The three fission yeast formins (Cdc12, For3, and Fus1) all nucleate actin assembly and remain continuously associated with the elongating actin filament barbed end, while incorporating thousands of actin monomers before dissociating. However, the specific rates for these reactions vary significantly and may therefore be functionally important.
Abstract
Fission yeast expresses three formins required for distinct actin cytoskeletal processes: Cdc12 (cytokinesis), For3 (polarization), and Fus1 (mating). We propose that in addition to differential regulation, key actin-assembly properties tailor formins for a particular role. In direct comparison to the well-studied Cdc12, we report the first in vitro characterization of the actin-assembly properties of For3 and Fus1. All three share fundamental formin activities; however, particular reaction rates vary significantly. Cdc12 is an efficient nucleator (one filament per approximately 3 Cdc12 dimers) that processively elongates profilin-actin at a moderate rate of 10 subunits s−1 μM−1, but lacks filament-bundling activity. Fus1 is also an efficient nucleator, yet processively elongates profilin-actin at one-half the rate of and dissociates 10-fold more rapidly than Cdc12; it also bundles filaments. For3 nucleates filaments 100-fold less well than Fus1, but like Cdc12, processively elongates profilin-actin at a moderate rate and lacks filament-bundling activity. Additionally, both the formin homology FH1 and FH2 domains contribute to the overall rate of profilin-actin elongation. We also confirmed the physiological importance of the actin-assembly activity of the fission yeast formins. Point mutants that disrupt their ability to stimulate actin assembly in vitro do not function properly in vivo.
INTRODUCTION
Formins are large multidomain cellular machines that employ a novel mechanism to assemble unbranched actin filaments from profilin-actin for fundamental eukaryotic cellular processes such as division, motility, polarization, morphology, and adhesion (Goode and Eck, 2007; Kovar et al., 2010). Formins contain two formin homology (FH) domains that nucleate and processively elongate actin filaments (Paul and Pollard, 2009b), which are flanked by less well-conserved regulatory regions. The FH2 domain forms a tethered head-to-tail dimer that nucleates actin assembly and then encircles and processively translocates toward the growing filament barbed end (Xu et al., 2004; Otomo et al., 2005b). The unstructured FH1 domain contains a variable number of proline-rich tracks that bind and deliver profilin-actin to the FH2-associated barbed end for elongation (Romero et al., 2004; Kovar, 2006; Kovar et al., 2010; Goode and Eck, 2007; Paul and Pollard, 2009b). Most organisms express multiple formin isoforms, from several in fungi to ∼15 in mammals and over 20 in plants (Higgs and Peterson, 2005; Rivero et al., 2005; Schonichen and Geyer, 2010), although the cellular roles of most isoforms have not been determined. Most studied formins have significantly different in vitro nucleation, elongation, and dissociation rates, as well as properties such as filament bundling and severing (Kovar, 2006; Kovar et al., 2010; Goode and Eck, 2007). A gratifying hypothesis is that both the regulatory and actin-assembly properties of different formin isoforms tailor each for a particular cellular process. However, with the exception of the two budding yeast formins Bni1 and Bnr1 (Pruyne et al., 2002; Sagot et al., 2002; Pring et al., 2003; Kovar and Pollard, 2004; Moseley et al., 2004; Moseley and Goode, 2005), the actin-assembly properties of all formins expressed by an individual organism have not been determined.
The fission yeast Schizosaccharomyces pombe is an ideal model to study how formins are tailored for specific cellular processes, because it expresses three isoforms with distinct roles: Cdc12 (cytokinesis), For3 (polarization), and Fus1 (mating; Chang et al., 1997; Petersen et al., 1998b; Feierbach and Chang, 2001; Nakano et al., 2002; Kovar et al., 2011). Multiple investigations have utilized genetic and cell biology approaches, such as time-lapse fluorescence microscopy, and mathematical theory to elucidate each formin's role in these processes (Petersen et al., 1995, 1998a, 1998b; Chang et al., 1997; Feierbach and Chang, 2001; Nakano et al., 2002; Martin and Chang, 2006; Wu et al., 2006; Martin et al., 2007; Vavylonis et al., 2008; Wang and Vavylonis, 2008; Yonetani et al., 2008; Yonetani and Chang, 2010; Coffman et al., 2009; Laporte et al., 2011). However, the in vitro actin-assembly properties have only been determined for the contractile ring formin Cdc12 (Kovar et al., 2003, 2006; Kovar and Pollard, 2004; Neidt et al., 2008, 2009; Yonetani et al., 2008). Therefore the actin-assembly properties of the actin cable formin For3 and mating formin Fus1 have been assumed rather than rigorously evaluated, which renders our mechanistic understanding of formin-mediated actin assembly for diverse cellular processes in fission yeast incomplete.
By directly comparing For3 and Fus1 to the well-characterized Cdc12, we show the first in vitro biochemical characterization of the actin-assembly abilities of these two isoforms. Like most other formins, all three stimulate nucleation, bind preassembled filament barbed ends tightly, remain processively associated with the elongating barbed end in both the absence and presence of profilin, add thousands of subunits before dissociating, and accelerate the rate of profilin-actin addition. However, particular rates vary significantly, suggesting that specific actin-assembly properties are functionally important. Cdc12 is an efficient nucleator that elongates profilin-actin at ∼10 subunits s−1 μM−1, dissociates slowly, and cannot bundle filaments. Fus1 is an efficient nucleator that elongates profilin-actin at only ∼5 subunits s−1 μM−1, dissociates 10-fold faster than Cdc12, and can bundle filaments. For3 is an extremely poor nucleator that, like Cdc12, elongates profilin-actin at ∼10 subunits s−1 μM−1 and dissociates slowly, and cannot bundle filaments. We also show that both the FH1 and FH2 domains contribute to the overall elongation rates of profilin-actin. Furthermore, the severity by which point mutations in the FH2 domain disrupts their ability to stimulate actin assembly in vitro correlates with the inability of all three formins to function in vivo.
RESULTS
The fission yeast formins stimulate actin assembly by different efficiencies
Our hypothesis is that although generally similar, the functionally diverse fission yeast formins Cdc12 (cytokinesis), Fus1 (mating), and For3 (polarization) have specific actin-assembly characteristics that are tailored for their particular cellular role. To elucidate and compare their actin-assembly properties, we expressed and purified from bacteria active fragment constructs containing the FH1 and FH2 domains (Figure 1A). We also examined the actin-assembly properties of the well-characterized budding yeast formin Bni1 as a comparative control (Pruyne et al., 2002; Sagot et al., 2002; Pring et al., 2003; Kovar and Pollard, 2004; Kovar et al., 2006 ; Moseley et al., 2004; Moseley and Goode, 2005; Paul and Pollard, 2008, 2009a).
FIGURE 1:
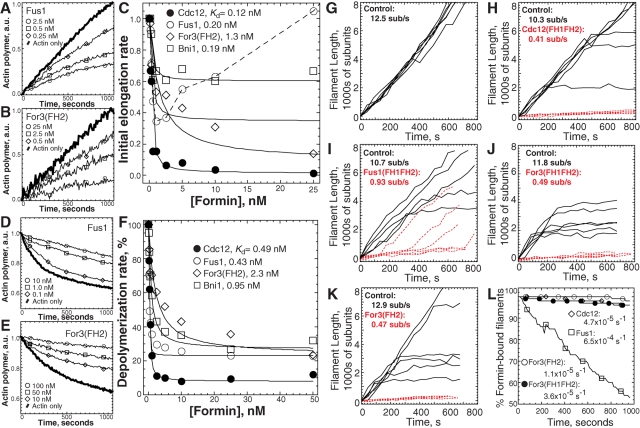
The fission yeast formins stimulate actin filament assembly with different nucleation efficiencies. (A) Domain organization of the three fission yeast formins Cdc12, Fus1, and For3, and budding yeast formin Bni1. Amino acid residues delineating FH1 and FH2 domain constructs are shown to the right. Each “P” signifies a putative profilin binding site, which is a proline-rich segment of 6–14 residues. (B to F) Spontaneous assembly of 2.5 μM Mg-ATP actin monomers (20% pyrene-labeled). (B and C) Time course of actin assembly in the absence (thick curve) or presence of the indicated concentrations of Fus1(FH1FH2), For3(FH1FH2), and For3(FH2). (D) Normalized plot of the dependence of actin-assembly rate (slope) on concentration of Cdc12(FH1FH2) (□), Fus1(FH1FH2) (▵), For3(FH1FH2) (•), For3(FH2) (◯), and Bni1(FH1FH2) (⋄). The inset shows For3(FH2) (◯) with an expanded x-axis. Dashed lines indicate the zero coordinate on the y-axis. (E) Nucleation efficiency: optimal number of formin dimers required to nucleate one filament. (F) Representative fluorescence micrographs of rhodamine-phalloidin–labeled actin filaments after spontaneous actin-assembly reactions reached plateau (2 h) in the absence (actin only) or presence of the indicated formin. Average filament lengths are reported in μm ± SD. Scale bar: 10 μm.
As expected the fission yeast formins promote actin monomer assembly, although their nucleation efficiencies vary significantly (Figure 1, B–F). Cdc12(FH1FH2) and Fus1(FH1FH2), as well as budding yeast Bni1(FH1FH2), stimulate actin polymerization by eliminating the lag phase (nucleation) and increasing the maximum rate (slope) of assembly over all formin concentrations (Figure 1, B and D). Curiously, For3(FH1FH2) has a biphasic effect on the spontaneous assembly rate (Figure 1, C and D). Low For3(FH1FH2) concentrations up to ∼50–100 nM increasingly inhibit actin assembly, whereas progressively higher concentrations stimulate actin assembly above the rate of actin alone. For3(FH2), which lacks the FH1 domain, behaves identically to For3(FH1FH2), with a biphasic effect that maximally promotes actin assembly at concentrations above 2 μM (Figure 1, C and D). Given the identical behavior of For3(FH1FH2) and For3(FH2), we primarily focused on the characterization of For3(FH2) in the absence of profilin.
