Abstract
The S locus, a single polymorphic locus, is responsible for self-incompatibility (SI) in the Brassicaceae family and many related plant families. Despite its importance, our knowledge of S-locus evolution is largely restricted to the causal genes encoding the S-locus receptor kinase (SRK) receptor and S-locus cysteine-rich protein (SCR) ligand of the SI system. Here, we present high-quality sequences of the genomic region of six S-locus haplotypes: Arabidopsis (Arabidopsis thaliana; one haplotype), Arabidopsis lyrata (four haplotypes), and Capsella rubella (one haplotype). We compared these with reference S-locus haplotypes of the self-compatible Arabidopsis and its SI congener A. lyrata. We subsequently reconstructed the likely genomic organization of the S locus in the most recent common ancestor of Arabidopsis and Capsella. As previously reported, the two SI-determining genes, SCR and SRK, showed a pattern of coevolution. In addition, consistent with previous studies, we found that duplication, gene conversion, and positive selection have been important factors in the evolution of these two genes and appear to contribute to the generation of new recognition specificities. Intriguingly, the inactive pseudo-S-locus haplotype in the self-compatible species C. rubella is likely to be an old S-locus haplotype that only very recently became fixed when C. rubella split off from its SI ancestor, Capsella grandiflora.
Self-incompatibility (SI) is an important mechanism used by many angiosperm species for preventing self-fertilization (selfing), which can be considered as an evolutionary dead end because it limits adaptive potential and causes inbreeding depression by the expression of recessive deleterious mutations (Lynch et al., 1995; Charlesworth et al., 2005; Newbigin and Uyenoyama, 2005; Busch and Schoen, 2008). SI is often controlled by a single polymorphic locus, the S locus. In the Brassicaceae, the S locus contains two specificity-determinant genes: S-LOCUS RECEPTOR KINASE (SRK), which encodes the stigmatic receptor kinase; and SP11/S-LOCUS CYSTEINE-RICH PROTEIN (SCR) (hereafter SCR), which encodes a small Cys-rich protein localized in the pollen coat, which is the ligand for the SRK receptor (Nasrallah, 2002). Pollen inhibition occurs when the same S-locus specificity is expressed by both pollen and pistil (Nasrallah, 2002). The S locus is exceptional, with its many transspecies polymorphisms (Charlesworth, 2006), providing a paradigm for frequency-dependent selection (Schierup and Vekemans, 2008; Leducq et al., 2011).
Transformation of Arabidopsis (Arabidopsis thaliana) with S-locus haplotypes from the self-incompatible Arabidopsis lyrata can restore SI in some accessions (Nasrallah et al., 2002). Therefore, it is likely that the initial inactivation of the S locus was a key step in the transition from SI to self-compatibility (SC) in this species. Arabidopsis is the model plant among SC species. The SI system is inactive in the selfing Arabidopsis reference strain Columbia (Col-0), and both SRK and SCR become pseudogenized (Kusaba et al., 2001). The inactivation of the S locus is correlated with the transition from outcrossing to selfing in both Arabidopsis and the Arabidopsis relative Capsella rubella (Tang et al., 2007; Guo et al., 2009; Tsuchimatsu et al., 2010). Transitions to SC are frequent in plants. This transition has occurred independently many times in several lineages (Barrett, 2002, 2010). In the Brassicaceae, most tribes include selfing species (Fobis-Loisy et al., 2004).
The molecular mechanism of action and functional polymorphisms at the S locus and in the SI system have been described extensively for Arabidopsis and related genera (Uyenoyama, 2000; Schierup et al., 2001, 2006; Kachroo et al., 2002; Sato et al., 2002; Takebayashi et al., 2003; Nasrallah et al., 2004; Prigoda et al., 2005; Bechsgaard et al., 2006; Paetsch et al., 2006; Liu et al., 2007; Mable and Adam, 2007; Sherman-Broyles et al., 2007; Tang et al., 2007; Busch et al., 2008; Shimizu et al., 2008; Guo et al., 2009; Castric et al., 2010; Tsuchimatsu et al., 2010).
Because of the difficulties with recovering the exceedingly diverse sequences at this locus by PCR, most analyses of the S locus have focused on SCR and SRK, and sometimes only on a partial S domain of the SRK gene. Other genes within the S-locus region have rarely been comprehensively examined (Kamau and Charlesworth, 2005; Hagenblad et al., 2006; Kamau et al., 2007; Ruggiero et al., 2008). There is, however, strong suppression of recombination and linked loci, which may influence the evolution of SI systems (Uyenoyama, 2005). Recently, it has been demonstrated that pseudo-SC can be caused by an S-locus-linked gene that can regulate SRK transcript levels (Liu et al., 2007). Detailed knowledge about S-locus sequences from additional relatives of Arabidopsis is also necessary to determine whether the S-locus polymorphisms in selfing species, such as Arabidopsis and C. rubella, reflect alleles that were present in a self-incompatible ancestor or whether they were produced by differential degradation of the S locus in different self-fertile populations (Sherman-Broyles et al., 2007). The age of S-locus allelic lineages in Arabidopsis relatives has thus far not been conclusively determined. Some allelic lineages at the S locus in A. lyrata are very old (Schierup et al., 2001; Paetsch et al., 2006; Edh et al., 2009): it is likely that they predate the divergence between the class I and class II allele lineages in Brassica, estimated to have occurred 40 million years ago (Uyenoyama, 1995). Finally, broader knowledge of S-locus sequence diversity is required for understanding how new specificities evolve in a two-gene system that relies on matching specificities between the two genes.
To address these questions, two factors must be considered. First, it is important to compare S-locus haplotypes between related SI and SC species. Second, both key genes and linked genes at the S locus should be analyzed. Therefore, we sequenced bacterial artificial chromosome clones (BACs) covering the S-locus region, including the core region from At4g21350 (PLANT U-BOX8 [PUB8]) to At4g21380 (ARK3), and several genes on either side, in two accessions of the selfing species Arabidopsis (Ath-Cvi) and C. rubella (Cru) and from four chromosomes of the outcrossing species A. lyrata (Aly-S16, Aly-S38, Aly-S50, and Aly-Sb). One additional S-locus haplotype each had already been sequenced and therefore was available from reference genome sequencing projects in Arabidopsis (Ath-Col; Arabidopsis Genome Initiative, 2000) and A. lyrata (Aly-Sa, from reference strain MN47; Hu et al., 2011). Our comparison of the eight S-locus haplotypes provides new insights into the evolution of the S locus in the Brassicaceae.
