Abstract
Exine, the outer plant pollen wall, has elaborate species-specific patterns, provides a protective barrier for male gametophytes, and serves as a mediator of strong and species-specific pollen-stigma adhesion. Exine is made of sporopollenin, a material remarkable for its strength, elasticity, and chemical durability. The chemical nature of sporopollenin, as well as the developmental mechanisms that govern its assembly into diverse patterns in different species, are poorly understood. Here, we describe a simple yet effective genetic screen in Arabidopsis (Arabidopsis thaliana) that was undertaken to advance our understanding of sporopollenin synthesis and exine assembly. This screen led to the recovery of mutants with a variety of defects in exine structure, including multiple mutants with novel phenotypes. Fifty-six mutants were selected for further characterization and are reported here. In 14 cases, we have mapped defects to specific genes, including four with previously demonstrated or suggested roles in exine development (MALE STERILITY2, CYP703A2, ANTHER-SPECIFIC PROTEIN6, TETRAKETIDE α-PYRONE REDUCTASE/DIHYDROFLAVONOL-4-REDUCTASE-LIKE1), and a number of genes that have not been implicated in exine production prior to this screen (among them, fatty acid ω-hydroxylase CYP704B1, putative glycosyl transferases At1g27600 and At1g33430, 4-coumarate-coenzyme A ligase 4CL3, polygalacturonase QUARTET3, novel gene At5g58100, and nucleotide-sugar transporter At5g65000). Our study illustrates that morphological screens of pollen can be extremely fruitful in identifying previously unknown exine genes and lays the foundation for biochemical, developmental, and evolutionary studies of exine production.
Plants have evolved remarkable and unique pollen walls. The pollen surface consists of inner (intine) and outer (exine) walls, coated with a lipid- and protein-rich extracellular matrix (the pollen coat; Fig. 1A). Exine provides several important functions to pollen grains: (1) it protects gametophytes in harsh environments and serves as a barrier against various physical and chemical factors and biological pathogens (Scott, 1994); (2) it helps with pollen dispersal and cross-pollination in some species (Faegri and van der Pijl, 1979); and (3) it enables stigma cells to capture the most appropriate pollen grains by providing species-specific factors for pollen-stigma recognition and adhesion (Zinkl et al., 1999).
Figure 1.
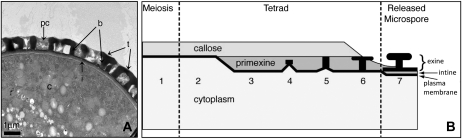
Plants have complex pollen walls. A, Transmission electron micrograph of a cross-section of Arabidopsis pollen. b, Baculae of exine; c, pollen grain cytoplasm; i, intine; pc, pollen coat; t, tectum of exine. B, Stages of exine assembly during microspore development (adapted from Scott [1994] and Paxson-Sowders et al. [1997]). Stage 1, pattern gene transcription and callose deposition; stage 2, patterning specifications; stage 3, primexine synthesis; stage 4, probaculae development; stage 5, protosporopollenin deposition; stage 6, callose removal; stage 7, sporopollenin strengthening and intine synthesis initiation.
There is an astonishing variety of exine patterns across different plant species, yet the overall pollen wall architecture is conserved among angiosperms. Often, the outer part of exine is elaborate, with radially directed rods (baculae) with expanded ends that often fuse to form patterned walls (muri) and a roof (tectum; Fig. 1A). Most pollen walls contain characteristic apertures or pores, thought to allow pollen tube emergence and changes in grain volume (Wodehouse, 1935; Heslop-Harrison, 1979a; Muller, 1979; Blackmore and Barnes, 1986; Katifori et al., 2010); these regions are often covered only by intine. The uniform appearance of apertures within a species indicates the presence of mechanisms that specify sites where exine is not deposited or is reduced. In fact, in many species, the aperture number and positions are genetically determined and tightly controlled: for example, lily (Lilium longiflorum) pollen has a single aperture that always develops at the tetrad perimeter (Heslop-Harrison, 1968a), and Arabidopsis (Arabidopsis thaliana) grains, like those of many other eudicots, contain three apertures arranged in a radially symmetrical fashion.
Exine is made of sporopollenin, a tough and chemically inert biopolymer. The chemical resistance of sporopollenin makes it a formidable challenge to deduce its structure. While its precise chemical structure remains unclear, chemical analysis suggested that sporopollenin is made of phenolics, fatty acids, and alkanes (Guilford et al., 1988; Ahlers et al., 1999, 2000; Wiermann et al., 2001). The synthesis and patterning of exine are generated through the combined activities of factors in both developing microspores that will become pollen grains (or in microspore mother cells) and in tapetum, a diploid secretory tissue that lines the anthers and surrounds the microspores. Tapetum is responsible for the synthesis of most of the sporopollenin materials (Piffanelli et al., 1998; Scott et al., 2004).
The precursor of exine (primexine) is synthesized just after meiosis II and is deposited onto the microspore plasma membrane, under a layer of callose (β-1,3 glucan) that temporarily separates microspores (Owen and Makaroff, 1995; Paxson-Sowders et al., 1997; for stages of exine assembly, see Fig. 1B). When callose wall deposition around the microspore mother cell and developing microspores is prevented either by premature activation of a β-1,3 glucanase that degrades callose (Worrall et al., 1992) or by mutations in a callose synthase (Dong et al., 2005; Nishikawa et al., 2005), the resulting pollen grains have severe defects in exine sculpting.
Primexine is made primarily of polysaccharides and acts as a loose scaffold for the attachment of sporopollenin monomers, resulting in partially polymerized protosporopollenin (Heslop-Harrison, 1968b). The genes that determine species-specific primexine patterns are likely transcribed within the diploid nucleus of a premeiotic microspore mother cell, and the pattern information is then inherited by microspores. Exine patterns are specified early in pollen development, with the accumulation of sporopollenin monomers over sites where baculae will appear (Heslop-Harrison, 1971; Sheldon and Dickinson, 1983). Ultimately, the number of baculae and the extent of their fusion determine exine patterns; for example, if the connections are minimal, the pattern may be reticulate, whereas additional connections can result in a smooth surface. Centrifugation experiments have shown that exine patterning can be perturbed during the early meiotic prophase (Sheldon and Dickinson, 1983) and that the patterning also requires coordination between the pollen plasma membrane and its underlying cytoskeleton, vesicles, and endoplasmic reticulum (ER; Dickinson and Sheldon, 1986; Takahashi and Skvarla, 1991; Fitzgerald and Knox, 1995; Perez-Munoz et al., 1995; Paxson-Sowders et al., 2001).
The study of pollen wall development is currently gaining momentum with the identification of a growing number of genes important for exine production and sporopollenin synthesis (Aarts et al., 1997; Paxson-Sowders et al., 2001; Ariizumi et al., 2003, 2004; Dong et al., 2005; Nishikawa et al., 2005; Morant et al., 2007; Guan et al., 2008; Suzuki et al., 2008; de Azevedo Souza et al., 2009; Dobritsa et al., 2009a, 2009b, 2010; Tang et al., 2009; Grienenberger et al., 2010; Kim et al., 2010). Among the phenotypes of the corresponding Arabidopsis mutants are absence of exine, abnormally low sporopollenin level, not fully polymerized sporopollenin, and changes in exine patterns. Several of the genes (MALE STERILITY2 [MS2], cytochromes P450 CYP703A2 and CYP704B1, ACYL-COENZYME A SYNTHETHASE5 [ACOS5], FACELESS POLLEN1 [FLP1], NO EXINE FORMATION1 [NEF1], LESS ADHESIVE POLLEN/POLYKETIDE SYNTHASE [LAP5/PKSB and LAP6/PKSA], and TETRAKETIDE α-PYRONE REDUCTASE [TKPR1/DRL1 and TKPR2/CCRL6]) participate in fatty acid modification or accumulation (Aarts et al., 1997; Ariizumi et al., 2003, 2004; Morant et al., 2007; de Azevedo Souza et al., 2009; Doan et al., 2009; Dobritsa et al., 2009b; Grienenberger et al., 2010; Kim et al., 2010), highlighting the role of this pathway in exine synthesis. The DEFECTIVE EXINE1 (DEX1) gene was suggested to contribute to exine assembly and encodes a predicted membrane-associated protein with similarity to animal integrins. DEX1 appears to interact with the plasma membrane to nucleate sporopollenin deposition (Paxson-Sowders et al., 2001), and in dex1 mutants, primexine deposition is significantly reduced, sporopollenin is randomly deposited on microspores, and the microspores are ultimately degraded. The RUPTURED POLLEN GRAIN1 (RPG1) gene encodes a seven-transmembrane-domain protein that localizes to the plasma membrane of microsporocytes (Guan et al., 2008). Mutations in this gene affect plasma membrane structure and primexine deposition and lead to irregular exine structure and microspore collapse (Guan et al., 2008).
Recently, the in silico approach was found to be a productive way for identifying new genes involved in sporopollenin synthesis or transport (Grienenberger et al., 2010; Kim et al., 2010; Quilichini et al., 2010): several of the genes implicated in this process have a very high coexpression correlation and are expressed together only in anthers at a particular stage of microspore development (Morant et al., 2007; Dobritsa et al., 2009b, 2010; Grienenberger et al., 2010; Kim et al., 2010; Quilichini et al., 2010).
Yet, despite the progress in the identification of several exine genes, it is clear that many other yet unknown players are involved in the complex process of exine production and its assembly into elaborate patterns. Some of the known exine genes do not have the distinct expression pattern associated with sporopollenin synthesis genes. Therefore, the traditional genetic approaches remain invaluable for the identification of members of the genetic network required for the biosynthesis, transport, and assembly of exine components. However, because genetic screens required visualization of minute exine details on tiny pollen grains and were expected to be very labor intensive, such screens have not been widely used in the past. Previously, a genetic screen has been performed using scanning electron microscopy to search for mutants (kaonashi [kns] mutants) with defective exine patterning (Suzuki et al., 2008). Twelve mutants that form at least five complementation groups were identified in this screen. One of them (kns2) affects exine pattern by increasing baculae density and has defects in a gene encoding a Suc phosphate synthase, the enzyme that catalyzes the rate-limiting step in Suc biosynthesis (Suzuki et al., 2008). It remains to be determined which genes are affected in the other kns mutants.
Here, we describe a large-scale genetic screen in Arabidopsis that aimed to identify genes involved in exine development using a novel, and simpler, screening approach. We screened for anthers and pollen that at low magnification (dissection microscope level) showed visible defects. We screened approximately 16,000 lines with random T-DNA insertions and used reverse genetics on a collection of mutations in 49 exine candidate genes. A secondary screen at a higher resolution (confocal microscope level) was then applied to the promising mutants to identify the ones with true exine defects. We isolated multiple mutants with abnormal exines that exhibited a large variety of phenotypes. These ranged from a complete lack of exine to a loss of the net-like structure, changes in the thickness of exine wall, irregular exine patterning, abnormal number and position of apertures, changes in the sizes of lacunae on the exine surface to exine that appeared morphologically normal yet demonstrated an altered autofluorescence spectrum, suggesting a change in its composition. We describe 49 mutants (some of which were found to be allelic) isolated through the forward genetic screen and seven mutants isolated through the reverse genetic screen. Through both approaches, we identified 14 genes, 10 of which to our knowledge have not been previously implicated in exine production.
RESULTS
Forward Genetic Screen
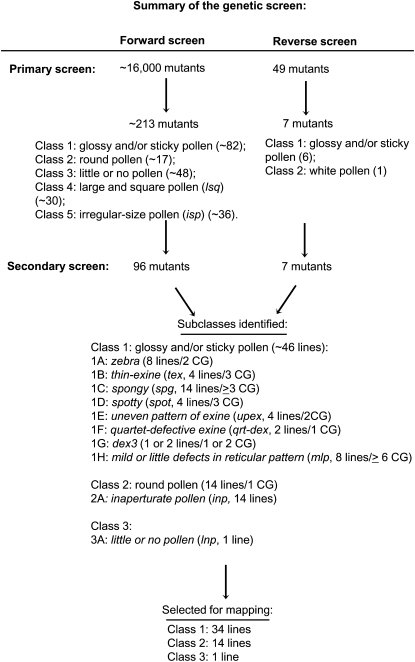
While characterizing and mapping several mutations that cause defects in exine structure (Nishikawa et al., 2005; Dobritsa et al., 2009a, 2010), we noticed that exine abnormalities, which can only be visualized using high-resolution microscopy, often also showed defects in anther and pollen morphology that can be easily detected using low-magnification standard dissecting stereomicroscopes. Therefore, we used low-magnification microscopy to perform a genetic screen for mutants with various morphological abnormalities in pollen and anthers. We used 179 pools of 100 lines from the Arabidopsis SALK T-DNA collection (Alonso et al., 2003) and screened approximately 16,000 different lines. The phenotypes that were selected in the primary screen included (1) abnormal anthers (e.g. brown, shriveled, glossy, or with little visible pollen), (2) pollen that exhibited unusually strong adherence to anthers and was not as easily shed as pollen of wild-type plants, (3) pollen that was released as aggregates and clumps and not as individual pollen grains, (4) pollen that had unusual size and/or shape (e.g. bigger or smaller than the wild type, round versus normally oblong), color (e.g. white or brown versus wild-type yellow), or light reflection (glossy or translucent versus wild-type matte), and (5) fertility defects that were combined with visible pollen or anther defects. As a result of this broadly defined primary screen, we identified 495 mutant candidates. Often, however, we found multiple mutants with identical or very similar phenotypes within a group of plants derived from a single set of pooled SALK lines, indicating that many of the mutant candidates from the same pool were likely siblings. Usually, only one or two of the lines with identical phenotypes derived from the same pool were subjected to further analysis. Therefore, we estimated that we had identified approximately 213 different mutant lines.