Since the length of filaments at the end of the reaction is inversely proportional to the number of filaments produced, we labeled filaments with rhodamine-phalloidin to compare relative formin nucleation efficiencies (Figure 1F). Control filaments without formin average 15.7 μm. Filaments produced by 50 nM Cdc12 or Fus1 (0.9–1.4 μm) are approximately five times shorter than filaments formed by 50 nM Bni1 (5.2 μm). For3(FH1FH2) and For3(FH2) require 10-fold more molecules (≥500 nM) to produce filaments 0.7 to 1.8 μm long. Given that the overall “bulk” spontaneous assembly rate is due to both the number of filaments and how fast they grow, we quantified the nucleation efficiency of each formin by accounting for the elongation rate (determined by total internal reflection fluorescence [TIRF] microscopy; see Figure 2). Cdc12 and Fus1 maximally produce one new filament per approximately three and two dimers, respectively (Figure 1E), whereas Bni1 produces one new filament per ∼26 dimers. For3 is a significantly less efficient nucleator, maximally producing one new filament per ∼172 dimers. Therefore the relative nucleation efficiency is Cdc12 = Fus1 > Bni1 > For3.
FIGURE 2:
The fission yeast formins bind to and reduce actin filament barbed-end dynamics by different amounts. (A to C) Seeded elongation: addition of 0.2 μM Mg-ATP-actin monomers (20% pyrene-labeled) to the barbed end of 0.5 μM preassembled actin filaments. (A and B) Time course of seeded assembly alone (thick curve) or in the presence of the indicated concentrations of Fus1(FH1FH2) (A) and For3(FH2) (B). (C) Dependence of the initial barbed-end assembly rate on formin concentration. Curve fits revealed equilibrium dissociation constants of 0.12 nM for Cdc12(FH1FH2) (•), 0.20 nM for Fus1(FH1FH2) (◯), 1.3 nM for For3(FH2) (⋄), and 0.19 nM for Bni1(FH1FH2) (□). Fus1(FH1FH2) concentrations ≥5.0 nM significantly nucleate actin monomer assembly under these conditions, so its curve fit was from 0 to 2.5 nM. (D to F) Filament disassembly: barbed-end loss of actin monomer from 5.0 μM preassembled filaments (50% pyrene labeled) upon dilution to 0.1 μM. (D-E) Depolymerization time-course in the absence (thick curve) or presence of the indicated concentrations of Fus1(FH1FH2) (D) and For3(FH2) (E). (F) Dependence of the depolymerization rate on formin concentration. Curve fits revealed equilibrium dissociation constants of 0.49 nM for Cdc12(FH1FH2) (•), 0.43 nM for Fus1(FH1FH2) (◯), 2.3 nM for For3(FH2) ⋄), and 0.95 nM for Bni1(FH1FH2) (□). (G to L) TIRF microscopy visualization of the spontaneous assembly of unlabeled 1.0 μM Mg-ATP-actin with 0.5 μM Mg-ATP-actin labeled with Oregon green. (G to K) Plots of the length of six individual filaments over time for control (solid lines) and formin-nucleated (red dashed lines) filaments. The average elongation rates are indicated (subunits s−1). (G) Actin only control (Video S1). (H) 1.0 nM Cdc12(FH1FH2) (Video S2). (I) 0.5 nM Fus1(FH1FH2) (Video S3). (J) 150 nM For3(FH1FH2) (Video S4). (K) 50 nM For3(FH2) (Video S5). (L) Dissociation rate. Percentage of formin-associated filaments over time. Curve fits revealed dissociation rates of 4.7 × 10−5 s−1 for Cdc12(FH1FH2) (⋄), 6.5 × 10−4 s−1 for Fus1(FH1FH2) (□), 3.6 × 10−5 s−1 for For3(FH1FH2) (•), and 1.1 × 10−5 s−1 for For3(FH2) (◯).
The weak nucleation activity of For3 is not due to poor dimerization. Cdc12 and For3 constructs containing translationally fused, tandem FH2 domains expected to “force” dimerization (Otomo et al., 2005b) retain the same overall activity as wild-type Cdc12(FH2) or For3(FH2) in spontaneous assembly and critical concentration assays (Supplemental Figure S1).
The fission yeast formins reduce barbed-end dynamics by different amounts in the absence of profilin
Most formins bind to actin filament barbed ends with low nanomolar affinity and decrease the rate of assembly and disassembly in the absence of profilin (Kovar et al., 2010). We measured the affinity of the fission yeast formins for barbed ends and determined their effects on barbed-end dynamics in the absence of profilin with multiple assays including seeded elongation, depolymerization, critical concentration, and TIRF microscopy (Figures 2, S2, and S3). All three fission yeast formins bind preassembled actin filament barbed ends and significantly lower the rate of both assembly (Figure 2, A–C) and disassembly (Figure 2, D–F), but they do so to different extents. Both Cdc12(FH1FH2) and For3(FH2) lower the barbed-end seeded assembly and depolymerization rates by ∼99 and ∼90%, whereas Fus1(FH1FH2) lowers the rates by ∼60 and ∼80%. Budding yeast Bni1(FH1FH2) lowers the assembly and depolymerization rates by ∼40 and ∼70%. Plots of the dependence of the initial seeded assembly rate (Figure 2C) or depolymerization rate (Figure 2F) on formin concentration revealed differences in their barbed-end affinities. Cdc12 and Fus1 bind barbed ends (Kd = ∼0.3 nM) ∼10-fold better than For3 (Kd = ∼2.0 nM). Bni1 binds barbed ends with a Kd of ∼0.6 nM.
For critical concentration assays, 1.0 μM Mg-ATP actin was allowed to reach steady state in the presence of different formins and total polymer concentration was measured by either pyrene fluorescence (Figure S2A) or high-speed centrifugation (Figure S2, B–C). Both strategies revealed that Cdc12(FH1FH2) and For3(FH2) shift the critical concentration from 0.1 μM to ∼0.6 μM, although ∼50-fold more For3(FH2) is required. Fus1(FH1FH2) shifts the critical concentration to only ∼0.25 μM. As for most other formins, the modest inhibitory effect of Bni1(FH1FH2) on barbed-end dynamics does not change the critical concentration. Cdc12 and For3 do not completely inhibit barbed-end dynamics, since a significantly lower amount of polymer is detected at steady state in the presence of capping protein, which completely blocks monomer addition and subtraction at the barbed end (Figure S2, B and C).
We directly observed formin-mediated actin assembly by TIRF microscopy to determine the specific elongation rates, processivity on the elongating barbed end, and barbed-end dissociation rate (Figures 2, G–L, and S3). We followed the assembly of individual filaments elongating from a mixture of 1.0 μM Mg-ATP unlabeled actin and 0.5 μM Mg-ATP actin labeled on Cys374 with Oregon green (Neidt et al., 2008). In the absence of formin, all filaments assembling from 1.5 μM Mg-ATP actin elongate their barbed ends at the same constant rate of ∼12.5 subunits s−1 (Figures 2G and S3, A–C, and Supplemental Video S1). As seen previously with Cdc12 (Kovar et al., 2003; Kovar and Pollard, 2004; Kovar, 2006; Neidt et al., 2008, 2009), two distinct filament populations that differ by elongation rate appear with all three fission yeast formins (Figures 2, H–K, and S3, D–R, and Videos S2–S6). The presence of two populations indicates processive association with the elongating barbed end, because if formins rapidly shuttle on and off the barbed end, it is expected that only one filament population will elongate at an intermediate rate (Kovar and Pollard, 2004; Kovar et al., 2006). The first population consists of internal control filaments that elongate from ∼10–13 subunits s−1. The second population consists of formin-associated filaments that elongate at a significantly lower rate: Cdc12(FH1FH2) = 0.4 subunits s−1 (Figures 2H and S3, D–F, and Video S2), Fus1(FH1FH2) = 0.9 subunits s−1 (Figures 2I and S3, G–I, and Video S3), For3(FH1FH2) = 0.5 subunits s−1 (Figures 2J and S3, J–L, and Video S4), and For3(FH2) = 0.5 subunits s−1 (Figures 2K and S3, M–R, and Videos S5–S6). Therefore, as predicted from “bulk” assays, the fission yeast formins significantly slow barbed-end elongation rates by ∼10- (Fus1) and 20-fold (Cdc12 and For3) in the absence of profilin. In summary, all three fission yeast formins bind actin filament barbed ends with different affinities and significantly reduce barbed-end dynamics in the absence of profilin.
Fus1 dissociates more rapidly than Cdc12 and For3 from the elongating barbed end
As visualized by TIRF microscopy, individual filaments that switch elongation rates from slow (formin-dependent) to fast (formin-independent) in the absence of profilin represent formin dissociation events (Figures 2I and S3, G–I, and Video S3). We measured the barbed-end dissociation rate of the fission yeast formins by plotting the percentage of formin-associated filaments (slow filaments) over time (Figure 2L). Cdc12, For3, and Fus1 all allow the addition of thousands of subunits before dissociating. However, Fus1(FH1FH2) dissociates 10-fold more rapidly than Cdc12(FH1FH2), For3(FH1FH2), and For3(FH2), which rarely dissociate on the timescale of our experiments: Fus1(FH1FH2) = 6.5 × 10−4 s−1, Cdc12(FH1FH2) = 4.7 × 10−5s−1, For3(FH1FH2) = 3.6 × 10−5s−1, and For3(FH2) = 1.1 × 10−5 s−1.
The fission yeast formins bind profilin and stimulate the assembly of profilin-actin
The major source of unassembled actin in fission yeast cells is associated with profilin (Lu and Pollard, 2001). By binding profilin through their proline-rich FH1 domain, formins are tailored to drive the rapid assembly of profilin-actin (Kovar et al., 2010). Formins vary considerably in the number and composition of profilin-binding, proline-rich tracks in their FH1 domains: For3 (four tracks), Cdc12 (two tracks), and Fus1 (one track; Figure 3A). The Fus1 FH1 domain is unconventional in that its single proline-rich region is interspersed with hydrophobic residues and contains only three consecutive prolines out of six thought to be ideal for profilin-binding (Perelroizen et al., 1994; Petrella et al., 1996). Although nonproline residues can contribute to profilin binding (Kursula et al., 2008), it is possible that Fus1 does not bind profilin efficiently or promote the assembly of profilin-actin.