RESULTS
Genomic Organization and Gene Content of the S Locus
For the scale of phylogenetic sampling, we covered geographic (A. lyrata from Europe and North America) and mating system (SC species Arabidopsis and C. rubella, SI species A. lyrata) diversity. There was only one BAC library of C. rubella available when we started this work a few years ago; thus, only a single C. rubella haplotype was sequenced. Six BACs covering the core S-locus region, from At4g21350 (PUB8) to At4g21380 (ARK3), were sequenced: Arabidopsis (one S-locus haplotype), A. lyrata (four S-locus haplotypes), and C. rubella (one S-locus haplotype). For SRK and SCR, the published SRKa/SCRa or SRKb/SCRb (Kusaba et al., 2001) is the same S-locus haplotype as Aly-Sa or Aly-Sb, respectively. Only for Aly-Sb, we could not completely fill all assembly gaps. Nevertheless, the four large contigs of Aly-Sb contained both SCR and SRK in addition to the other flanking genes of the S locus. All six sequenced S-locus haplotypes share the region from At4g21326 to At4g21430, except Aly-S50, where the sequence from exon 1 to part of exon 6 of At4g21326 is missing (Fig. 1C). A phylogenetic tree (Supplemental Fig. S1) indicated that the recovered S-locus haplotypes span several S-locus lineages across Arabidopsis and Capsella relatives.
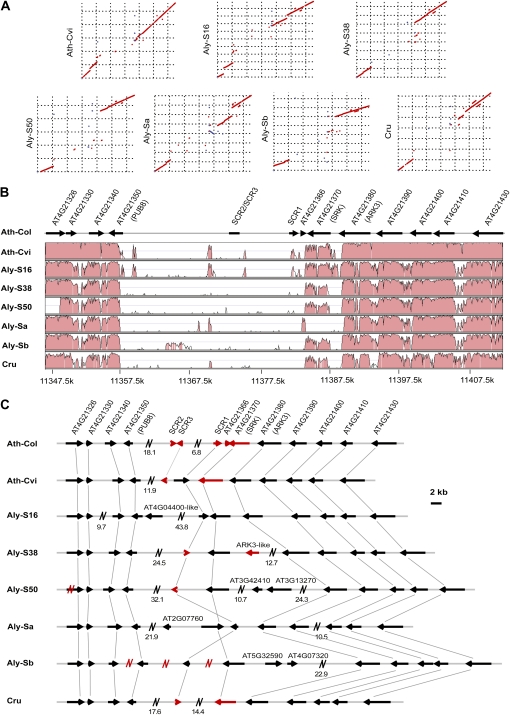
Figure 1.
Comparison of S-locus haplotypes. A, Dot plot highlighting sequence similarity and rearrangement between each S-locus haplotype and Ath-Col. B, A VISTA plot indicating similarity between Ath-Col and the other S-locus haplotypes. C, A comparison of gene content among different S-locus haplotypes. Red arrows indicate the SRK-, SCR-, and ARK3-related truncated sequences. Red N symbols represent gaps, and black N symbols represent large fragments not drawn to scale (actual length in kb is given below). [See online article for color version of this figure.]
Comparing the six sequenced S-locus haplotypes and the Arabidopsis and A. lyrata reference genome S-locus haplotypes, the length of the eight S-locus haplotypes (from At4g21326 to At4g21430) ranges from 56.1 kb (Ath-Cvi) to 116.5 kb (Aly-Sb; Supplemental Table S1). Sequences on either side of the core S-locus region are highly conserved and syntenic, while the core region is highly diverged (Fig. 1, A and B). Gene order and orientation are conserved (Fig. 1, B and C), except for Aly-Sa, which has an inversion including SCR and SRK (Fig. 1), as well as Aly-S50 and Aly-Sb, which have an inversion of the SCR gene (Fig. 1C). It is likely that in the majority of S-locus haplotypes, there is one SCR gene and one SRK gene in head-to-head orientation.
The 11 protein-coding genes found in the Arabidopsis reference S-locus haplotype Ath-Col are conserved in all S-locus haplotypes. Aly-S16, Aly-S50, Aly-Sa, and Aly-Sb, however, have one or two extra genes, which are not shared with any of the other S-locus haplotypes (Fig. 1C; Supplemental Table S1). In addition, there are some partially duplicated genes, including SCR-like sequences (SCR2 and SCR3) and an SRK-like gene (At4g21366) in Ath-Col, an SCR3-like fragment in Ath-Cvi, and an ARK3-like fragment in Aly-S38.
The content of transposable elements in different S-locus haplotypes varies greatly, from 5.45% in Ath-Cvi to 31.27% in Aly-Sb (Supplemental Tables S2 and S3). Arabidopsis S-locus haplotypes have the fewest transposable elements, while Cru is intermediate. The number and insertion times of long terminal repeat (LTR) retrotransposons also differ (Supplemental Table S2).
Transspecific Allele Sharing and Coevolution
In Ath-Col, Aly-S38, Aly-S50, and Cru (Fig. 2; Table I), SCR is represented by a truncated open reading frame (ORF), as inferred from deduced amino acid sequences. In Ath-Col, Ath-Cvi, and Cru, the SRK genes also have truncated ORFs (Kusaba et al., 2001; Tang et al., 2007; Shimizu et al., 2008; Guo et al., 2009). The amino acid sequences of the SCR gene are highly diverged, but the eight conserved Cys residues (Kusaba et al., 2001) were found in each S-locus haplotype with complete ORF (Fig. 2).
Figure 2.
Alignment of SCR-related sequences.
Table I. The SCR direction, intron length, and distance from SRK.
