrates that regulation of swarming during phase variation is genetically separable from the pigment pathway. Subsequent qRT-PCR analysis confirmed this result because a subset of genes up-regulated in the WT tan variant, which is swarm deficient, were also up-regulated in the dkx− swarm deficient mutant but not the dkx− swarm proficient mutant. Finally, disruption of a putative DNA binding protein whose expression was increased in yellow variants has identified a candidate master regulator of phase variation. Taken together, these data provide new insights into the yellow-tan phase process and reveal fundamental links with several known pathways in M. xanthus.
RESULTS
Microarray analysis of yellow and tan phase variant populations
M. xanthus undergoes phase variation forming either yellow, swarm proficient (Swr++) or tan, swarm deficient (Swr(+)) colonies that differ in their abilities to develop. To identify genes whose products likely contribute to differences in pigment production, swarming, and development, we compared the transcriptomes of two independent yellow and tan variants using DNA microarray analysis. Yellow and tan variants were naturally occurring phase variants derived from M. xanthus WT strain DK1622. RNA harvested from the two yellow and two tan CTPM broth-grown cultures was used to synthesize Cy3-labeled cDNA to probe a M. xanthus microarray containing 7335 genes of the predicted and annotated 7457 M. xanthus DK1622 genes. Data compiled from the microarrays were normalized and tested for differential expression. Genes whose hybridization signal resulted in an adjusted p value ≤ 0.05 were considered statistically significant and were selected for further study.
qRT-PCR was performed to confirm the results obtained from the microarray analysis. Sets of oligonucleotides were designed for (1) genes with significantly increased expression in yellow or tan variants, (2) genes that were just above the adjusted P-value cutoff, (3) genes that did not show a significant difference between tan and yellow variants, and (4) 16S rRNA, the endogenous control. The WT yellow strain was used as the calibrator. Microarray results that were not in agreement with qRT-PCR results are not subjected to further analysis.
Thirty-seven genes whose expression was up-regulated in yellow variant populations relative to tan variants were identified using the combined data from two microarray experiments. A subset of these genes was subjected to further analysis using qRT-PCR. Values presented are the average of at least six qRT-PCR results for each gene assayed in triplicate on at least two independent occasions.
These genes are listed in Table 1A where log increase values are given; values in the text are given as fold increases. Expression of multiple genes involved in synthesis of the yellow pigment, DKX, which is absent in tan variants, was increased in yellow variants relative to the tan variant (see Table 1A for MXAN4292, MXAN4295, MXAN4298, and MXAN4301). Expression of a nearby hypothetical gene, MXAN4288, was also increased and may identify another player in the DKX pathway.
Table 1.
| A. Yellow variants show increased expression of 37 genes | ||||
|---|---|---|---|---|
| p.value.adj | array log increase |
qRT-PCR* | MXAN # | Annotation |
| 0.02103 | 2.12 | MXAN0123 | conserved hypothetical protein | |
| 0.00811 | 2.79 | 1.6 | MXAN0228 | Putative HTH-Xre DNA-binding protein |
| 0.00108 | 4.43 | 5.3 | MXAN0504 | hypothetical protein |
| 0.00707 | 3.41 | 2.9 | MXAN1286 | ABC transporter, ATP-binding/permease protein |
| 0.01494 | 3.65 | MXAN1288 | carbamoyltransferase family protein, putative | |
| 0.00164 | 3.08 | 2.4 | MXAN1289 | phytanoyl-CoA dioxygenase family protein |
| 0.01401 | 2.36 | MXAN1650 | peptidase, S1A (chymotrypsin) subfamily | |
| 0.01758 | 3.38 | MXAN1724 | lipoprotein, putative | |
| 0.03972 | 2.23 | 1.2 | MXAN2675 | glutamyl-tRNA synthetase |
| 0.01494 | 2.57 | MXAN2791 | protease B | |
| 0.03648 | 1.97 | MXAN2817 | triosephosphate isomerase | |
| 0.02561 | 3.10 | MXAN3296 | ribosomal protein S7 | |
| 0.02666 | 3.99 | 1.5 | MXAN3299 | ribosomal protein S10 |
| 0.04257 | 3.73 | MXAN3306 | ribosomal protein L16 | |
| Expression of genes for synthesis of polyketide myxovirescin/TA, a peptidase inhibitor | ||||
| 0.02543 | 2.90 | 1.4 | MXAN3943 | Cytochrome P450 family protein (hydroxylase TaH) |
| 0.02881 | 2.30 | 1.8 | MXAN3949 | acyl carrier protein, putative (TaB) |
| Putative role in non-ribosomal synthesis | ||||
| 0.04493 | 2.67 | 1.3 | MXAN4084 | cyclic nucleotide-binding domain protein |
| Expression of genes for synthesis of polyketide DKxanthene, the yellow pigment | ||||
| 0.02881 | 2.21 | MXAN4288 | hypothetical protein | |
| 0.01293 | 2.97 | MXAN4292 | dkxM; polyketide synthase | |
| 0.00025 | 2.55 | 2.5 | MXAN4294 | dkxK; arsenical-transporting ATPase family |
| 0.03155 | 3.25 | 2.4 | MXAN4295 | dkxJ; patatin-like phospholipase family |
| 0.03445 | 2.50 | 1.4 | MXAN4298/9 | dkxG; polyketide synthase type 1 |
| 0.04014 | 2.44 | MXAN4301 | dkxE; polyketide synthase type 1 | |
| 0.01758 | 2.44 | MXAN4746 | TonB-dependent receptor | |
| 0.01401 | 2.59 | MXAN4770 | 3-oxoacyl-(acyl-carrier-protein) reductase | |
| 0.00108 | 3.30 | MXAN4924 | Hypothetical GC-trans-RRR | |
| 0.04493 | 1.83 | 1.2 | MXAN5138 | site-specific DNA methyltransferase |
| 0.02543 | 2.31 | MXAN5261 | lipoprotein, putative | |
| 0.01293 | 3.12 | MXAN5392 | lipoprotein, putative | |
| 0.02561 | 2.29 | 1.2 | MXAN5478 | LysM domain protein |
| 0.02543 | 3.26 | 2.1 | MXAN5703 | PspA (phage shock protein A) |
| 0.00811 | 3.96 | MXAN5970 | peptidase, S8 (subtilisin) family | |
| 0.01401 | 2.66 | 1.4 | MXAN5974 | hydrolase, alpha/beta fold family |
| 0.02881 | 2.61 | MXAN6494 | conserved protein (M23 peptidase-like C-terminus) | |
| 0.01494 | 2.76 | MXAN6704 | GNAT family; ribosomal protein alanine N-acetyltransferase-like protein | |
| 0.03648 | 2.47 | MXAN6734 | response regulator/sensory box histidine kinase | |
| 0.05616 | 2.49 | MXAN6862 | MotA/TolQ/ExbB proton channel family protein | |
| B. Expression of four genes is increased in tan variants compared with yellow variants. | ||||
|---|---|---|---|---|
| p.value.adj | fold increase |
MXAN # | Annotation | |
| 0.02881 | 3.75 | 4.6 | MXAN4371 | serine/threonine kinase Pkn14 |
| 0.02615 | 2.69 | 1.5 | MXAN4479 | serine/threonine protein kinase, putative |
| 0.016 | 3.42 | 3.9 | MXAN6911 | TonB-dependent siderophore receptor |
| 0.016 | 5.04 | 1.9 | MXAN4389 | KatB; catalase |
Genes showing significant expression increases in M. xanthus DK1622 yellow (1A) and tan (1B) phase variants after two independent microarray studies. Column headings: p value adj is the adjusted p value, a BH multiple testing correction; log increase represents the change in expression and is derived from coefficient value; RT-PCR value represents the average of two independent experiments for a subset of genes that were the focus of this work; MXAN # and Annotation are derived from the M. xanthus genome (Goldman et al., 2006); in some cases, annotation has been updated to reflect recent additions to the databases, such as naming of dkx genes (Meiser et al., 2008).
Expression of other polyketide gene operons was also up-regulated in yellow compared to tan variants. Yellow variants showed increased expression of MXAN3943, a gene that encodes a cytochrome P450 family protein, and MXAN3949, a gene that encodes a putative acyl carrier protein. Both MXAN3943 and MXAN3949 are situated in the polyketide synthesis operon for antibiotic myxovirescin, also known as TA, targets a signal peptidase that is required for lipoprotein processing (Dan Wall, personal communication). Myxovirescin/TA has been characterized genetically (Paitan et al., 1999b, Tolchinsky et al., 1992) and chemically (Furstner et al., 2007) because of its useful pharmacological properties. The purified product of one of these genes, MXAN3943, encoding a P450 hydroxylase TaH, was shown to previously to be involved in synthesis of myxovirescin/TA (Paitan et al., 1999a). Some of the genes identified in the microarray may correspond to genes involved in regulation of myxovirescin (Varon et al., 1992). MXAN4084, which is adjacent to another large operon encoding polyketide synthase components, may be involved in regulation of myxovirescin-like macrolide or a related compound. The C-terminus of MXAN4084, annotated as a cyclic dinucleotide binding protein, shares similarity with the effector domain of the CAP family of transcriptional activators such as FNR and CRP. Expression of MXAN4084 was increased 6-fold in yellow cells.
There was a >10-fold increase in expression of an ABC transporter and (ornithine) carbamoyltransferase family protein encoded as the first and last genes respectively of what appears to be a three-gene operon shown previously to be essential for developmental aggregation (Ward et al., 1998). This ABC transporter was identified as a partner for FrzZ from a yeast two-hybrid enrichment. A TonB-dependent receptor, MXAN4746, which appears to form an operon with a second gene encoding a putative lipoprotein, was up 5 fold in yellow variants.
The 6-fold increased in 3-oxoacyl-(acyl carrier protein) reductase (MXAN4770) in yellow cells was intriguing because this enzyme is needed for synthesis of the Pseudomonas aeruginosa autoinducer 3-oxo-C(12)-homoserine lactone (Nigaud et al., 2010). Although HSL-like molecules from M. xanthus have not been identified by bioassay, it is possible that yellow variants produce a novel quorum-sensing-like molecule.
MXAN6862, which encodes a member of the MotA/TolQ proton channel family, was more highly expressed in yellow variants; its role may be to energize proteins in the membrane or participate in transport. Expression of ribosomal protein genes, S7, S10, and L16, and genes encoding three putative lipoproteins was increased in yellow variants as well as several genes whose products likely contribute to synthesis of fatty acids and phospholipids.
A gene (MXAN5478) encoding a LysM-like cell wall degrading protein was increased in yellow variants and may play a role in cell wall metabolism or lysis. At least two putative peptidases (one chymotrypsin subfamily and one subtilisin family), a protease B homolog, and a GNAT-family acetyltransferase showed 5-, 15-, 6- and 7-fold increases, respectively in yellow variants.
Several potential DNA and effector-binding proteins, including an adenine-specific DNA methyltransferase, a member of the helix turn helix-xenobiotic response element (HTH-Xre) family, a CAP-like cyclic NMP-binding protein (mentioned above), a hybrid sensor kinase/response regulator, and a gene whose product is related to PspA (phage-shock protein), which is known to suppress sigma-54 dependent transcription, were found to be increased in yellow relative to tan variants.
A number of genes encoding hypothetical and conserved hypothetical proteins were increased in yellow variants. MXAN0504, whose expression was increased over 20-fold in yellow variants, is an isolated gene encoding a small protein that weakly resembles DsbA. The gene for the hypothetical protein MXAN0123 is part of an operon that includes a uracil phosphoribosyltransferase and a GTP cyclohydrolase-like enzyme that together may synthesize a cofactor or signal molecule. MXAN4924 is predicted to encode a 50 kDa membrane protein that contains a C-terminal GC-trans-RRR domain (Gly-Cys -transmembrane helix-Arg-rich cluster), that is thought to serve as a protein-targeting domain.
The serine-threonine kinase Pkn14 is increased in tan variants
Expression of four genes, shown in Table 1B, was increased in tan variants relative to yellow variants. Of these, expression of genes encoding two protein kinases, MXAN4371 (Pkn14) and MXAN4479, was increased 13 and 6-fold, respectively. MXAN6911, which is predicted to encode a TonB-dependent siderophore receptor, was enriched about 11-fold in tan variants. Although expression of MXAN4389 (katB) encoding catalase was increased in only one microarray experiment, the levels were increased in both qRT-PCR experiments. Furthermore, aliquots of tan variants produced copious amounts of oxygen indicative of catalase activity in a hydrogen peroxide assay (Accessexcellence, 2010) compared with modest gas production from a similar aliquot of yellow cells.