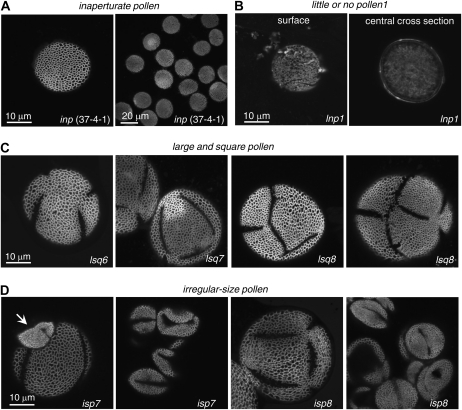
Based on the most obvious morphological defects visible at low magnification, we placed mutants into five primary-screen classes: (1) glossy and/or sticky pollen that is not easily released from the anthers (approximately 82 lines; Supplemental Fig. S1); (2) round pollen (approximately 17 lines); (3) little or no pollen produced (this phenotype was usually associated with sterility or reduction in fertility; approximately 48 lines); (4) large and square pollen (approximately 30 lines); and (5) irregular-size pollen produced by the same anther (approximately 36 lines; Fig. 2).
Figure 2.
Summary of the genetic screen. CG, Number of genetic complementation groups formed by mutants in a phenotypic subclass.
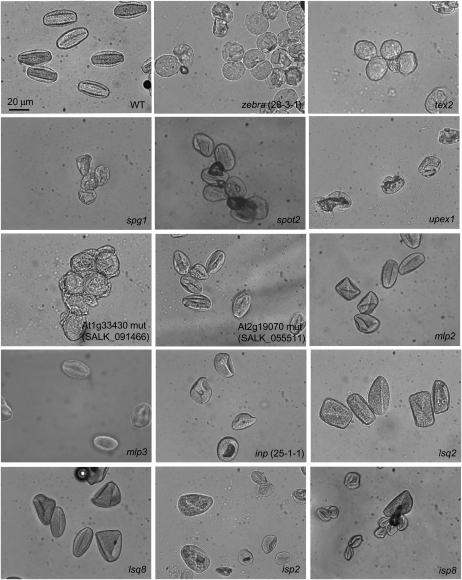
Initially, the screen also included assessment of the morphology of unhydrated pollen at higher magnification. This revealed that shapes of the mutant pollen grains were often significantly distorted (Fig. 3). It also indicated that mutants grouped together in the same primary-screen class (e.g. glossy and/or sticky pollen) sometimes had noticeably different pollen phenotypes when observed with a higher magnification (Fig. 3).
Figure 3.
Examples of shape and size distortion in pollen grains observed in the mutants identified in the primary screen. Unhydrated pollen grains were visualized with Zeiss Axioskop. Magnification is the same for all images. WT, Wild type. The mutants shown were assigned to the following primary-screen classes based on their dissecting microscope pollen phenotypes: (1) glossy and/or sticky pollen (zebra [28-3-1], tex2, spg1, spot2, upex1, SALK_091466 [At1g33430], SALK_055511 [At2g19070], mlp2, and mlp3); (2) round pollen (inp [25-1-1]); (3) large and square pollen (lsq2 and lsq8); (4) irregular-size pollen (isp2 and isp8). Note that more details of pollen morphology are visible in the images obtained with this method than with the dissecting microscopes used for the primary screen. These images, therefore, do not serve to illustrate phenotypic differences used for mutant placement into five classes during the primary screen.
Reverse Genetic Screen
Prior to the start of our screen, the chemical analysis of sporopollenin (Guilford et al., 1988; Ahlers et al., 1999, 2000; Wiermann et al., 2001) and a small set of available exine mutants (Aarts et al., 1997; Ariizumi et al., 2003; Dong et al., 2005; Nishikawa et al., 2005; Morant et al., 2007) provided evidence that biosynthetic pathways that produce fatty acids, phenylpropanoids, and some other small molecules are important for exine synthesis. Therefore, we also screened mutants disrupted in 49 candidate genes using the same visual approach as described above (Fig. 2). We targeted genes encoding enzymes in the two pathways with the strong chemical and/or genetic evidence for a role in exine formation (fatty acid and phenylpropanoid), genes encoding enzymes from the pathways with controversial roles in exine synthesis (flavonoid and carotenoid), as well as a number of genes that are predominantly expressed in developing Arabidopsis anthers or are homologs of anther-specific genes from other plants. The mutants came primarily from the SALK and SAIL (Sessions et al., 2002) T-DNA collections (Table I; Supplemental Tables S1 and S2). Our analysis was limited by the availability of mutations in regions where they were likely to affect gene functions.
Table I.
Genes identified in the reverse genetics screen that, when mutated, affect pollen and anther morphology
| Gene | Primary-Screen Mutant Phenotype | Exine Mutant Phenotypea | Annotation/Function | Putative Pathway | Expression Patternb | Insertion Lines Tested |
| At1g33430 | Glossy/sticky pollen | upex (allelic to upex1 and upex2)c | Glycosyl transferase | Buds (stages 9–12), not in ap3 and ag | SALK_091466 | |
| At1g65060 | Glossy/sticky pollen | mlp5 | 4-Coumarate-CoA ligase (4CL3) | Phenolics | Buds (stages 9–10), petals | SALK_017894, WiscDsLox503E07 |
| At1g69500 | Glossy/sticky pollen | zebra (allelic to lines 28-3-1, 37-2-3, 119-1-1, 131-3-1, 155-4-1, 174-154-2)c | CYP704B1, fatty acid ω-hydroxylase | Fatty acid | Buds (stages 9–10), not in ap3 and ag | SAIL_1149_B03 |
| At2g19070 | Glossy/sticky pollen | mlp6 | Spermidine hydroxycinnamoyl transferase | Phenolics (spermidine conjugates) | Buds (stages 9–10), not in ap3 and ag | SALK_055511 |
| At3g28780 | Glossy/sticky pollen | mlp7 | Gly-rich protein | Stamen | GT_5_86366 | |
| At4g14080 | Glossy/sticky pollen | mlp (allelic to mlp4)c | β-1,3-Glucanase/glycosyl hydrolase (A6/MEE48) | Callose degradation | Buds (stages 9–10), not in ap3 and ag | WiscDSLox285F04 |
| At5g13930 | White pollen | mlp8 | Chalcone synthase TT4 | Phenolics (flavonoids) | Multiple tissues and stages | SALK_020583 |
Phenotypic categories developed in the secondary screen with confocal microscopy. For descriptions of the different categories, see “Results.”
Based on expression data from the microarray (e.g. GENEVESTIGATOR [Zimmerman et al., 2004] and/or Massively Parallel Signature Sequencing [Meyers et al., 2004]) databases.
Indicates additional alleles that were found during the forward genetic screen.
We found seven genes in which mutations affected pollen or anther morphology (At1g33430, At1g65060, A1g69500, At2g19070, At3g28780, At4g14080, and At5g13930; Table I). For 19 genes, no morphological changes in the anthers or pollen were observed, despite the presence of homozygous insertions in their open reading frames or 5′ untranslated regions (Supplemental Table S1). For the rest of the genes (Supplemental Table S2), no obvious morphological changes were found. However, the results were inconclusive, usually because the insertion locations were in noncoding regions.
Of the genes that affected pollen or anther morphology, six of the seven mutants produced pollen that looked glossy and/or adhered more tightly to anthers (glossy and/or sticky pollen). These six genes have predicted anther-specific expression or are expressed in the buds at the stages of exine development (Table I). The seventh, an insertion in At5g13930/TT4, a gene for chalcone synthase (CHS) that plays a key role in the flavonoid biosynthesis and is not anther specific, did not have the glossy and/or sticky pollen phenotype but resulted in the absence of pollen pigments and pollen that was white, unlike the yellow-colored wild-type pollen and similar to the pollen from chs mutants in maize (Zea mays) and petunia (Petunia hybrida; Coe et al., 1981; Mo et al., 1992; Napoli et al., 1999).
Secondary Screen: Mutant Phenotypes and Gene Isolation
We used auramine O staining and laser scanning confocal microscopy (LSCM) to examine exine in the identified mutants. Exine structure was examined in representatives of different mutant classes that produced mature pollen. While we briefly describe here phenotypes of several primary-screen mutant classes, our further study concentrated primarily on the members of class 1 (glossy and/or sticky pollen) and class 2 (round pollen), because they exhibited phenotypes more directly related to exine formation. As a result of the secondary screen, 34 mutants from the glossy and/or sticky pollen category, 14 mutants from the round pollen category (inaperturate [inp]; see below), and one mutant from the little or no pollen category were selected for mapping (Fig. 2).
We used a number of techniques to identify genes responsible for the abnormal exine phenotypes. Because the T-DNA mutagenesis collection was used for the screen, we utilized two different PCR-based methods (McElver et al., 2001; Alonso et al., 2003) to identify positions of the T-DNA insertions. Results of the T-DNA insertion mapping for the lines in which insertions were identified are summarized in Supplemental Table S3. In several cases, the identified T-DNA insertions were responsible for the exine defects. However, in many instances, they did not cosegregate with the observed phenotypes (Dobritsa et al., 2009b; Supplemental Table S3; see below), and for a few lines, we could not identify the positions of the T-DNA insertions. Thus, on a case-by-case basis, we used additional approaches, such as linking the defects to molecular markers through bulked segregant analysis (Michelmore et al., 1991), checking the pollen phenotypes of the sequence-indexed T-DNA insertion lines available for the candidate genes, placing the mutations into complementation groups, comparing the phenotypes of the mutants found in the forward genetic screen with those found through reverse genetics or with previously known exine mutants, followed by PCR amplification and sequencing of these genes in the mutant lines. Table II summarizes the results of the mapping. Below, we describe the secondary-screen exine phenotypes of the mutants and isolation of the affected genes.
Table II.
Mapping of lesions in the mutants with defective exine isolated through the forward genetic screen
| Secondary-Screen Phenotypic Category | Lines Forming a Complementation Group | T-DNA Insertion Site (if Relevant) | Genetic Mappinga | Gene Affected/Candidate Gene |
| zebra | SAIL_1149_B03, 28-3-1, 37-2-3, 119-1-1, 131-3-1, 155-4-1, 174-154-2 | SAIL_1149_B03: At1g69500; other lines, various sites | 28-3-1 and 37-2-3: bottom of chromosome 1 (markers NGA111 and NGA692) | At1g69500b-g (CYP704B1), anther-specific fatty acid ω-hydroxylase |
| zebra | 135-2-3 | Top of chromosome 3 (markers NGA162 and GAPAB) | At3g11980b-f (MS2), anther-specific fatty acyl reductase | |
| thin-exine | tex1 | Top of chromosome 1 (marker F21M12) | At1g01280c-f (CYP703A2), anther-specific fatty acid in-chain hydroxylase | |
| thin-exine | tex2 | Bottom of chromosome 5 (markers CIW10 and MBK5) | At5g65000c-g, NST | |
| thin-exine | tex3, tex4 | tex4: At4g35420 | tex3: NGA1107 marker on chromosome 4; tex4: CIW7 marker on chromosome 4 | At4g35420ce (TKPR1/DRL1), anther-specific tetraketide α-pyrone reductase |
| spongy | spg1 | Bottom of chromosome 1 (markers NGA111 and NGA692) | ||
| spongy | spg2 | At1g27600 | Top of chromosome 1 (marker ZFPG) | At1g27600ce, putative glycosyl transferase |
| spongy | spg3, spg4, spg5, spg6, spg7, spg8, spg9 | spg3, spg4, spg7: chromosome 4 (CIW7 marker) | ||
| spotty | spot1 | At5g58100 | At5g58100cf, novel protein | |
| spotty | spot2 | Top of chromosome 1 (marker F21M12) and bottom of chromosome 5 (markers CIW10 and MBK5) | ||
| spotty | spot3, spot4 | Bottom of chromosome 1 (markers NGA111 and NGA692) | ||
| uneven pattern of exine | upex1, upex2, SALK_091466 | At1g33430 | At1g33430cf, putative glycosyl transferase, anther specific | |
| uneven pattern of exine | upex3 | Chromosome 3 (marker GAPAB) | ||
| quartet-defective exine | qrt-dex1, qrt-dex2 | At4g20050 | At4g20050cf (QRT3), polygalacturonase | |
| defective exine3 | dex3 | Chromosome 4 (marker CIW7) | ||
| mild or little defects in reticulate pattern | mlp1 | Chromosome 3 (markers GAPAB, F1P2-TGF, and NGA112) | ||
| mild or little defects in reticulate pattern | mlp2 | At2g30710 | At2g30710c, RabGAP/TBC domain protein | |
| mild or little defects in reticulate pattern | mlp3 | At2g30760 | At2g30760c, novel protein | |
| mild or little defects in reticulate pattern | mlp4 | At4g14080 | At4g14080cf (A6/MEE48), putative β-1,3-glucanase/glycosyl hydrolase, anther specific | |
| inaperturate pollen | 25-1-1, 31-1-2, 37-4-1, 41-4-1, 42-4-1, 47-2-1, 52-1-1, 60-4-2, 77-93-1, 83-122-1, 105-1-4, 114/116-3-3, 117-3-7, 118-2-2 | 25-1-1, 31-1-2, 37-4-1, 52-1-1: chromosome 4 (marker CIW7) | ||
| little or no pollen | lnp1 | Top of chromosome 3 (marker NGA162) |
Linkage to the indicated markers was established through bulked segregant analysis.
For details, see Dobritsa et al. (2009b).
Contains a mutation in the gene.
Does not complement other mutant alleles of this gene.
Bulked segregant analysis demonstrates linkage of the phenotype to the chromosomal region where this gene is located.
Other lines with insertions in this gene have identical phenotypes.
The mutant phenotype is rescued by a full-size genomic construct.
Class 1: Glossy and/or Sticky Pollen
This primary-screen class included the largest number of mutants with different abnormalities in the exine structure. Forty-six mutant lines, many of them with dramatic changes in their exines, are described below. These mutants were placed in eight subclasses (secondary-screen categories) based on their exine phenotypes. The number of lines indicated in parentheses below refers to the number of isolates with a particular phenotype that were recovered from different sets of pooled seeds in the forward genetics screen and/or in the reverse genetics screen. Also in parentheses, we indicate the number of established complementation groups (CG) formed by mutants from a given subclass, which suggests the number of affected exine loci.
1A: zebra (Eight Lines/Two CG)
These mutants lacked exine, which was replaced by a thin layer of a material that did not stain well with the exine-specific dye auramine O, and contained rare bits of auramine O-positive aggregates irregularly distributed on the grain surface. We recently described these mutants in detail elsewhere (Dobritsa et al., 2009b). The zebra mutants fell into two complementation groups that corresponded to the fatty acid hydroxylase CYP704B1 and the fatty acyl reductase MS2 (Dobritsa et al., 2009b).