−1 indicates that only the first exon or part of it was found. NA indicates that the distance from SCR to SRK cannot be estimated for the gaps within Aly-Sb.
| BAC | Direction | Intron Length | Distance from SRK |
| bp | |||
| Ath-Col | + | 782 | 710 |
| Ath-Cvi | + | 2,036 | 227 |
| Aly-S16 | + | 1,044 | 255 |
| Aly-S38 | + | −1 | 3,812 |
| Aly-S50 | − | −1 | 5,249 |
| Aly-Sa | − | 1,519 | 1,751 |
| Aly-Sb | − | 78 | NA |
| Cru | + | −1 | 14,422 |
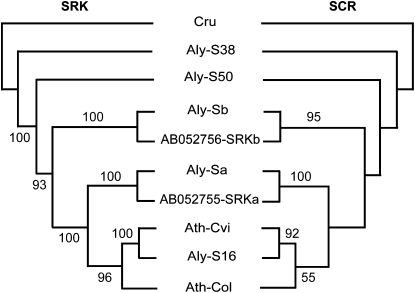
The topologies of the phylogenetic trees for SCR and SRK match. Ath-Cvi and Aly-S16, as well as Aly-S38 and Cru, show close relationships, supporting a pattern of coevolution for SCR and SRK (Fig. 3; Supplemental Figs. S1 and S2). None of the other genes at the S locus showed the same transspecies pattern (Supplemental Fig. S3), not even At4g21380 (ARK3), which is less than 3 kb away from the SRK gene in most S-locus haplotypes. Thus, linkage disequilibrium is apparently weak, and limited to a narrow region, from SCR to SRK.
Figure 3.
Matching phylogenetic trees of the SCR and SRK sequences.
The length of the single intron in SCR can be very different, even between closely related S-locus haplotypes such as Ath-Cvi (2,036 bp) and Aly-S16 (1,044 bp; Table I). The length of the intergenic region between SRK and SCR, for SCR with only the first exon, which was calculated based on the distance from the end of the first exon of SCR to SRK, is also highly variable, from 227 bp in Ath-Cvi to 14,422 bp in Cru (Table I).
Gene Conversion
There are frequent duplications in the core S-locus region, including duplications of SCR, SRK, and ARK3. SRK and ARK3 are closely related in sequence, and five potential gene conversion events were detected, with the size of the converted fragment ranging from 31 to 391 bp (Table II). Gene conversion also appears to have occurred between SRK and ARK3 in both Aly-Sb and Ath-Col as well as between SRK-like (At4g21366) and SRK and between SRK-like (At4g21366) and ARK3 in Ath-Col. Affected were exon 7 of SRK and ARK3, except for the gene conversion event between At4g21366 and ARK3 in Ath-Col, which included exons 6 and 7.
Table II. Gene conversions in SRK- and ARK3-related sequences.
| Species | Sequence 1 | Sequence 2 | Pa | Beginb | Endc | Lengthd |
| Arabidopsis | At4g21366 | SRK_Ath-Col | 0.0000 | 4,145 | 4,560 | 391 |
| ARK3_Ath-Col | At4g21366 | 0.0034 | 4,392 | 4,422 | 31 | |
| ARK3_Ath-Col | At4g21366 | 0.0450 | 4,516 | 4,546 | 31 | |
| ARK3_Ath-Col | SRK_Ath-Col | 0.0407 | 4,392 | 4,422 | 31 | |
| A. lyrata | ARK3_Aly-Sb | SRK_Aly-Sb | 0.0015 | 4,490 | 4,532 | 43 |
Simulated P values of the global inner fragments obtained from 10,000 permutations.
First nucleotide of the converted region.
Last nucleotide of the converted region.
Length of the converted region without alignment gaps.
Evidence for Positive Selection
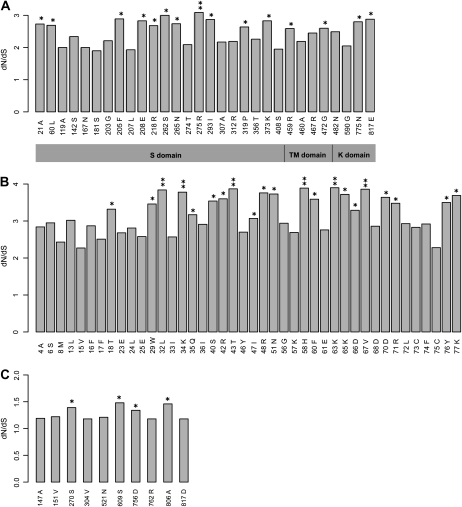
We tested whether the evolutionary divergence of the S locus has been affected by positive selection. Because pseudogenes confound such tests and show different evolutionary patterns than functional genes, we excluded them from our analyses. The ratio of nonsynonymous and synonymous mutation (dN/dS) averaged across all sites and lineages ranged from 0.11 (At4g21350) to 1.21 (SCR; Table III). For three genes, SCR, SRK, and ARK3, the positive selection model fit the data significantly better than the neutral model (Table III), although the SCR gene had a much higher proportion of codons under positive selection than SRK and ARK3 (Fig. 4).
Table III. Evidence for adaptive evolution of genes located in the S-locus region.
M0 is the dN/dS ratio averaged across all sites and lineages. M7 versus M8 indicates the outcome of the likelihood test. −1 means that the value of 2Δℓ (twice the log likelihood difference between two models) is 0 or less than 0.0001 and there is no significant difference between the two models. Estimates of parameters indicate the proportion of amino acids predicted to be under adaptive evolution (and their corresponding dN/dS ratio) from model M8. NA indicates not applicable because there is no significant difference between the positive selection model and the neutral model. Positively selected sites are amino acids under positive selection with Bayesian posterior P > 0.95 based on model M8 using the Bayes Empirical Bayes method. Ns is the number of sequences in the analysis. LA indicates the codon in the alignment. For the SRK gene, Aly-S16 is the reference sequence, and for the SCR gene, Ath-Cvi is the reference sequence. For ARK3 and other genes, Ath-Col is the reference. * P < 0.05, ** P < 0.001, *** P < 0.0001.
| Gene | M0 | M7 Versus M8 (2Δℓ) | Estimates of Parameters | Positively Selected Sites | Ns | LA |
| At4g21326 | 0.198 | −1 | NA | NA | 8 | 700 |
| At4g21330 | 0.217 | 3.894 | NA | NA | 8 | 210 |
| At4g21340 | 0.312 | 0.069 | NA | NA | 8 | 309 |
| At4g21350 (PUB8) | 0.106 | −1 | NA | NA | 8 | 374 |
| SCR | 1.210 | 7.896* | 44.2% (ω = 2.955) | 32L, 34K, 43T, 48R, 58H, 63K, 67V | 4 | 95 |
| At4g21370 (SRK) | 0.340 | 26.137*** | 4.3% (ω = 3.300) | 275R | 5 | 868 |
| At4g21380 (ARK3) | 0.198 | 11.882* | 0.3% (ω = 9.127) | None with P > 0.95 | 8 | 854 |
| At4g21390 | 0.113 | −1 | NA | NA | 8 | 851 |
| At4g21400 | 0.325 | −1 | NA | NA | 7 | 718 |
| At4g21410 | 0.185 | 0.195 | NA | NA | 7 | 680 |
| At4g21430 | 0.314 | 2.965 | NA | NA | 8 | 940 |
Figure 4.