A dkxG mutant undergoes phase variation
The finding that expression of a set of genes encoding for DKX biosynthesis proteins was elevated in yellow variants relative to tan variants agreed with the observed phenotype of these two variants. We initiated a phenotypic and molecular comparison between a dkx− pigment mutant and yellow and tan variants to better understand the relationship between pigment production and swarming during phase variation and to begin to elucidate the functions of other genes identified in the microarray. To obtain a dkx− mutant for these studies, we screened among a pool of KanR M. xanthus DK1622 colonies that had been mutagenized with the transposon Himar (Lampe et al., 1996) for tan mutants that swarmed on 0.3% agar.
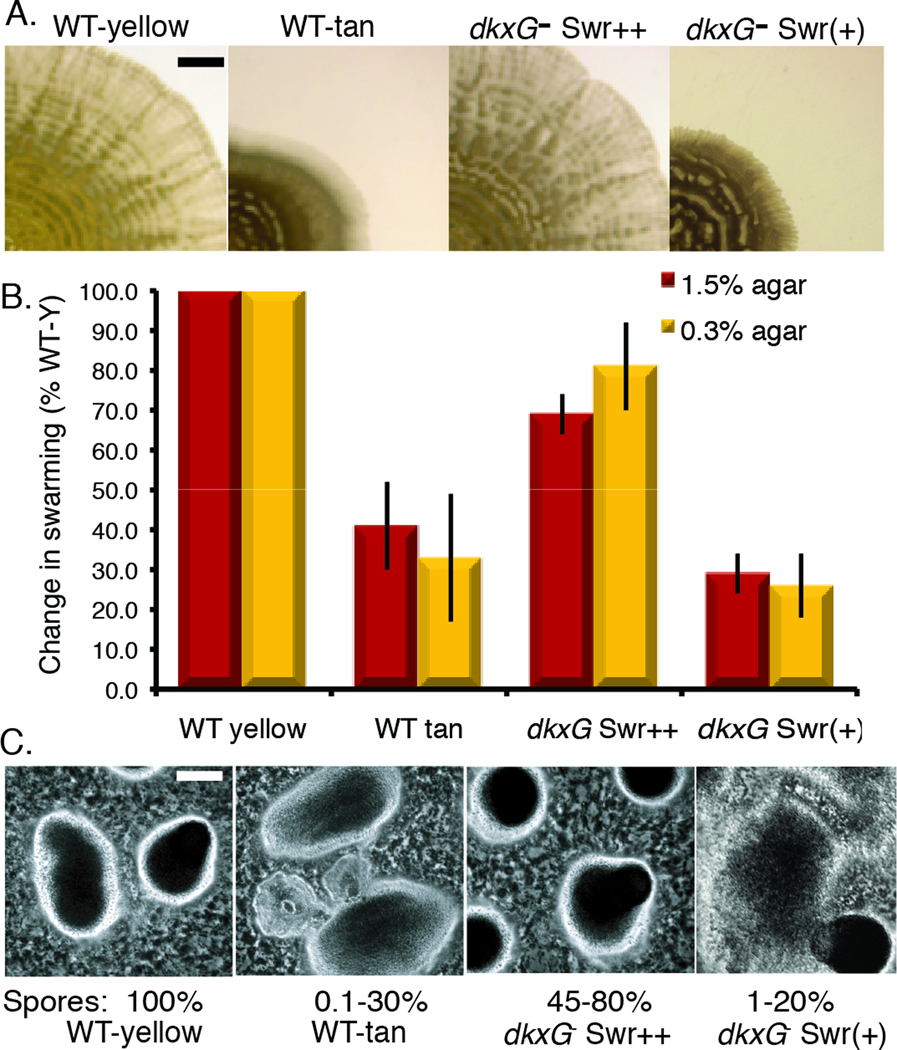
Sequence analysis showed that one of the tan mutants was due to insertion of the transposon in MXAN4299 (dkxG), a gene encoding a non-ribosomal peptide synthase/polyketide synthase needed for DKX production (Meiser et al., 2008). Importantly, upon transfer of this mutant from 0.3% to 1.5% agar and during routine handling, we observed two distinct colony morphologies for the dkxG− mutant. Spectral analysis at 400 nm showed that both colony types were tan due to loss of DKX, but one morphotype, MxH2535, produced large colonies similar to the WT yellow variant while the other morphotype, named MxH2576, produced small colonies similar to the WT tan variant (Fig 1). Swr(+) colonies arose from the dkxG Srw++ mutants (3×10−3 n= 2500 colonies) at about the same rate as Swr(+) arose from the WT Srw++ (8×10−3, n= 2200). The switch from Swr(+) to Swr++ was slightly higher at 2×10−2 and 1×10−2 for the WT and dkxG mutant, respectively.
Fig 1.
The dkxG− mutant produces Swr++ and Swr(+) phase variants. (A) Swarming is reduced significantly in tan variants and in dkxG− Swr(+) mutants. Microphotos of WT yellow, WT tan, MxH2535 (dkxG− Swr++ mutant), and MxH2576 (dkxG− Swr(+)) swarms on 0.3% agar after 72 h. Bar = 2.5 mm. Photos were taken with a Nikon SMZ-U stereoscope. (B). Swarming on 1.5 and 0.3% agar provides an estimate of the function of A- and S-gliding motility systems, respectively. Results are the average of triplicate samples from three independent assays and are presented relative to the WT yellow culture (100%). Swarming was measured as described in Experimental procedures. (C). Development of fruiting bodies was induced on starvation agar. Photos were taken after 96 h with a Nikon FXA microscope at 30X. Bar = 100 µm
Small colony size is usually an indication of a problem with a motility component, so we used swarm and videomicroscopy assays to quantify gliding. M. xanthus used two mechanisms to glide over surfaces. Adventurous (A)-motility allows cells to move independently on agar surfaces; the mechanism of A-gliding motility is still being investigated (Mauriello et al., 2010). The genetically independent social (S)-motility system, which is powered by extension and retraction of type IV pili, requires that cells be in close contact for gliding to occur. Studies of swarming have revealed that a functioning A-motility system is needed for swarming on 1.5% (hard) agar whereas a functioning S-motility system is needed for swarming on 0.3% (soft) agar (Shi & Zusman, 1993). As previous studies measured swarming only on 1.5% agar surfaces, we examined swarming capabilities of yellow and tan variants alongside the dkxG− mutants on 1.5% and 0.3% agar surfaces to determine if swarming was affected under both of these conditions. After 5 days, the pattern and texture of the MxH2535 swarm was nearly identical to that of the WT yellow variant (Fig 1A) and the change in swarm area was 70% to 80% that of WT yellow variant on 1.5% and 0.3% agar respectively (Fig 1B). Based on these results, MxH2535 was designated dkxG− Swr++. In contrast, MxH2576, designated dkxG− Swr(+), produced significantly smaller swarms after 5 days (Fig 1A) and the change in swarm area was less than 30% that of the WT yellow variant and 66% that of the WT tan variant on both swarm media (Fig 1B). These results suggest that DKX production does not govern M. xanthus phase variation but represents one pathway that is regulated by phase variation.
Swarming differences are magnified at high cell densities
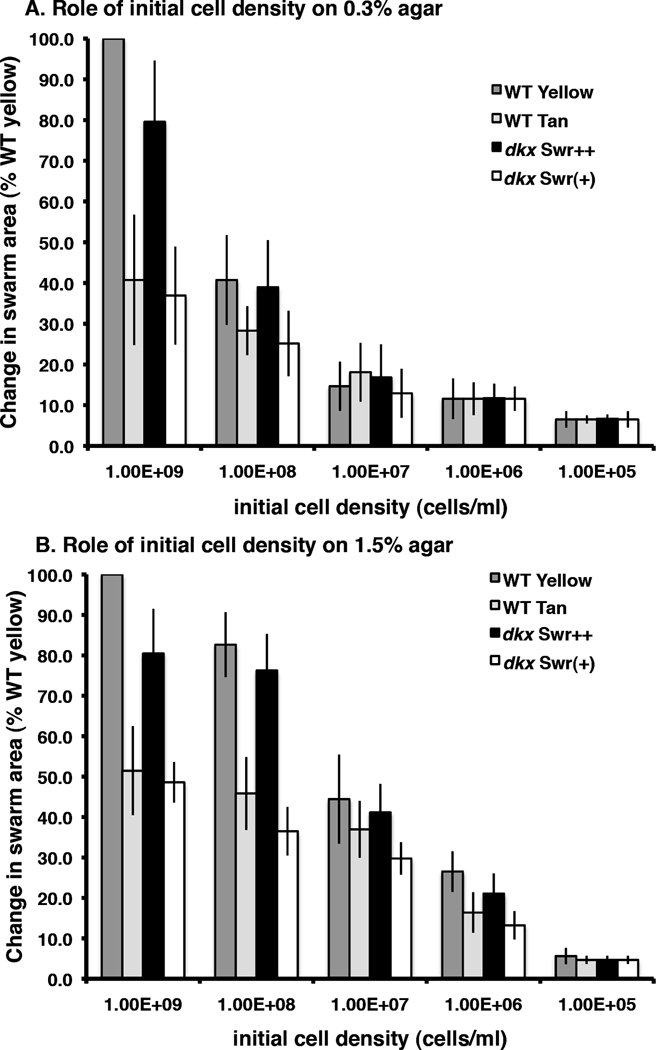
It has been reported that yellow and tan variants exhibit different density-dependent growth and swarming rates (Rosenberg et al., 1977). To examine contributions of DKX in density-dependent swarming, WT yellow, WT tan, and both dkx− morphotypes were assayed on soft and hard agar surfaces at different cell densities. Swarming on soft agar was robust at high cell densities (109 cells ml−1), but even a 10-fold decrease in initial cell density resulted in a significant decrease in swarming (Fig 2A). This affect was quite dramatic on 0.3% agar where S-gliding predominates. This agrees with previous reports showing that the efficiency of S-gliding, but not A-gliding, requires a high density of cells. Swarming of the dkxG− Swr++ was 80% that of the WT yellow variant when the initial cell density was high but this difference disappeared at lower initial cell densities on both 0.3 and 1.5% agar surfaces. Similarly, swarming of the dkxG− Swr(+) mutant was consistently less than the WT tan variant. Hence DKX, directly or indirectly, may stimulate swarming at high cell densities.
Fig. 2. Swarming on soft agar is particularly sensitive to initial cell density.
(A) The change in swarm area on 0.3% agar was determined after 5 d at 32°C for one yellow (WT, grey bars) and three tan (WT tan, light grey bars; dkxG− Swr++, black bars; dkxG−, Swr(+) white bars) strains. Each strain was grown in liquid medium, adjusted to the cell density indicated in the legend and spotted on 0.3% CTPM agar plates that were incubated at 32° for 5 d. Assays were performed in triplicate. Data shown are the average of two experiments. (B) Identical to (A) except that each cell density sample was spotted on 1.5% CTPM agar. All values are relative to the WT yellow swarm value at 1E+09 initial cell density (100%).
Swarming was reduced significantly for all four strains at low cell densities on both agar surfaces (Fig 2A, B). Swarms did not expand even when prolonged incubation (5–7 d) should have allowed for increased cell density due to growth. After 5 days, the size of a swarm was smaller than the size of a typical isolated colony for each strain on a streak plate. Cells within the swarm were still viable, but they did not move beyond the boundary of the swarm. This result was surprising because it suggests that swarming can override (or even suppress) gliding since even a low density of cells interfered with the ability of cells to gliding away from the swarm.
Time-lapse videomicroscopy, which provides a measure of gliding motor function and reversal frequency, was performed to determine if changes in motor or taxis functions might contribute to the dramatic swarming defects. Time-lapse assays did not reveal significant differences in the rate of gliding (10.5 µm min−1 WT yellow versus 8.9 µm min−1 WT tan; see supplemental movies WTyellow.avi and WTtan.avi) or reversal period (9.8 min rev−1 WT yellow and 11.2 min rev−1 WT tan) on 1.5% agar pads. Rates were similar for the dkx− mutants. Hence, the reduced ability of the tan variant and the dkx− Swr(+) mutant to swarm at high cell density on 1.5% agar cannot be accounted for by changes in the A-motility motor.