1B: thin-exine (tex; Four Lines/Three CG)
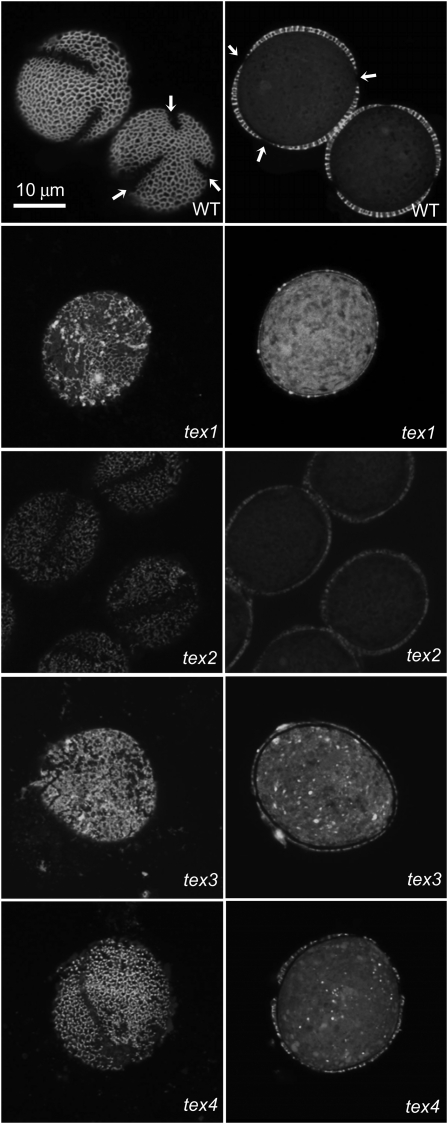
In these lines, exine was very thin (approximately 0.2–0.4 μm versus 1 μm for the wild-type pollen; Fig. 4). In tex1, the reticulate pattern of exine was retained, although large areas of the surface had bare patches where exine was absent. However, in tex2 to tex4, the regular reticulate pattern was no longer present (Fig. 4). Pairwise crosses placed tex mutants into three complementation groups composed of tex1, tex2, and tex3/tex4, respectively. Because bulked segregant analysis linked tex1 to the top of chromosome 1 (Table II) near the exine gene CYP703A2 (Morant et al., 2007), we sequenced this gene in tex1 (referred to as line 40-4-1 by Dobritsa et al. (2009b). tex1 had a single nucleotide change (G to C) in the last exon of CYP703A2 that caused a Gly-405→Arg conversion. The tex1 thin-exine phenotype was very similar to that of the lap4-1 allele (Dobritsa et al., 2009b) of CYP703A2. A cross between tex1 and lap4-1 demonstrated that they do not complement each other: all 24 F1 progeny had the glossy and sticky pollen phenotype. Thus, tex1 is an allele of CYP703A2.
Figure 4.
Confocal images of mutant pollen with thin exine. Grains were stained with auramine O. Optical sections through the surface (left) and center (right) of pollen grains demonstrate differences in exine patterns and thickness between the wild type (WT) and four mutants. Magnification is the same for all images. Arrows point to three apertures in the wild-type pollen grains.
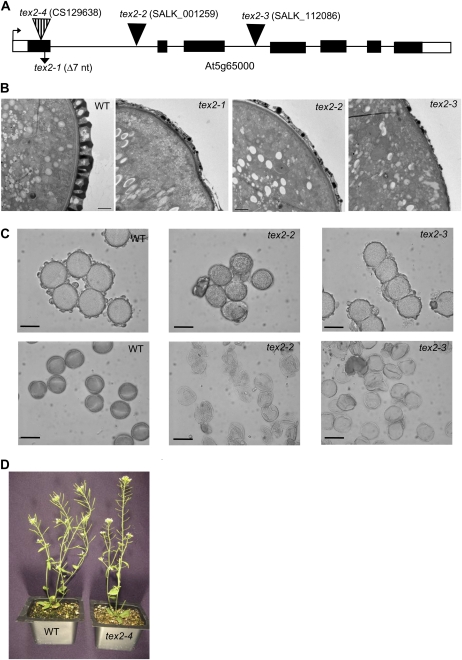
The tex2 mutation was linked to the bottom of chromosome 5 (Table II). Additional fine-mapping allowed us to map the mutation to an 82-kb region between 25.926 and 26.008 Mb on chromosome 5. Sequencing of the genes in this interval identified a seven-nucleotide deletion early in the coding region of At5g65000, which encodes a putative nucleotide-sugar transporter (NST). This mutation was termed tex2-1 and was predicted to lead to protein truncation after the first 29 amino acids. Three insertion lines available for this gene (SALK_001259, SALK_1120886, and CS129638) did not complement the tex2-1 defects and were referred to as tex2-2, tex2-3, and dex2-4, respectively. tex2-2 and tex2-3 had T-DNA insertions in introns 1 and 3, respectively, and tex2-4 had a transposon inserted in exon 1 at the beginning of the open reading frame (Fig. 5A). All three lines exhibited pollen defects. The first two lines, similar to tex2-1, produced fertile plants with glossy and sticky acetolysis-sensitive pollen (Fig. 5C) and the thin-exine phenotype identical to that of tex2-1 pollen (Fig. 5B). The third line, tex2-4, resulted in sterile plants with no pollen produced (Fig. 5D). To confirm the identity of the TEX2 gene, a complementation construct containing the entire At5g65000 gene under the native promoter was introduced into tex2-1 and tex2-4/+ plants. Wild-type anther and pollen phenotypes were restored in all BASTA-resistant homozygous tex2-1 (20 lines) and tex2-4 (seven lines) T1 progeny, confirming At5g65000 as the TEX2 gene and indicating that the tex2-4 pollen phenotype and sterility were due to the defects in TEX2. Both tex2-1 and tex2-4 mutations are located close to each other in the beginning of the coding sequence and are expected to generate null alleles. The reason for the differences in pollen viability and mutant phenotypes between these two alleles is currently unknown. Possibly, the different ecotype backgrounds (Columbia [Col-0] and Landsberg erecta [Ler], respectively) affect the phenotypes of tex2-1 and tex2-4.
Figure 5.
Pollen defects in the tex2 alleles. A, The At5g65000 gene region. One of the two transcripts supported by the existence of ESTs is shown. Exons are shown as rectangles, with black boxes representing coding regions. Positions of mutations in tex2-1 to tex2-4 alleles are shown. The second predicted transcript differs from this one after exon 5 and, therefore, should also be affected in all the mutant alleles. nt, Nucleotides. B, Transmission electron micrographs of pollen grain sections from the wild type (WT), tex2-1, tex2-2, and tex2-3. Exine is abnormally thin in the tex2 mutants. Bars = 1 μm. C, tex2 pollen grains are sensitive to acetolysis. Top panels, pollen grains of the wild type, tex2-2, and tex2-3 mock treated with 50 mm Tris-HCl, pH 7.5, have similar morphology. Bottom panels, wild-type pollen grains treated with the acetolysis mixture remain intact, whereas tex2-2 and tex2-3 pollen grains are lysed after acetolysis. Bars = 20 μm. D, tex2-4 plants are sterile and have short siliques compared with the wild type. [See online article for color version of this figure.]
Sections of the developing tex2-4 anthers revealed normal pollen development until stage 9 of anther development (Sanders et al., 1999), when abnormalities were noticed in microspores exhibiting cytoplasm retraction from cell walls (Supplemental Fig. S2). By late stage 11, the tex2-4 pollen completely degenerated, with cell debris left in the anther locules (Supplemental Fig. S2, E and F). The cytoplasmic shrinkage, such as the one observed in tex2-4 microspores, is one of the hallmarks of programmed cell death (Buckner et al., 2000; Häcker, 2000) and has previously been described for some mutants with defects in microsporogenesis (Polowick and Sawhney, 1995; Yang et al., 2003; Coimbra et al., 2009), including the ms1 mutant, in which pollen wall formation is abnormal (Vizcay-Barrena and Wilson, 2006). It could also be caused by plasmolysis (e.g. due to changing osmotic conditions in the locular cavity and compromised cell walls). Interestingly, exine deposition around the developing tex2-4 microspores was visible in stage 9 anthers (Supplemental Fig. S2C). This was in contrast to the lap5 lap6 and acos5 exine mutants, which also demonstrated microspore defects beginning at the same stage and resulted in sterile plants with no mature pollen, but they showed no exine production around the developing microspores (de Azevedo Souza et al., 2009; Dobritsa et al., 2010).
The tex4 mutant, another member of the thin-exine class, harbored a T-DNA insertion in the last exon of At4g35420, a recently identified tetraketide α-pyrone reductase (TKPR1/DRL1; Grienenberger et al., 2010). The importance of the anther-specific TKPR1/DRL1 for exine production was recently demonstrated (Tang et al., 2009; Grienenberger et al., 2010). The noncomplementing tex3 mutation mapped to the bottom of chromosome 4 in the vicinity of TKPR1/DRL1. However, sequencing of TKPR1/DRL1 from the tex3 mutant revealed no nucleotide changes in its coding or noncoding regions, including introns and the 240 bp upstream and 140 bp downstream of the gene. It remains to be seen if tex3 affects the function of TKPR1/DRL1.
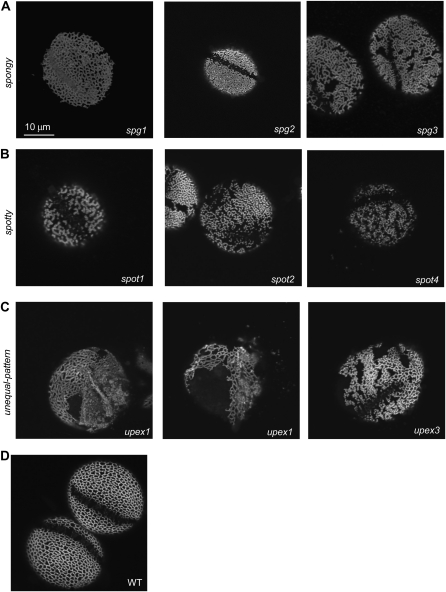
1C: spongy (spg; 14 Lines/Three or More CG)
In these mutants, the regular reticulate pattern of exine was not maintained, and on many occasions, instead of a regular net-like structure, exine had irregularly distributed elongated tectum elements and lacunae of variable sizes and shapes, resembling pollen surfaces from the species with rugulate or perforate exine patterns (Fig. 6A). One of the lines (spg2) also demonstrated reduced ability to hydrate normally, as many pollen grains in this line retained oval shape upon placement into the aqueous environment (Fig. 6A).
Figure 6.
Confocal images of mutant exine surfaces from several subclasses of glossy and/or sticky pollen. A, Examples of mutants with spongy exine. spg2 has problems with pollen hydration. B, Examples of mutants with spotty exine. C, Lines upex1 and upex3 have pollen grains with only portions of exine maintaining reticulate structure. Note the easy shedding of exine in upex1. D, The wild type (WT). Grains were stained with auramine O, and magnification is the same for all images.
Complementation crosses performed on nine of the spg lines placed them into three complementation groups: spg1, spg2, and spg3 to spg9, respectively. spg2 harbored a T-DNA insertion in the first intron of At1g27600, a putative glycosyl transferase. Consistent with this, genetic mapping also localized spg2 to the general vicinity of At1g27600 (Table II). In addition, SALK_037323, a line with an annotated T-DNA insertion at an almost identical position, had the identical pollen exine phenotype. spg1 was linked to the markers NGA111 and NGA692 at the bottom of chromosome 1. In the case of the third spg complementation group, all three alleles used for mapping (spg3/spg4/spg7) had linkage to the CIW7 marker on chromosome 4.
1D: spotty (spot; Four Lines/Three CG)
In these lines, exine elements appeared to be largely disconnected, indicating possible problems with tectum formation (Fig. 6B).
For spot1, both PCR-based methods found an insertion in At5g58100, encoding a novel protein. Three independent sequence-indexed lines (SALK_061320, SALK_041228, and SALK_079847) with T-DNA inserted into different regions of this gene generated plants exhibiting the same pollen exine phenotype, strongly suggesting that mutations in At5g58100 were responsible for the abnormally developed exine.
The rest of the spot mutants formed two complementation groups: spot2 and spot3/spot4. For spot3/spot4, the defect mapped to the bottom of chromosome 1 (Table II). For spot2, surprisingly, linkage to two areas, the top of chromosome 1 (markers F21M12 and ZFPG) and the bottom of chromosome 5 (markers CIW10 and MBK5), was found by bulked segregant analysis. Mapping with additional markers in these regions using 57 individual F2 mutant progeny confirmed this result. We observed a segregation ratio of 208 wild type to 57 mutants for the spot2 phenotype in F2, consistent with simple Mendelian 3:1 phenotypic segregation (χ2 = 1.64, 0.5 > P > 0.1) and with the hypothesis that a single recessive locus was affected. Thus, it remains to be seen what kind of a genomic rearrangement caused this phenotype and this inheritance pattern.
1E: uneven pattern of exine (upex; Four Lines/Two CG)
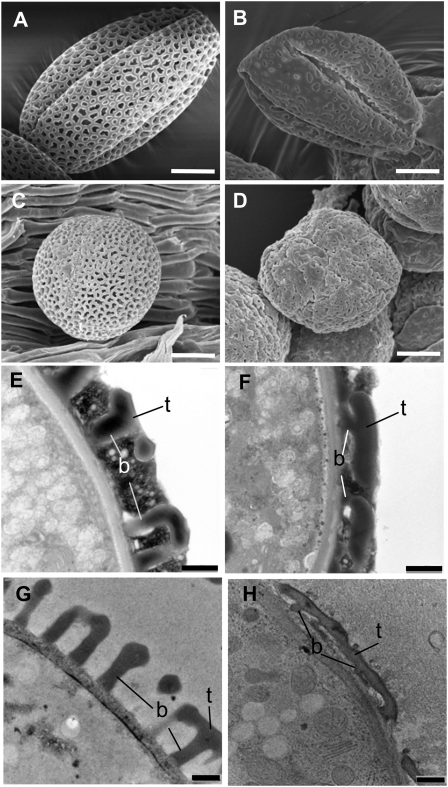
Three lines (upex1–upex3) identified through a forward genetics approach had a grossly disorganized exine pattern, with only portions of the pollen surface areas maintaining reticulate structure. upex1 (Fig. 6C) and upex2 (data not shown) had very similar types of defects: whereas some surface regions retained reticulate exine, other areas lacked exine or had exine that appeared practically smooth and had small lacunae. In addition, their exine dissociated easily from the pollen surface (Fig. 6C, middle), leaving some grains in the microscopy samples exineless. Another line (SALK_091466) with an identical phenotype was discovered via reverse genetics and had a mutation in At1g33430, a putative glycosyl transferase (Table I). Additional imaging of upex1 with scanning electron microscopy confirmed the abnormal patterning of exine and the presence of lacunae that were fewer in number and smaller in size than in the wild type (Fig. 7, A–D). Transmission electron microscopy (TEM) of upex1 pollen demonstrated that its exine had very short baculae and overdeveloped tectum (Fig. 7, E–H). In upex3, portions of the surface that were lacking the reticulate exine pattern appeared spotty, giving an impression that the normal tectum connections were missing between baculae in these areas (Fig. 6C).