FIGURE 3:
The fission yeast formins stimulate the assembly of profilin-actin. (A) Amino acid sequence of the FH1 domains; For3(718-837), Cdc12(882-972), and Fus1(792-868). Putative profilin-binding segments are outlined with black boxes over white letters. (B) Dependence of profilin's intrinsic tryptophan fluorescence on the concentration of the indicated FH1 domains. Curve fits revealed equilibrium dissociation constants of 1.7 μM for Cdc12(FH1), 1.3 μM for Fus1(FH1), and 6.5 μM for For3(FH1). (C to F) Spontaneous assembly of 2.5 μM Mg-ATP actin (20% pyrene-labeled). (C and D) Time-course of actin assembly in the absence (thick curve) or presence of formin ± 2.5 μM profilin. (C) 25 nM Cdc12(FH1FH2) (•), Cdc12 with profilin (▴), 7.5 nM Fus1(FH1FH2) (◯), and Fus1 with profilin (▵). (D) 15 nM Cdc12(FH1FH2) (•), Cdc12 with profilin (▴), 500 nM For3(FH1FH2) (◯), and For3 with profilin (▵). (E) Dependence of the normalized actin-assembly rate (slope) on the concentration of profilin for 25 nM Cdc12(FH1FH2) (•), 7.5 nM Fus1(FH1FH2) (◯), 500 nM For3(FH1FH2) (▵), and 1000 nM For3(FH2) (□). (F) Representative fluorescence micrographs of actin filaments labeled with rhodamine-phalloidin 5 min after initiation of spontaneous assembly reactions in the presence of formin with and without 2.5 μM profilin. Average filaments lengths ± SD are indicated. Scale bar: 10 μm. (G to J) TIRF microscopy visualization of the spontaneous assembly of unlabeled 0.9 μM Mg-ATP-actin with 0.1 μM Mg-ATP-actin labeled on lysines with Alexa green and 2.5 μM fission yeast profilin. Plots of the length of six individual filaments over time for control (solid lines) and/or formin-nucleated (red dashed lines) filaments are shown. The average elongation rates are indicated (subunits s−1). Control and formin-associated filaments could not be differentiated for Cdc12 and For3. (G) Actin and profilin control (Video S7). (H) 2.5 nM Cdc12(FH1FH2) and profilin (Video S8). (I) 5.0 nM Fus1(FH1FH2) and profilin (Video S9). (J) 150 nM For3(FH1FH2) and profilin (Video S10).
The affinity of fission yeast profilin for all three FH1 domains was determined by measuring the change in profilin's intrinsic tryptophan fluorescence upon binding FH1 (Figure 3B). This assay likely reveals the binding of profilin to the highest-affinity, proline-rich site within each FH1 domain. We found that profilin binds to Cdc12(FH1) (Kd = 1.7 μM) and Fus1(FH1) (Kd = 1.3 μM) with similar affinities. For3(FH1) increases profilin's intrinsic tryptophan fluorescence by only one-half as much as Cdc12 or Fus1, potentially indicating a different binding conformation, and has a slightly lower affinity (Kd = 6.5 μM).
We then examined the ability of fission yeast formin(FH1FH2) constructs to stimulate the assembly of actin over a range of profilin concentrations in the pyrene actin assay (Figure 3, C–E). Both Cdc12(FH1FH2) and Fus1(FH1FH2) stimulate the assembly 2.5 μM Mg-ATP-actin in the presence of optimal profilin concentrations (∼2.5–5.0 μM) faster than actin alone, although Cdc12(FH1FH2) assembles profilin-actin faster than Fus1(FH1FH2) (Figure 3, C and E). Profilin's effect on both Cdc12(FH1FH2) and Fus1(FH1FH2) is biphasic (Figure 3E). Low profilin concentrations (up to ∼5.0 μM) increase the spontaneous assembly rate, whereas higher profilin concentrations reduce assembly, because profilin inhibits nucleation and excess free profilin competes with profilin-actin for the FH1 domain (Kovar et al., 2003, 2006; Paul and Pollard, 2008; Vavylonis et al., 2008).
For3(FH2), which lacks the profilin-binding FH1 domain, does not assemble profilin-actin well (Figure 3E). Surprisingly, we found that profilin also reduces pyrene fluorescence in the presence of For3(FH1FH2) (Figure 3, D and E). Therefore either For3(FH1FH2) cannot utilize profilin-actin or For3(FH1FH2) does not incorporate actin labeled on Cys374 with pyrene in the presence of profilin. To differentiate between these possibilities, we fixed and stained actin filaments with rhodamine-phalloidin 300 s after reactions were initiated (Figure 3F). Since profilin inhibits nucleation, few filaments are formed by 300 s in the absence of formin. Many long filaments (∼10.0 μm) are assembled by Cdc12(FH1FH2) with 2.5 μM profilin. As expected, For3(FH2) does not assemble long filaments in the presence of profilin. However, For3(FH1FH2) assembles many long filaments (∼8.0 μm) with 2.5 μM profilin. Therefore, although For3(FH1FH2) can stimulate profilin-actin assembly, For3(FH1FH2) does not efficiently incorporate actin labeled with pyrene on Cys374 in the presence of profilin. Characterization of chimeras mixing the FH1 and FH2 domains from Cdc12 and For3 (FH1cdc12FH2For3 and FH1For3FH2cdc12), suggest that both the For3 FH1 and FH2 domains contribute to selecting against profilin bound to pyrene-actin (Figure S4).
We utilized TIRF microscopy to determine elongation rates in the presence of profilin (Figures 3, G–J, and S5 and Videos S7–S10). Reactions with 2.5 μM fission yeast profilin were assembled from a mixture of 0.9 μM Mg-ATP unlabeled actin and 0.1 μM Mg-ATP actin labeled on lysines with Alexa Fluor 488 isothiocyanate (Alexa green). In control reactions without formin, all filaments elongate their barbed ends at the same constant rate of ∼6.0 subunits s−1 (Figures 3G and S5, A–C, and Video S7). All three formins stimulate the assembly of profilin-actin as indicated by a ∼10-fold increase in the density of filaments per 10 μm2, 450 s after reactions were initiated (Figure S5, A, D, G, and J). Two distinct filament populations are present in reactions with profilin and Fus1(FH1FH2) (Figures 3I and S5, G–I, and Video S9), whereas only one population is detected for Cdc12(FH1FH2) (Figures 3H and S5, D–F, and Video S8) and For3(FH1FH2) (Figures 3J and S5, J–L, and Video S10), because the formin-associated filaments elongate at approximately the same rate as control filaments. Fission yeast profilin increases the elongation rate of filaments nucleated by Cdc12, Fus1, and For3 to ∼7.0, 2.0, and 6.0 subunits s−1, respectively. Therefore, based on normalized actin concentrations, fission yeast formin nucleated filaments elongate approximately threefold (Fus1) to 20-fold (Cdc12 and For3) faster in the presence of profilin.
The barbed-end elongation rate in the presence of both Cdc12 and Fus1 has a biphasic dependence on the concentration of profilin (Figure S5M). Lower profilin concentrations increase the elongation rate of 1.5 μM actin (0.5 μM Oregon Green 488 iodoacetamide [Oregon green]–labeled), with a maximal effect in the range of 2.5–5.0 μM profilin. Elongation is increasingly inhibited at higher concentrations, due to competition between profilin-actin and excess profilin for the formin FH1 domain (Kovar et al., 2006; Vavylonis et al., 2006). Cdc12-associated barbed ends elongate at least two- to threefold faster than Fus1-associated barbed ends over a range of optimal profilin concentrations from 2.5 to 10 μM.
Both the FH1 and FH2 domains contribute to the elongation rate of profilin-actin
Although all fission yeast formin FH1FH2 domains facilitate the addition of profilin-actin faster than actin alone, the specific elongation rates do not strictly correlate with the number of putative profilin-binding sites in the FH1 domain (Figure 3). Therefore each proline-rich track may not contribute equally to the assembly of profilin-actin, and the FH2 domain may play a significant role in the addition of profilin-actin to the barbed end. To evaluate the contribution of the fission yeast formin FH1 and FH2 domains to the barbed-end addition of profilin-actin, we utilized TIRF microscopy to compare the elongation rates of a series of Cdc12 and Fus1 FH1FH2 domain chimeras (Figure 4). We found that the FH1 domain contributes more than simply binding to profilin-actin, and there is an important interaction between the FH1 and FH2 domains that affects the elongation rate.
FIGURE 4:
Comparison of formin chimera constructs reveals that both the FH1 and FH2 domains contribute to barbed-end elongation of profilin-actin. (A) Schematic of formin chimera constructs. Colors indicate origin of FH1 (light) and FH2 (dark) domains where Fus1 is blue and Cdc12 is red. Each FH1 domain “P” signifies an independent proline-rich track predicted to bind profilin. (B) Bar graph of the average elongation rates (subunits/s) of formin-associated filaments in the absence and presence of 2.5 μM profilin, determined by TIRF microscopy visualization of the assembly of 1.0 μM unlabeled Mg-ATP-actin with 0.5 μM Oregon green–labeled Mg-ATP-actin. Rates are adjusted based on normalization of internal control filaments to 10.0 subunits s−1 μM−1.
In the absence of profilin, the FH2 domain alone determines the elongation rate of all FH1FH2 chimera-associated filaments. Chimeras containing the Fus1 FH2 domain elongate at ∼0.5 subunits s−1, whereas chimeras containing the Cdc12 FH2 domain elongate at ∼0.25 subunits s−1 (Figure 4).
Both the Cdc12 and Fus1 FH1 domains facilitate the addition of profilin-actin to FH2-bound barbed ends faster than actin alone (Figure 4). However, the particular combination of FH1 and FH2 domains contributes differently to the specific elongation rate, which reveals four general principles of fission yeast formin(FH1FH2)-mediated elongation of profilin-actin. First, elongation rates increase proportional to the number of proline-rich tracks when the origin of the FH1 and FH2 domains are the same. Fus1(FH11PFH2)-associated filaments elongate one-half the rate of Fus1(FH12PFH2)-associated filaments, and Cdc12(FH11PFH2)-associated filaments elongate one-half the rate of Cdc12(FH12PFH2)-associated filaments (Figure 4). Second, the number of proline-rich tracks in the FH1 domain does not increase the elongation rate indefinitely. Cdc12(FH12PFH2)- and Cdc12(FH14PFH2)-associated filaments elongate at approximately the same rate, as do Cdc12(FH12P)Fus1(FH2)- and Cdc12(FH14P)Fus1(FH2)-associated filaments (Figure 4). Third, each proline-rich track does not contribute equally. For example, the single proline-rich track in the FH1 domain of Fus1 contributes less than the first proline-rich track in the FH1 domain of Cdc12. Fus1(FH11PFH2)-associated filaments elongate approximately twofold slower than Cdc12(FH11P)Fus1(FH2)-associated filaments, and Fus1(FH11P)Cdc12(FH2)-associated filaments elongate approximately threefold slower than Cdc12(FH11PFH2)-associated filaments (Figure 4). Fourth, the FH2 domain contributes significantly to the rate of profilin-actin assembly. For example, Fus1(FH11PFH2)-associated filaments elongate approximately threefold faster than Fus1(FH11P)Cdc12(FH2)-associated filaments (Figure 4).