Affects of phase variation and pigment production on development
Defects in swarming can have grave consequences for the ability of cells to form aggregates needed for fruiting body formation. When starved for nutrients, the Swr++ yellow variants formed aggregates within 16–20 hs, which were consistently transformed into dark fruiting mounds (Fig 1C). For reasons that are not entirely clear, independent cultures of tan variants arrested early in development (ridge stage) about 50% of the time and progressed to mound stage about 50% of the time. A tan variant mound with partial darkening in the center is shown in Fig 1C. Production of heat-resistant spores was low (0.1–1% of WT yellow levels) when morphogenesis did not progress beyond the ridge aggregation stage and higher (1–30% of WT levels) when mounds formed. MxH2535 produced dark fruiting bodies containing roughly the same number of heat-resistant spores (60 ± 20%) as the WT yellow variant (Fig 1C). The fruiting body developmental phenotypes of the dkxG− Swr(+) derivative paralleled that of the tan WT variants (Fig 1C). Using the Kroos-Kaiser developmental assay (high cell density on starvation medium (Kroos et al., 1986)), the dkxG− Swr(+) mutant produced roughly 1–20% the WT yellow complement of heat-resistant spores by day 5.
We note that the dkx− strain described by Meiser et al produced spores that were visually indistinguishable from the dark refractile spores produced by the WT strain yet the spores of their mutant failed to germinate after heat and sonication treatment (Meiser et al., 2006). Their development assay was performed in submerged culture at a much lower cell density, which might explain our differing results. To test this, we assayed our dkxG- strains using the submerged culture method. After days 3 and 5, samples were subjected to sonication with and without heat treatment. We found no significant difference in spore yield between WT yellow, WT tan, dkxG− Swr++ and dkxG− Swr(+) after sonication treatment only. However, samples treated with sonication plus heat gave significantly different results. When adjusted for the differences in initial cell density, survival of spores from WT yellow and dkxG− Swr++ strains was proportional to the levels measured from plate assays. However, spore levels for the WT tan and dkxG− Swr(+) strains after development in submerged culture were reduced >100-fold (Fig S1). We conclude that swarm proficient strains form spores in high and low cell density assays, whereas the swarm deficient strains form spores efficiently only at high cell densities.
The dkxG− Srw++ mutant shows enhanced cohesion
The differences we observed in swarming and development might be attributed to changes to gliding components during phase variation. Although no known gliding genes were identified in the microarray assay, increased levels of enzymes involved in post-translational modification might contribute to the efficiency of swarming in yellow cells. The abilities of M xanthus cells to agglutinate (aka cell cohesion) and bind dyes, such as Congo red (CR) and trypan blue (TB), are correlated positively with production of extracellular fibrils and pili needed for S-gliding motility. Mutations in difA, difC and difE genes abolish production of fibrils and reduce cohesion and CR binding while mutations in difD and difG overproduce fibrils (Black & Yang, 2004, Dana & Shimkets, 1993). Mutations in pilA and aberrant localization of PilA affect production of fibrils and hence diminish cohesion and dye binding (Black et al., 2006, Yang et al., 2010).
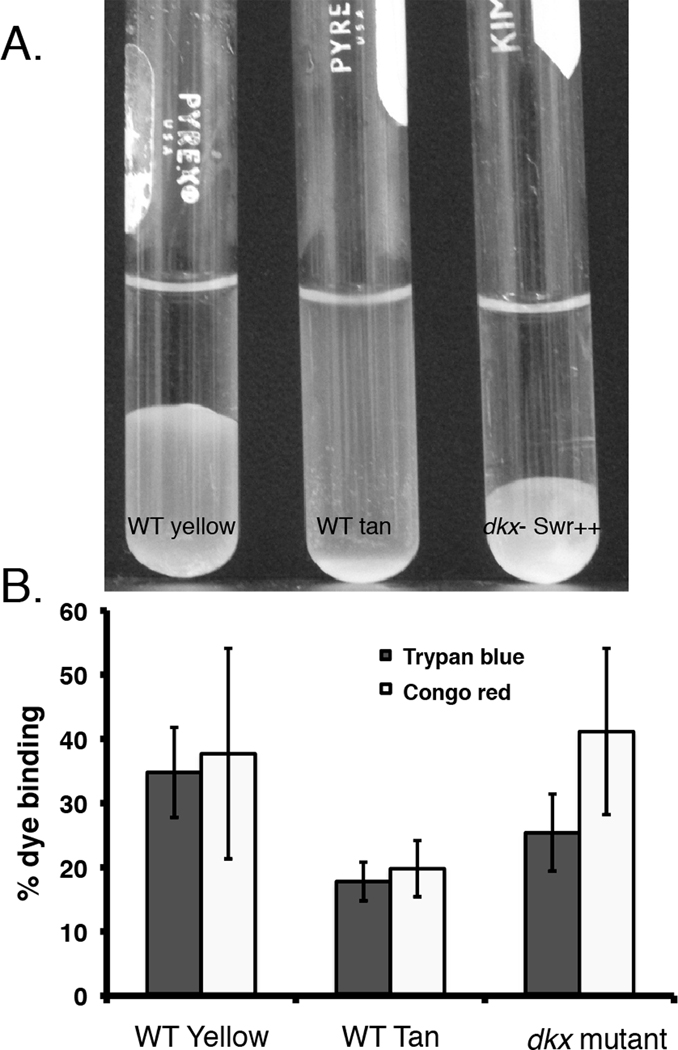
Cohesion and dye-binding assays were used to measure fibril production in WT yellow, WT tan and dkx− strains. As shown in Fig 3, the WT yellow cells agglutinated quickly to form a yellow colored mass at the bottom of the tube (endpoint shown in Fig 3A). The tan dkxG− Swr++ mutant aggregated at about the same rate (not shown) as the WT yellow resulting in a mass of cells at the base of the tube (Fig 3A). Surprisingly, the aggregate formed by the dkxG− Swr++ mutant was more compact than the WT yellow despite the fact that the assay started at the same initial cell density for both samples. In contrast, the tan variant failed to agglutinate in the cohesion assay and cells remained dispersed in buffer (Fig 3A, middle tube). The agglutination phenotype of the dkxG− Swr(+) mutant was indistinguishable from the WT tan variant (data not shown).
Fig 3. Tan variants are defective in cell cohesion.
(A) The ability of cells to agglutinate was performed at 32 °C for 2 h. Tan variants are less cohesive than WT and cell cohesion is increased in tan dkxG− Swr++ mutants. (B) Decreased agglutination in tan variants is associated with a reduced ability to bind Congo red and trypan blue. Dye binding of the dkxG− mutant was reproducibly higher than the tan variant. The absorbance of each sample was measured at 490 nm to detect Congo red and 585 nm for trypan blue. Values represent the average of triplicate samples from two independent assays.
The dye binding results, shown in Fig 3B, followed a similar trend. CR binding by the dkxG− Swr++ mutant was slightly higher than the yellow variant. TB binding, which is thought to be more specific for fibrils (Black et al., 2006) was 72% that of the WT yellow variant. The results with the dkxG− Swr++ mutant implicate a CR-binding component, other than fibrils, in the cohesion process. Alternatively a surface component critical for cohesion might be more accessible in the dkxG− Swr++ mutant. The factor that contributed to increased cohesion might be responsible for the slight reduction in swarming by the dkxG− Swr++ mutant compared with the WT yellow variant. Dye binding of the tan variant was about 60% (for both CR and TB) that of the yellow variant.
Tan phase variants have a survival advantage
Decreases in swarming might be caused by changes in surface components, as described above, gliding motor defects, or differences between the growth rates of variants. To determine if differences in growth rate were affecting swarming, we measured growth of strains in liquid medium at 32°C with initial cell densities ranging from 103 to 107 cells ml−1. Tan cultures doubled every 3.6 ± 0.4 h and 3.4 ± 0.3 h, whereas yellow cultures doubled every 3.9 ± 0.3 h and 3.6 ± 0.9 h in liquid medium when the initial cell densities were high (107 and 106 cells ml−1 respectively)(See graphs in Fig S2). At lower cell densities, the doubling times were very similar: 4.1 h and 4.4 h at 107 cells ml−1, 4.0 h and 4.1 h at 106 cells ml−1 initial density and 4.6 h and 4.5 h at 105 cells ml−1 initial cell densities for yellow and tan cultures respectively. Both yellow and tan cultures had comparable lag periods in liquid Casitone medium of ≈10 hr (±2 h) for every 10-fold change in density. The doubling time for MxH2535 (dkxG− Swr++) was 3.9 h when the initial cell density was 107 cells ml−1, similar to WT yellow variant. Hence, the reduction in swarming in the tan variant is not due to a slower growth rate because our results showed that tan variants grew faster than yellow variants.
The growth rate studies described above were performed by monitoring cell scattering in a Klett meter in conjunction with viable plate counts at regular intervals. The results of plate counts revealed consistent differences between yellow and tan variants in post-exponential phase survival. Yellow and tan variants both reached a density of about 109 cells ml−1 20–30 h post inoculation (Fig 4 and Fig S2 growth curve at 32°C). About 24 h after reaching the maximum density in broth, the Klett values of yellow cultures began to decrease rapidly. In contrast, the Klett values of tan cultures decreased at a slower rate and reached a stationary phase density that was consistently higher than yellow cultures. Stationary phase tan cultures yielded 8–10 fold higher CFUs than comparable aliquots from yellow cultures.
Fig 4. Survival of tan variants was superior to yellow variants in liquid medium.
Yellow (black bars) and tan (grey bars) variants were cultured at 32° for 24 h, then shifted to 22° for the duration of the experiment. Aliquots were removed at times indicated and assayed for CFU; CFUs were screened visually for phase (yellow or tan). (A) The WT yellow variant was used as inoculum. At 24 h, the culture was at Klett 110 and contained ≈ 5 × 108 yellow variants and 7 × 106 tan variants (1.4%). Growth (determined by Klett reading) peaked at 24–26 h. With increasing time of incubation, viability of both variant types decreased with the rate of decrease being steeper for yellow variants (solid line) that tan variants (dashed line). (B) Protocol as in (A) except that the initial inoculum was the WT tan variant.
The survival effect was more dramatic at 22°C, a temperature range that mimics the natural habitat of M. xanthus habitat. Similar as the results shown in Fig S2, the post-exponential viability of yellow cultures decreased at a rate of ≈106 cells per day while viability of tan cultures decreased at a rate of ≈ 105 cells per day (Fig 4 WT yellow = solid line; WT tan = dashed line). Stationary phase survival favored tan variants as the incubation time increased. As shown in Fig 4, cultures that initially were predominantly yellow yielded almost exclusively tan variant colonies (Fig 4A, grey bars) after 168 h. The quantity of tan cells within the yellow cultures was stable over time which suggests that the survivors were pre-existing tan cells, not yellow to tan convertants. Cultures that were tan at the time of inoculation continued to yield high numbers of tan CFU for > 168 h (Fig 4B). Enhanced survival appeared to be an intrinsic property of tan cells and not the result of feeding off necrotic yellow cells because the density and viability of the tan variant cultures was the same regardless of the initial number of yellow cells. Survival results for the dkxG− Swr++ and Swr(+) mutants were similar to those of the WT-yellow and tan variants respectively (not shown).
Ultraviolet light stimulates tan to yellow phase variation
Previous studies showed that the ratio of yellow to tan variants was higher after exposure to UV (Burchard et al., 1977). However it was not clear if UV was causing a increase in the frequency of tan to yellow conversion or if tan variants were simply more sensitive to UV damage due to lack of pigment because investigators lacked strains with stable yellow/tan phenotypes. The role of UV is significant as it may provide insight into the mechanism of phase variation. The stable tan phenotype of the dkxG− mutants allowed us to determine if lack of pigment contributed to UV sensitivity. Equivalent amounts of washed cells were exposed to UV for 0–4 min and then plated on rich medium to quantify survival and look for evidence of phase variation. After exposure to 1 min of UV, the survival numbers of the yellow variant, tan variant, and dkxG mutants were nearly identical (range 68–72% CFU relative to CFUs at t= 0). However, after exposure for 3 min, survival of cells from yellow WT cultures was >10-fold higher than that of the tan WT culture. Moreover, the survivors from the tan variant population, albeit lower in number, were enriched for yellow variants compared to the t=0 control, suggesting that UV treatment stimulated phase variation. The loss of pigment did not contribute to UV sensitivity, because the tan dkxG− mutant, when assayed in parallel, produced the same number of CFUs after 2, 3, and 4 min UV exposure as the WT yellow culture. The results for the dkxG Swr++ mutant show that loss of pigment does not make cells more sensitive to UV. Rather, the lower overall survival of cells from tan cultures suggests that sensitivity of tan variants to UV is due to an aspect of phase variation that is independent of DKxanthene production.