Positive selection and variation in dN/dS among codons. A, SRK. B, SCR. C, ARK3. Single asterisks represent codons with a Bayesian posterior probability of positive selection greater than 75%, and double asterisks represent codons with a Bayesian posterior probability of positive selection greater than 95%.
DISCUSSION
Genomic Organization and Gene Content of the S Locus in the Arabidopsis/Capsella Lineage
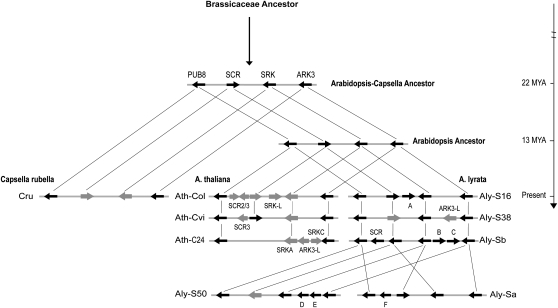
Although the core S-locus region between the PUB8 gene and ARK3 is very fluid and entails many independent insertions, duplications, and inversions, genes flanking the outermost PUB8 and ARK3 genes were found to be conserved with respect to gene orientation and order in the eight analyzed S-locus haplotypes. The more common head-to-head orientation of SCR and SRK, however, is changed in Aly-S50, Aly-Sb, and Ath-C24 (Fig. 5). Aly-Sa has an inversion including the SCR-SRK region, but the relative orientation is similar to the other S-locus haplotypes. We conclude that the ancestral organization of the S locus in the Arabidopsis/Capsella lineage appears to be one SCR gene and one SRK gene in head-to-head orientation, flanked by the PUB8 and ARK3 genes transcribed from the same strand as SRK (Fig. 5). The extra genes, which are not shared with any of the other S-locus haplotypes, must have originated independently in specific lineages.
Figure 5.
Proposed history of the S locus. Divergence times (million years ago [MYA]) on the right are calculated based on the synonymous substitutions among the ADH gene orthologs extracted from the reference genomes (Col-0 and MN47) and GenBank (AF110435; C. rubella), using the spontaneous mutation rate estimated in Arabidopsis (Ossowski et al., 2010). The divergence time between A. lyrata and Arabidopsis is consistent with a recent study using fossil evidence (Beilstein et al., 2010). Gray arrows indicate truncated ORFs. Letters A to F represent inserted genes (relative to Ath-Col): A, AT4G04400-like; B, AT5G32590-like; C, AT4G07320-like; D, AT3G42410-like; E, AT3G13270-like; F, AT2G07760-like.
Transspecies Polymorphism and Coevolution
Transspecies patterns of polymorphism are typical for plant SI systems (Sato et al., 2002; Bechsgaard et al., 2006; Paetsch et al., 2006), a fungal SI system (May et al., 1999), and the vertebrate animal major histocompatibility complex system (Adams et al., 2000). In the Brassica genus, some S-locus haplotypes have been estimated to be about 20 to 40 million years old (Uyenoyama, 1995). In the Solanaceae, the S-RNase, which is the female specificity determinant of SI in this family, has been estimated to be nearly 70 million years old (Xue et al., 1996). Some allelic lineages of the major histocompatibility complex system of animals appear to have persisted for at least 20 million years (Klein et al., 1998). Because the S-locus genes appear to be under selection, one cannot easily determine the age of the S-locus haplotype with a molecular clock. The close relationship of the Ath-Cvi and Aly-S16 haplotypes between Arabidopsis and A. lyrata as well as of the Aly-S38 and Cru haplotypes between A. lyrata and C. rubella (Paetsch et al., 2006) suggest that these S-locus haplotypes are at least 13 and 22 million years old, respectively (Fig. 5).
In the Brassicaceae, recombination events disrupting the linkage of matched SRK and SCR alleles are expected to result in the breakdown of SI. The pattern of coevolution between SRK and SCR suggests that they are in strong linkage disequilibrium, consistent with the previous population genetics study in A. lyrata (Kamau et al., 2007). In Brassica, the tree topology of SRK and SCR is basically similar (Sato et al., 2002; Edh et al., 2009). The structural and sequence heterogeneity between SCR and SRK sequences is likely the most important mechanism suppressing recombination and maintaining SI. This conclusion is strongly supported by results from Castric et al. (2010), who found that recombination could occur when individuals were homozygous for recessive alleles, indicating that there is no general suppression of recombination in this region. It is interesting that coevolution and linkage are limited to a really small region around SCR and SRK.
Duplication, Gene Conversion, Positive Selection, and Evolution of New Specificities
There are several hypotheses for the generation of new SI specificities (Uyenoyama et al., 2001; Chookajorn et al., 2004; Charlesworth et al., 2005). In one hypothesis (Uyenoyama et al., 2001), a self-compatible intermediate is generated in which partial breakdown of SI occurs because of a mutation in one of the SI specificity determinants. Subsequent compensatory mutations in the other specificity determinant then cause the pistil to reject the new pollen type, thus restoring the SI system. In the outcrossing species A. lyrata, many self-compatible accessions have been found in North America. SC, therefore, is likely to have originated multiple times (Mable et al., 2005; Mable and Adam, 2007; Mable, 2008; Foxe et al., 2010), and these self-compatible haplotypes might also be precursors of new SI haplotypes. Another model suggests that new SI specificities arise through gradual modification of SRK-SCR affinities (Chookajorn et al., 2004). We found that SCR appears to be under much stronger diversifying selection than SRK, consistent with a structure-function analysis of SCR that showed this protein to be unusually tolerant to sequence changes (Chookajorn et al., 2004), which suggests that SCR is the major engine for the modification of SRK-SCR affinities.