Figure 7.
Abnormalities in exine patterning of upex1 pollen. A to D, Scanning electron micrographs of the surface structure of pollen grains from the wild type (A and C) and the upex1 mutant (B and D). Samples were prepared without fixation (A and B) or with fixation (C and D). E to H, Transmission electron micrographs of pollen grain sections from the wild type (E and G) and upex1 (F and H). Note short baculae (b) and overdeveloped tectum (t) of the mutant exine. The TEM images are of the mature pollen (E and F) and of the developing microspores from stage 11 buds (G and H). Bars = 5 μm (A–D) and 500 nm (E–H).
The upex subclass was composed of three noncomplementing mutants (upex1/upex2/SALK_091466) and of upex3, which complemented the other mutations. For upex3, linkage to the GAPAB marker on chromosome 3 was established. The noncomplementing upex1/upex2 lines, as well as SALK_091466, had T-DNA insertions in At1g33430, a putative glycosyl transferase. An unrelated T-DNA insertion in At1g33430 (SAIL_544_C02) had an identical pollen phenotype. According to the public expression databases, such as GENEVESTIGATOR (Zimmermann et al., 2004) and Massively Parallel Signal Sequencing (Meyers et al., 2004), At1g33430 has anther-specific expression in stage 9 to 10 buds (at the time of exine development) and is not expressed in the stamen-lacking ap3 and ag mutants. We confirmed this expression pattern with reverse transcription-PCR and by expressing GUS under the control of the At1g33430 promoter (Supplemental Fig. S3).
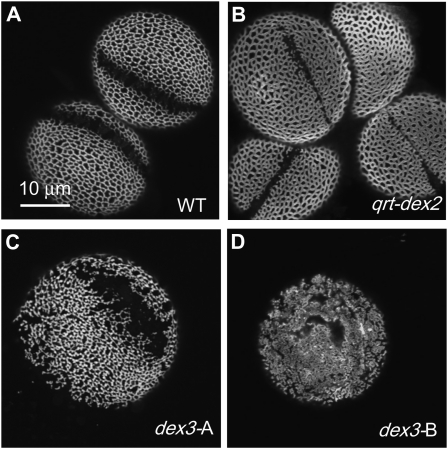
1F: quartet-defective exine (qrt-dex; Two Lines/One CG)
Two lines had the glossy and sticky pollen phenotype, which cosegregated with the qrt phenotype manifested as defects in microspore separation and the resulting release of four products of meiosis as tetrads and not monads of pollen. Pollen exine from these two lines had broader muri and smaller lacunae than the wild type (Fig. 8B; data not shown). Similar exine patterning defects and glossy and sticky pollen phenotypes were previously observed in the mutant alleles of QRT3 (S. Nishikawa, personal communication). However, these types of exine defects are not generally associated with the qrt phenotype, as mutations in the other two QRT genes, QRT1 and QRT2, do not cause additional anther- or pollen-associated phenotypes (A.A Dobritsa, unpublished data).
Figure 8.
Examples of mutants with qrt and dex3 phenotypes. A, The wild type (WT). B, qrt-dex2 has a qrt phenotype as well as larger exine surface coverage, with broader muri and smaller lacunae. C and D, Two exine phenotypes were observed in the progeny of dex3. C, Phenotype of the original isolate (dex3-A). D, Novel phenotype segregated in the next generations (dex3-B). Grains were stained with auramine O, and magnification is the same for all images.
In qrt-dex1, a T-DNA insertion was located to the QRT3 gene. A cross between qrt-dex1 and qrt-dex2 did not rescue the mutant phenotypes, indicating that the same gene is likely affected in both lines. QRT3 was previously identified as a polygalacturonase and suggested to degrade the pectic polysaccharides in the primary cell wall of the microspore mother cell (Rhee et al., 2003). Exine phenotypes of qrt-dex1 and qrt-dex2 indicate that, in addition to degradation of the primary cell well, polygalacturonase activity is important for proper exine patterning.
1G: defective exine3 (dex3; One or Two Lines)
The original isolate (dex3-A) had an abnormal but still largely reticulate exine pattern with irregular aperture margins (Fig. 8C). In the next generation, however, the progeny of this self-fertilized plant segregated phenotypes: whereas some progeny had the parental exine phenotype and normal fertility, the rest of the plants displayed reduced fertility and produced few pollen grains. In the latter case, the pollen grains were deformed and had a novel exine phenotype with smaller lacunae and larger surface coverage (dex3-B; Fig. 8D).
1H: mild or little defects in reticulate pattern (mlp; Eight Lines/Six or More CG)
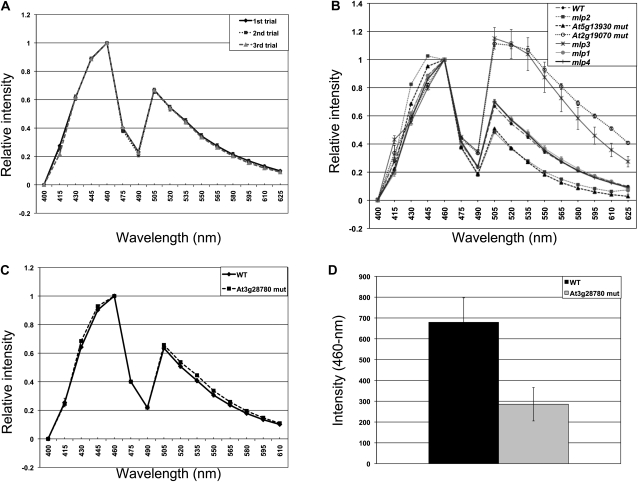
Eight mutants had obvious glossy and/or sticky pollen phenotypes when assessed using dissecting microscopes but had few, if any, changes in their exine morphology when visualized using LSCM (Supplemental Fig. S4). In mlp1, exine retained the regular reticulate structure but had a finer mesh pattern than the wild-type exine. mlp2 and mlp3 had an abnormally shaped glossy pollen (Fig. 3), but their exine had an overall preserved reticulate pattern (Supplemental Fig. S4). Similarly, regular reticulate structure was also present in the glossy pollen of mlp4 (Supplemental Fig. S4). In addition, four lines identified through reverse genetics (mlp5–mlp8, corresponding to At1g65060, At2g19070, At3g28780, and At5g13930, respectively) had very little changes in their exine patterns (Supplemental Fig. S4), despite having glossy and/or sticky pollen phenotypes easily distinguishable at the dissecting microscope level (Supplemental Fig. S1).
Exine is an autofluorescing structure, likely due to the presence of phenolics and, possibly, some other compounds (Roshchina, 2003). To find if exines of the mutants with almost no pattern defects differ from the wild-type exine in their autofluorescence characteristics, we performed LSCM λ scans on pollen surfaces using 405-nm light for excitation. Emission spectra of exines were recorded in the interval of 400 to 625 nm. Under these conditions, the wild-type exine fluorescence spectrum consisted of two peaks: a higher one, with the maximum at 460 nm, and a lower one, at 505 nm (Fig. 9A). To account for individual differences in fluorescence intensity between pollen grains, all the data were normalized to the intensity of the 460-nm peaks. The method generated highly reproducible results: Figure 9A shows three separate trials on the wild-type Col-0 pollen grains (n = 10 grains per trial). When spectra of the mutants were analyzed, we noticed changes in the spectral fingerprints of several lines. For instance, both mlp3 and mlp6 (At2g19070) showed dramatic increases in the 505-nm peak, which became higher than the 460-nm peak (Fig. 9B). Interestingly, while all the grains in mlp3 demonstrated a tendency to have a larger 505-nm peak, they exhibited much higher variation in the emission spectra at the longer wavelengths than their wild-type counterparts or other mutants, perhaps reflecting greater differences among the individual mlp3 pollen in the quantity or quality of materials from which their exine is made. Pollen from mlp2, mlp5 (At1g65060), a different insertion in At1g65060 (WiscDsLox503E07), and mlp8 (At5g13930) had reductions in the 505-nm peak as well as changes in the shape of the 460-nm peak (Figs. 9B and 10A). For mlp7 (At3g28780), we detected no changes in the autofluorescence spectrum of exine when the data were normalized to the 460-nm peak (Fig. 9C); however, the absolute fluorescence intensity of the mutant was reduced (illustrated as size of the 460-nm peak in Fig. 9D). Two mutants in this category (mlp1 and mlp4) did not differ from the wild type in their exine fluorescence patterns (Fig. 9B).
Figure 9.
Some mutants with mild or little defects in exine morphology have abnormal exine autofluorescence. Λ scans of exine autofluorescence were recorded in the 400- to 625-nm interval (n = 10 grains per genotype). The data were normalized to 460-nm peak intensity, and means and se values were calculated. Wild-type exine generates two peaks, the higher one at 460 nm and the lower one at 505 nm. A, The spectra are highly reproducible. Three separate trials were performed on the wild-type exine; n = 10 grains per trial. B, Examples of autofluorescence spectral fingerprints of several mutants. The mlp1 and mlp4 lines do not demonstrate noticeable changes in their exine spectra compared with the wild type (WT), while the spectra of the other mutants shown here are significantly different from the wild type. C and D, Although the exine spectrum of the mutant of At3g28780 is not different from the wild type when data are normalized to the 460-nm peaks (C), the absolute fluorescence intensity of the mutant is greatly reduced, demonstrated as intensity of the 460-nm peaks (D).
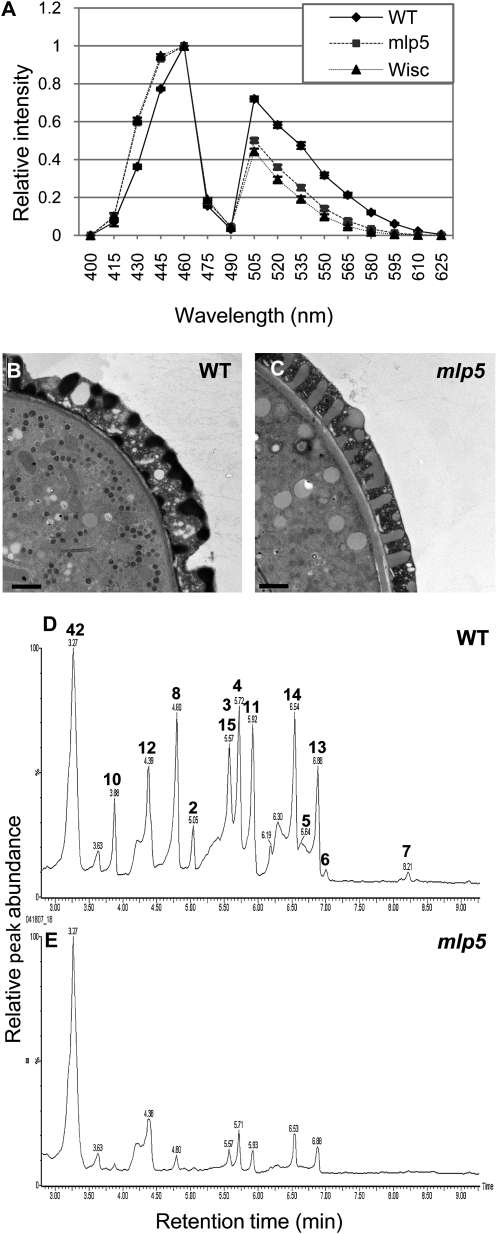
Figure 10.
mlp5 has abnormal pollen exine properties and defects in the production of flavonoids during anther development. A, Exine autofluorescence spectra of the wild-type (WT) and two mutants in the 4CL3 gene (mlp5 [SALK_017894] and Wisc [WiscDsLox503E07]). B and C, Transmission electron micrographs of pollen grain sections from the wild type (B) and mlp5 (C). Exine in mlp5 is less electron dense compared with the wild type. Bars = 1 μm. D and E, Levels of flavonoids produced in stage 9/10 anthers are strongly reduced in mlp5. UPLC-quadrupole time-of-flight-MS chromatograms of flavonoids in the wild type (D) and mlp5 (E) are shown. The peaks in D are annotated in boldface according to the identifier numbers in Supplemental Table S4. Proposed annotations of the peaks are as follows: 2, unknown; 3, kaempherol 7-rhamnoside; 4, kaempherol 3-galactoside-7-rhamnoside; 5, unknown; 6, isorhamnetin 3-glucoside; 7, kaempheride 3-glucoside; 8, quercetin 3-glucoside-7-rhamnoside; 10, unknown; 11, kaempheride 3,7-diglucoside; 12, kaempherol 3-glucoside-7,4′-dirhamnoside; 13, luteolin 4′-methyl ether 7-rutinoside; 14, kaempherol 3-rhamnoside-7-rhamnoside; 15, kaempherol 3-O-rutinoside; 42, 8-methylthio-octyl glucosinolate.