RT-PCR reveals different expression profiles between tan variants and dkxG− mutants
Assays for swarming, dye binding, UV sensitivity and development revealed significant similarities between WT yellow and dkxG Swr++ strains that were distinct from those of the tan variant and the dkxG Swr(+) strains. To determine if these patterns were based on changes at the molecular level, the expression profiles of a subset of genes identified from the microarray were determined. As shown in Table 2 (and Fig S3) expression of genes for synthesis of DKX, represented here by MXAN4295 (dkxJ) and MXAN4294 (dkxK; ATPase) was reduced in the tan variant (due to phase variation) and both of the dkxG− mutants (due to gene disruption of a gene in the pathway). dkxG was decreased in the tan variant and the dkxG Swr++ strain, but surprisingly was increased in the Swr(+) variant of the dkx mutant. The dkxG− Swr(+) mutant, which is phenotypically very similar o the WT tan variant, presented a slightly different expression profile than the tan variant and its dkxG Swr++ relative. For example expression of genes involved in production of the antibiotic myxovirescin was increased in the dkxG− Swr(+) mutant but decreased in the WT tan variant and tan variants. Similarly, expression of MXAN0228, which encodes a potential regulator (HTH-Xre) and MXAN5138 (methyltransferase) was increased in the dkxG− Swr(+) mutant and decreased in the tan variant.
Table 2.
WT phase variants share patterns of gene expression that are similar to dkx mutants
| dkxJ | dkxK | dkxG | hypothetical protein | |||||
|---|---|---|---|---|---|---|---|---|
| RQ value | range (−/+) | RQ value | range (−/+) | RQ value | range (−/+) | RQ value | range (−/+) | |
| WT Yellow | 1 | 0.72/2.62 | 1 | 0.60/1.52 | 1 | 0.36/0.57 | 1 | 0.16/0.20 |
| DKX Swr+ | 0.17 | 0.09/0.21 | 0.40 | 0.19/0.37 | 0.95 | 0.32/0.8 | 1.44 | 0.83/1.96 |
| WT Tan | 0.07 | 0.02/0.03 | 0.34 | 0.11/0.15 | 0.65 | 0.29/0.52 | 0.54 | 0.15/0.22 |
| DKX Swr− | 0.21 | 0.087/0.15 | 0.62 | 0.15/1.17 | 2.56 | 0.86/1.49 | 2.15 | 0.86/1.43 |
| myxovirescin 1 | myxovirescin 2 | cNMP-binding | DNA methyltransferase | |||||
|---|---|---|---|---|---|---|---|---|
| RQ value | range (−/+) | RQ value | range (−/+) | RQ value | range (−/+) | RQ value | range (−/+) | |
| WT Yellow | 1 | 0.22/0.35 | 1 | 0.6/1.48 | 1 | 0.6/1.47 | 1 | 0.31/0.46 |
| DKX Swr+ | 0.89 | 0.54/1.37 | 1.07 | 0.6/1.34 | 1.13 | 0.59/1.51 | 1.12 | 0.45/0.87 |
| WT Tan | 0.55 | 0.23/0.38 | 0.55 | 0.29/0.62 | 1.18 | 0.23/1.48 | 0.52 | 0.34/1.26 |
| DKX Swr− | 1.78 | 0.91/1.85 | 1.77 | 1.13/3.14 | 3.53 | 2.62/1.42 | 3.12 | 1.74/4.97 |
| HTH-Xre | lysM | phage integrase-like | pkn14 | |||||
|---|---|---|---|---|---|---|---|---|
| RQ value | range (−/+) | RQ value | range (−/+) | RQ value | range (−/+) | RQ value | range (−/+) | |
| WT Yellow | 1 | 0.29/0.41 | 1 | 0.19/0.23 | 1 | 0.23/0.30 | 1 | 0.48/1.09 |
| DKX Swr+ | 0.96 | 0.57/0.75 | 1.29 | 0.46/0.71 | 0.85 | 0.32/0.50 | 0.97 | 0.33/0.49 |
| WT Tan | 0.34 | 0.01/0.75 | 1.21 | 0.33/0.46 | 1.59 | 0.33/0.41 | 8.25 | 3.18/6.16 |
| DKX Swr− | 1.31 | 0.35/1.67 | 2.01 | 0.85/1.48 | 3.36 | 1.42/2.46 | 13.02 | 5.09/9.61 |
| TonB-dependent receptor | catalase | |||
|---|---|---|---|---|
| RQ value | range (−/+) | RQ value | range (−/+) | |
| WT Yellow | 1 | 0.08/0.08 | 1 | 0.31/0.46 |
| DKX Swr+ | 1.19 | 0.46/0.76 | 0.91 | 0.37/0.62 |
| WT Tan | 5.58 | 2.42/4.27 | 1.87 | 0.5/0.68 |
| DKX Swr− | 7.20 | 3.54/6.97 | 3.54 | 1.45/2.47 |
Expression of a subset of genes known to be differentially regulated during phase variation was compared for WT yellow, WT tan, dkx Swr++ and dkx Srw(+) strains. Expression of dkxJ and dkxK is decreased in all three tan strains due to phase variation or dkxG mutation. Expression of genes encoding the TonB-dependent receptor, serine-threonine kinase Pkn14 and catalase, which are indicated by the rightmost gold, green, and blue-green bars, respectively, was increased in the tan variant and the dkxG− Swr(+) mutant only. Others are discussed in the text. dkxG− Swr++, WT tan, and dkxG− Swr(+) expression levels were relative to the WT yellow (calibrator, set to 1) plotted on a log scale as decreased (< 1) or increased expression (>1). The endogenous control for qRT-PCR was 16S RNA. Legend: dkxJ (MXAN4295), dkxK (MXAN4294), dkxG, (MXAN4299), dkx adenyltransferase (MXAN4305), hypothetical (MXAN0504), myxovirescin 1 (MXAN3943), myxovirescin 2 (MXAN3949), cNMP binding (MXAN4084), DNA MT (methyltransferase; MXAN5138), HTH-Xre (MXAN0228), lysM (MXAN5478), phage integrase-like (MXAN1200), pkn14 (MXAN4371), TonB-dependent siderophore receptor (MXAN6911), and catalase (MXAN4389). Bars represent the range RQ min and RQ max for each sample.
Significantly, expression of genes encoding Pkn14 (gold bar), the putative TonB-dependent receptor (green bar) and catalase (blue green bar), which was shown by microarray to be increased in the tan variant was also increased in the dkxG− Swr(+) mutant, but not in the dkxG− Swr++ mutant. These data show that the change in swarming between dkxG Swr++ and dkxG Swr(+) was accompanied by changes in gene expression for pkn14, MXAN6911 and katB that parallel the differences we found between the WT yellow and tan variants by microarray. Hence the functions of the TonB-dependent receptor, KatB, and Pkn14 are uniquely associated with swarm pathway.
Disruption of MXAN0228 abolishes production of DKX and blocks the swarm switch
MXAN0228, which is predicted to encode a 13 kDa DNA binding protein belonging to the HTH-xenobiotic response element (Xre) superfamily of proteins, was up-regulated in yellow cells relative to tan variants. Members of the Xre family include bacterial methylases and phage regulatory proteins Cro and C1. We were intrigued by the fact that MXAN0228 also contains a conserved HipB domain. HipB is the neutralizing component of a toxin-antitoxin system whose function is associated with persistence in bacterial cells, a phenotype that is reminiscent of the tan variant survival phenotype. HipB interacts with the toxin HipA, which contains a serine-threonine kinase-like fold and phosphorylates EF-Tu (Schumacher et al., 2009). MXAN0097 is predicted to encode a 46 kDa protein that shares 25%/43% identity/similarity with the E coli HipA; expression of MXAN0097 was not differentially regulated in yellow and tan variants in our study.
To determine if MXAN0228 regulates phase variation, we engineered a disruption of this gene. MXAN0228 is a single gene operon; hence construction of a disruption using an internal fragment of the gene would not be expected to confer polar effects on adjacent genes. Plasmid pHS2, was electroporated into WT yellow cells and KanR transformants were recovered. All 42 transformants recovered exhibited a stable tan color. Surprisingly, disruption of MXAN0228 (HTH-Xre) in an otherwise WT tan Swr(+) cells yielded only tan Swr++ derivatives (n= 54). Since all of the transformants (from two independent electroporations) were Swr++, this suggests that the xre− Swr++ phenotype is epistatic to that of the Swr(+) phenotype of the tan variant.
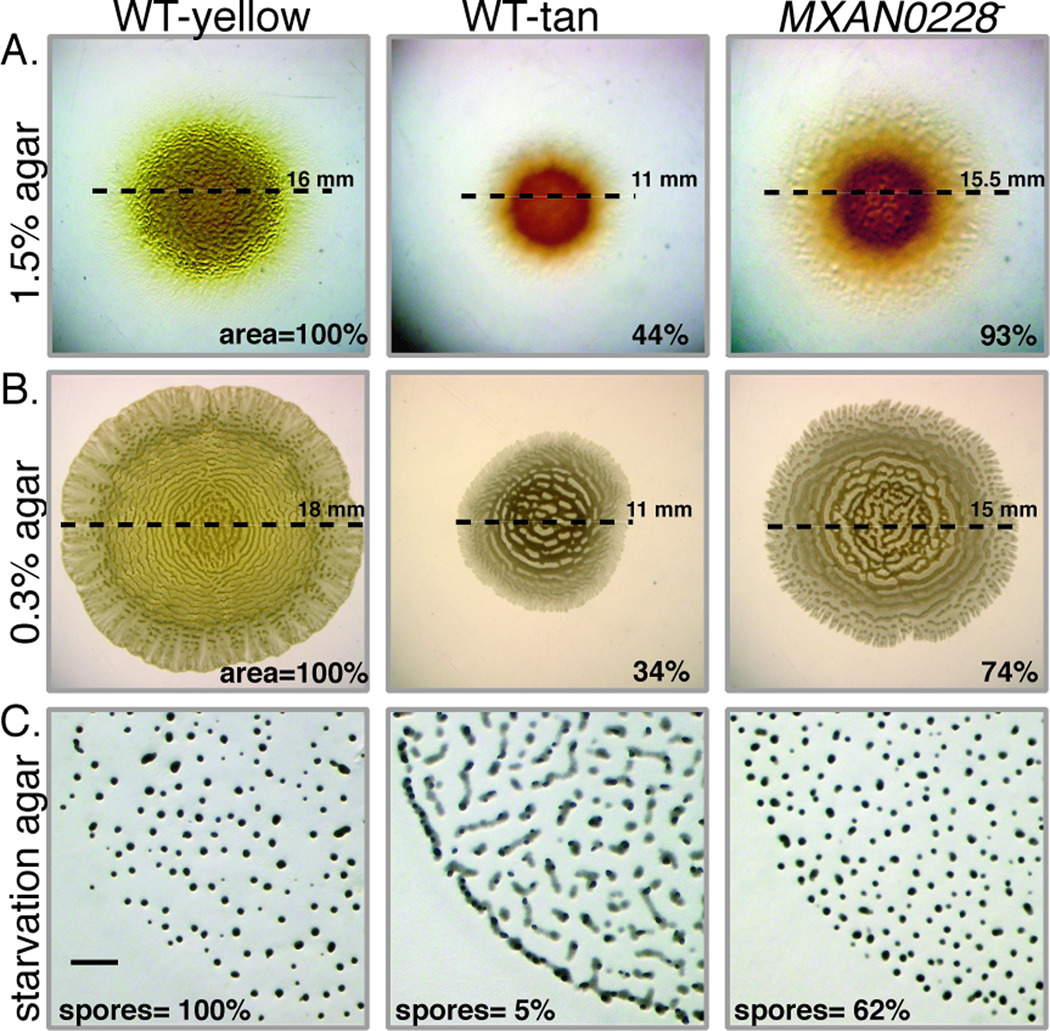
The phenotype of the MXAN0228 disruption mutant, MxH2546, was nearly identical to that of the dkx− Swr++ mutant. As shown in Fig 5, MxH2546 exhibited near normal abilities to swarm on hard (Fig 5A, 93% of WT yellow) and soft (Fig 5B; 74% of WT yellow) agar surfaces and produced fruiting bodies that were similar is size, density, and distribution to the WT yellow variant (Fig 5C).
Fig 5.