Fus1, but not Cdc12 or For3, bundles actin filaments
Actin filaments in the contractile ring (Cdc12) and actin cables (For3) are bundled (Kamasaki et al., 2005, 2007), whereas filament organization during sexual conjugation (Fus1) is less clear (Petersen et al., 1998a). Bundling in fission yeast is thought to be primarily mediated by “traditional” cross-linkers such as fimbrin and α-actinin (Nakano et al., 2001; Wu et al., 2001; Takaine et al., 2009; Skau and Kovar, 2010; Skau et al., 2011). However, some formin FH2 domains bundle actin filaments (Moseley and Goode, 2005; Harris et al., 2006; Michelot et al., 2006), albeit at significantly higher concentrations than required for processive barbed-end elongation.
By low-speed cosedimentation at 10,000 × g, we found that Fus1(FH1FH2) and fimbrin Fim1 cross-link filaments preassembled from 3.0 μM Mg-ATP-actin monomers, whereas Cdc12(FH1FH2) and For3(FH2) do not (Figure 5A). Saturation of cross-linking occurs at ∼2.0 μM Fus1(FH1FH2), compared with ∼0.5 μM Fim1 (Figure 5B). Both Fim1 and Fus1(FH1FH2) organize filaments into dense bundles composed of multiple, aligned filaments (Figure 5C). High-speed sedimentation at 100,000 × g revealed that Fus1(FH1FH2) binds preassembled filaments with a Kd of ∼0.1 μM (Figure 5D).
FIGURE 5:
Fus1, but not Cdc12 or For3, binds to and cross-links actin filaments. (A and B) Low-speed sedimentation. (A) Coomassie Blue–stained gels of supernatants (SUP) and pellets (PEL) after 3.0 μM preassembled Mg-ATP-actin filaments were incubated with 0.75, 1.5, and 3.0 μM fission yeast fimbrin (Fim1), or formins Cdc12(FH1FH2), MBP-For3(FH2), and Fus1(FH1FH2), and spun at 10,000 × g. Actin and Fim1/Formin are marked to the left. (B) Dependence of actin remaining in the low-speed supernatant on the concentration of Fim1 (•) and Fus1(FH1FH2) (◯). (C) Representative fluorescence micrographs of rhodamine-phalloidin–labeled actin filaments in the absence (actin only) or presence of 500 nM Fim1 or Fus1(FH1FH2). Scale bar: 5 μm. (D) High-speed sedimentation. Fraction of 0.75 μM Fus1(FH1FH2) bound (% in pellet) to actin filaments following sedimentation at 100,000 × g. A curve fit revealed a dissociation constant of 0.10 μM.
FH2 domain mutations cause mild to severe defects in actin assembly in vitro and function in vivo
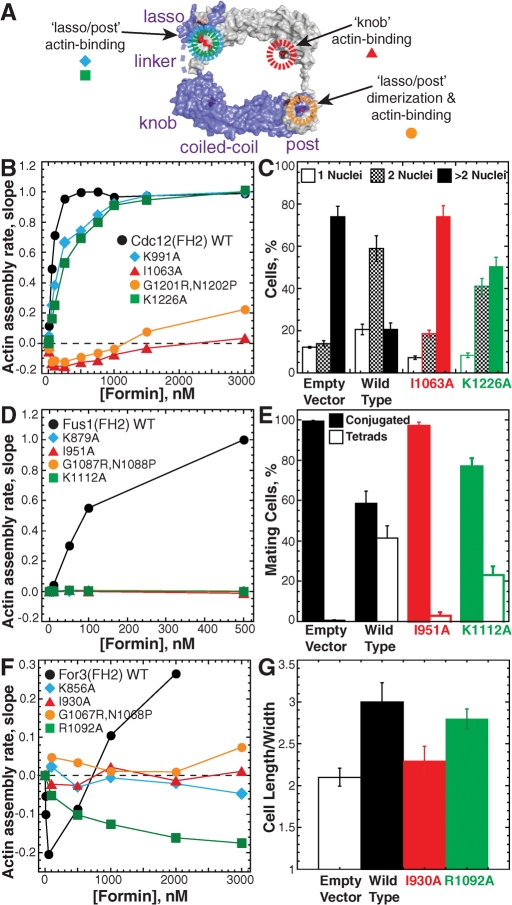
Although phylogenetically distinct (Higgs and Peterson, 2005; Rivero et al., 2005; Schonichen and Geyer, 2010), fission yeast formin FH2 domains align with other FH2 domains by sequence similarity (∼40–60%) and predicted secondary structure (Figure S6). The fission yeast FH2 domains are predicted to form a flexible “tethered dimer” architecture similar to budding yeast formin Bni1, for which actin-free and actin-bound structures have been solved (Xu et al., 2004; Otomo et al., 2005b). The Bni1 FH2 domain forms a head-to-tail homodimer composed of bundles of α-helices that fall into four subregions from the N- to C-terminus: lasso, knob, coiled-coil, and post. A short (∼17) amino acid linker joins the lasso and knob regions. Each half of the dimer contains two conserved actin-binding sites, one in the knob and one in the lasso–post region (Otomo et al., 2005b). Additionally, the dimerization interface consists of highly conserved residues that include the core GNF/YMN motif originally used to identify FH2 domains (Castrillon and Wasserman, 1994). We investigated the importance of the predicted fission yeast formin actin-binding and dimerization regions by testing the in vitro actin-assembly properties and in vivo function of a series of FH2 domain mutations (Figures 6 and S7–S9).
FIGURE 6:
Mutations of conserved residues in the FH2 domain impair formin activity in vitro and in vivo. (A) Schematic of the budding yeast Bni1(FH2) “tethered dimer” structure (pdb ly64; Otomo et al., 2005b). The lasso, linker, knob, coiled-coil, and post from one hemidimer (purple) are labeled. Mutations in conserved residues in the lasso–post actin-binding (blue and green), lasso–post dimerization and actin-binding (orange), and knob actin-binding (red) regions are indicated. (B, D, and F) Spontaneous assembly of 20% pyrene-labeled 2.5 μM Mg-ATP actin monomers. Dependence of the actin-assembly rate (slope) on the concentration of the indicated (B) MBP-Cdc12(FH2), (D) MBP-Fus1(FH2), or (F) MBP-For3(FH2) constructs. (C, E, and G) Complementation of mutant fission yeast formin strains by expression of medium-strength p572-41X-formin(full length)-GFP constructs containing the indicated FH2 domain point mutations. Error bars specify SD of triplicate experiments. (C) Percent of temperature-sensitive mutant cdc12-112 cells (strain KV427) with one, two, or more than nuclei following 16 h at 36°C in Edinburgh minimal media (EMM). (E) Percentage of h90 fus1Δ cells (strain EG999; Petersen et al., 1998b) that have mated and then formed tetrads (Tetrads) or not (Conjugated) following 36 h in ME mating medium at 25°C. (G) Average morphology (cell length/width) of for3Δ cells (strain BFY9; Feierbach and Chang, 2001), following 20 h at 25°C in EMM.
Cytokinesis formin Cdc12
Gel filtration revealed that mutations in the predicted actin-binding residues of Cdc12(FH2) (Figure S7A), K991A (lasso), I1063A (knob), and K1226A (post), do not disrupt dimerization (Figure S7B). Cdc12(FH2) mutations G1201R, N1202P (post) also do not disrupt dimerization (Figure S7B), suggesting that other residues within the extensive lasso–post interface also contribute to dimerization. Cdc12(FH2) mutations cause defects in the ability to stimulate actin monomer assembly that range from mild to severe: K991A < K1226A < G1201R, N1202P < I1063A (Figures 6B and S7C). Cdc12(FH2) mutants I1063A and G1201R, N1202P also have a reduced affinity for preassembled barbed ends (Figure S7D). Wild-type Cdc12(FH2), K991A, and K1226A have an equilibrium dissociation constant Kd of ∼0.3 nM, whereas mutants I1063A and G1201R, N1202P have a Kd of 10.6 and a Kd of 24.1 nM, respectively.
We assessed the importance of Cdc12-mediated actin filament assembly in vivo by testing the ability of full-length Cdc12-GFP plasmids containing point mutations in the FH2 domain to complement the temperature-sensitive mutant cdc12-112 strain at the restrictive temperature of 36°C (Figures 6C and S7E–G). Cdc12-112 cells carrying an empty vector: 1) fail to form contractile rings (visualized with the myosin regulatory light chain RLC-RFP); 2) deposit new cell wall material sparingly around a broad region of the cell cortex (calcofluor); and 3) contain more than two nuclei in ∼75% of their population (as seen by 4′,6-diamidino-2-phenylindole [DAPI] staining; Figures 6C and S7, E–G; Chang et al., 1997). Cdc12-112 cells expressing wild-type Cdc12-GFP are largely corrected, forming contractile rings that direct the formation of focused septa oriented perpendicular to the long axis of the cell (∼70% normal septa), and only ∼20% contain more than two nuclei. Although wild-type and mutant constructs are expressed at similar levels, there is considerable variation from cell to cell, which explains incomplete complementation by wild-type Cdc12-GFP (Figure S7F). Examination of cdc12-112 cells expressing FH2 mutant Cdc12-GFP constructs revealed that the ability of Cdc12 to drive cytokinesis correlates with its ability to stimulate actin assembly in vitro. All Cdc12-GFP mutants deposit septal material through the formation of contractile ring-like assemblages, although the normalcy of rings and septa varies considerably (Figure S7, E–G). Weaker actin-assembly mutants K1226A (∼35% normal septa, ∼50% with more than two nuclei) and K991A (∼10% normal septa, ∼65% with more than two nuclei) are more similar to wild-type than cdc12-112 cells expressing stronger actin-assembly mutants I1063A (∼3% normal septa, ∼75% with more than two nuclei) and G1201A, N1202P (∼5% normal septa, ∼75% with more than two nuclei).