An additional factor might be gene conversion, which has been found in Brassica between SLG and SRK (Sato et al., 2002; Fujimoto et al., 2006) and between SRK and its possible paralogs in A. lyrata (Charlesworth et al., 2003). We have identified gene conversion events among SRK, ARK3, and duplicated SRK-like (At4g21366) genes. Interestingly, we found that gene conversion was largely restricted to the portion of the region encoding the kinase domain. Although the kinase domain of the SRK protein does not seem to directly contribute to the specificity of pollen/pistil recognition (Rea et al., 2010), it might indirectly influence the specificity of SRK by affecting protein conformation. In addition, we found that only three out of 11 genes (SCR, SRK, and ARK3) in the S-locus region show evidence of positive selection. Previous studies have found many sites in the S domain of SRK to be under positive selection in these species (Sainudiin et al., 2005; Castric and Vekemans, 2007). We also found that ARK3, which is the S-domain receptor-like kinase that is most closely related to SRK, has a history of gene duplication, gene conversion, and positive selection. These features of ARK3 could be a motor for the evolution of SRK, given the apparent gene conversion between ARK3 and SRK that we found here. In summary, there is a strong indication that duplication, gene conversion, and positive selection all contribute to the evolution of new S-locus specificities, as has been suggested previously.
Relationship between the S Locus and Selfing
One hypothesis for the labile nature of SI is that the transition from outcrossing to selfing can provide a selective advantage at a given time and place (Stebbins, 1957; Busch et al., 2010; Goldberg et al., 2010; Wright and Barrett, 2010). A single inactivated S-locus haplotype was fixed in C. rubella, representing the key step in the recent transition to selfing for this species (Guo et al., 2009). In contrast, SC has apparently originated several times independently in Arabidopsis (Bechsgaard et al., 2006; Sherman-Broyles et al., 2007; Tang et al., 2007; Shimizu et al., 2008; Boggs et al., 2009b; Tsuchimatsu et al., 2010). In accessions with a Col-0-like SRK allele, SCR became pseudogenized after the gene was disrupted by an inversion (Boggs et al., 2009b; Tsuchimatsu et al., 2010). In the accession Cvi-0, SCR is intact but SRK has a truncated ORF (Tang et al., 2007; Shimizu et al., 2008; this work). Both SCR and SRK have truncated ORFs in Col-0 (Kusaba et al., 2001). C24, which also has a truncated SRK ORF, lacks SCR altogether (Sherman-Broyles et al., 2007). In C. rubella, the ORFs of all SCR alleles and some SRK alleles are truncated (Guo et al., 2009). Interestingly, one A. lyrata haplotype, Aly-S38, is very similar to C. rubella, with a closely related SCR with a truncated ORF and an SRK with a complete ORF (Fig. 3; Supplemental Fig. S1). This indicates that the truncated S-locus haplotype in C. rubella could be very old, despite it having been fixed only very recently. Although the simultaneous maintenance of both nonfunctional and functional S-locus haplotypes is rare (Uyenoyama et al., 2001; Porcher and Lande, 2005), two of the S-locus haplotypes we recovered for A. lyrata appear to contain a SCR allele with a truncated ORF, which spans two different S-locus lineages across Arabidopsis and Capsella relatives (Supplemental Fig. S1). It will be interesting to investigate the activity of those S-locus haplotypes with truncated ORFs (such as Aly-S38 and Aly-S50), given that the SCR gene of Cvi may not be functional even though it has a complete ORF (Boggs et al., 2009a).
Unfortunately, the sequences of Aly-S38 and Aly-S50 come from a BAC library generated a decade ago, and the individual plants with these haplotypes are no longer available. Because of the great diversity of S-locus haplotypes, finding the same haplotypes in extant samples would be very difficult, if not impossible. On the other hand, the distinction of functional/nonfunctional S-locus haplotypes was not a main focus of our study. The strength of our work is that it is, to our knowledge, the first sequencing study of the S-locus region in such a large number of Arabidopsis relatives. With additional high-quality sequences of the S-locus region from A. lyrata and other related species, one can address the mechanisms of the evolution of the S-locus region, determine whether S-locus haplotypes with truncated ORFs are functional or not, and also address the question of the age of inactive S-locus haplotypes with more confidence.
CONCLUSION
Our results are consistent with previous studies of the S locus in that we found that the two SI-determining genes, SCR and SRK, show patterns of coevolution. Duplication, gene conversion, and positive selection have been important factors in the evolutionary history of these two genes and appear to contribute to the generation of new recognition specificities. An unexpected result is that the inactive pseudo-S-locus haplotype in the SC species C. rubella might be an old S-locus haplotype that only very recently became fixed when C. rubella split off from its SI ancestor, Capsella grandiflora. Finally, our data provide not only an important resource for understanding S-locus evolution but can also guide future functional studies of the SI system.
MATERIALS AND METHODS
BAC Screen, Sequencing, and Assembly
We generated a 400-bp probe derived from PUB8 (At4g21350) by PCR amplification of Arabidopsis (Arabidopsis thaliana) Col-0 using primers G10720 (5′-TTAATCTCCACTCTCATCTCTC-3′) and G10721 (5′-CTGATTCCTTTCTCTCCCTATC-3′) and used it to screen BAC libraries of Arabidopsis Cvi-0, Arabidopsis lyrata, and Capsella rubella by filter hybridization. The A. lyrata individuals used in constructing the BAC library were from the Pech area in Bavaria (Germany), and the C. rubella individuals were from Gargano, Italy. End sequencing was used to map the borders of positive BAC clones, and clones that include the region from At4g21326 to At4g21430 were analyzed by shotgun sequencing.
For Arabidopsis Cvi-0, BAC35E6 (Ath-Cvi) was analyzed. For the outcrossing A. lyrata, we first sequenced part of the SRK locus (using G12957 primer; 5′-CTTCGAGCTTGGTTTCTTCA-3′) to differentiate different S-locus haplotypes. We then used these SRK sequences and other available SRK sequences from GenBank to construct a phylogenetic tree for the different S-locus haplotypes. Among 16 positive BAC clones, there were three distinct S-locus haplotypes (named after the similarity to well-known SRK sequences of A. lyrata) besides Aly-Sb, located on the SRKb BAC of A. lyrata reported by Kusaba and colleagues (2001), hereafter represented as Aly-Sb, and Aly-Sa from the reference strain MN47 (Supplemental Fig. S1). Both Aly-Sa and Aly-Sb are derived from accessions of A. lyrata collected in Michigan (Kusaba et al., 2001). In the end, four independent A. lyrata BAC clones, BAC6K22 (Aly-S16), BAC39O17 (Aly-S38), BAC27M5 (Aly-S50), and Aly-Sb (Kusaba et al., 2001), were selected for sequencing. From the C. rubella library, we selected BAC15P16 (Cru).