The phenotype of the MXAN0228 (HTH-Xre) mutant is similar to the dkxG Swr++ mutant. Swarming of the tan MXAN0228 mutant was similar to the WT yellow culture on 1.5% agar plate (panel A) contrast was enhanced to show the extent of swarming at the edge) and 0.3% agar (panel B) whereas the WT tan variant showed reduced swarming on both surfaces. Dashed lines represent the swarm diameter; the area in the lower right corner represents the average final change in swarm area over 5 d relative to the WT yellow variant. (C) WT tan variants formed long translucent mounds, in contrast to the dark fruiting bodies formed by the WT yellow variant and the MXAN0228 mutant on TPM starvation agar. The average number of heat-resistant spores (three independent experiments) is shown in the lower left corner. Bar = 100 µm.
The phenotype of the MXAN0228 mutant suggests that the product of this gene, the HTH-Xre protein, plays a key role in regulation of phase variation. Not only was DKX synthesis abolished but a systematic examination of several thousand colonies failed to reveal a yellow variant or a Swr(+) morphotype from MXAN0228 mutants. No Swr(+) colonies were detected among over 5000 colonies screened so the rate is less than 10−4. Hence, pigment production and the ability to vary the swarm phase is blocked or significantly reduced in the MXAN0228 mutant.
The transcriptomes of WT yellow, WT tan and the MXAN0228 mutant were analyzed by qRT-PCR. As shown in Fig 6, expression of dkxJ, dkxK and dkxG was reduced dramatically in both the WT tan and MXAN0228 mutant. In contrast, expression of myxovirescin genes was similar in the MXAN0228 mutant to the WT yellow variant whereas these genes are down-regulated in the WT tan variant. This suggests that expression of myxovirescin is regulated by a mechanism that is independent of MXAN0228 and distinct from DKX regulation. Expression of pkn14 and MXAN6911 (TonB-dependent siderophore receptor), which were significantly increased in the WT tan variant and in the dkxG− Swr(+) mutant, were increased only slightly in the MXAN0228 mutant background, which suggests that regulates these genes directly or indirectly. Finally, increased expression of the MXAN0228 gene hints that MXAN0228 is subject to autoregulation.
Fig. 6.
The transcriptome of MxH2546 (MXAN0228−) mutant suggests that HTH-Xre is a positive regulator of dkx operons. WT tan and MxH2546 (MXAN0228::pHS2) expression levels were relative to the WT yellow (calibrator, set to 1) plotted on a log scale as decreased (< 1) or increased expression (>1). The endogenous control for qRT-PCR was 16S RNA. Legend: dkxJ (MXAN4295), dkxK (MXAN4294), dkxG, (MXAN4299), dkx adenyltransferase (MXAN4305), hypothetical (MXAN0504), myxovirescin 1 (MXAN3943), myxovirescin 2 (MXAN3949), cNMP binding (MXAN4084), DNA methyltransferase (MXAN5138), HTH-Xre (MXAN0228), lysM (MXAN5478), phage integrase-like (MXAN1200), pkn14 (MXAN4371), TonB-dependent receptor (MXAN6911), and catalase (MXAN4389).
DISCUSSION
Microbes employ complicated regulatory circuits to integrate a multitude of signals needed to adapt to changing environmental conditions. One such regulatory process is phase variation, a term used to describe the ability of a subset of microbes in a population to specifically alter the expression of components that often confer some selective advantage. In M. xanthus, phase variation results in cells that express a different set of genes observable as colonies with differences in pigment, texture, and swarming capabilities. In this study, we identified a core set of 41 genes that are differentially regulated during phase variation. Thirty-seven genes were up-regulated in yellow variants (core yellow genes) relative to tan variants and four genes were up-regulated in tan variants (core tan genes) relative to yellow variants. Hence, less than 1% of the M. xanthus genome is devoted to this important process. This set of genes provides a focal point to begin a detailed molecular analysis of phase variation.
In the Myxococcus social system, the role of yellow variants, with their superior swarming capabilities, might be to seek out food and produce components that protect the population as a whole. Several clusters of core yellow genes encode proteins for non-ribosomal biosynthetic pathways whose products might function as signaling molecules to regulate swarming and development or antibiotics that provide protection. For example, multiple genes previously shown to be essential for synthesis of the polyketide DKxanthene were highly expressed in yellow variants as opposed to tan variants (Fig 7A). DKxanthene probably is important for survival of M. xanthus in nature as it has been shown to play a critical role in the formation of heat-resistant spores in the laboratory (Meiser et al., 2006). Genes for components involved in synthesis of the antibiotic myxovirescin (aka TA) (MXAN3943 and MXAN3949) also were expressed more highly in yellow variants and release more active myxovirescin into the medium than tan cells (data not shown). Increased production of myxovirescin might explain why yellow variants have a greater tendency to lyse during stationary phase than do tan variants. Myxovirescin was initially reported to interfere with cell wall synthesis (Zafriri et al., 1981) because it stimulates cell lysis, but has been shown to inhibit a lipoprotein peptidase. During growth, M. xanthus cells appear to be protected from self-destruction by this antibiotic because enzymes involved in synthesis of myxovirescin are located in the membrane and presumably export the antibiotic out of the cell (Simunovic et al., 2003). However, changes in cell architecture during stationary phase or when cells are starving may make yellow variants sensitive to their own myxovirescin, thus triggering lysis. On the other hand, the low levels of myxovirescin in tan variants should contribute to their enhanced survival during stationary phase. Although we did not quantify expression of these genes in starving cells, we imagine that the expression pattern might be conserved during development. If true, then lysis of yellow variants could allow release of DKX, which Meiser et al have shown promotes sporulation of tan variants (Meiser et al., 2006).
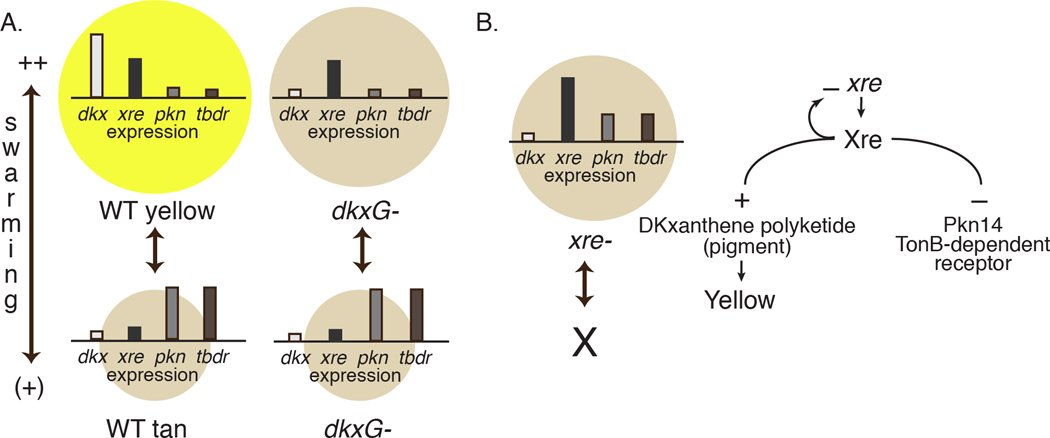
Fig 7. HTH-Xre regulates multiple phenotypes associated with phase variation.
A. Expression patterns of dkx, xre, pkn, and tnbr illustrated against the backdrop of their WT and dkxG− phase variation phenotype. The relative expression levels are shown as bars on the line within each circle. Each circle represents the swarm size and color. dkx and xre are expressed highly and pkn and tbdr are expressed poorly in yellow swarm ++ cells (left panel top circle). The pattern is reversed in tan swarm(+) variants (left panel bottom circle). The switch to Swr(+) is accompanied by activation of pkn14 and MXAN6911, encoding a threonine-serine kinase and TonB-dependent receptor respectively, both of which may contribute to the enhanced survival of the tan Swr(+) variants. Loss of dkxG affects expression of dkx but mutants can phase vary between Swr++ (right panel top circle) and Swr(+)(right panel bottom circle) because MXAN0228 is active in the dkxG− background. dkxJ: DKxanthene gene MXAN4295; xre: HTH-Xre MXAN0228; pkn14: protein kinase; tnbr: TonB-dependent receptor MXAN6911.
B. The MXAN0228 encodes a putative HTH-Xre protein with a tan Swr++ locked phenotype. Left panel shows the expression profile of dkx, xre, pkn, and tnbr in the xre228 mutant which produces a tan swarm that is about 75% the size of the WT yellow parent (indicated by the size and position of the tan circle). Disruption of MXAN0228 blocks expression of DKX polyketide and prevents switching (indicated with X) from Swr++ to Swr (+). Expression of pkn and tbdr is slightly elevated in the xre228 mutant compared with the WT yellow strain, which might account for the slight reduction in swarm size. Expression of xre228 is elevated in the Δxre228 background suggesting that Xre228 regulates its own expression directly or indirectly (right panel).
Some of the phenotypic differences between yellow and tan variants during development can be explained from the data presented in this study. Elevated levels of MXAN1286 and MXAN1288 likely contribute to the abilities of yellow cells to aggregate and form fruiting bodies. MXAN1286 and MXAN1288 are the first and third genes of a three-gene operon that encodes an ABC transporter and ornithine-like carbamoyltransferase, respectively. This ABC transporter was identified previously because it interacts with the CheY-like response regulator FrzZ in the yeast-two hybrid assay (Ward et al., 1998). Disruption of the MXAN1286 transporter blocked developmental aggregation. The defects of the mutant could be complemented by mixing with WT cells, which led the authors to suggest that ABC transporter exports a compound needed for autochemotaxis during development. Although Ward and co-workers found that disruption of MXAN1288 did not produce the same aggregation defect as the MXAN1286 mutant, the phenotype of a carbamoyltransferase mutant might have been missed depending on the type of assay used. Elevated levels of a carbamoyl transferase in yellow M. xanthus variants might protect these cells from a component that is analogous to phaseolotoxin, a toxin produced by Pseudomonas that targets carbamoyltransferase. E. coli and Salmonella are inhibited by phaseolotoxin but Pseudomonas is immune to its own toxin because at 18° it produces an ornithine carbamoyltransferase that is insensitive to phaseolotoxin (Staskawicz et al., 1980). Although a phaseolotoxin has not been reported from M. xanthus, a potential substitute might be a heme-like component derived from products of the operon adjacent to the MXAN1286 operon. MXAN1289, which is predicted to encode a phytanoyl-CoA dioxygenase, was up-regulated in yellow cells. MXAN1289 is the last gene in a large operon that includes a non-ribosomal peptide synthetase and a protoporphyrinogen oxidase.
Yellow variants exhibited more vigorous swarming than tan variants, but this difference was not dependent on pigment production because as summarized in Fig 7A, the dkxG mutant produced both Swr++ and Swr(+) phenotypes that could phase vary with respect to swarming. Since swarming, particularly swarming on 0.3% agar, required a high cell density, we considered that some of the genes expressed in Swr+ strains, such as MXAN1289 (above) and MXAN4084 (cyclic NMP binding protein), might be involved in production or reception of a signal molecule that promotes swarming in a density dependent manner. MXAN6862, a MotA/TolQ homolog adjacent to aglS, an A-gliding motility gene (Youderian et al., 2003) may contribute to the robust swarming on 1.5% agar enjoyed by yellow, but not tan, variants.
The differences in texture, cohesiveness and dye binding between Swr++ and Swr (+) variants are indicative of changes in the cell surface and extracellular matrix (ECM). Although genes known to be involved in production of ECM material were not among the core genes that we identified in this search, genes whose products might modify existing surface components during phase variation were identified. Several putative lipoproteins, transferases, peptidases, and proteases may contribute to restructuring the envelope or redirecting protein processing. Increased expression of MXAN6704, a relative of GNAT-family acetyltransferase RimI in yellow variants, but not in tan variants, argues that modifications in the translation machinery contribute to phase variation. Similarly, expression of a subset of ribosomal proteins was increased in yellow variants; this increase was not due to growth rate because yellow variants had a slightly longer doubling time than tan variants. Instead, some ribosomal proteins might play specific roles in phase variation, an idea that has been suggested for Mycoplasma, where simple sequence repeats that contribute to phase variation have been found in genes for many ribosomal proteins (Rocha & Blanchard, 2002).