Mating formin Fus1
Fus1(FH2) mutations (Figure S8A) K879A (lasso), I951A (knob), and G10876R, N1088P (post), and K112A (post) all cause severe defects in the ability to stimulate actin monomer assembly in vitro (Figures 6D and S8B). We investigated the ability of full-length Fus1-GFP constructs containing point mutations in the FH2 domain to allow homothallic mating type h90 fus1Δ mutant cells to conjugate and form tetrads following 36 h in malt extract (ME) mating media (Figures 6E and S8, C–E; Petersen et al., 1998b). Fus1Δ cells carrying an empty vector are able to conjugate, but < 2% form tetrads. Expression of wild-type Fus1-GFP allows ∼40% of conjugated cells to form tetrads, whereas significantly fewer conjugated fus1Δ cells expressing Fus1-GFP mutants I1951A (∼3%) and K1112A (∼20%) form tetrads. Therefore the ability of Fus1 to stimulate actin assembly is important for mating.
Polarity formin For3
For3(FH2) mutations (Figure S9A) K856A (lasso), I930A (knob), and G1067R, N1068P (post) completely abolish the ability to both inhibit and stimulate actin monomer assembly in vitro, whereas R1092A (post) retains some inhibitory activity (Figures 6F and S9B). We tested the importance of For3-mediated actin filament assembly in vivo by assessing whether full-length For3-GFP constructs containing point mutations in the FH2 domain polarize for3Δ cells (Figures 6G and S9, C–E). Asynchronous for3Δ cells with an empty vector are depolarized (Feierbach and Chang, 2001; Nakano et al., 2002), resulting in an average length divided by width (μm/μm) of only a little over 2.0. Polarity is largely restored in for3Δ cells expressing wild-type For3-GFP, with an average length/width of ∼3.0. Polarization in for3Δ cells expressing FH2 mutant For3-GFP constructs I930A (length/width = ∼2.3) and R1092A (length/width = ∼2.8) correlates nicely with their relative ability to affect actin assembly in vitro.
DISCUSSION
Fission yeast expresses three formin isoforms required for different cellular processes: Cdc12 (cytokinesis), For3 (polarization), and Fus1 (mating). Although Cdc12 has been well characterized (Kovar et al., 2003, 2006; Kovar and Pollard, 2004; Neidt et al., 2008, 2009; Yonetani et al., 2008; Skau et al., 2009), for the first time we report the actin-assembly properties of For3 and Fus1. By direct comparison, we found that Cdc12, For3, and Fus1 have general actin-assembly properties similar to those of other formins (Figure 7). All three stimulate actin assembly; remain processively associated with the elongating barbed end in both the absence and presence of profilin, while incorporating thousands of actin monomers before dissociating; reduce the barbed-end elongation and disassembly rates in the absence of profilin; and drive the assembly of profilin-actin faster than actin alone. However, the specific rates for these reactions vary significantly (Figure 7).
FIGURE 7:
Fission yeast formin actin-assembly properties. Top, cartoon diagram of the general actin filament structures assembled by the three fission yeast formins. Bottom, comparison of the major actin-assembly properties of the fission yeast formins, determined for the first time for Fus1 and For3 in this study.
The cytokinesis formin Cdc12 is an efficient nucleator (one filament per three Cdc12 dimers), binds with high affinity to preassembled barbed ends (Kd = 0.1 nM), is highly processive (Koff = 4.7 × 10−5 s−1) and elongates profilin-actin at a moderate rate (11.0 subunits s−1) but lacks actin filament-bundling activity. The mating formin Fus1 is also an efficient nucleator (one filament per two Fus1 dimers) and binds with high affinity to barbed ends (Kd = 0.2 nM), is processive, and efficiently bundles actin filaments but elongates profilin-actin at one-half the rate of (5.0 subunits s−1) and dissociates an order of magnitude more quickly than Cdc12 (Koff = 6.5 × 10−4 s−1). The polarity formin For3 is an extremely poor nucleator (one filament per 170 For3 dimers) and binds with lower affinity to barbed ends (Kd = 1.3 nM), but is highly processive (Koff = 3.6 × 10−5 s−1) and elongates profilin-actin at a moderate rate similar to that of Cdc12 (10.0 subunits s−1); it also lacks bundling activity. Furthermore, mutations that disrupt the actin-assembly properties of the fission yeast formins in vitro abrogate their function in vivo (Figures 6 and S7 to S9).
The atypical actin-assembly properties of For3
We and other researchers have communicated unpublished results that For3 constructs purified from either bacteria or yeast do not stimulate actin assembly (Goode and Eck, 2007), which our new findings suggest is due to a combination of factors. First, For3 constructs express poorly and form higher-order aggregates that need to be removed by gel filtration (Figure S10). Second, For3 inhibits actin assembly at low concentrations, while stimulating actin assembly only at higher concentrations. This biphasic behavior is caused by a combination of activities, including inefficient nucleation, a high affinity for preassembled filament barbed ends, and significant inhibition of barbed-end elongation. Lower concentrations of For3 do not create many new filaments but bind to spontaneously assembled barbed ends and slow their elongation ∼20-fold. Substantial new filament formation (nucleation) at higher For3 concentrations ultimately increases the “bulk” polymerization rate despite their slow elongation rate. Fission yeast actin capping protein has a similar biphasic activity (Kovar et al., 2005). It is possible that the nucleation activity of For3 is better with fission yeast actin (Takaine and Mabuchi, 2007; Ti and Pollard, 2011), or with For3 constructs that include the C-terminus (Gould et al., 2011). However, preliminary investigations of Cdc12, Fus1, and For3 constructs that extend past the FH2 domain have not revealed increased actin-assembly properties (Scott and Kovar, unpublished data). It is also possible that native For3 is posttranslationally modified.
Most formins favor unlabeled actin over Cys374-labeled actin in the presence of profilin, but do so by varying amounts (Kovar et al., 2006). Cdc12 and Fus1 are rare examples of formins that do not differentiate between unlabeled and Cys374-labeled actin in the presence of profilin (Kovar and Pollard, 2004; Kovar et al., 2006), whereas For3 almost exclusively utilizes unlabeled actin. We have recently discovered other formins that completely select against Cys374-labeled actin in the presence of profilin, including Drosophila diaphanous and Chlamydomonas CrFor1 (J. Sees, J. Winkelman, M. Glista, and D. Kovar, unpublished data). Therefore interpreting pyrene assays with formin and profilin requires additional assays that allow filament visualization.
The poor nucleation activity of For3 may be crucial for polarized actin cable assembly, where multiple rounds of asynchronous nucleation and elongation of new filaments “push/carry” older filaments within the bundle to the cell interior (Martin and Chang, 2006; Wang and Vavylonis, 2008). Simultaneous nucleation of numerous filaments by activated For3 molecules might lead to an extremely short (0.6 μm), thick bundle that does not extend inward. Second, it possible that For3 does not nucleate new filaments but instead binds and processively elongates preassembled filaments nucleated by the Arp2/3 complex in adjacent actin patches, analogous to the “convergent elongation” model proposed for the assembly of filopodia (Svitkina et al., 2003). On nucleation and/or barbed-end association, the actin filament elongation properties of For3 align well with current models. Actin cables assemble from cell tips at 0.3 μm s−1 (100 subunits s−1), which pushes the pointed end into the cell interior (Martin and Chang, 2006). For3 processively elongates profilin-actin in vitro at ∼10 subunits s−1 μM−1 of actin, which corresponds to a reasonable ∼150 subunits/s/15 μM actin in the cell. Following the addition of only 100–200 subunits, mechanisms must be in place that inhibit For3-mediated elongation but that apparently do not remove For3 from the filament.
Roles of the FH2 domain in profilin-actin assembly
Formins enhance the elongation rate of profilin-actin by increasing the local actin concentration and directing actin monomers onto the barbed end (Romero et al., 2004; Kovar et al., 2006; Vavylonis et al., 2006; Paul and Pollard, 2008); the FH1 and FH2 domains are therefore thought to play complementary roles. The FH2 domain remains processively associated with the barbed end, whereas the FH1 domain binds profilin-actin and transfers it to the barbed end. Profilin must then dissociate from the barbed end to allow additional rounds of assembly.
We have proposed that profilin and/or profilin-actin makes a critical interaction with the FH2 domain during transfer to the barbed end (Neidt et al., 2009), which is supported by three lines of evidence. First, analysis of chimeras between formins that prefer different profilin isoforms revealed that both the FH1 and FH2 domains contribute to profilin specificity (Neidt et al., 2009). Second, as discussed in the preceding section, certain formin and profilin combinations select against the addition of actin labeled on Cys374. Results from chimeras mixing the FH1 and FH2 domains from Cdc12 and For3 suggest that selection against Cys374-labeled by For3 occurs at both the association of profilin-actin with the FH1 domain, as well as association of FH1-profilin-actin with the FH2-associated barbed end (Figure S4). Third, reported here (Figure 4) and elsewhere (Paul and Pollard, 2008; Neidt et al., 2009; Vidali et al., 2009), the profilin-actin elongation rate of chimeras between the FH1 and FH2 domains of diverse formins is not strictly proportional to the FH1 domain and its number of proline-rich, profilin-binding sites. Thus the FH2 domain, rather than profilin binding to the FH1 domain, is often the rate-limiting step in formin-mediated elongation.