For one of the mlp mutants, mlp5, we also performed TEM analysis of pollen. Interestingly, we found that in TEM images, the mutant exine was consistently less electron dense than the wild-type exine (samples were prepared twice with the same results; trial 1, n = 19 pollen for mlp5, n = 14 pollen for the wild type; trial 2, n = 25 pollen for mlp5, n = 19 pollen for the wild type [Fig. 10, B and C]). This is consistent with mlp5 exine having chemical changes that cause it to bind less of the electron-dense stains used in sample preparations.
mlp5 has defects in 4CL3, a 4-coumarate-CoA ligase, which was previously proposed to be involved in one branch of phenylpropanoid metabolism, production of flavonoids (Ehlting et al., 1999). To see what chemical changes mlp5 exine might have, we performed a metabolomics analysis on extracts from developing (stage 9/10) anthers. Ultra-HPLC coupled to mass spectrometry (UPLC-MS) and gas chromatography-mass spectrometry were used to separate and detect metabolites. The sample preparation and analysis were done as described before (Dobritsa et al., 2010).
Consistent with the proposed role for 4CL3, the most notable change in the metabolic profile of mlp5, in comparison with the wild type, was a very strong reduction in the amount of flavonoids (Fig. 10, D and E; Supplemental Table S4). When anther metabolites from mlp5 and wild-type plants were compared by UPLC-MS, reduction in the levels of multiple glycosides of the flavonoid kaempferol was apparent in the mutant (Supplemental Table S4). Several other kaempherol glycosides (kaempferol 7-rhamnoside, kaempferol 3-galactoside-7-rhamnoside, kaempferide 3-glucoside) were absent in mlp5. Additionally, several other flavonoids were missing or had reduced levels, including luteolin 4′-methyl ether 7-rutinoside, quercetin 3-glucoside-7-rhamnoside, and isorhamnetin 3-glucoside (Supplemental Table S4). Associated with the decrease in flavonoids was a concomitant 5-fold increase in Phe, which serves as the entry point into phenylpropanoid pathways (Supplemental Table S5). Interestingly, we also found a 5-fold increase in the amount of another phenolic compound, ferulic acid (Supplemental Table S5), commonly recognized as a precursor in the biosynthesis of the strong plant polymer lignin and suggested to be one of the phenolic precursors of sporopollenin. Based on these results, we hypothesize that changes in exine autofluorescence and TEM staining in mlp5 may be due to reductions in the amounts of the aromatic constituents of sporopollenin.
Another mlp mutant, mlp4, harbors a T-DNA insertion in the gene At4g14080, encoding an anther-specific glycosyl hydrolase family 17 protein, A6/MEE48. An independent transposon insertion into this gene (WiscDSLox285F04) had an identical glossy and sticky pollen phenotype, providing evidence that mutations of the A6/MEE48 gene are responsible for pollen defects. It has been suggested that A6 is a β-1,3-glucanase that may be important for the dissolution of callose walls around the microspore tetrads (Hird et al., 1993). mlp2 harbors a T-DNA insertion in At2g30710 that encodes a RabGAP/TBC domain-containing protein, and mlp3 harbors an insertion in At2g30760 encoding a novel protein. Further studies will be necessary to determine if these genes are in fact required for pollen development. The mlp1 mutation was localized to chromosome 3 (Table II).
Class 2: Round Pollen (inp)
Plants with round pollen and no other pollen/anther defects (e.g. no glossy and/or sticky pollen phenotype) visible at the dissection microscope level were identified among 17 pools of SALK lines and had a very uniform phenotype. The pollen grains of these mutants had a normal reticulate pattern of exine but completely lacked apertures (Fig. 11A) and were dubbed inp. Because apertures are often used as exit points for pollen tubes, the fertility of the inp plants was assessed. They exhibited well-developed siliques and a normal number of seeds per silique (e.g. 54.2 ± 3.5 for the inp line 25-1-1 versus 51.3 ± 2.5 for the wild type [n = 12 siliques in both cases]), indicating that pollen tubes in Arabidopsis are capable of breaking through exine walls.
Figure 11.
Confocal images of exine from several mutant classes. A, Examples of mutant lines with pollen completely lacking apertures (inp phenotype). B, Exine-lacking line lnp1, which generates very few pollen grains. C, Examples of mutant pollen that belongs to the large and square pollen (lsq) category. Note the abnormal number and positions of apertures. D, Examples of mutant pollen that belong to the irregular-size pollen (isp) category. The arrow points to a small grain next to the larger grain (isp7 [left]); both are derived from the same anther. Larger-than-normal grains sometimes have abnormal number and positions of apertures (isp8 [left]). Smaller-than-normal grains have smaller lacunae and broader walls, resulting in more excessively covered pollen surface (isp7, left and right; isp8, right). Grains were stained with auramine O. Magnification is the same as in A (left) for all images except A (right). For reference to the wild-type exine, see Figures 4 and 7.
Complementation crosses performed on 14 inp lines derived from different seed pools revealed that they all belonged to the same complementation group and, therefore, were most likely allelic. All four of the inp lines that were tested with bulked segregant analysis had mutations linked to the CIW7 marker on chromosome 4 (Table II).
Class 3: Little or No Pollen
Mutations in several genes previously implicated in exine production cause the collapse of pollen grains during development, resulting in reduction or complete absence of pollen on dehisced anthers (Aarts et al., 1997; Paxson-Sowders et al., 1997; Wilson et al., 2001; Ito and Shinozaki, 2002; Ariizumi et al., 2004; Dong et al., 2005; Morant et al., 2007; Guan et al., 2008; Tang et al., 2009). In fact, identifying mutants with defects in male fertility was until recently the primary way to discover genes involved in exine development (Aarts et al., 1993; Paxson-Sowders et al., 1997; Ariizumi et al., 2003, 2004; Guan et al., 2008). Thus, during our screen, we have also focused on lines that generated little or no pollen. Approximately 48 such lines were identified. A male-sterile line, little or no pollen1 (lnp1), was found to produce very few glossy pollen grains, which, when examined using LSCM, were found to lack exine (Fig. 11B). A mutation in lnp1 was linked to the marker NGA162 on chromosome 3. Since two known exine genes, MS2 and DEX1, reside in this region, we performed complementation crosses with ms2 mutants and also PCR amplified and sequenced both genes from the lnp1 plants. We found that the lnp1 mutation complemented ms2, and the sequences of both MS2 and DEX1 genes from this line were indistinguishable from the wild type, indicating that a third gene required for exine production likely resides in this area.
Class 4: Large and Square Pollen
Approximately 30 lines (large and square pollen1 [lsq1]–lsq30) were identified. Several lines from this class had pollen with apertures that were abnormal in number or position (Fig. 11C). Grains were significantly larger than those from the wild type. For at least some members of this class (lsq2, lsq3, and lsq4), phenotypes were likely dominant, with F1 plants exhibiting the lsq phenotype after a backcross with Col-0. In the F2 generation for these three lines, segregation of three types of progeny was observed: wild-type-looking plants with wild-type pollen; plants with larger flowers and large and square pollen; and finally, dwarf plants with a variety of phenotypic defects, such as bent pistils, anthers with little or no pollen, and sterility. It is known that abnormal meiotic divisions of the microspore mother cells affect the number and placement of apertures in microspores. For instance, when divisions are blocked by colchicine, the whole mother cell acts as a single spherical spore and either does not form apertures or forms one irregular, randomly placed aperture (Sheldon and Dickinson, 1986). Similarly, genetic disruption of male meiotic cytokinesis by mutations in the kinesin-encoding TETRASPORE/STUD gene results in the development of large pollen grains with an abnormal number and position of apertures (Hulskamp et al., 1997; Spielman et al., 1997). We hypothesize that similar types of defects may cause the phenotypes that were observed in this class of mutants. This class has not been further pursued.
Class 5: Irregular-Size Pollen
Approximately 36 lines (irregular-size pollen1 [isp1]–isp36) were identified. Lines that had this type of defect generated pollen of variable size in the same anther (Fig. 11D; Supplemental Fig. S4). In several cases, the larger than wild-type pollen grains had extra apertures, resembling the lsq class of mutants (Fig. 11D; isp8). Pollen grains that were smaller than the wild type had exine with a reticulate pattern but smaller than normal lacunae and broader walls or muri, resulting in more excessively covered pollen surface (Fig. 11D). We stained pollen grains in several of those lines with 4′,6-diamino-phenylindole (DAPI) to visualize DNA and discovered that larger grains had two compact generative nuclei and a diffusely stained vegetative nucleus, similar to the wild type (Supplemental Fig. S5). At the same time, all of the smaller grains lacked DAPI staining (Supplemental Fig. S5). Thus, they were not viable pollen but rather empty exine shells, either generated by the disrupted cleavage during meiotic divisions or representing collapsed pollen grains, similar to the ones produced by the mutant defective in the REVERSIBLY GLYCOSYLATED POLYPEPTIDE (RGP1) and RGP2 gene functions. Small and collapsed pollen grains in this mutant are products of normal meiotic divisions but later have defects in intine production and pollen mitotic divisions (Drakakaki et al., 2006). This class has not been analyzed further.
DISCUSSION
A Novel Screen Identifies Multiple Pollen Mutants That Have Defective Exine
The field of exine and sporopollenin research, which was at an impasse for a long time, is now at a place similar to where cutin and suberin studies were until a few years ago (for review, see Pollard et al., 2008), when joint chemical, biochemical, and, especially, genetic approaches helped to greatly advance the research in these areas. Whereas the chemical properties of exine’s sporopollenin had prevented straightforward characterization of its composition, recent discoveries of several exine genes have reignited interest in this structure and will likely help to gain better understanding of the nature of this unique compound, of the pathways that lead to its production, and of the developmental mechanisms that control exine’s assembly into intricate and diverse three-dimensional patterns.
We have conducted a novel genetic screen in Arabidopsis to discover genes involved in pollen exine development. While many of the previously identified genes required for exine development were discovered in screens concentrating on male sterility (van der Veen and Wirtz, 1968; Aarts et al., 1993; Paxson-Sowders et al., 1997; Wilson et al., 2001; Ariizumi et al., 2003, 2004, 2005, 2008; Guan et al., 2008), our studies (Nishikawa et al., 2005; Dobritsa et al., 2009b, 2010) have demonstrated that male sterility or even reduction in male fertility are not the obligate characteristics of mutants with defects in exine-specific genes: many mutants with defects in exine structure have normal fertility. This suggests that male-sterility screens are biased against finding some of the exine mutants. In fact, the results of the screen presented here, as well as the results of the recent kns screen that also used a fertility-independent approach (Suzuki et al., 2008), support this notion: most of the mutant lines that we chose for more detailed characterization (53 out of 56) and 11 out of the 12 kns mutants have normal fertility, despite sometimes very prominent exine abnormalities. These include, for instance, the zebra mutants, in which the exine layer was missing (Dobritsa et al., 2009b).
We used a simple visual screen in which we looked at the state of anthers and pollen grains using standard dissecting microscopes, relying on changes in pollen shape, light reflection, color, and ease of release from anthers to identify potential exine mutants. This method, although labor intensive compared with sterility screens, is nevertheless much simpler than the kns screen (Suzuki et al., 2008) performed using scanning electron microscopy or a screen for pollen-stigma adhesion, in which the lap mutants affecting exine production were identified (Zinkl and Preuss, 2000; Nishikawa et al., 2005). The strategy proved to be effective: we discovered approximately 213 different mutant lines, which were placed into five phenotypic categories recognized with dissecting microscopes.
A secondary screen among multiple representatives from different categories, which utilized confocal microscopy and concentrated on changes in exine, allowed us to prioritize work with these categories. The glossy and/or sticky pollen category contained mutants with the most striking defects in exine structure and pattern, including those with the zebra, thin-exine, spongy, spotty, uneven-pattern, and defective exine phenotypes, and screening for the glossy and/or sticky pollen phenotype appears to be a reliable way to isolate exine mutants. Most of the mutants (34 identified through the forward screen and six identified through reverse genetics) that we chose to pursue belonged to this category. How could exine abnormalities lead to changes in pollen luster or the tendency of pollen grains to form aggregates? The effects may be direct, caused by changes in the regularity of the net-like tectum or by the aberrant quantity or quality of sporopollenin constituents, which lead to changes in light interaction with the exine surface. Alternatively, they may be indirect, caused, for instance, by changes in the amount of pollen coat that defective exines can hold. This, in turn, may lead to the abnormal light reflection or may mediate clump formation.
The second class of the discovered mutants included those that did not have glossy and/or sticky pollen phenotypes but appeared round using dissecting microscopes. All of these mutants had exines with normal reticulate patterning but lacked apertures.
Finally, because of the negative effect that exine defects have on pollen viability in some mutants (Aarts et al., 1997; Paxson-Sowders et al., 2001; Ariizumi et al., 2004; Guan et al., 2008; de Azevedo Souza et al., 2009; Dobritsa et al., 2009a, 2010), we also kept the lines that produced little or no pollen. Approximately 48 such lines were identified. While only one of these lines (lnp1) was pursued here, the rest can be rescreened, looking at the exine formation in these mutants during microspore development.
Disruption of the Exine Gene Network Can Have Many Morphological Outcomes
Mutants discovered in our screen had a variety of exine phenotypes, ranging from very mild (mlp) to severe (zebra, tex2, lnp1). Based on the morphology of their exines, we grouped the identified mutants into different subclasses, resulting in zebra, tex, spg, spot, upex, qrt-dex, dex3, mlp, inp, and lnp categories. The phenotypic differences in the mutants underscore the fact that exine development is a complex and highly regulated process and that disruption of this process at various points leads to morphological differences in exines.
Although our understanding of the genetic network that facilitates exine synthesis and patterning is still very limited, the identification of some players important for exine development has highlighted the roles of several processes required for exine formation. These include the formation of a temporary callose wall around the pollen mother cell and tetrads of microspores (CALLOSE SYNTHASE5 gene), primexine deposition (DEX1, NEF1, RPG1), and the production and modification of fatty acid-derived components of sporopollenin (CYP704B1, CYP703A2, MS2, ACOS5, LAP5/PKSB, LAP6/PKSA, TKPR1/DRL1, TKPR2/CCRL6).
Exine phenotypes of the mutants discovered in our study provide some clues to the roles of corresponding genes. Mutants that have almost no exine or a very thin exine (zebra, tex) often have defects in genes participating in sporopollenin synthesis or transport. We hypothesize that the currently unknown gene disrupted in lnp1 may also be involved in this process. The specification of exine reticulate pattern appears to be genetically separate from sporopollenin production, as hypomorphic mutations (tex1 and lap4-1) in a gene involved in sporopollenin biosynthesis (CYP703A2) result in very thin exines, which, nevertheless, have quite regular reticulate patterns.