Significantly, we found increased expression of pkn14 and MXAN6911, which encode a serine-threonine kinase discovered by S. Inouye (Nariya & Inouye, 2005) and putative TonB-dependent siderophore receptor, respectively was specifically associated with the Swr(+) strains, the tan variant and the dkxG− Swr(+) mutant. While it is premature to speculate on the function of the TonB-receptor protein, the function of Pkn14 in growth and development provides valuable insight to the molecular advantage of being a tan variant. During vegetative growth, Pkn8 phosphorylates Pkn14, which in turn phosphorylates MrpC, the antitoxin portion of the MrpC-MazF toxin-antitoxin system (Nariya & Inouye, 2008). MrpC phosphorylation prevents the build-up of free MrpC, which also regulates expression of MazF. Pkn14 is not active in starving cells and as a consequence, MrpC appears to be degraded resulting in release of MazF. The endoribonuclease activity of MazF ultimately results in cell death (Nariya & Inouye, 2008). The link between tan variants, Pkn14, and Mrp begins to provide insight to the phenotype of asg mutants, one of which, asgA, is needed for expression of the mrp operon during development (Sun & Shi, 2001). The asg mutants are unable to produce an essential extracellular signal early in the development of fruiting bodies and spores (Kuspa et al., 1992). The asg mutants exhibit a dramatic tan phenotype during vegetative growth which suggests that these components, which include an essential DNA binding protein (Plamann et al., 1994), a response regulator (Plamann et al., 1995) and the sigma factor RpoD, may play a role in regulating at least some aspects of the complex phase variation pathway.
Our results suggest that the dynamic of the Pkn14 MrpC pathway is different in tan variants than in yellow variants. Elevated levels of Pkn14 in tan variants would be expected to protect MrpC and delay the onset of MazF-mediated autolysis. This protection likely persists during development when higher levels of Pkn14 would give tan variants the edge over yellow variants, whose lack of Pkn14 would launch the MazF programmed cell death response. Although we did not find evidence that Pkn8, the Pkn14 modifier, was expressed in tan variants, expression of MXAN4479, an alternative pSTK, was increased and may regulate the activity of Pkn14 in tan variants.
A long-range goal of our work is to elucidate the mechanism that regulates the phase variation switch. The discovery that expression of an adenine methyltransferase was increased in yellow cells is tantalizing since DNA methylation is known to generate epigenetic signals that regulate phase variation (Broadbent et al., 2010, Jakomin et al., 2008, Peterson & Reich, 2008, Zaleski et al., 2005). Phase may also be regulated or maintained by sensor proteins that respond to environmental signals. At least one signal transduction component was identified in this study as being up-regulated in yellow variants. MXAN6734 encodes a 74 kDa PAS/PAC sensor signal transduction histidine kinase predicted to reside in the cytoplasmic membrane. Since MXAN6734 contains a predicted heme-binding site, oxygen, light, or an unknown ligand may be critical for maintenance of yellow variants or may activate the master switch that initiates phase variation.
A key regulator of the phase variation process was identified during this study. As shown in Fig 7B, the putative HTH-Xre protein encoded by MXAN0228 (xre228) plays a significant role in the phase variation regulatory hierarchy because deletion of xre228 abolishes production of the pigment DKXanthene and reduces production of myxovirescin. Moreover, loss of Xre228 greatly reduces the frequency at which cells switch from swarm proficient to a swarm deficient state because we were unable to detect any Swr (+) derivatives of this mutant. Xre228 exhibited characteristics associated with other master or bistable switch proteins that regulate their own expression, directly or indirectly. If Xre228 behaves as a bistable switch, we propose that moderate levels of Xre228 would favor expression of DKxanthene and factors that facilitate swarming. However, xre expression is inhibited if the levels of Xre get too high, resulting in decreased expression of dkx and increased expression of pkn and tbdr which favor the tan Swr(+) phenotype. The identification of a key regulator of phase variation sets the stage for refining the roles that this complex process plays in Myxococcus swarming, survival, and development.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
The Myxococcus xanthus strains included wild-type strain DK1622 (Wall et al., 1999). M. xanthus was grown routinely at 32°C in CTPM liquid medium (1% Casitone, 10 mM Tris, 1 mM potassium phosphate, 5 mM MgSO4, final pH 7.6). CTPM was supplemented with kanamycin (Kan; 40 µg/ml) when applicable. Restriction endonucleases and DNA modifying enzymes were from New England Biolabs. The mariner transposon Himar (Lampe et al., 1996) was used to mutagenize the WT yellow variant and tan derivatives were isolated. Chromosomal DNA recovered from one of the stable tan mutants was digested with SacII, treated with ligase, and KanR plasmids named pGF88 were recovered in E. coli. A 1.7 kbp MluI - XbaI fragment from pGF88 was cloned into pSMART®LC Kan vector for sequencing. Sequence of the resulting plasmid, named pGF89 showed that the Himar transposon had inserted at bp 637 of the 8922 bp MXAN4299 gene, which encodes a non-ribosomal peptide synthase/polyketide synthase needed for production of the yellow pigment DKxanthene (Meiser et al., 2006).
Analysis of phase variants and mutants
A wild type strain (DK1622) that shows characteristic normal gliding motility, swarming, fruiting body morphogenesis, sporulation, and phase variation was used as the source for yellow and tan phase cells. The DKxanthene mutants, MxH2535 (dkgG Swr++) and MxH2576 (dkxG Swr(+)), are devoid of the yellow pigment and were assayed in parallel with the WT yellow and tan variants. The four strains were compared using assays for swarming, aggregation, dye-binding, UV sensitivity, fruiting body development and production of heat-resistant spores.
Growth assays were performed on three separate occasions to determine the doubling time, doubling time as a function of initial cell density, density-dependent swarming on hard and soft agar, and long-term survival of cells in liquid culture. 50 µl aliquots of cells were transferred from log phase cultures to fresh medium and cell density was monitored at 3–6 h intervals for 48 h, then every 12 h up for up to 16 days. To determine how Klett readings were correlated with viable cell numbers, aliquots were removed from broth cultures at 24 h intervals to monitor cell viability and screen visually for phase changes. Colony forming units (CFU) were determined by removing aliquots of cells at times indicated in figures and diluting in CTPM broth. After an initial 1:10 dilution (20 µl cell into 180 µl broth), six sequential 10-fold serial dilutions were made and aliquots of each dilution were plated on CTPM agar medium. After 4 days growth at 32°C, plates were scored for total number of yellow and tan colonies.
Phase switching values were determined by visual inspection of over 2000 colonies that arose from single cells. For each strain to be assayed, a single phase type colony (for example, yellow Swr++) was used to inoculate CTPM medium, grown to a cell density of 4× 108 cells ml−1, and then serially diluted in CTPM broth. Aliquots of several dilutions were plated on CTPM agar (± Kan where applicable) and incubated ay 32°C for 4–5 days. To compensate for potential differences in recovery of different cell types, aliquots of diluted samples were plated using two methods – one aliquot was spread on the agar using a glass rob and another aliquot was added to 50°C CTPM soft agar (0.7% agar) and poured on the agar. Colonies recovered from soft agar were transferred to fresh CTPM agar using toothpicks to determine the phenotype. Colony morphologies were evaluated by color (yellow vs tan) and swarm morphology (large matte-textured colonies = Swr++ and small, shiny surfaced colonies = Swr(+)). Although recovery of colonies was at least 10-fold lower on spread plates, we did not find a difference in the ratio of yellow vs tan or Swr++ vs Swr(+) morphologies between spread plates and those recovered from soft agar overlap plates.
UV sensitivity was quantified by exposing 10–20 µl aliquots of cells suspended in TPM buffer to a density of 5 × 108 cells ml−1 to 254 nm light (10,000 µJoules) for up to 4 min. with gentle mixing by pipette at 1 min intervals. At designated times, cells were removed and diluted 1:10 six times. 3 µl aliquots of each dilution were plated on CTPM medium in triplicate and incubated at 32°C. Growth was visible after 3–5 days; plates were examined for growth for 10 days. Catalase activity was determined by visual analysis of gas bubbles upon addition of 15 µl 0.06 M H2O2 to 50 µl of 5 × 109 M. xanthus cells (yellow and tan) washed with 50 mM potassium phosphate at pH 7.
The swarming rate of each mutant was compared with the WT strain on CTPM medium with 0.3% and 1.5% high grade agar as described elsewhere (Shi & Zusman, 1993). Cells were grown to a density of about 5×108 cells ml−1 in CTPM medium at 32°C, harvested by centrifugation and suspended in TPM medium to a density of about 5×109 cells ml−1 and spotted in 3 µl aliquots on 0.3% and 1.5% CTPM agar and incubated at 32°C or 18°C. The diameter was measured every 24 h for up to 5 days.
The rate of gliding of individual cells was measured by time-lapse videomicroscopy of cells on 1.5% CTPM agar or in a 50:50 mixture of 2x CTPM broth and 1% methylcellulose (fc 0.5% methylcellulose) at 20 sec intervals for 20 min as described elsewhere (Patryn et al., 2010). Samples were incubated at 32°C for 1 h prior to taking measurements. Metamorph tracking of at least 30 motile cells was used to generate velocity rates, but only reversing cells factored into cell reversal frequency by the Motility Macro v2.2 (Higgs & Merlie, 2008). Cells were considered to reverse if they progressed one cell length then paused and moved in a new direction at least 110 degrees from the original direction of motion.
Development of fruiting bodies and spores was initiated by centrifugation of 1 ml of 5×108 cells and suspension in 0.1 vol TPM buffer (CTPM medium lacking Casitone) as described (Kroos et al., 1986). Aliquots (20µl) were spotted on TPM agar and incubated at 32°C for 5 days. Developing samples were examined every 12 to 24 hrs under a stereomicroscope to monitor the progress of rippling, mound formation, and darkening of the fruiting body. On days 2 and 5, a sample was removed and examined by diffraction interference contrast microscopy to determine if rod-shaped M. xanthus cells had differentiated to spheres and to monitor the appearance of refractile spores. On day 5, plates containing developing cultures were incubated at 50°C for 2 hs, scraped into TPM buffer, and sonicated briefly to disperse the clumps of cells within the fruiting body. Samples were serially diluted in TPM and then spotted on CTPM agar to stimulate germination.
The protocol of Jacoksen (Jakobsen et al., 2004) was used to monitor development in submerged culture. Briefly, 1 ml of 5×108 cells was harvested by centrifugation and the cell pellet was resuspended in 0.1 vol MC7 buffer. Aliquots (25 µl) were mixed with 375 µl of MC7 buffer and transferred to 24-wells microtiter plates and incubated at 32°C for 3 and 5 days. The suspensions were sonicated and incubated for 2 h in a 50°C waterbath and dilutions of the suspensions were plated on CTPM agar plates to quantify germination of spores.
Methods to compare relative levels of exopolysaccharide (EPS) using Congo red and trypan blue have been published (Lancero et al., 2004) and were modified slightly for this study. Briefly, mid-log phase CTPM broth-grown M. xanthus cells, were harvested by centrifugation and the cell pellet was suspended to 5 × 108 cells ml−1 in TPM-CR buffer (10 mM Tris, 1 mM potassium phosphate, 5 mM MgSO4, 15 µg ml−1 Congo red (or 10 µg ml−1 trypan blue), final pH 7.6). Cell-free samples containing TPM+dye buffer were used as controls. All samples were vortexed briefly and incubated at 25°C in the dark. After 30 min, the cells were removed by centrifugation and 900 µl of each supernatant was transferred to a cuvette. The absorbance of each sample was measured at 490 nm for Congo red (585 nm for trypan blue) and compared to the absorbance of the cell-free control. Each test sample and control sample was analyzed in triplicate.
Cell aggregation was assayed by harvesting 3 ml of mid-log phase cells by centrifugation at 10,000 × g for 5 min, washed once in 10 mM MOPS (morpholinepropanesulfonic acid) buffer at pH 6.8, and suspended by gentle pipetting in aggregation buffer (10 mM MOPS pH 6.8 1 mM MgCl2, 1mM CaCl2) to ≈ 5 ×108 cells/ml. The cells suspensions were incubated without shaking at 32 °C, and the OC600 was measured at 10-intervals for at least 2 h.
The amount of DKX was estimated by spectral analysis of 5 ×108 log-phase cell harvested by centrifugation and suspended in 1ml of TPM. DKX was measured at 400 nm using a Spectonic 21D spectrophotometer (Milton Roy).