These results strongly suggest that the FH2 domain has an important role in profilin-actin elongation. We propose that, in addition to binding the proline-rich FH1 domain and actin, profilin also contains a region important for interacting with the formin FH2 domain. Although this interaction could be important for addition of profilin-actin to the FH2 domain-associated barbed end, we favor the hypothesis that interaction between profilin and the FH2 domain is important for dissociation of profilin from the barbed end following the incorporation of its actin monomer. A crystal structure of the budding yeast formin Bni1 FH2 domain in complex with actin revealed that the FH2 “knob” region makes a critical contact in the hydrophobic groove of actin subdomain 1, which is immediately next to the site that binds profilin (Otomo et al., 2005b; Vavylonis et al., 2006). Steric clash between profilin and the FH2 “knob” region is likely to occur in the “stair-stepping” model (Otomo et al., 2005a), whereas steric clashes may not occur in a more recently proposed “stepping second” model of processive elongation (Paul and Pollard, 2009a, 2009b).
Formin specialization in fungi, animals, and plants
Eukaryotes have multiple formin genes, including two in budding yeast, three in fission yeast, 15 in mammals, and more than 20 in plants, that perform diverse cellular functions from cell migration and adhesion, to cell division and intracellular trafficking (Higgs and Peterson, 2005; Goode and Eck, 2007; Kovar et al., 2010; Schonichen and Geyer, 2010). Given that these actin filament-based structures have different temporal, spatial, mechanical, and dynamic requirements, it is quite likely that different processes require formins with specific properties. Consistent with this hypothesis, the intrinsic in vitro actin-assembly properties, including nucleation efficiency, barbed-end elongation rate, barbed-end dissociation rate, and ability to bundle and/or sever filaments, vary widely among diverse formins (Higgs, 2005; Kovar, 2006; Goode and Eck, 2007; Kovar et al., 2010).
Differences in actin-assembly properties appear to be physiologically important. For example, a fast formin-mediated elongation rate is critical for cell polarization in the moss Physcomitrella patens (Vidali et al., 2009). The slow formin For1 cannot substitute for the fast formin For2 in polarization of moss plants. Similarly, overexpression of a constitutively active Cdc12 construct lacking its C-terminus produces robust ectopic interphase contractile rings in fission yeast (Yonetani and Chang, 2010). However, overexpression of a chimeric Cdc12 construct containing the inefficient nucleating FH1FH2 domains of For3 produces significantly weaker contractile rings with severe defects (Yonetani and Chang, 2010). Determining the full extent to which the specific actin-assembly properties of formins are physiologically important will require examination of a range of different cell types that exclusively express formins with altered properties.
MATERIALS AND METHODS
S. pombe strains and cell microscopy
Formin mutant strains: (1) cdc12 temperature-sensitive strain KV427; cdc12-112, rlc1-mRFP::kanMX6; (2) fus1 null strain EG999; h90, fus1Δ::leu2+ (Petersen et al., 1998b); (3) for3 null strain BFY9; for3Δ::kanMX6 (Feierbach and Chang, 2001). Cell morphology and organization were observed by differential interference contrast (DIC) and epifluorescence microscopy. Images were collected on an Orca-ER camera (Hamamatsu, Bridgewater, NJ) on an IX-81 microscope (Olympus, Tokyo, Japan), with a 60×, 1.4 numerical aperture Plan Apo objective. Nuclei and septa were visualized with DAPI (nuclei) and calcofluor (septa), as described previously (Kovar et al., 2003), and quantified for at least 200 cells. The expression levels of full-length formin-GFP constructs were determined by measuring the mean GFP fluorescence using ImageJ (http://rsb.info.nih.gov/ij/download.html).
DNA constructs for expression in bacteria and S. pombe
Three types of formin constructs were prepared for bacterial expression: 1) FH1, FH2, and FH1FH2 domain constructs for Cdc12, Fus1, and For3; 2) tandem FH2 domain constructs for Cdc12 and For3; and 3) chimera FH1::FH2 domain constructs between Cdc12, Fus1, and For3 containing the FH1 domain of one formin linked to the FH2 domain of the other. Cdc12(FH1) and Cdc12(FH1FH2) constructs have been described previously (Neidt et al., 2008). All constructs were prepared by standard cloning procedures involving PCR amplification (iProof, Bio-Rad, Hercules, CA) from S. pombe genomic DNA with a 6x His sequence included in the reverse primer. PCR products were cloned into either pET21a (EMD4Biosciences, San Diego, CA), pET21a-MBP(TEV) (Neidt et al., 2008), or pGEX-KText-GST(Thrombin) (Guan and Dixon, 1991) using restriction enzymes. Constructs contained the following FH1 and FH2 regions, numbers indicating the specific amino acids (Figure 1A): Cdc12(FH1FH2) [882–1390], Cdc12(FH2) [973–1390], Cdc12(FH1) [882–972], Fus1(FH1FH2) [792–1277], Fus1(FH2) [869–1277], Fus1(FH1) [792–862], For3(FH1FH2) [718–1265], For3(FH2) [838–1265], For3(FH1) [718–837]. Inserts were confirmed by sequencing. Budding yeast formin Bni1(FH1FH2) pQE70-Bni1(1227–1766) and fission yeast profilin pMW172-SpPRF bacterial expression constructs have been described (Lu and Pollard, 2001; Kovar and Pollard, 2004).
Full-length, medium-strength fission yeast expression constructs that include a C-terminal GFP for visualization were also prepared by standard PCR and restriction digest cloning procedures: p572-41Xnmt1-cdc12(1–1841)-GFP, p572-41Xnmt1-fus1(1–1372)-GFP, and p572-41Xnmt1-for3(1–1461)-GFP.
Mutagenesis of plasmid expression constructs
FH2 mutations were made using QuikChange XL site-directed mutagenesis kit (Stratagene, Agilent, Santa Clara, CA). Residues important for actin binding and dimerization were selected based on the three-dimensional structure and mutational analysis of Bni1 (Otomo et al., 2005b), and alignment of fission yeast to budding yeast FH2 amino acid sequences (Figure S6). FH2 residues important for actin binding, K1359 (lasso), I1431 (knob), and K1601 (post), were mutated to alanine residues: 1) Cdc12-K991A (lasso), I1063A (knob), and K1226A (post); 2) Fus1-K879A (lasso), I951A (knob), and K1112A (post); and 3) For3-K856A (lasso), I930A (knob), and R1092A (post). FH2 residues possibly important for both actin binding and dimerization G1576, N1577 (post) were mutated to arginine and proline: 1) Cdc12-G1201R, N1202P; 2) Fus1- G1087R, N1088P; and 3) For3-G1067R, N1068P.
Protein purification
Cdc12(FH1FH2)-HIS and Cdc12(FH2)-HIS (Neidt et al., 2008), Bni1(FH1FH2)-HIS (Kovar and Pollard, 2004), mouse capping protein (Palmgren et al., 2001), fission yeast profilin (Lu and Pollard, 2001), and fission yeast fimbrin Fim1 (Skau and Kovar, 2010), were purified from bacteria as described. MBP-Cdc12(FH2)-HIS, Fus1(FH1FH2)-HIS, Fus1(FH2)-HIS, For3(FH1FH2)-HIS, MBP-For3(FH2)-HIS, GST-For3(FH2)-HIS, For3(FH2)-HIS, MBP-Cdc12(FH2)::Cdc12(FH2)-HIS, MBP-For3(FH2)::For3(FH2)-HIS, GST-For3(FH2)::Cdc12(linker)-HIS, MBP-Cdc12(FH2)-HIS mutants, MBP-Fus1(FH2)-HIS mutants, and MBP- and GST-For3(FH2)-HIS mutants were purified by expressing constructs in Escherichia coli strain BL21-Codon Plus (DE3)-RP (Stratagene) with 0.5 mM isopropyl β-d-thiogalactopyranoside for 16 h at 16°C. Harvested cells were resuspended in extraction buffer (50 mM NaH2PO4, pH 8.0, 500 mM NaCl, 10% glycerol, 10 mM imidazole, 10 mM β-mercaptoethanol (BME) supplemented with protease inhibitors, and homogenized in an EmulsiFlex-C3 (Avestin, Ottawa, Canada). The homogenate was clarified at 30,000 and 50,000 × g for 15 and 30 min, respectively, and the extract was incubated with Talon Metal Affinity Resin (Clontech, Mountain View, CA) for 1–2 h at 4°C, and then loaded onto a disposable column. After a 50-ml wash with extraction buffer, formins were eluted with elution buffer (50 mM NaH2PO4, pH 8.0, 500 mM NaCl, 10% glycerol, 10 mM BME, and 250 mM imidazole) and dialyzed overnight in either HiTrap Q buffer A (Fus1: 20 mM Tris HCl, pH 8.5, 50 mM NaCl, 5% glycerol, 0.01% NaN3, and 1 mM dithiothreitol [DTT]), or formin buffer (Cdc12 and For3: 20 mM HEPES, pH 7.4, 1 mM EDTA, 200 mM KCl, 0.01% NaN3, and 1 mM DTT) in the absence or presence of 3 μM tobacco etch virus (TEV) protease to remove maltose-binding protein (MBP) or thrombin protease to remove glutathione S-transferase (GST). Dialyzed Fus1 was loaded on a 1.0-ml HiTrap Q column (GE Healthcare, Waukesha, WI) and eluted with a linear gradient from 50 to 500 mM NaCl. Pure Fus1 was dialyzed overnight in formin buffer, concentrated using Centriprep YM-30 (Millipore, Billerica, MA) to >10 μM, flash frozen in liquid nitrogen, and stored at −80°C. Unfrozen and flash-frozen Fus1 both maintained activity over time. Dialyzed Cdc12, For3, tethered dimers, and Cdc12 mutants were concentrated using Centriprep YM-30 and loaded on a 24-ml Superdex 200 10/300 gel-filtration column (GE Healthcare; Figure S10). Protein in gel-filtration fractions suggestive of aggregates and dimers was pooled separately and concentrated using aquacide (Calbiochem, San Diego, CA) or Centriprep YM-30 (Millipore) to > 10 μM. For3 aggregates exclusively inhibit actin assembly, whereas stable For3 dimers inhibit actin assembly at lower concentrations and enhance actin assembly at higher concentrations (Figure S10). All For3 constructs slowly lost activity over time, samples were therefore kept on ice and used within 1 mo.