Phenotypes of some other mutants (spg, spot, upex, qrt-dex, dex3) suggest that the affected genes are responsible for exine patterning. This process, which in Arabidopsis ultimately results in the development of a regular array of baculae of a certain height and the formation of tectum with regularly distributed lacunae of a certain size, is likely complex. Some of the developmental elements possibly contributing to exine patterning are the formation of primexine of the right thickness and the undulation of plasma membrane at regular intervals: the distribution of membrane protrusions may provide spatial information for the development of future baculae (Takahashi, 1995; Suzuki et al., 2008). At1g33430 disrupted in upex1 and upex2 appears to control the height of baculae and the proper fusion of their distal ends to form patterned tectum, and genes affected in upex3 and the spot mutants are likely involved in tectum development. The spg mutants resemble the type 3 kns mutants (Suzuki et al., 2008), which have incomplete or irregular tectum development. Suzuki et al. (2008) hypothesized that the type 3 kns mutants are involved in creating pretectum or in the biosynthesis or deposition of sporopollenin on a growing tectum. We suggest that abnormalities in baculae distribution may also contribute to the phenotypes of the spg or type 3 kns mutants.
Some of the discovered genes are novel and would have been difficult to identify without a forward genetic screen. For instance, At5g58100 affected in spot1 encodes a protein with no obvious functional motifs and does not exhibit anther-specific expression. Yet, the existence of three independent lines with insertions in this gene, all exhibiting the same phenotype, indicates that this gene is in fact important for exine development.
Similarly, At5g65000, encoding a putative NST affected in tex2, is not anther specific and could be difficult to identify as important for exine development through other approaches. The TEX2 protein is well conserved in the plant kingdom (sharing 73% amino acid sequence identity with orthologs in maize and sorghum [Sorghum bicolor], 71% identity with orthologs in rice [Oryza sativa] and pine [Pinus spp.], and 62% identity with an ortholog in the moss Physcomitrella patens). NSTs constitute a family of transmembrane proteins that transport nucleotide-activated sugars from cytoplasm to ER and Golgi lumens, where activated sugars are used as substrates for various glycosyl transferases that synthesize polysaccharides and modify proteins and lipids (Kawakita et al., 1998; Berninsone and Hirschberg, 2000). Although its own substrate specificity is unknown at present, in a multispecies phylogenetic tree of the NST family, TEX2 clusters together with another Arabidopsis protein, At5g41760 (Bakker et al., 2005), capable of transporting CMP-sialic acid into the Golgi apparatus (Bakker et al., 2008). Interestingly, two putative glycosyl transferases (At1g33430/UPEX1/UPEX2 and At1g27600/SPG2) were also identified in our screen. It remains to be determined if these proteins depend on TEX2 for their sugar substrates. At1g27600 was recently described as IRREGULAR XYLEM9-LIKE and shown to participate as a minor player, along with its closest homolog IRREGULAR XYLEM9, in the biosynthesis of the hemicellulose glucuronoxylan, a major component of plant secondary cell walls (Wu et al., 2010). In addition to these enzymes indicating a possible link between carbohydrate metabolism and exine production, the product of the KNS2 gene, discovered in the kns screen, encodes a Suc phosphate synthase (Suzuki et al., 2008). Primexine, the precursor of exine, whose presence is critical for proper sporopollenin deposition and exine formation (Paxson-Sowders et al., 2001; Guan et al., 2008), is made of polysaccharides such as cellulose (Rowley and Dahl, 1977). Therefore, a possibility exists that the NST TEX2, two glycosyl transferases, and the Suc phosphate synthase KNS2 contribute to the synthesis of polysaccharides required for primexine generation.
Mutants with Little Changes in Their Exine Patterns May Have Abnormal Exine Composition
Mutants belonging to one of the subclasses (mlp) found in our screen were easily distinguishable as glossy and/or sticky pollen at the dissecting microscope level yet had little, if any, morphological changes in their exine when viewed by confocal microscopy. For some of these mutants, changes in the exine autofluorescence spectrum or in the emission intensity were detected, indicating likely changes in exine composition; for others, no defects were found. Although it is possible that the glossy and/or sticky pollen lines with no apparent changes in exine morphology or fluorescence have defects in the pollen coat rather than in exine per se, to determine if there are some structural changes in the exine of these mutants, other methods may have to be applied. For example, atomic force microscopy can be used to measure the stiffness of the exine material in those mutants. One of those mutants is mlp4, affecting the anther-specific A6 gene. A6 is a glycosyl hydrolase family 17 protein proposed to be important for the timely dissolution of callose walls around tetrads of microspores (Hird et al., 1993). Although the function of A6 has not been formally proven, its expression is strongly correlated with the genes involved in sporopollenin synthesis, such as ACOS5, CYP704B1, MS2, and others. Interestingly, another glycosyl hydrolase family 17 protein (At3g23770) that shares significant homology with A6 has strong coexpression with A6 and the genes in the predicted sporopollenin biosynthesis network. It is possible, therefore, that A6 and At3g23770 have redundant functions during microspore and exine development and that a double mutant may have more revealing exine phenotypes.
Autofluorescence in plants is often caused by the presence of phenolic compounds, and phenolics in sporopollenin are likely strong contributors to exine autofluorescence. Further studies are necessary to clarify if the mlp mutants with abnormalities in the exine autofluorescence spectrum have exine with abnormal phenolic composition. Our study suggested this as a possibility. We found that two mlp mutants affecting the flavonoid pathway (mlp5 and mlp8) had pollen exine with lower 550-nm autofluorescence peaks, whereas a mutant affecting the production of hydroxycinnamoyl spermidine (mlp6) had a relative increase in the 505-nm peak. At the moment, it is unclear if changes in autofluorescence are caused by changes in these particular phenolic compounds or if they are caused by pleiotropic effects of mutations that affect different metabolites. As more genes required for defined steps in the sporopollenin synthesis pathway are discovered and characterized, their mutants can be used to determine the contribution of the respective components of sporopollenin to exine autofluorescence. It is conceivable that better understanding of exine autofluorescence can lead in the future to novel ways of mutant isolation (e.g. by screening pollen with a fluorescence-activated cell sorter).
For one of the mlp lines, mlp5, in addition to changes in exine autofluorescence, we also found decreased exine staining in TEM samples. This further supported the notion that although exine patterning was normal in this mutant, its exine composition was altered.
mlp5 has defects in 4CL3, a 4-coumarate-CoA ligase. 4-Coumarate-CoA ligases (EC 6.2.1.12) play roles in early steps of the phenylpropanoid pathway. They channel carbon flow from primary metabolism to different branches of secondary phenolic metabolism, such as the synthesis of flavonoids or lignin. This process is done by converting 4-coumaric acid and other hydroxycinnamic acids into the corresponding CoA esthers (Hahlbrock and Scheel, 1989; Douglas, 1996). Based on the in vitro properties of the enzyme, its expression patterns, and evolutionary relationships, it has been suggested that 4CL3 in Arabidopsis is likely involved in flavonoid metabolism and not in the production of lignin or other phenolic compounds (Ehlting et al., 1999). Consistent with the involvement of 4CL3 in flavonoid production, our metabolite profiling analysis of the extracts from developing mlp5 anthers at the time of exine production revealed a dramatic reduction in the amount of flavonoids. Additionally, we found a 5-fold increase in the accumulation of Phe, a precursor of phenolic compounds, and, interestingly, a similar 5-fold increase in the accumulation of ferulic acid, which is known as a precursor in lignin production. Previously, the in vitro studies of 4CL3 enzymatic activities demonstrated that although the enzyme had the strongest substrate specificity toward 4-coumarate, it also possessed a noticeable activity toward ferulate (Ehlting et al., 1999; Costa et al., 2005). Results of chemical analyses of sporopollenin suggested that both ferulic and 4-coumaric acids are possible constituents of sporopollenin (Schulze Osthoff and Wiermann, 1987; Wehling et al., 1989; Rozema et al., 2001; de Leeuw et al., 2006). Based on our results, we hypothesize that among the roles of 4CL3 in anthers may be the synthesis of phenolic components of sporopollenin and that ferulate-CoA and 4-coumarate-CoA may either be directly incorporated in sporopollenin or act as precursors for sporopollenin’s phenolic components. At present, it is unknown if a reduction in the amount of flavonoids in mlp5 anthers indicates that they are also used as precursors in sporopollenin synthesis or, more likely, reflects 4CL3 participation in another pathway (e.g. the production of flavonoids for pollen coat). The 4CL3 products may also contribute to the recently discovered pathway leading to the formation of hydroxycinnamoyl spermidines in pollen (Fellenberg et al., 2008, 2009; Grienenberger et al., 2009; Matsuno et al., 2009). These spermidine conjugates are components of pollen coat (Grienenberger et al., 2009), yet there is some evidence that they may also be present in exines (Gubatz et al., 1986).
Aperture Development in Exines Is Specified Separately from the Reticulate Pattern
The round pollen category contained mutants with pollen entirely lacking apertures (inp phenotype) but with a normal reticulate pattern, indicating that these types of exine elements are specified separately. All 14 inp lines that were tested for complementation were noncomplementing, suggesting that these mutations were allelic. Identification of the mutants with inaperturate pollen raises interesting developmental and evolutionary questions. In pollen of most plant species, apertures represent a well-defined and tightly controlled element of exine patterning, and they are species specific in number and morphology. This indicates that the exine deposition machinery reliably recognizes some areas on the pollen surface as different from others and does not deposit exine into these areas. How pollen generates and defines these functionally distinct cellular domains and then translates cellular asymmetries into extracellular differences is an exciting developmental and cell biological problem. The product of the INP gene is likely involved either in asymmetry establishment or in its interpretation. Given the variety of aperture morphologies across the phylogenetic spectrum of plants, understanding of the cellular mechanism for generating extracellular asymmetries will eventually allow investigation of the evolutionary malleability of these types of programs in a well-defined system that provides an enormous diversity of structures to probe.
In Arabidopsis, apertures are formed at the points of contact between neighboring microspores in a tetrahedral tetrad generated through simultaneous microsporogenesis (Owen and Makaroff, 1995; Furness and Rudall, 2004). It would be interesting to see if the shape of microspore tetrads and their development in inp mutants (e.g. simultaneous versus successive microsporogenesis) differ from those of the wild type. It was demonstrated in other plant species that proper aperture positioning is microtubule dependent (Dover, 1972; Sheldon and Dickinson, 1983, 1986). Also, the formation of an ER shield (the colpal shield), which blocks the deposition of primexine and sporopollenin at the sites of future apertures, was suggested to account for the development of apertures, at least in some species (Heslop-Harrison, 1963, 1968b). It would be important to check if microtubule formation, meiotic spindle orientation, and ER positioning are normal in these mutants. Identifying the gene defective in this complementation group will likely provide additional information about the mechanism that determines the number and positions of apertures.
The inp mutants are also ideal for addressing the functional roles of apertures. Traditionally, apertures have been regarded as points of exit for germinating pollen tubes. In fact, for pollen tubes of many plant species for which detailed studies of pollen germination have been performed, apertures are the only way out (Heslop-Harrison, 1979b; Cresti et al., 1985; Heslop-Harrison and Heslop-Harrison, 1985, 1992). However, in Arabidopsis, pollen tubes do not always use apertures and often break out through the exine walls (Edlund et al., 2004). Additionally, inaperturate pollen, although much less common, does exist among all major plant groups (many gymnosperms, some monocots [e.g. members of Orchidaceae, Liliaceae, and Zingiberaceae], and a few eudicots [e.g. some taxa in Solanaceae, Salicaceae, and Ranunculaceae]) and has independently evolved numerous times (Furness, 2007). Most members of the Brassicaceae family, to which Arabidopsis belongs, have fairly uniformly patterned pollen with reticulate exine and three slit-like apertures (Rollins and Banerjee, 1979). However, examples of plants with inaperturate pollen also exist in Brassicaceae (e.g. Matthiola incana has spheroidal pollen with reticulate exine and no apertures; Furness, 2007). It has been suggested that the evolution of pollen with apertures of the type that Arabidopsis possesses (tricolpate pollen), which is very widespread among eudicots, is a potential key innovation ensuring selective advantage and underlying eudicot success (Furness and Rudall, 2004). Our recovery of the inp mutants in Arabidopsis will provide a means to test this hypothesis. The inp mutants displayed apparently normal fertility. Careful in vitro and in vivo studies of pollen germination, as well as competitive crosses using wild-type and inaperturate pollen, will be required to determine if having three apertures gives pollen a significant advantage in germination.
Difficulties in the Isolation of Exine Genes Found through Forward Genetic Screening of the SALK Collection
We chose to screen the T-DNA-mutagenized SALK collection (Alonso et al., 2003) with a goal of fast recovery of the mutated sequences that cause defective exine phenotypes. However, we found that for a significant number of mutants that were pursued, mutations causing abnormal exine phenotypes were not directly associated with T-DNA insertions (Table II; Supplemental Table S3). This made the isolation of the responsible genes far from straightforward. For example, for all seven zebra isolates identified in the forward screen, the T-DNA insertions were not localized to the genes later found to be responsible for the exine phenotypes (CYP704B1 and MS2). When the nature of the mutations was finally determined for the zebra lines, we found examples of short deletions (one and 42 nucleotides long), a larger genomic rearrangement, and single-nucleotide substitutions (Dobritsa et al., 2009b). Similarly, for the thin-exine lines tex1 and tex2, the T-DNAs were mapped to positions completely different from the ones found to be responsible for the mutant phenotypes, and the mutations causing the phenotypes were identified as a single-nucleotide substitution and a seven-nucleotide deletion, respectively. In addition, for tex3, spg5, upex2, lnp1, and the four lines belonging to the inp complementation group, the T-DNA insertions were mapped to chromosomes different from the one to which the mutations were linked through bulked segregation analysis. In spot2, bulked segregation analysis linked the defect to the regions of both chromosomes 1 and 5, indicating the possibility of a translocation or another genomic rearrangement.