Microarray analysis
Published protocols that have been demonstrated to work with M. xanthus and related bacteria (Kadam et al., 2008, Pham et al., 2006, Jakobsen et al., 2004, Call et al., 2003, Britton et al., 2002) were used to generate microarray data. Printed Corning slides were generated at the Research Technology Support facility at Michigan State University and contained single prints of 70-mer single stranded DNA oligomers representing 7335 of the predicted annotated 7457 M. xanthus genes. Two independent microarrays experiments were performed. In each case, triplicate slides were probed with cDNA from RNA harvested from yellow and tan derivatives of two WT (DK1622) stocks – one that had recently been removed from the freezer and one that came from a different freezer stock of the WT strain and had been in use for at least one month. To adjust for the possibility of variation due to changing ratios of yellow and tan variants in our WT populations, the qRT-PCR experiments were performed on separate occasions from independent isolates. An aliquot of the sample used to prepare mRNA was plated right before harvest to ensure that the cells were still predominantly yellow (or tan). Superscript III First strand was used to synthesize cDNA, which was labeled with Alexafluor 555 carboxlic acid succinimidyl ester (Invitrogen, LaJolla, CA). Samples with adjusted p values of <0.05 from the two independent array runs were considered statistically significant. All analyses were done in the R programming language (Team, 2010), raw intensity data were read in and checked for quality using the Limma package (Smyth, 2005). The data were then background corrected using background subtraction and the arrays were normalized with quantile normalization (Bolstad et al., 2003). A simple linear model was fit to the data and variances were corrected using an empirical Bayes approach (Smyth, 2004) from the bioconductor limma package. Results were corrected for multiple testing using the false discovery rate procedure of Benjamini and Hochberg (BH) and considered significant if their BH adjusted p-values were < 0.05 (Benjamini & Hochberg, 1995).
Real-time PCR
WT yellow and tan variants were grown to a density of 5 × 108 cells ml−1 in CTPM medium at 32°C. A 1 ml aliquot was harvested by centrifugation at 13,700 × g and the cell pellet was suspended in 0.5 ml PBS buffer. Total mRNA was extracted using RNAprotect Bacterial reagent (with DNase) and RNeasy minikit according to manufacturers instruction (Qiagen, Valencia CA). 500 ng total RNA was used to produce cDNA using a Hexanucleotide Mix (Roche, Indianapolis, IN) as primer for the reaction using the Superscriptase II kit (Invitrogen). The resulting cDNA was diluted 1:25 or 1:1250 for probing target genes and 16s rRNA templates respectively. 5 µl of diluted cDNA sample was added to 20 µl of a PCR mixture prepared from Power SYBR® Green PCR Master Mix (Applied Biosystems, Carlsbad, CA), which contained each primer at a concentration of 160 nM. Primers, listed in Table 2, were chosen using Primer Express 3.0 software (Applied Biosystems) and were designed to amplify a region of about 150–200 bp within each transcript.
Supplementary Material
Table 3.
Strains and molecular reagents
|
M. xanthus strains |
Genotype | Phenotype | Construction | Reference |
|---|---|---|---|---|
| DK1622 | wild type | A+S+ KanS | (Wall & Kaiser, 1999) | |
| MxH2535 | DK1622 MXAN4299::miniHimar-GFP | KanR tan Swr++ | mini-Himar-GFP transposon into DK1622 | This study |
| MxH2546 | MXAN0228::pHS2 | KanR tan | pHS2 into DK1622 | This study |
| MxH2576 | DK1622 MXAN4299::miniHimar-GFP | KanR tan Swr(+) | mini-Himar-GFP transposon into DK1622 | This study |
| Plasmid | Construction | Reference | ||
| pCR-Blunt II-TOPO | cloning vector KanR | Invitrogen | ||
| pSMART® LCKan | cloning vector KanR | Lucigen | ||
| Pmini-Himar-GFP | Mini-Himar transposon mutagenesis vector; generates transcriptional gfp fusions | Gift from Heidi Kaplan | ||
| pGF88 | Ligation of 5.0 kbp of SacII fragment from MxH2535 chromosomal DNA | This Study | ||
| pGF89 | 1.7 kbp MluI XbaI fragment from pGF88 into pSMART® LCKan | This Study | ||
| pHS2 | 337 bp PCR product of MXAN0228 D-For by D-Rev cloned into pCR-Blunt II-TOPO | This Study | ||
| Oligonucleotides for PCR | ||||
| Gene | D-For | D-Rev | ||
| MXAN0288 | 5’ GCATGGACAAGAAACTCGCAACCA | 5’ AACAGCTTCACCTTCGCGGG | ||
| Oligonucleotides for RT-PCR | ||||
| Gene | RT-PCR Forward | RT-PCR Reverse | ||
| MXAN0185 | 5' GGCTTGGGTTTCGTCACATC | 5' GCGGACAACAGCAAAGGAA | ||
| MXAN0228 | 5' AGAAACTCGCAACCACAATCG | 5' CTCGGTGGCCACATCGA | ||
| MXAN0504 | 5' AAGGCTGGCAGGGAAGGTA | 5' GCACGCGTAGTGGAAGAACTG | ||
| MXAN0682 | 5' AAGGCTGGCGCGTGAAG | 5' CGGGTCGGTCTGCTCGTA | ||
| MXAN1200 | 5' CCGAGCCAGTTAAGGCATAGC | 5' CCGCACAAGCTCTCCGTACT | ||
| MXAN1289 | 5' CGACGGCGTTCTGAATGACT | 5' GCATGTTCCAGCGGTTGAA | ||
| MXAN1525 | 5' ACCATTGGGCGGGATCA | 5' GTTGGTGGAGCCCAGGTCTT | ||
| MXAN1926 | 5' ACATGACCATCGTCGGCA | 5' CCAGCTCCAGGACTGTCAGTC | ||
| MXAN2675 | 5' ACGCACGCGACCTGGAT | 5' TCCATCAGGTGGTCATCAATGA | ||
| MXAN2921 | 5' GATCGTTTCATCGCCAACAGT | 5' CTCGCGGCCCCAGAA | ||
| MXAN2967 | 5' GTCGCAGAAGCGGCAGAT | 5' CGTTACCCTCGAAGTCGTAGATG | ||
| MXAN2991 | 5' CCGCACGTTCCAGTCCAT | 5' CGGGCGTGCCCATCA | ||
| MXAN3084 | 5' CGACGCCATCAAGCTCTATGA | 5' TTTCCCTGGGCGATGAAG | ||
| MXAN3299 | 5' ACGCGCATCAACAAGTTCAC | 5' CTGCTCACGGCTCTTCTTGTC | ||
| MXAN3306 | 5' AAGCGTGGCGGCAAGA | 5' CTTCACCACCGCGACGTAGT | ||
| MXAN3450 | 5' GACTACCTGGTGAAGCCCTTTG | 5' GCCGGGCATCGATGTG | ||
| MXAN3574 | 5' ACGGTCCAGACTCCTACGGG | 5' AAGCCGTTGGATGTTAGCCA | ||
| MXAN3943 | 5' TCCCTCCCTGGACGTGTTC | 5' CGGTCCTGAATCTCCTGGTAGA | ||
| MXAN3949 | 5’ TGTTGAGCGCCAGCTTCTC | 5’ GGTACCCGAACTCGAATCACA | ||
| MXAN4084 | 5’ GAGCGACAGCTCTCCGAAGA | 5’ GGCCGAGGCCGTCATT | ||
| MXAN4294 | 5' CCGGTTGGGCGAGTACCT | 5' GGTGGACGGGCAATCCA | ||
| MXAN4295 | 5' GGCTGCATGTCGTCGTACCT | 5' GGCCTCCAGCATCATCTTCA | ||
| MXAN4299 | 5' GCTTCGCGTCTCACTCCAA | 5' TGGTAGACAATCTGCAGGTCGAT | ||
| MXAN4305 | 5’ GACCTGGCGTACATCCTCTACAC | 5’ CATGGCACCACTCGATGAAG | ||
| MXAN4371 | 5' TGGTGGAGGAAGTGCAACTG | 5' CCCCGGTGAATCTTGAAGTAGT | ||
| MXAN4389 | 5' GGCATCCCGGCGAACTA | 5' CGGCGGTGAGGCTCTTC | ||
| MXAN4479 | 5' GGCCGGTCCGATATCTTCTC | 5' GGCACCTCCTCCGTCTTCA | ||
| MXAN4862 | 5' TGATGAGCCCGGAAATGC | 5' CATCCAGACGAGTGGCAAGAC | ||
| MXAN5138 | 5' CGGCTTCGCGATGCA | 5' CTTGAACCAGGTGACGGTGTT | ||
| MXAN5445 | 5' ATCTTCAGCAACGGGTGCTT | 5' GCAGGTCCCGATCTTGATGA | ||
| MXAN5478 | 5' CGGCCTGTACACCGTCAAGT | 5' GATTTTGTCCGGGTCCTTGAG | ||
| MXAN5646 | 5' GTTCTTGTCGAGCCTCTTCCA | 5' GCTGCCCGGCAATCC | ||
| MXAN5703 | 5' GAGAAGATCCAGGCCAACGA | 5' TGACGAGCCCCATCTCCAT | ||
| MXAN5783 | 5' CACCGCGCAGAAGTCGTT | 5' CGTTGGCGAAGTCGGAGTA | ||
| MXAN5974 | 5' CGGCTTCGCCTTCACCTT | 5' CACTGGGCCTCGTCATAACC | ||
| MXAN6046 | 5' CTACCTCATGGCTCGCAACTT | 5' TTCATCCCATCCATGTGGAA | ||
| MXAN6298 | 5' GCAACGAGCTGTGGAGCAA | 5' GTCGAACGAGCGGTTGAAG | ||
| MXAN6704 | 5' GCCCTGACGCAGCTCAAG | 5' CCGACAAGCTGGCTTTGC | ||
| MXAN6911 | 5' GACGGCAGCGTCGTCAA | 5' GCCGTACTCGGCGGTAATC | ||
| MXAN7181 | 5' GGTACGCACCTTGAAATTCGA | 5' GGAGGGCCCAGTGGAAGT | ||
| MXAN7441 | 5' TCCGTGACGTCTACAAGCTGAT | 5' ATGGCGGACGCGTTCTC | ||
Abbreviations: Kan, kanamycin. A, adventurous gliding motility; S, social gliding motility; tan, tan variant; yellow, yellow variant; Swr, swarming; GFP, green fluorescent protein; for, forward; rev, reverse. M. xanthus genes are given the designation MXANxxxx [http://cmr.jcvi.org/tigr-scripts/CMR/GenomePage.cgi?org=gmx].
Acknowledgements
The authors thank Dr. S. Minnich for critical reading of the manuscript and Jeff Landgraf (Michigan State University) for printing of DNA microarray slides. This research is based in part upon work supported by grants MCB1052525 from the NSF to PLH and Cooperative Agreement DBI-0939454 from the NSF. GF and HS were supported by grant P20RR016454 (NIH-INBRE).
Abbreviations
- DKX
DKxanthene
References
- Accessexcellence. Catalase lab website. 2010 http://www.accessexcellence.org/AE/ATG/data/released/0074-GenNelson/index.php.