Following elution from Talon resin, MBP-Fus1(FH1)-HIS and MBP-Cdc12(FH1)-HIS were concentrated with Centriprep YM-3 (Millipore) and dialyzed overnight in HiTrap Q buffer A (pH 8.5) with 3 μM TEV protease to remove MBP. Protein was loaded on a 1.0-ml HiTrap Q column, eluted, and dialyzed overnight in amylose buffer (20 mM Tris HCl, pH 7.4, 500 mM NaCl, 1 mM EDTA, 10% glycerol, 0.01% NaN3, and 1 mM DTT). Protein was incubated with Amylose Resin (New England Biolabs, Ipswich, MA) for 1 h at 4°C and loaded onto a disposable column. After a 50-ml wash with amylose buffer, proteins were eluted with amylose elution buffer (amylose buffer + 10 mM maltose) and dialyzed overnight in extraction buffer. Proteins were run over a second Talon column and dialyzed overnight in formin(FH1) buffer (20 mM Tris, pH 7.5, 150 mM KCl, and 0.2 mM DTT). Pure formin(FH1) was concentrated using Centriprep YM-3 or aquacide to >100 μM, stored on ice, and used within 1 mo.
MBP-For3(FH1) was purified similarly. Following elution from Talon resin, protein was dialyzed overnight in extraction buffer with 3 μM TEV protease to remove MBP. Protein was run over a second Talon column, then dialyzed into HiTrap Q buffer A. Protein was loaded on a 1.0-ml HiTrap Q column, eluted as above, and dialyzed overnight in formin(FH1) buffer. Protein was concentrated using aquacide, loaded on a Superdex 200 gel-filtration column, and pure formin(FH1) fractions were pooled and concentrated using aquacide to >100 μM.
Ca-ATP actin was purified from chicken skeletal muscle (Trader Joe's) and rabbit skeletal muscle (Pel-Freez, Rogers, AR), as previously described (Spudich and Watt, 1971). Gel-filtered actin was labeled on Cys-374 with pyrenyl iodoacetamide (pyrene) or Oregon green (Invitrogen) and labeled on lysines with Alexa green (Invitrogen; Kuhn and Pollard, 2005; Neidt et al., 2008). Immediately prior to each experiment, 5–15 μM Ca-ATP actin was converted to Mg-ATP actin by adding 0.1 vol of 2 mM EGTA and 0.5 mM MgCl2 for 2 min at 25°C.
Protein concentrations were determined with extinction coefficients as follows: unlabeled actin, A290 = 26,600 M−1 cm−1 (Houk and Ue, 1974); pyrene-actin, (A290 – [A344 × 0.127]) × 38.5 μM (Cooper et al., 1983); Oregon-green actin, [Total Ca-actin] = (A290 – [A491 × 0.16991])/26,600 M−1 cm−1, [Ca-OG-actin] = A491/77,800 M−1 cm−1 (Kovar et al., 2003); fission yeast profilin, A280 = 20,065 M−1 cm−1 (Lu and Pollard, 2001); mouse capping protein, A280 = 76,300 M−1 cm−1 (Palmgren et al., 2001). Extinction coefficients (A280 in units M−1 cm−1) were estimated with ProtParam (http://us.expasy.org/tools) from the amino acid composition: 1) 6x His-tagged constructs: Cdc12(FH1FH2) = 51,255 (Kovar et al., 2003); Fus1(FH1FH2) = 34,880; Fus1(FH2) = 33,265; For3(FH2) = 39,560; Bni1(FH1FH2) = 42,985; 2) 6x His and MBP- or GST-tagged constructs: MBP-Cdc12(FH2) = 119,095; MBP-Cdc12(FH2)::Cdc12(FH2) = 170,350; MBP-For3(FH1FH2) = 110,380; MBP-For3(FH2) = 107,400; MBP-For3(FH2)::For3(FH2) = 146,960; GST-For3(FH2) = 82,670. Point mutations did not change extinction coefficients. Protein concentrations of 6x His-tagged FH1 constructs Cdc12(FH1), Fus1(FH1), and For3(FH1) were determined by A205 in water ([(A205FH1− A205buffer)/30]/mol wt).
Fluorescence spectroscopy
Actin assembly was measured from the fluorescence of a trace of pyrene-actin (excitation at 364 nm and emission at 407 nm) with Spectramax Gemini XPS (Molecular Devices, Sunnyvale, CA) and Safire2 (Tecan, Durham, NC) fluorescence plate readers. Final protein concentrations are indicated in the figure legends.
For spontaneous assembly assays, 15 μM of 20% pyrene-labeled Mg-ATP-actin was placed in an upper row of a 96-well nonbinding black plate (Corning, Corning, NY). Other proteins to be assayed (formin, profilin, etc.) and 10X KMEI (500 mM KCl, 10 mM MgCl2, 10 mM ethylene glycol tetraacetic acid [EGTA], and 100 mM imidazole, pH 7.0) were placed in a lower row of the plate. For seeded assembly assays, 5.0 μM unlabeled Mg-ATP-actin was preassembled in the upper row of a 96-well plate, which was followed by addition of other proteins to be assayed (formin, profilin, etc.). Twenty percent pyrene-labeled Mg-ATP-actin (2.0 μM) was placed in a lower row. For depolymerization assays, a 5.0 μM mixture of 20% pyrene-labeled Mg-ATP-actin monomers was preassembled in the upper row of the plate for 2 h. Formin and 10X KMEI were placed in the lower row. Reactions for all assays were initiated by mixing contents of the lower wells with the actin monomers in the upper wells. The critical concentration for actin assembly was determined by assembling 1.0 μM of 20% pyrene-labeled Mg-ATP-actin monomers in the presence of a range of concentrations of formin in a 96-well black plate. The final amount of filamentous actin was determined after a 16-h incubation in the dark at 25°C.
The affinity of profilin for formin(FH1) was determined by measuring profilin's intrinsic tryptophan fluorescence by excitation at 295 nm and emission at 323 nm (Perelroizen et al., 1994; Petrella et al., 1996). Profilin (1.0 μM) was incubated with a range of formin(FH1) concentrations for 30 min, then profilin fluorescence was read in a Safire2 fluorescence plate reader and plotted versus formin(FH1) concentration. The fluorescence of formin(FH1) alone was subtracted from the fluorescence in the presence of profilin. Affinities (Kd) were determined by fitting a quadratic function to the dependence of the concentration of bound profilin on the concentration of formin(FH1).
Initial polymerization and depolymerization rates, barbed-end affinity, and nucleation efficiency
The rates of spontaneous assembly reactions were determined by measuring the linear slopes from 10 to 50% of actin assembly. Seeded assembly rates were measured from the linear fit of the first 500s. Depolymerization slopes were fit by linear curves over the first 200–250 s. Depolymerization rates were expressed as a percent normalized to the rate of actin alone. The affinity of formin for actin filament barbed ends was determined by fitting plots of the dependence of either the initial assembly rate or the initial depolymerization rate on the concentration of formin, with the equation
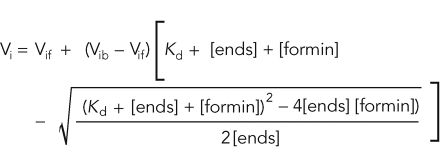 |
Vi is the observed elongation or depolymerization rate, Vif is the elongation or depolymerization rate when barbed ends are free, Vib is the elongation or depolymerization rate when barbed ends are bound, [ends] and [formin] are barbed-end and formin concentrations. Nucleation efficiency was calculated by dividing the spontaneous assembly rate (slope) by the elongation rate (k+) in the absence and presence of profilin and dividing by the formin concentration.
Microscopy of fluorescently labeled filaments
Products of spontaneous assembly reactions were examined by fluorescence microscopy as previously described (Blanchoin et al., 2000; Kovar et al., 2003). Following the spontaneous assembly of unlabeled Mg-ATP-actin, filaments were incubated for 5 min with 1 μM TRITC-Phalloidin (Fluka Biochemika, Switzerland). Reactions were terminated by a 250-fold dilution in fluorescence buffer (50 mM KCl, 1 mM MgCl2, 100 mM DTT, 20 μg/ml catalase, 100 μg/ml glucose oxidase, 3 mg/ml glucose, 0.5% methylcellulose, and 10 mM imidazole, pH 7.0) and absorbed to coverslips coated with 0.05 μg/μl poly-l-lysine. Fluorescence images were collected with a cooled CCD camera (Orca-ER, Hamamatsu) on an Olympus IX-81 microscope.
Total internal reflection fluorescence (TIRF) microscopy
Images of Oregon green–labeled or Alexa green–labeled actin filaments excited by total internal reflection (Olympus IX-71 microscope; fit with through-the-objective TIRF illumination) were collected at 15-s intervals with an iXon EMCCD camera (Andor Technology, Belfast, UK). As described in detail previously (Kovar et al., 2003, 2006; Kovar and Pollard, 2004; Kuhn and Pollard, 2005; Neidt et al., 2008), a mixture of 1.0 μM unlabeled Mg-ATP actin and 0.5 μM (33%) Mg-ATP Oregon-green actin or 0.9 μM unlabeled Mg-ATP actin and 0.1 μM (10%) Mg-ATP Alexa-green actin was mixed with 2X TIRF buffer (1X: 10 mM Imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM DTT, 0.2 mM ATP, 50 μM CaCl2, 15 mM glucose, 20 μg/ml catalase, 100 μg/ml glucose oxidase and 0.5% [500 centipoise] methylcellulose) and formin and/or profilin and transferred to a flow cell for imaging.
Low- and high-speed actin filament sedimentation
The ability of fission yeast formins to cross-link actin filaments was determined from low-speed cosedimentation assays. Mg-ATP-actin (15 μM) was preassembled for 2.0 h at 25°C, then 3.0 μM was aliquoted into Eppendorf tubes and incubated with a range of concentrations of fission yeast fimbrin (Fim1; Skau and Kovar, 2010) or formins Cdc12(FH1FH2), MBP-For3(FH2), and Fus1(FH1FH2) for 20 min at 25°C. Samples were spun at 10,000 × g for 20 min at 25°C. Pellets and supernatants were separated by 12.5% SDS–PAGE and stained with Coomassie Blue. The intensity of protein bands (densitometry) was then determined with an Odyssey Infrared Imager (LI-COR Biosciences, Lincoln, NE).