BAC DNA was prepared using the Large Construct DNA Preparation Kit (Qiagen), physically sheared, and shotgun libraries were constructed using the TOPO Shotgun Subcloning Kit (Invitrogen). Shotgun clones were sequenced on an ABI 3730XL DNA Analyzer using T7 and SP6 primers. Sequences were processed using Phred for base calling, CROSS_MATCH for removing vector sequences, and Phrap and Consed for assembly (Ewing and Green, 1998; Ewing et al., 1998; Gordon et al., 1998). Primer walking and PCR were used to fill or polish gaps and ambiguous regions. The sequences and the assembled contigs were in accordance with the Bermuda standards of sequencing (http://www.genome.gov/10001812). The S-locus region of Col-0 was extracted from the Arabidopsis reference genome (http://www.arabidopsis.org/), and the S-locus region of the MN47 strain of A. lyrata was extracted from the reference genome as well (http://genome.jgi-psf.org/Araly1/Araly1.home.html).
Sequence Annotation and Phylogenetic Analysis
Annotation of the BAC sequences was performed using FGENESH (http://linux1.softberry.com/berry.phtml). The annotation was confirmed by comparison with Arabidopsis (The Arabidopsis Information Resource 8). The SCR gene was annotated by comparing it with SCR1, SCR2, and SCR3 of Arabidopsis (Col-0), SCRa (AB052753) and SCRb (AB052754) of A. lyrata, SP11-6 (AF195625) of Brassica oleracea, and SP11-9 of Brassica rapa (AB022078). Spidey (http://www.ncbi.nlm.nih.gov/spidey/) was used to confirm the annotation based on the alignment to Arabidopsis mRNA when needed. VISTA (http://genome.lbl.gov/vista/index.shtml) and Mummer version 3.19 (Kurtz et al., 2004) were used to compare and view BAC sequences.
Sequences were aligned using ClustalX version 1.81 (Thompson et al., 1997) and then refined manually. PAUP* version 4.0b10 (Swofford, 2003) was used to reconstruct the phylogenetic tree. For the maximum parsimony tree, a heuristic search was performed with the MULPARS option, tree bisection-reconnection branch swapping. The neighbor-joining trees were constructed using the two-parameter model (Kimura, 1980; Saitou and Nei, 1987). Topological robustness was assessed by bootstrapping with 1,000 replicates (Felsenstein, 1985).
Transposable Element Analysis
RepeatMasker version 3.2.5 (http://www.repeatmasker.org/) was used to detect the content of transposable elements using the Arabidopsis library of Repbase (RM database version 20080611). LTR_STRUC (McCarthy and McDonald, 2003) was used to identify full-length LTR retrotransposons. Each LTR retrotransposon was classified (gypsy-like, copia-like, others) based on BLASTN and T-BLAST searches of the Brassicaceae repeat database (http://tigrblast.tigr.org/euk-blast/index.cgi?project=plant.repeats). The sequence divergence between the two LTRs of each intact LTR retrotransposon was used to estimate the insertion time (T) using the formula T = K/(2)(r), where K is the number of substitutions per site between two LTRs, estimated with the two-parameter model using the distmat program implemented in the EMBOSS package (http://emboss.sourceforge.net/), and r is the spontaneous mutation rate estimated in Arabidopsis, 7 × 10−9 mutations per site per generation (year; Ossowski et al., 2010).
Detection of Gene Conversion
Gene conversion was analyzed using GENECONV version 1.81a (Sawyer, 1989) with default settings. Based on an alignment of DNA sequences, GENECONV detects gene conversion by looking for sufficiently similarly aligned segments between a pair of sequences. A global value of P ≤ 0.05 was used as a criterion for further analysis of the conversion tracts detected by GENECONV. Given that GENECONV could not distinguish between gene conversion and unequal crossing over, significant GENECONV results are indicated as conversion events or tracts (Mondragon-Palomino and Gaut, 2005).
Positive Selection Detection Using Likelihood Ratio Tests
Potentially positive selection was analyzed with the program codeml of PAML version 3.15 (Yang, 1997). The first step was to find sites with ω (dN/dS) > 1 by comparing a null model (M7) that does not allow for sites with ω > 1 and a general model that does (M8) by performing a likelihood ratio test using codeml. If model M7 was rejected in favor of model M8, we took this as evidence against the ω ratio being confined to the interval (0, 1) for all sites. The second step was to use a likelihood ratio test to identify residues (with Bayesian posterior probability) under positive selection with the Bayes Empirical Bayes tool (Yang et al., 2005). We assumed that sites with ω > 1 and a high probability (P ≥ 95%) were likely to be under positive selection.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers HQ376928-HQ376932 and EF637083.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Selection of BAC clones for shotgun sequencing based on the phylogeny of SRK using the most parsimonious method.
Supplemental Figure S2. A single most parsimonious tree based on SCR gene amino acid sequences.
Supplemental Figure S3. Phylogenetic trees using the neighbor-joining method with two-parameter model based on the coding regions of nine protein-coding genes flanking the S locus, from At4g21326 to At4g21430, and the SRK gene to detect transspecies polymorphism.
Supplemental Table S1. BAC information and gene density.
Supplemental Table S2. Repeat content and full-length LTR retrotransposon content and insertion times.
Supplemental Table S3. Transposable element content in S-locus haplotypes.
Acknowledgments
We thank June B. Nasrallah for giving the A. lyrata Aly-Sb BAC clone; Maarten Koornneef for providing access to the Cvi-0 BAC library; Norman Warthmann, Kirsten Bomblies, Stefan Henz, and Namita Tripathi for technical advice; Stanley A. Sawyer for helpful suggestions on the usage of GENECONV for gene conversion analysis; Beth Rowan for greatly improving the manuscript; and Carmen Melchers for proofreading.