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- Berleman JE, Kirby JR. Multicellular development in Myxococcus xanthus is stimulated by predator-prey interactions. J Bacteriol. 2007;189:5675–5682. doi: 10.1128/JB.00544-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleman JE, Scott J, Chumley T, Kirby JR. Predataxis behavior in Myxococcus xanthus. Proc Natl Acad Sci U S A. 2008;105:17127–17132. doi: 10.1073/pnas.0804387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black WP, Xu Q, Yang Z. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol Microbiol. 2006;61:447–456. doi: 10.1111/j.1365-2958.2006.05230.x. [DOI] [PubMed] [Google Scholar]
- Black WP, Yang Z. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J. Bacteriol. 2004;186:1001–1008. doi: 10.1128/JB.186.4.1001-1008.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, Grossman AD. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J Bacteriol. 2002;184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent SE, Davies MR, van der Woude MW. Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation-dependent mechanism. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchard RP, Burchard AC, Parish JH. Pigmentation phenotype instability in Myxococcus xanthus. Can J Microbiol. 1977;23:1657–1662. doi: 10.1139/m77-238. [DOI] [PubMed] [Google Scholar]
- Burchard RP, Dworkin M. Light-induced lysis and carotinogenesis in Myxococcus xanthus. J. Bacteriol. 1966;91:535–545. doi: 10.1128/jb.91.2.535-545.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call DR, Bakko MK, Krug MJ, Roberts MC. Identifying antimicrobial resistance genes with DNA microarrays. Antimicrob Agents Chemother. 2003;47:3290–3295. doi: 10.1128/AAC.47.10.3290-3295.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvio C, Celandroni F, Ghelardi E, Amati G, Salvetti S, Ceciliani F, Galizzi A, Senesi S. Swarming differentiation and swimming motility in Bacillus subtilis are controlled by swrA, a newly identified dicistronic operon. J Bacteriol. 2005;187:5356–5366. doi: 10.1128/JB.187.15.5356-5366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvio C, Osera C, Amati G, Galizzi A. Autoregulation of swrAA and motility in Bacillus subtilis. J Bacteriol. 2008;190:5720–5728. doi: 10.1128/JB.00455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Chu F, Kolter R, Losick R. Bistability and biofilm formation in Bacillus subtilis. Molecular Microbiology. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana JR, Shimkets LJ. Regulation of Cohesion-Dependent Cell Interactions in Myxococcus xanthus. J. Bacteriol. 1993;175:3636–3647. doi: 10.1128/jb.175.11.3636-3647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- Furstner A, Bonnekessel M, Blank JT, Radkowski K, Seidel G, Lacombe F, Gabor B, Mynott R. Total synthesis of myxovirescin A1. Chemistry. 2007;13:8762–8783. doi: 10.1002/chem.200700926. [DOI] [PubMed] [Google Scholar]
- Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, et al. Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci U S A. 2006;103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris BZ, Kaiser D, Singer M. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 1998;12:1022–1035. doi: 10.1101/gad.12.7.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs PI, Merlie JP., Jr . Myxococcus xanthus: Cultivation, Motility and Development. In: Whitworth D, editor. Myxobacteria: Multicellularity and Differentiation. Washington DC: ASM Press; 2008. pp. 465–478. [Google Scholar]
- Jakobsen JS, Jelsbak L, Jelsbak L, Welch RD, Cummings C, Goldman B, Stark E, Slater S, Kaiser D. Sigma54 enhancer binding proteins and Myxococcus xanthus fruiting body development. J Bacteriol. 2004;186:4361–4368. doi: 10.1128/JB.186.13.4361-4368.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakomin M, Chessa D, Baumler AJ, Casadesus J. Regulation of the Salmonella enterica std fimbrial operon by DNA adenine methylation, SeqA, and HdfR. J Bacteriol. 2008;190:7406–7413. doi: 10.1128/JB.01136-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam SV, Wegener-Feldbrugge S, Sogaard-Andersen L, Velicer GJ. Novel transcriptome patterns accompany evolutionary restoration of defective social development in the bacterium Myxococcus xanthus. Mol Biol Evol. 2008;25:1274–1281. doi: 10.1093/molbev/msn076. [DOI] [PubMed] [Google Scholar]
- Kaiser D. Coupling cell movement to multicellular development in myxobacteria. Nat Rev Microbiol. 2003;1:45–54. doi: 10.1038/nrmicro733. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Rudner R, Losick R. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol Microbiol. 2004;52:357–369. doi: 10.1111/j.1365-2958.2004.03996.x. [DOI] [PubMed] [Google Scholar]
- Kim SK, Kaiser D. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell. 1990;6:19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- Kroos L, Kuspa A, Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- Kuspa A, Plamann L, Kaiser D. Identification of heat-stable A-factor from Myxococcus xanthus. J. Bacteriol. 1992;174:3319–3326. doi: 10.1128/jb.174.10.3319-3326.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe DJ, Churchill ME, Robertson HM. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- Lancero H, Caberoy NB, Castaneda S, Li Y, Lu A, Dutton D, et al. Characterization of a Myxococcus xanthus mutant that is defective for adventurous motility and social motility. Microbiology. 2004;150:4085–4093. doi: 10.1099/mic.0.27381-0. [DOI] [PubMed] [Google Scholar]
- Laue BE, Gill RE. Use of a phase variation-specific promoter of Myxococcus xanthus in a strategy for isolation a phase-locked mutant. J. Bacteriol. 1994;176:5341–5349. doi: 10.1128/jb.176.17.5341-5349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue BE, Gill RE. Using a phase-locked mutant of Myxococcus xanthus to study the role of phase variation in development. J. Bacteriol. 1995;177:4089–4096. doi: 10.1128/jb.177.14.4089-4096.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauriello EM, Mignot T, Yang Z, Zusman DR. Gliding motility revisited: how do the myxobacteria move without flagella? Microbiol Mol Biol Rev. 2010;74:229–249. doi: 10.1128/MMBR.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiser P, Bode HB, Muller R. The unique DKxanthene secondary metabolite family from the myxobacterium Myxococcus xanthus is required for developmental sporulation. Proc Natl Acad Sci U S A. 2006;103:19128–19133. doi: 10.1073/pnas.0606039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiser P, Muller R. Two functionally redundant Sfp-type 4'-phosphopantetheinyl transferases differentially activate biosynthetic pathways in Myxococcus xanthus. Chembiochem. 2008;9:1549–1553. doi: 10.1002/cbic.200800077. [DOI] [PubMed] [Google Scholar]
- Meiser P, Weissman KJ, Bode HB, Krug D, Dickschat JS, Sandmann A, Muller R. DKxanthene biosynthesis--understanding the basis for diversity-oriented synthesis in myxobacterial secondary metabolism. Chem Biol. 2008;15:771–781. doi: 10.1016/j.chembiol.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- Nariya H, Inouye S. Identification of a protein Ser/Thr kinase cascade that regulates essential transcriptional activators in Myxococcus xanthus development. Mol Microbiol. 2005;58:367–379. doi: 10.1111/j.1365-2958.2005.04826.x. [DOI] [PubMed] [Google Scholar]
- Nigaud Y, Cosette P, Collet A, Song PC, Vaudry D, Vaudry H, Junter GA, Jouenne T. Biofilm-induced modifications in the proteome of Pseudomonas aeruginosa planktonic cells. Biochim Biophys Acta. 2010;1804:957–966. doi: 10.1016/j.bbapap.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Paitan Y, Orr E, Ron EZ, Rosenberg E. Cloning and characterization of a Myxococcus xanthus cytochrome P-450 hydroxylase required for biosynthesis of the polyketide antibiotic TA. Gene. 1999a;228:147–153. doi: 10.1016/s0378-1119(98)00609-x. [DOI] [PubMed] [Google Scholar]
- Paitan Y, Orr E, Ron EZ, Rosenberg E. Genetic and functional analysis of genes required for the post-modification of the polyketide antibiotic TA of Myxococcus xanthus. Microbiology. 1999b;145(Pt 11):3059–3067. doi: 10.1099/00221287-145-11-3059. [DOI] [PubMed] [Google Scholar]
- Patryn J, Allen K, Dziewanowska K, Otto R, Hartzell PL. Localization of MglA, an essential gliding motility protein in Myxococcus xanthus. Cytoskeleton. 2010 doi: 10.1002/cm.20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SN, Reich NO. Competitive Lrp and Dam assembly at the pap regulatory region: implications for mechanisms of epigenetic regulation. J Mol Biol. 2008;383:92–105. doi: 10.1016/j.jmb.2008.07.086. [DOI] [PubMed] [Google Scholar]
- Pham VD, Shebelut CW, Jose IR, Hodgson DA, Whitworth DE, Singer M. The response regulator PhoP4 is required for late developmental events in Myxococcus xanthus. Microbiology. 2006;152:1609–1620. doi: 10.1099/mic.0.28820-0. [DOI] [PubMed] [Google Scholar]
- Plamann L, Davis JM, Cantwell B, Mayor J. Evidence that asgB encodes a DNA-binding protein essential for growth and development of Myxococcus xanthus. J. Bacteriol. 1994;176:2013–2020. doi: 10.1128/jb.176.7.2013-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann L, Li Y, Cantwell B, Mayor J. The Myxococcus xanthus asgA gene encodes a novel signal transduction protein required for multicellular development. J. Bacteriol. 1995;177:2014–2020. doi: 10.1128/jb.177.8.2014-2020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP, Blanchard A. Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res. 2002;30:2031–2042. doi: 10.1093/nar/30.9.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolbetzki A, Ammon M, Jakovljevic V, Konovalova A, Sogaard-Andersen L. Regulated secretion of a protease activates intercellular signaling during fruiting body formation in M. xanthus. Dev Cell. 2008;15:627–634. doi: 10.1016/j.devcel.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Keller KH, Dworkin M. Cell density-dependent growth of Myxococcus xanthus on casein. J Bacteriol. 1977;129:770–777. doi: 10.1128/jb.129.2.770-777.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science. 2009;323:396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Zusman DR. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc. Natl. Acad. Sci. USA. 1993;90:3378–3382. doi: 10.1073/pnas.90.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets LJ. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Ann. Rev. Microbiol. 1999;53:525–549. doi: 10.1146/annurev.micro.53.1.525. [DOI] [PubMed] [Google Scholar]
- Simunovic V, Gherardini FC, Shimkets LJ. Membrane localization of motility, signaling, and polyketide synthetase proteins in Myxococcus xanthus. J Bacteriol. 2003;185:5066–5075. doi: 10.1128/JB.185.17.5066-5075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Sogaard-Andersen L. Cell polarity, intercellular signalling and morphogenetic cell movements in Myxococcus xanthus. Curr Opin Microbiol. 2004;7:587–593. doi: 10.1016/j.mib.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Sogaard-Andersen L, Overgaard M, Lobedanz S, Ellehauge E, Jelsbak L, Rasmussen AA. Coupling gene expression and multicellular morphogenesis during fruiting body formation in Myxococcus xanthus. Mol Microbiol. 2003;48:1–8. doi: 10.1046/j.1365-2958.2003.03399.x. [DOI] [PubMed] [Google Scholar]
- Staskawicz BJ, Panopoulos NJ, Hoogenraad NJ. Phaseolotoxin-insensitive ornithine carbamoyltransferase of Pseudomonas syringae pv. phaseolicola: basis for immunity to phaseolotoxin. J Bacteriol. 1980;142:720–723. doi: 10.1128/jb.142.2.720-723.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Shi W. Analyses of mrp genes during Myxococcus xanthus development. Journal of Bacteriology. 2001;183:6733–6739. doi: 10.1128/JB.183.23.6733-6739.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RDC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- Tolchinsky S, Fuchs N, Varon M, Rosenberg E. Use of Tn5lac to study expression of genes required for production of the antibiotic TA. Antimicrob Agents Chemother. 1992;36:2322–2327. doi: 10.1128/aac.36.10.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude MW. Re-examining the role and random nature of phase variation. FEMS Microbiol Lett. 2006;254:190–197. doi: 10.1111/j.1574-6968.2005.00038.x. [DOI] [PubMed] [Google Scholar]
- van der Woude MW, Baumler AJ. Phase and antigenic variation in bacteria. Clin Microbiol Rev. 2004;17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon M, Fuchs N, Monosov M, Tolchinsky S, Rosenberg E. Mutation and mapping of genes involved in production of the antibiotic TA in Myxococcus xanthus. Antimicrob Agents Chemother. 1992;36:2316–2321. doi: 10.1128/aac.36.10.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D, Kaiser D. Type IV pili and cell motility. Mol. Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- Wall D, Kolenbrander PE, Kaiser D. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for Type IV pilus biogenesis, social motility and development. J. Bacteriol. 1999;181:24–33. doi: 10.1128/jb.181.1.24-33.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MJ, Mok KC, Astling DP, Lew H, Zusman DR. An ABC Transporter Plays a Developmental Aggregation Role in Myxococcus xanthus. J. Bacteriol. 1998;180:5697–5703. doi: 10.1128/jb.180.21.5697-5703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Lux R, Hu W, Hu C, Shi W. PilA localization affects extracellular polysaccharide production and fruiting body formation in Myxococcus xanthus. Mol Microbiol. 2010;76:1500–1513. doi: 10.1111/j.1365-2958.2010.07180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youderian P, Burke N, White DJ, Hartzell PL. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol Microbiol. 2003;49:555–570. doi: 10.1046/j.1365-2958.2003.03582.x. [DOI] [PubMed] [Google Scholar]
- Zafriri D, Rosenberg E, Mirelman D. Mode of action of Myxococcus xanthus antibiotic TA. Antimicrob Agents Chemother. 1981;19:349–351. doi: 10.1128/aac.19.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaleski P, Wojciechowski M, Piekarowicz A. The role of Dam methylation in phase variation of Haemophilus influenzae genes involved in defence against phage infection. Microbiology. 2005;151:3361–3369. doi: 10.1099/mic.0.28184-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.