Affinity of Fus1 for actin filaments was determined by high-speed cosedimentation. Mg-ATP-actin (15 μM) was preassembled for 2.0 h at 25°C. Fus1(FH1FH2) (1.5 μM) was incubated for 20 min at 25°C with a range of concentrations of preassembled actin filaments. Samples were spun for 20 min at 100,000 × g at 25°C. Pellets and supernatants were separated by 12.5% SDS–PAGE and stained with Coomassie Blue, and the intensity of protein bands was determined with an Odyssey Infrared Imager. The affinity (Kd) of Fus1 for actin filaments was determined by fitting a quadratic function to the dependence of the concentration of bound Fus1 dimers (loss of Fus1 from supernatants) on the concentration of actin.
Supplementary Material
Acknowledgments
We thank Wenjing Zong for initial purification of For3, Mike Glista for filament images shown in Figure 5C, Elizabeth Kovar for critical reading of the manuscript, and members of the Kovar lab for technical assistance and helpful comments. We also thank Fred Chang (Columbia University, New York) and Olaf Nielsen (University of Copenhagen, Denmark) for strains. This work was supported by grants from the Edward Mallinckrodt, Jr., Foundation and National Institutes of Health (R01 GM-079265 to D.R.K.).
Abbreviations used:
- Alexa green
Alexa Fluor 488 isothiocyanate
- BME
β-mercaptoethanol
- DAPI
4′,6-diamidino-2-phenylindole
- DIC
differential interference contrast
- DTT
dithiothreitol
- EGTA
ethylene glycol tetraacetic acid
- EMM
Edinburgh minimal media
- FH
formin homology
- GST
glutathione S-transferase
- MBP
maltose-binding protein
- ME
malt extract
- Oregon green
Oregon Green 488 iodoacetamide
- TEV
tobacco etch virus
- TIRF
total internal reflection fluorescence
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0492) on August 24, 2011.
REFERENCES
- Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Wasserman SA. diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Nile AH, Lee IJ, Liu H, Wu JQ. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol Biol Cell. 2009;20:5195–5210. doi: 10.1091/mbc.E09-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Walker SB, Pollard TD. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J Muscle Res Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- Feierbach B, Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr Biol. 2001;11:1656–1665. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Gould CJ, Maiti S, Michelot A, Graziano BR, Blanchoin L, Goode BL. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr Biol. 2011;21:384–390. doi: 10.1016/j.cub.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem. 2006;281:14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houk TW, Jr, Ue K. The measurement of actin concentration in solution: a comparison of methods. Anal Biochem. 1974;62:66–74. doi: 10.1016/0003-2697(74)90367-4. [DOI] [PubMed] [Google Scholar]
- Kamasaki T, Arai R, Osumi M, Mabuchi I. Directionality of F-actin cables changes during the fission yeast cell cycle. Nat Cell Biol. 2005;7:916–917. doi: 10.1038/ncb1295. [DOI] [PubMed] [Google Scholar]
- Kamasaki T, Osumi M, Mabuchi I. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J Cell Biol. 2007;178:765–771. doi: 10.1083/jcb.200612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR. Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol. 2006;18:11–17. doi: 10.1016/j.ceb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Bestul AJ, Li Y, Scott BJ, Carlier MF. Formin-mediated actin assembly. In: Carlier MF, editor. Actin-based Motility. New York: Springer; 2010. pp. 279–316. [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Sirotkin V, Lord M. Three's company: the fission yeast actin cytoskeleton. Trends Cell Biol. 2011;21:177–187. doi: 10.1016/j.tcb.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Wu JQ, Pollard TD. Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol Biol Cell. 2005;16:2313–2324. doi: 10.1091/mbc.E04-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursula P, Kursula I, Massimi M, Song YH, Downer J, Stanley WA, Witke W, Wilmanns M. High-resolution structural analysis of mammalian profilin 2a complex formation with two physiological ligands: the formin homology 1 domain of mDia1 and the proline-rich domain of VASP. J Mol Biol. 2008;375:270–290. doi: 10.1016/j.jmb.2007.10.050. [DOI] [PubMed] [Google Scholar]
- Laporte D, Coffman VC, Lee IJ, Wu JQ. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Pollard TD. Profilin binding to poly-l-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Chang F. Dynamics of the formin for3p in actin cable assembly. Curr Biol. 2006;16:1161–1170. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- Martin SG, Rincon SA, Basu R, Perez P, Chang F. Regulation of the formin for3p by cdc42p and bud6p. Mol Biol Cell. 2007;18:4155–4167. doi: 10.1091/mbc.E07-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot A, Derivery E, Paterski-Boujemaa R, Guerin C, Huang S, Parcy F, Staiger CJ, Blanchoin L. A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr Biol. 2006;16:1924–1930. doi: 10.1016/j.cub.2006.07.054. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J Biol Chem. 2005;280:28023–28033. doi: 10.1074/jbc.M503094200. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Imai J, Arai R, Toh EA, Matsui Y, Mabuchi I. The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J Cell Sci. 2002;115:4629–4639. doi: 10.1242/jcs.00150. [DOI] [PubMed] [Google Scholar]
- Nakano K, Satoh K, Morimatsu A, Ohnuma M, Mabuchi I. Interactions among a fimbrin, a capping protein, and an actin-depolymerizing factor in organization of the fission yeast actin cytoskeleton. Mol Biol Cell. 2001;12:3515–3526. doi: 10.1091/mbc.12.11.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidt EM, Scott BJ, Kovar DR. Formin differentially utilizes profilin isoforms to rapidly assemble actin filaments. J Biol Chem. 2009;284:673–684. doi: 10.1074/jbc.M804201200. [DOI] [PubMed] [Google Scholar]
- Neidt EM, Skau CT, Kovar DR. The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J Biol Chem. 2008;283:23872–23883. doi: 10.1074/jbc.M803734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell. 2005a;18:273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005b;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- Palmgren S, Ojala PJ, Wear MA, Cooper JA, Lappalainen P. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J Cell Biol. 2001;155:251–260. doi: 10.1083/jcb.200106157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Energetic requirements for processive elongation of actin filaments by FH1FH2 formins. J Biol Chem. 2009a;284:12533–12540. doi: 10.1074/jbc.M808587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton. 2009b;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelroizen I, Marchand JB, Blanchoin L, Didry D, Carlier MF. Interaction of profilin with G-actin and poly(L-proline) Biochemistry. 1994;33:8472–8478. doi: 10.1021/bi00194a011. [DOI] [PubMed] [Google Scholar]
- Petersen J, Nielsen O, Egel R, Hagan IM. F-actin distribution and function during sexual differentiation in Schizosaccharomyces pombe. J Cell Sci. 1998a;111:867–876. doi: 10.1242/jcs.111.7.867. [DOI] [PubMed] [Google Scholar]
- Petersen J, Nielsen O, Egel R, Hagan IM. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J Cell Biol. 1998b;141:1217–1228. doi: 10.1083/jcb.141.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Weilguny D, Egel R, Nielsen O. Characterization of fus1 of Schizosaccharomyces pombe: a developmentally controlled function needed for conjugation. Mol Cell Biol. 1995;15:3697–3707. doi: 10.1128/mcb.15.7.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella EC, Machesky LM, Kaiser DA, Pollard TD. Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry. 1996;35:16535–16543. doi: 10.1021/bi961498d. [DOI] [PubMed] [Google Scholar]
- Pring M, Evangelista M, Boone C, Yang C, Zigmond SH. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 2003;42:486–496. doi: 10.1021/bi026520j. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Rivero F, Muramoto T, Meyer AK, Urushihara H, Uyeda TQ, Kitayama C. A comparative sequence analysis reveals a common GBD/FH3-FH1-FH2-DAD architecture in formins from Dictyostelium, fungi and metazoa. BMC Genomics. 2005;6:28. doi: 10.1186/1471-2164-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Schonichen A, Geyer M. Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim Biophys Acta. 2010;1803:152–163. doi: 10.1016/j.bbamcr.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Skau CT, Courson DS, Bestul AJ, Winkelman JD, Rock RS, Sirotkin V, Kovar DR. Actin filament bundling by fimbrin is important for endocytosis, cytokinesis, and polarization in fission yeast. J Biol Chem. 2011;286:26964–77. doi: 10.1074/jbc.M111.239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skau CT, Kovar DR. Fimbrin and tropomyosin competition regulates endocytosis and cytokinesis kinetics in fission yeast. Curr Biol. 2010;20:1415–1422. doi: 10.1016/j.cub.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skau CT, Neidt EM, Kovar DR. Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol Biol Cell. 2009;20:2160–2173. doi: 10.1091/mbc.E08-12-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaine M, Mabuchi I. Properties of actin from fission yeast Schizosaccharomyces pombe and interaction with fission yeast profilin. J Biol Chem. 2007;282:21683–21694. doi: 10.1074/jbc.M611371200. [DOI] [PubMed] [Google Scholar]
- Takaine M, Numata O, Nakano K. Fission yeast IQGAP arranges actin filaments into the cytokinetic contractile ring. EMBO J. 2009;28:3117–3131. doi: 10.1038/emboj.2009.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti SC, Pollard TD. Purification of actin from fission yeast Schizosaccharomyces pombe and characterization of functional differences from muscle actin. J Biol Chem. 2011;286:5784–5792. doi: 10.1074/jbc.M110.199794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Kovar DR, O'Shaughnessy B, Pollard TD. Model of formin-associated actin filament elongation. Mol Cell. 2006;21:455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Wu JQ, Hao S, O'Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- Vidali L, van Gisbergen PAC, Guerin C, Franco P, Li M, Burkart G, Augustine RC, Blanchoin L, Bezanilla M. Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc Natl Acad Sci USA. 2009;106:13341–13346. doi: 10.1073/pnas.0901170106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Vavylonis D. Model of For3p-mediated actin cable assembly in fission yeast. PLoS One. 2008;3:e4078. doi: 10.1371/journal.pone.0004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Bahler J, Pringle JR. Roles of a fimbrin and an α-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol Biol Cell. 2001;12:1061–1077. doi: 10.1091/mbc.12.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- Yonetani A, Chang F. Regulation of cytokinesis by the formin cdc12p. Curr Biol. 2010;20:561–566. doi: 10.1016/j.cub.2010.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonetani A, Lustig RJ, Moseley JB, Takeda T, Goode BL, Chang F. Regulation and targeting of the fission yeast formin cdc12p in cytokinesis. Mol Biol Cell. 2008;19:2208–2219. doi: 10.1091/mbc.E07-07-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.