References
- Adams EJ, Cooper S, Thomson G, Parham P. (2000) Common chimpanzees have greater diversity than humans at two of the three highly polymorphic MHC class I genes. Immunogenetics 51: 410–424 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Barrett SC. (2002) The evolution of plant sexual diversity. Nat Rev Genet 3: 274–284 [DOI] [PubMed] [Google Scholar]
- Barrett SC. (2010) Understanding plant reproductive diversity. Philos Trans R Soc Lond B Biol Sci 365: 99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechsgaard JS, Castric V, Charlesworth D, Vekemans X, Schierup MH. (2006) The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol Biol Evol 23: 1741–1750 [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. (2010) Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 18724–18728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs NA, Dwyer KG, Shah P, McCulloch AA, Bechsgaard J, Schierup MH, Nasrallah ME, Nasrallah JB. (2009a) Expression of distinct self-incompatibility specificities in Arabidopsis thaliana. Genetics 182: 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs NA, Nasrallah JB, Nasrallah ME. (2009b) Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PLoS Genet 5: e1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch JW, Joly S, Schoen DJ. (2010) Does mate limitation in self-incompatible species promote the evolution of selfing? The case of Leavenworthia alabamica. Evolution 64: 1657–1670 [DOI] [PubMed] [Google Scholar]
- Busch JW, Schoen DJ. (2008) The evolution of self-incompatibility when mates are limiting. Trends Plant Sci 13: 128–136 [DOI] [PubMed] [Google Scholar]
- Busch JW, Sharma J, Schoen DJ. (2008) Molecular characterization of Lal2, an SRK-like gene linked to the S-locus in the wild mustard Leavenworthia alabamica. Genetics 178: 2055–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric V, Bechsgaard JS, Grenier S, Noureddine R, Schierup MH, Vekemans X. (2010) Molecular evolution within and between self-incompatibility specificities. Mol Biol Evol 27: 11–20 [DOI] [PubMed] [Google Scholar]
- Castric V, Vekemans X. (2007) Evolution under strong balancing selection: how many codons determine specificity at the female self-incompatibility gene SRK in Brassicaceae? BMC Evol Biol 7: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. (2006) Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet 2: e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Bartolomé C, Schierup MH, Mable BK. (2003) Haplotype structure of the stigmatic self-incompatibility gene in natural populations of Arabidopsis lyrata. Mol Biol Evol 20: 1741–1753 [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Vekemans X, Castric V, Glémin S. (2005) Plant self-incompatibility systems: a molecular evolutionary perspective. New Phytol 168: 61–69 [DOI] [PubMed] [Google Scholar]
- Chookajorn T, Kachroo A, Ripoll DR, Clark AG, Nasrallah JB. (2004) Specificity determinants and diversification of the Brassica self-incompatibility pollen ligand. Proc Natl Acad Sci USA 101: 911–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edh K, Widén B, Ceplitis A. (2009) The evolution and diversification of S-locus haplotypes in the Brassicaceae family. Genetics 181: 977–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194 [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Fobis-Loisy I, Miege C, Gaude T. (2004) Molecular evolution of the S locus controlling mating in the Brassicaceae. Plant Biol (Stuttg) 6: 109–118 [DOI] [PubMed] [Google Scholar]
- Foxe JP, Stift M, Tedder A, Haudry A, Wright SI, Mable BK. (2010) Reconstructing origins of loss of self-incompatibility and selfing in North American Arabidopsis lyrata: a population genetic context. Evolution 64: 3495–3510 [DOI] [PubMed] [Google Scholar]
- Fujimoto R, Sugimura T, Nishio T. (2006) Gene conversion from SLG to SRK resulting in self-compatibility in Brassica rapa. FEBS Lett 580: 425–430 [DOI] [PubMed] [Google Scholar]
- Goldberg EE, Kohn JR, Lande R, Robertson KA, Smith SA, Igić B. (2010) Species selection maintains self-incompatibility. Science 330: 493–495 [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. (1998) Consed: a graphical tool for sequence finishing. Genome Res 8: 195–202 [DOI] [PubMed] [Google Scholar]
- Guo YL, Bechsgaard JS, Slotte T, Neuffer B, Lascoux M, Weigel D, Schierup MH. (2009) Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc Natl Acad Sci USA 106: 5246–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenblad J, Bechsgaard J, Charlesworth D. (2006) Linkage disequilibrium between incompatibility locus region genes in the plant Arabidopsis lyrata. Genetics 173: 1057–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A, Nasrallah ME, Nasrallah JB. (2002) Self-incompatibility in the Brassicaceae: receptor-ligand signaling and cell-to-cell communication. Plant Cell (Suppl) 14: S227–S238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamau E, Charlesworth B, Charlesworth D. (2007) Linkage disequilibrium and recombination rate estimates in the self-incompatibility region of Arabidopsis lyrata. Genetics 176: 2357–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamau E, Charlesworth D. (2005) Balancing selection and low recombination affect diversity near the self-incompatibility loci of the plant Arabidopsis lyrata. Curr Biol 15: 1773–1778 [DOI] [PubMed] [Google Scholar]
- Kimura M. (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120 [DOI] [PubMed] [Google Scholar]
- Klein J, Sato A, Nagl S, O'hUigin C. (1998) Molecular trans-species polymorphism. Annu Rev Ecol Syst 29: 1–21 [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. (2004) Versatile and open software for comparing large genomes. Genome Biol 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Dwyer K, Hendershot J, Vrebalov J, Nasrallah JB, Nasrallah ME. (2001) Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13: 627–643 [PMC free article] [PubMed] [Google Scholar]
- Leducq JB, Llaurens V, Castric V, Saumitou-Laprade P, Hardy OJ, Vekemans X. (2011) Effect of balancing selection on spatial genetic structure within populations: theoretical investigations on the self-incompatibility locus and empirical studies in Arabidopsis halleri. Heredity 106: 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Sherman-Broyles S, Nasrallah ME, Nasrallah JB. (2007) A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr Biol 17: 734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery J, Bürger R. (1995) Mutation accumulation and the extinction of small populations. Am Nat 146: 489–518 [Google Scholar]
- Mable BK. (2008) Genetic causes and consequences of the breakdown of self-incompatibility: case studies in the Brassicaceae. Genet Res (Camb) 90: 47–60 [DOI] [PubMed] [Google Scholar]