T-DNA or transposon integrations in Arabidopsis have been previously associated with chromosomal rearrangements, large and small deletions, and translocations (Forsbach et al., 2003; Boavida et al., 2009), and examples of the recovery of point mutations through T-DNA screens have been described (Dieterle et al., 2001; Takeda et al., 2004; Marrocco et al., 2006). Yet, the frequency of such events in our collection of isolates appears to be quite high, making the SALK collection less attractive for use in forward genetic screens than we expected. These features might be more common than is usually anticipated for T-DNA-mutagenized collections, because a recent forward genetic screen of the T-DNA Versailles collection followed by the mapping of the recovered mutations revealed a very similar trend in terms of a high number of non-T-DNA-tagged mutated genes that instead displayed single-nucleotide changes, small deletions, and other genomic rearrangements (De Muyt et al., 2009).
MATERIALS AND METHODS
Forward Genetic Screen
We screened pooled T-DNA insertion lines derived from the SALK collection (Alonso et al., 2003). The Col-0 ecotype of Arabidopsis (Arabidopsis thaliana) served as the wild-type control. The entire content of each vial containing pooled T3 and T4 seeds from 100 individual lines was planted onto four flats on a 1:1 mix of C2 (Conrad Fafard) and Metro-Mix (Sun Gro Horticulture) or Schultz Potting Soil Plus (Spectrum Brands) soil. Plants from 179 seed pools were screened (individual sets of pools CS75001–CS75003, CS75007, and CS75011–CS75185 from the combined sets of pools CS76502 and CS76504 were obtained from the Arabidopsis Biological Resource Center [ABRC]). Plants were grown either in a greenhouse (in an air-conditioned room at 22°C) under ambient light conditions or in growth chambers with 12 h of 100 μE m−2 s−1 fluorescent lighting at 22°C. Six hundred to 800 plants were grown and screened from most vials, and we considered this an equivalent of screening approximately 100 individual lines. Zeiss Stemi-2000C, Olympus SZ51, or equivalent dissecting stereomicroscopes with magnification of 50× to 80× were used. Open flowers were collected, pollen was brushed on a microscope slide, and anthers were examined for abnormalities in anther or pollen morphology. Neither anthers nor pollen underwent any treatment for primary screening. More than 123,000 plants were screened. We estimate that we screened an equivalent of at least 16,000 individual lines. Mutants with potential exine defects were backcrossed with Col-0 at least once.
Reverse Genetic Screen
Available insertion mutants for 49 candidate exine genes were obtained from the ABRC or the Nottingham Arabidopsis Stock Center. One or two insertion lines were used for each gene, with a preference given to those with insertion positions within predicted protein-coding regions. Thirty-six plants were grown for each line. We confirmed the presence of insertions in the lines using gene-specific primers developed through the SIGnAL iSect Primer Design program (http://signal.salk.edu/isects.html). For a list of primers used to confirm insertions, see Supplemental Table S6. Screening methods were identical to those used for the forward genetic screen. Once potential exine mutants were identified, their phenotypes were confirmed in the next generation, and plants were backcrossed with a background ecotype (Col-0 or Ler) at least once.
Phenotypic Analysis during the Secondary Screen
Shapes of unhydrated pollen were assessed by placing pollen grains in 100% glycerol and examining with a 100× objective on a Zeiss Axioskop compound microscope (Nomarski optics). To visualize the exine structure, pollen grains were removed from anthers onto a microscope slide (by gently brushing an anther on a slide or by removing pollen with thin forceps), and a small drop of auramine O solution was added to pollen. Auramine O was dissolved in 50 mm Tris-HCl, pH 7.5, and diluted to 0.001% in 17% Suc. Pollen grains were simultaneously hydrated and stained with auramine O and then examined with a laser scanning confocal microscope (Olympus Fluoview FV1000, 100× objective, fluorescein isothiocyanate settings). In addition to images of the exine surface, we took optical sections by LSCM through the center of pollen grains to determine exine thickness. Multiple pollen grains were observed for each line (n ≥ 20). For lines exhibiting interesting abnormal exine phenotypes, pollen from multiple flowers (and multiple plants, after the lines were established) was assessed. For most mutant lines, the phenotypes of all grains in each line were very similar. The exceptions to this were lsq and isp lines, which produced pollen of different sizes, shapes, or aperture placement, progeny of the dex3 isolate, which segregated two different mutant exine phenotypes, and upex lines, in which all pollen grains had abnormal exine yet particular patterning defects and the placement of reticulate patches varied from grain to grain. Scanning electron microscopy, TEM, and acetolysis were performed as described (Dobritsa et al., 2009b). Sectioning of developing anthers and toluidine blue staining were performed as described (Dobritsa et al., 2010).
Spectral Analysis of Exine Autofluorescence
Pollen grains were mounted in 100% glycerol, illuminated using a 405-nm laser (14% laser power), and adjusted for focus and photomultiplier tube voltage using AlexaFluor488 settings. Lambda scans were performed on exine surfaces (n ≥ 10 grains) using a laser scanning confocal microscope (Olympus Fluoview FV1000, 100× objective, scan interval of 400–625 nm, step size of 15 nm, bandwidth of 15 nm). Background intensity was subtracted, then intensity data were normalized to the intensity of the 460-nm peaks and mean and se values were calculated.
Identification of Exine Genes
Two PCR methods were used to map T-DNA insertions in the lines discovered via forward screen: the adapter-based method used by Alonso et al. (2003) for mapping positions of SALK T-DNA insertions, and thermal asymmetric interlaced-PCR (McElver et al., 2001). Whenever possible, after determining the positions of T-DNA insertions, we confirmed that the disrupted gene was responsible for the observed phenotype by checking the phenotypes of additional insertion alleles available from the ABRC. For lines with causal mutations unlinked to the T-DNA, we crossed the lines with defective exine with the Ler ecotype and performed bulked segregant analysis to identify the genomic intervals in which the mutations were located. We used five to six molecular markers per chromosome available through The Arabidopsis Information Resource (http://www.arabidopsis.org/). Complementation crosses were performed on mutants with similar exine phenotypes. In several cases, we identified an affected gene by comparing exine phenotypes of the mutants isolated via the forward genetic screen with mutants identified through reverse genetics, performing complementation analysis, and sequencing the gene. To complement the tex2 defects, the At5g65000 gene (from 516 nucleotides upstream of the start codon to 251 nucleotides downstream of the stop codon) was amplified by PCR (primers SacI-At5g65000-FF [5′-GAGCTCTTCAACCCGAGGACATGAGAAG-3′] and AgeI-At5g65000-IR [5′-ACCGGTATTCAAAACTGTATACATCATGTG-3′]), digested with SacI/AgeI, and introduced into the modified binary vector pGreenII02229 (Hellens et al., 2000; von Besser et al., 2006). Homozygous tex2-1 and heterozygous tex2-4/+ plants were transformed by the floral dip method (Clough and Bent, 1998), and transgenic plants were selected with BASTA. Homozygous tex2-4 progeny were identified among the transgenic T1 plants using a diagnostic PCR (primers CS129638-LP [5′-TTGTAACTCGATCCACCCTTG-3′], CS129638-RP [5′-GATAACGCGAAAGGTTACACG-3′], and DS3-1 [5′-ACCCGACCGGATCGTATCGGT-3′]).
The Arabidopsis Genome Initiative locus numbers for the genes analyzed in this article are as follows: CYP703A2 (At1g01280), At1g27600, At1g33430, 4CL3 (At1g65060), CYP704B1 (At1g69500), SHT (At2g19070), MS2 (At3g11980), At3g28780, A6/MEE48 (At4g14080), QRT3 (At4g20050), TKPR1/DRL1 (At4g35420), TT4 (At5g13930), At5g58100, At5g65000.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Examples of anthers from mutants that have the glossy and/or sticky pollen phenotype.
Supplemental Figure S2. Microspores produced by the tex2-4 mutant degenerate during development.
Supplemental Figure S3. At1g33430 is expressed in the developing anthers.
Supplemental Figure S4. Examples of glossy and/or sticky pollen mutants that exhibit a normal reticulate pattern under LSCM.
Supplemental Figure S5. DAPI staining of irregular-size pollen (isp).
Supplemental Table S1. Candidate genes that did not affect pollen and anther morphology, when mutated.
Supplemental Table S2. Candidate genes in which the analysis did not produce conclusive results.
Supplemental Table S3. Positions of the T-DNA insertions mapped for various mutants identified in the screen.
Supplemental Table S4. UPLC-MS analysis of metabolite levels in the extracts from wild-type and mlp5 developing anthers.
Supplemental Table S5. Gas chromatography-mass spectrometry analysis of metabolite levels in the extracts from wild-type and mlp5 developing anthers.
Supplemental Table S6. Primers used to verify the positions of T-DNA insertions in candidate genes.
Acknowledgments
We thank undergraduates Veder Garcia, Carol Casey, Gatoya Jones, Alex Hoover, Andrew Mutka, and Jacob Roshanmanesh for help with the forward genetic screen and bulked segregant analysis, Jean Greenberg and Aretha Fiebig for critical reading of the manuscript, the ABRC and the Nottingham Arabidopsis Stock Center for mutant resources, and John Zdenek, Judy Coswell, and Sandra Suwanski at the University of Chicago greenhouse for excellent plant growth facilities.
References
- Aarts MG, Dirkse WG, Stiekema WJ, Pereira A. (1993) Transposon tagging of a male sterility gene in Arabidopsis. Nature 363: 715–717 [DOI] [PubMed] [Google Scholar]
- Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A. (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J 12: 615–623 [DOI] [PubMed] [Google Scholar]
- Ahlers F, Bubert H, Steuernagel S, Wiermann R. (2000) The nature of oxygen in sporopollenin from the pollen of Typha angustifolia L. Z Naturforsch C 55: 129–136 [DOI] [PubMed] [Google Scholar]
- Ahlers F, Thom L, Lambert J, Kuckuk R, Wiermann R. (1999) 1H NMR analysis of sporopollenin from Typha angustifolia. Phytochemistry 5: 1095–1098 [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Inatsugi R, Nishida I, Sato S, Kato T, Tabata S, Toriyama K. (2004) Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J 39: 170–181 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K. (2003) A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol Biol 53: 107–116 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K. (2005) The HKM gene, which is identical to the MS1 gene of Arabidopsis thaliana, is essential for primexine formation and exine pattern formation. Sex Plant Reprod 18: 1–7 [Google Scholar]
- Ariizumi T, Kawanabe T, Hatakeyama K, Sato S, Kato T, Tabata S, Toriyama K. (2008) Ultrastructural characterization of exine development of the transient defective exine 1 mutant suggests the existence of a factor involved in constructing reticulate exine architecture from sporopollenin aggregates. Plant Cell Physiol 49: 58–6718045813 [Google Scholar]
- Bakker H, Routier F, Ashikov A, Neumann D, Bosch D, Gerardy-Schahn R. (2008) A CMP-sialic acid transporter cloned from Arabidopsis thaliana. Carbohydr Res 343: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Bakker H, Routier F, Oelmann S, Jordi W, Lommen A, Gerardy-Schahn R, Bosch D. (2005) Molecular cloning of two Arabidopsis UDP-galactose transporters by complementation of a deficient Chinese hamster ovary cell line. Glycobiology 15: 193–201 [DOI] [PubMed] [Google Scholar]
- Berninsone PM, Hirschberg CB. (2000) Nucleotide sugar transporters of the Golgi apparatus. Curr Opin Struct Biol 10: 542–547 [DOI] [PubMed] [Google Scholar]
- Blackmore S, Barnes SH. (1986) Harmomegathic mechanisms in pollen grains. Blackmore S, Ferguson IK, , Pollen and Spores: Form and Function, Vol 12. Academic Press, London, pp 137–149 [Google Scholar]
- Boavida LC, Shuai B, Yu H-J, Pagnussat GC, Sundaresan V, McCormick S. (2009) A collection of Ds insertional mutants associated with defects in male gametophyte development and function in Arabidopsis thaliana. Genetics 181: 1369–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner B, Johal GS, Janick-Buckner D. (2000) Cell death in maize. Physiol Plant 108: 231–239 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coe EH, McCormick SM, Modena SA. (1981) White pollen in maize. J Hered 72: 318–320 [Google Scholar]
- Coimbra S, Costa M, Jones B, Mendes MA, Pereira LG. (2009) Pollen grain development is compromised in Arabidopsis agp6 agp11 null mutants. J Exp Bot 60: 3133–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MA, Bedgar DL, Moinuddin SGA, Kim K-W, Cardenas CL, Cochrane FC, Shockey JM, Helms GL, Amakura Y, Takahashi H, et al. (2005) Characterization in vitro and in vivo of the putative multigene 4-coumarate:CoA ligase network in Arabidopsis: syringyl lignin and sinapate/sinapyl alcohol derivative formation. Phytochemistry 66: 2072–2091 [DOI] [PubMed] [Google Scholar]
- Cresti M, Ciampolini DL, Mulcahy DL, Mulcahy GB. (1985) Ultrastructure of Nicotiana alata pollen, its germination and early tube formation. Am J Bot 72: 719–727 [Google Scholar]
- de Azevedo Souza C, Kim SS, Koch S, Kienow L, Schneider K, McKim SM, Haughn GW, Kombrink E, Douglas CJ. (2009) A novel fatty acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw JW, Versteegh GJM, van Bergen PF. (2006) Biomacromolecules of algae and plants and their fossil analogues. Plant Ecol 182: 209–233 [Google Scholar]
- De Muyt A, Pereira L, Vezon D, Chelysheva L, Gendrot G, Chambon A, Lainé-Choinard S, Pelletier G, Mercier R, Nogué F, Grelon M. (2009) A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet 5: e1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson HG, Sheldon JM. (1986) The generation of patterning at the plasma membrane of the young microspore of Lilium. Blackmore S, Ferguson IK, , Pollen and Spores: Form and Function, Vol 12. Academic Press, London, pp 1–17 [Google Scholar]