- Mable BK, Adam A. (2007) Patterns of genetic diversity in outcrossing and selfing populations of Arabidopsis lyrata. Mol Ecol 16: 3565–3580 [DOI] [PubMed] [Google Scholar]
- Mable BK, Robertson AV, Dart S, Di Berardo C, Witham L. (2005) Breakdown of self-incompatibility in the perennial Arabidopsis lyrata (Brassicaceae) and its genetic consequences. Evolution 59: 1437–1448 [PubMed] [Google Scholar]
- May G, Shaw F, Badrane H, Vekemans X. (1999) The signature of balancing selection: fungal mating compatibility gene evolution. Proc Natl Acad Sci USA 96: 9172–9177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy EM, McDonald JF. (2003) LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics 19: 362–367 [DOI] [PubMed] [Google Scholar]
- Mondragon-Palomino M, Gaut BS. (2005) Gene conversion and the evolution of three leucine-rich repeat gene families in Arabidopsis thaliana. Mol Biol Evol 22: 2444–2456 [DOI] [PubMed] [Google Scholar]
- Nasrallah JB. (2002) Recognition and rejection of self in plant reproduction. Science 296: 305–308 [DOI] [PubMed] [Google Scholar]
- Nasrallah ME, Liu P, Nasrallah JB. (2002) Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science 297: 247–249 [DOI] [PubMed] [Google Scholar]
- Nasrallah ME, Liu P, Sherman-Broyles S, Boggs NA, Nasrallah JB. (2004) Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proc Natl Acad Sci USA 101: 16070–16074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbigin E, Uyenoyama MK. (2005) The evolutionary dynamics of self-incompatibility systems. Trends Genet 21: 500–505 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Lucas-Lledó JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M. (2010) The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetsch M, Mayland-Quellhorst S, Neuffer B. (2006) Evolution of the self-incompatibility system in the Brassicaceae: identification of S-locus receptor kinase (SRK) in self-incompatible Capsella grandiflora. Heredity 97: 283–290 [DOI] [PubMed] [Google Scholar]
- Porcher E, Lande R. (2005) Loss of gametophytic self-incompatibility with evolution of inbreeding depression. Evolution 59: 46–60 [PubMed] [Google Scholar]
- Prigoda NL, Nassuth A, Mable BK. (2005) Phenotypic and genotypic expression of self-incompatibility haplotypes in Arabidopsis lyrata suggests unique origin of alleles in different dominance classes. Mol Biol Evol 22: 1609–1620 [DOI] [PubMed] [Google Scholar]
- Rea AC, Liu P, Nasrallah JB. (2010) A transgenic self-incompatible Arabidopsis thaliana model for evolutionary and mechanistic studies of crucifer self-incompatibility. J Exp Bot 61: 1897–1906 [DOI] [PubMed] [Google Scholar]
- Ruggiero MV, Jacquemin B, Castric V, Vekemans X. (2008) Hitch-hiking to a locus under balancing selection: high sequence diversity and low population subdivision at the S-locus genomic region in Arabidopsis halleri. Genet Res (Camb) 90: 37–46 [DOI] [PubMed] [Google Scholar]
- Sainudiin R, Wong WS, Yogeeswaran K, Nasrallah JB, Yang Z, Nielsen R. (2005) Detecting site-specific physicochemical selective pressures: applications to the class I HLA of the human major histocompatibility complex and the SRK of the plant sporophytic self-incompatibility system. J Mol Evol 60: 315–326 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sato K, Nishio T, Kimura R, Kusaba M, Suzuki T, Hatakeyama K, Ockendon DJ, Satta Y. (2002) Coevolution of the S-locus genes SRK, SLG and SP11/SCR in Brassica oleracea and B. rapa. Genetics 162: 931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S. (1989) Statistical tests for detecting gene conversion. Mol Biol Evol 6: 526–538 [DOI] [PubMed] [Google Scholar]
- Schierup MH, Bechsgaard JS, Nielsen LH, Christiansen FB. (2006) Selection at work in self-incompatible Arabidopsis lyrata: mating patterns in a natural population. Genetics 172: 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup MH, Mable BK, Awadalla P, Charlesworth D. (2001) Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics 158: 387–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup MH, Vekemans X. (2008) Genomic consequences of selection on self-incompatibility genes. Curr Opin Plant Biol 11: 116–122 [DOI] [PubMed] [Google Scholar]
- Sherman-Broyles S, Boggs N, Farkas A, Liu P, Vrebalov J, Nasrallah ME, Nasrallah JB. (2007) S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell 19: 94–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu KK, Shimizu-Inatsugi R, Tsuchimatsu T, Purugganan MD. (2008) Independent origins of self-compatibility in Arabidopsis thaliana. Mol Ecol 17: 704–714 [DOI] [PubMed] [Google Scholar]
- Stebbins GL. (1957) Self-fertilization and population variability in the higher plants. Am Nat 41: 337–354 [Google Scholar]
- Swofford DL. (2003) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- Takebayashi N, Brewer PB, Newbigin E, Uyenoyama MK. (2003) Patterns of variation within self-incompatibility loci. Mol Biol Evol 20: 1778–1794 [DOI] [PubMed] [Google Scholar]
- Tang C, Toomajian C, Sherman-Broyles S, Plagnol V, Guo YL, Hu TT, Clark RM, Nasrallah JB, Weigel D, Nordborg M. (2007) The evolution of selfing in Arabidopsis thaliana. Science 317: 1070–1072 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimatsu T, Suwabe K, Shimizu-Inatsugi R, Isokawa S, Pavlidis P, Städler T, Suzuki G, Takayama S, Watanabe M, Shimizu KK. (2010) Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature 464: 1342–1346 [DOI] [PubMed] [Google Scholar]
- Uyenoyama MK. (1995) A generalized least-squares estimate for the origin of sporophytic self-incompatibility. Genetics 139: 975–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama MK. (2000) Evolutionary dynamics of self-incompatibility alleles in Brassica. Genetics 156: 351–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama MK. (2005) Evolution under tight linkage to mating type. New Phytol 165: 63–70 [DOI] [PubMed] [Google Scholar]
- Uyenoyama MK, Zhang Y, Newbigin E. (2001) On the origin of self-incompatibility haplotypes: transition through self-compatible intermediates. Genetics 157: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Barrett SC. (2010) Evolution: the long-term benefits of self-rejection. Science 330: 459–460 [DOI] [PubMed] [Google Scholar]
- Xue Y, Carpenter R, Dickinson HG, Coen ES. (1996) Origin of allelic diversity in Antirrhinum S locus RNases. Plant Cell 8: 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13: 555–556 [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. (2005) Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22: 1107–1118 [DOI] [PubMed] [Google Scholar]