- Dieterle M, Zhou YC, Schäfer E, Funk M, Kretsch T. (2001) EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev 15: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan TTP, Carlsson AS, Hamberg M, Bülow L, Stymne S, Olsson P. (2009) Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli. J Plant Physiol 166: 787–796 [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Lei Z, Nishikawa S, Urbanczyk-Wochniak E, Huhman DV, Preuss D, Sumner LW. (2010) LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol 153: 937–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, Nishikawa S, Preuss D, Urbanczyk-Wochniak E, Sumner LW, Hammond A, Carlson AL, Swanson RJ. (2009a) LAP3, a novel plant protein required for pollen development, is essential for proper exine formation. Sex Plant Reprod 22: 167–177 [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Shrestha J, Morant M, Pinot F, Matsuno M, Swanson R, Møller BL, Preuss D. (2009b) CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol 151: 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DPS. (2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J 42: 315–328 [DOI] [PubMed] [Google Scholar]
- Douglas CJ. (1996) Phenylpropanoid metabolism and lignin biosynthesis: from weeds to trees. Trends Plant Sci 1: 171–178 [Google Scholar]
- Dover GA. (1972) The organization and polarity of pollen mother cells of Triticum aestivum. J Cell Sci 11: 699–711 [DOI] [PubMed] [Google Scholar]
- Drakakaki G, Zabotina O, Delgado I, Robert S, Keegstra K, Raikhel N. (2006) Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiol 142: 1480–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund AF, Swanson R, Preuss D. (2004) Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell (Suppl) 16: S84–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, Büttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E. (1999) Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J 19: 9–20 [DOI] [PubMed] [Google Scholar]
- Faegri K, van der Pijl L. (1979) The Principles of Pollination Ecology. Pergamon, Oxford [Google Scholar]
- Fellenberg C, Böttcher C, Vogt T. (2009) Phenylpropanoid polyamine conjugate biosynthesis in Arabidopsis thaliana flower buds. Phytochemistry 70: 1392–1400 [DOI] [PubMed] [Google Scholar]
- Fellenberg C, Milkowski C, Hause B, Lange PR, Böttcher C, Schmidt J, Vogt T. (2008) Tapetum-specific location of a cation-dependent O-methyltransferase in Arabidopsis thaliana. Plant J 56: 132–145 [DOI] [PubMed] [Google Scholar]
- Fitzgerald MA, Knox RB. (1995) Initiation of primexine in freeze-substituted microspores of Brassica campestris. Sex Plant Reprod 8: 99–104 [Google Scholar]
- Forsbach A, Schubert D, Lechtenberg B, Gils M, Schmidt R. (2003) A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol Biol 52: 161–176 [DOI] [PubMed] [Google Scholar]
- Furness CA. (2007) Why does some pollen lack apertures? A review of inaperturate pollen in eudicots. Bot J Linn Soc 155: 29–48 [Google Scholar]
- Furness CA, Rudall PJ. (2004) Pollen aperture evolution: a crucial factor for eudicot success? Trends Plant Sci 9: 154–158 [DOI] [PubMed] [Google Scholar]
- Grienenberger E, Besseau S, Geoffroy P, Debayle D, Heintz D, Lapierre C, Pollet B, Heitz T, Legrand M. (2009) A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J 58: 246–259 [DOI] [PubMed] [Google Scholar]
- Grienenberger E, Kim SS, Lallemand B, Geoffroy P, Heintz D, Souza CdeA, Heitz T, Douglas CJ, Legrand M. (2010) Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22: 4067–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan YF, Huang XY, Zhu J, Gao JF, Zhang HX, Yang ZN. (2008) RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol 147: 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubatz S, Herminghaus S, Meurer B, Strack D, Wiermann R. (1986) The location of hydroxycinnamic acid amides in the exine of Corylus pollen. Pollen Spores 28: 347–354 [Google Scholar]
- Guilford WJ, Schneider DM, Labovitz J, Opella SJ. (1988) High resolution solid state C NMR spectroscopy of sporopollenins from different plant taxa. Plant Physiol 86: 134–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häcker G. (2000) The morphology of apoptosis. Cell Tissue Res 301: 5–17 [DOI] [PubMed] [Google Scholar]
- Hahlbrock K, Scheel D. (1989) Physiology and molecular biology of phenylpropanoid metabolism. Annu Rev Plant Physiol Plant Mol Biol 40: 347–369 [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J. (1963) An ultrustructural study of pollen wall ontogeny in Silene pendula. Grana Palynol 4: 1–24 [Google Scholar]
- Heslop-Harrison J. (1968a) Pollen wall development: the succession of events in the growth of intricately patterned pollen walls is described and discussed. Science 161: 230–237 [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J. (1968b) Wall development within the microspore tetrad of Lilium longiflorum. Can J Bot 46: 1185–1192 [Google Scholar]
- Heslop-Harrison J. (1971) Wall pattern formation in angiosperm microsporogenesis. Symp Soc Exp Biol 25: 277–300 [PubMed] [Google Scholar]
- Heslop-Harrison J. (1979a) An interpretation of the hydrodynamics of pollen. Am J Bot 66: 737–743 [Google Scholar]
- Heslop-Harrison J. (1979b) Aspects of the structure, cytochemistry and germination of the pollen of rye. Ann Bot (Lond) 44: 1–47 [Google Scholar]
- Heslop-Harrison J, Heslop-Harrison Y. (1985) Germination of stress-tolerant Eucalyptus pollen. J Cell Sci 73: 135–157 [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison Y, Heslop-Harrison J. (1992) Germination of monocolpate angiosperm pollen evolution of the actin cytoskeleton and wall during hydration activation and tube emergence. Ann Bot (Lond) 69: 385–394 [Google Scholar]
- Hird DL, Worrall D, Hodge R, Smartt S, Paul W, Scott R. (1993) The anther-specific protein encoded by the Brassica napus and Arabidopsis thaliana A6 gene displays similarity to beta-1,3-glucanases. Plant J 4: 1023–1033 [DOI] [PubMed] [Google Scholar]
- Hulskamp M, Parekh NS, Grini P, Schneitz K, Zimmermann I, Lolle SJ, Pruitt RE. (1997) The STUD gene is required for male-specific cytokinesis after telophase II of meiosis in Arabidopsis thaliana. Dev Biol 187: 114–124 [DOI] [PubMed] [Google Scholar]
- Ito T, Shinozaki K. (2002) The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol 43: 1285–1292 [DOI] [PubMed] [Google Scholar]
- Katifori E, Alben S, Cerda E, Nelson DR, Dumais J. (2010) Foldable structures and the natural design of pollen grains. Proc Natl Acad Sci USA 107: 7635–7639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakita M, Ishida N, Miura N, Sun-Wada G-H, Yoshioka S. (1998) Nucleotide sugar transporters: elucidation of their molecular identity and its implication for future studies. J Biochem 123: 777–785 [DOI] [PubMed] [Google Scholar]
- Kim SS, Grienenberger E, Lallemand B, Colpitts CC, Kim SY, Souza CdeA, Geoffroy P, Heintz D, Krahn D, Kaiser M, et al. (2010) LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22: 4045–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrocco K, Zhou Y, Bury E, Dieterle M, Funk M, Genschik P, Krenz M, Stolpe T, Kretsch T. (2006) Functional analysis of EID1, an F-box protein involved in phytochrome A-dependent light signal transduction. Plant J 45: 423–438 [DOI] [PubMed] [Google Scholar]
- Matsuno M, Compagnon V, Schoch GA, Schmitt M, Debayle D, Bassard JE, Pollet B, Hehn A, Heintz D, Ullmann P, et al. (2009) Evolution of a novel phenolic pathway for pollen development. Science 325: 1688–1692 [DOI] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al. (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S. (2004) The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res 14: 1641–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV. (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88: 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y, Nagel C, Taylor LP. (1992) Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc Natl Acad Sci USA 89: 7213–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant M, Jørgensen K, Schaller H, Pinot F, Møller BL, Werck-Reichhart D, Bak S. (2007) CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19: 1473–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J. (1979) Form and function in angiosperm pollen. Ann Miss Bot Gard 66: 593–632 [Google Scholar]
- Napoli CA, Fahy D, Wang HY, Taylor LP. (1999) white anther: a petunia mutant that abolishes pollen flavonol accumulation, induces male sterility, and is complemented by a chalcone synthase transgene. Plant Physiol 120: 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S, Zinkl GM, Swanson RJ, Maruyama D, Preuss D. (2005) Callose (beta-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biol 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen HA, Makaroff CA. (1995) Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma 185: 7–21 [Google Scholar]
- Paxson-Sowders DM, Dodrill CH, Owen HA, Makaroff CA. (2001) DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol 127: 1739–1749 [PMC free article] [PubMed] [Google Scholar]
- Paxson-Sowders DM, Owen HA, Makaroff CA. (1997) A comparative ultrastructural analysis of exine pattern development in wild-type Arabidopsis and a mutant defective in pattern formation. Protoplasma 198: 53–65 [Google Scholar]
- Perez-Munoz CA, Webster BD, Jernstedt JA. (1995) Spatial congruence between exine pattern, microtubules and endomembranes in Vigna pollen. Sex Plant Reprod 8: 147–151 [Google Scholar]
- Piffanelli P, Ross JHE, Murphy DJ. (1998) Biogenesis and function of the lipidic structures of pollen grains. Sex Plant Reprod 11: 65–80 [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB. (2008) Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Polowick PL, Sawhney VK. (1995) Ultrastructure of the tapetal cell wall in the stamenless-2 mutant of tomato (Lycopersicon esculentum): correlation between structure and male-sterility. Protoplasma 189: 249–255 [Google Scholar]
- Quilichini TD, Friedmann MC, Samuels AL, Douglas CJ. (2010) ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol 154: 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Osborne E, Poindexter PD, Somerville CR. (2003) Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol 133: 1170–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RC, Banerjee UC. (1979) Pollens of the Cruciferae. Bussey Institution, Harvard University, Cambridge, MA [Google Scholar]
- Roshchina VV. (2003) Autofluorescence of plant secreting cells as a biosensor and bioindicator reaction. J Fluoresc 13: 403–420 [Google Scholar]
- Rowley JR, Dahl AO. (1977) Pollen development in Artemisia vulgaris with special reference to glycocalix material. Pollen Spores 19: 169–284 [Google Scholar]
- Rozema J, Broekman RA, Blokker P, Meijkamp BB, de Bakker N, van de Staaij J, van Beem A, Ariese F, Kars SM. (2001) UV-B absorbance and UV-B absorbing compounds (para-coumaric acid) in pollen and sporopollenin: the perspective to track historic UV-B levels. J Photochem Photobiol B 62: 108–117 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu Y-C, Lee PY, Truong MT, Beals TP, Goldberg RB. (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Schulze Osthoff K, Wiermann R. (1987) Phenols as integrated compounds of sporopollenin from Pinus pollen. J Plant Physiol 131: 5–15 [Google Scholar]
- Scott RJ. (1994) Pollen exine: the sporopollenin enigma and the physics of pattern. Scott RJ, Stead AD, , Molecular and Cellular Aspects of Plant Reproduction. University Press, Cambridge, UK, pp 49–81 [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG. (2004) Stamen structure and function. Plant Cell (Suppl) 16: S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon JM, Dickinson HG. (1983) Determination of patterning in the pollen wall of Lilium henryi. J Cell Sci 63: 191–208 [DOI] [PubMed] [Google Scholar]
- Sheldon JM, Dickinson HG. (1986) Pollen wall formation in Lilium: the effect of chaotropic agents, and the organisation of the micro-tubular cytoskeleton during pattern development. Planta 168: 11–23 [DOI] [PubMed] [Google Scholar]
- Spielman M, Preuss D, Li FL, Browne WE, Scott RJ, Dickinson HG. (1997) TETRASPORE is required for male meiotic cytokinesis in Arabidopsis thaliana. Development 124: 2645–2657 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Masaoka K, Nishi M, Nakamura K, Ishiguro S. (2008) Identification of kaonashi mutants showing abnormal pollen exine structure in Arabidopsis thaliana. Plant Cell Physiol 49: 1465–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M. (1995) 3-Dimensional aspects of exine initiation and development in Lilium longiflorum (Liliaceae). Am J Bot 82: 847–854 [Google Scholar]
- Takahashi M, Skvarla JJ. (1991) Exine pattern formation by plasma membrane in Bougainvillea spectabilis Willd. (Nyctaginaceae). Am J Bot 78: 1063–1069 [Google Scholar]
- Takeda S, Tadele Z, Hofmann I, Probst AV, Angelis KJ, Kaya H, Araki T, Mengiste T, Mittelsten Scheid O, Shibahara K, et al. (2004) BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev 18: 782–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LK, Chu H, Yip WK, Yeung EC, Lo C. (2009) An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in Arabidopsis. New Phytol 181: 576–587 [DOI] [PubMed] [Google Scholar]
- van der Veen JH, Wirtz P. (1968) EMS-induced genic male sterility in Arabidopsis thaliana: a model selection experiment. Euphytica 17: 371–377 [Google Scholar]
- Vizcay-Barrena G, Wilson ZA. (2006) Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J Exp Bot 57: 2709–2717 [DOI] [PubMed] [Google Scholar]
- von Besser K, Frank AC, Johnson MA, Preuss D. (2006) Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133: 4761–4769 [DOI] [PubMed] [Google Scholar]
- Wehling K, Niester C, Boon JJ, Willemse MTM, Wiermann R. (1989) p-Coumaric acid: a monomer in the sporopollenin skeleton. Planta 179: 376–380 [DOI] [PubMed] [Google Scholar]
- Wiermann R, Ahlers F, Schmitz-Thom I. (2001) Sporopollenin. Stenbüchel A, Hofrichter M, , Biopolymers, Vol 1. Wiley-VCH Verlag, Weinheim, Germany, pp 209–227 [Google Scholar]
- Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ. (2001) The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J 28: 27–39 [DOI] [PubMed] [Google Scholar]
- Wodehouse RP. (1935) Pollen Grains: Their Structure, Identification and Significance in Science and Medicine. McGraw-Hill, New York [Google Scholar]
- Worrall D, Hird DL, Hodge R, Paul W, Draper J, Scott R. (1992) Premature dissolution of the microsporocyte callose wall causes male sterility in transgenic tobacco. Plant Cell 4: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A-M, Hörnblad E, Voxeur A, Gerber L, Rihouey C, Lerouge P, Marchant A. (2010) Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol 153: 542–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Makaroff CA, Ma H. (2003) The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15: 1281–1295 [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkl GM, Preuss D. (2000) Dissecting Arabidopsis pollen-stigma interaction reveals novel mechanisms that confer mating specificity. Ann Bot (Lond) 85: 15–21 [Google Scholar]
- Zinkl GM, Zwiebel BI, Grier DG, Preuss D. (1999) Pollen-stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 126: 5431–5440 [DOI] [PubMed] [Google Scholar]