Abstract
Understanding the fundamentals of G-quadruplex formation is important both for targeting G-quadruplexes formed by natural sequences and for engineering new G-quadruplexes with desired properties. Using a combination of experimental and computational techniques, we have investigated the effects of site-specific substitution of a guanine with C8-modified guanine derivatives, including 8-bromo-guanine, 8-O-methyl-guanine, 8-amino-guanine, and 8-oxo-guanine, within a well-defined (3 + 1) human telomeric G-quadruplex platform. The effects of substitutions on the stability of the G-quadruplex were found to depend on the type and position of the modification among different guanines in the structure. An interesting modification-dependent NMR chemical-shift effect was observed across basepairing within a guanine tetrad. This effect was reproduced by ab initio quantum mechanical computations, which showed that the observed variation in imino proton chemical shift is largely influenced by changes in hydrogen-bond geometry within the guanine tetrad.
Introduction
Guanine-rich nucleic acid sequences can fold into four-stranded structures called G-quadruplexes (1–3). Mounting evidence supports the role of G-quadruplex structures in telomere biology (4–8) and gene regulation (7–10). These structures are considered to be potential therapeutic targets (11–13). In addition, engineered G-quadruplex-forming sequences with anticoagulant (14–17), anti-HIV (18–21), and anti-cancer (22–25) activities have been discovered.
G-quadruplex structures (1–3) are formed by the stacking of multiple layers of G⋅G⋅G⋅G tetrads (26) (Fig. 1, a and b). The diverse folding topologies observed for G-quadruplexes are related to specific structural features including strand orientation, connecting-loop type, and the glycosidic bond angle of guanines within G-tetrads (1–3,27). The preferred folding topology of a G-quadruplex-forming nucleic acid sequence can be altered by introducing a guanine like aromatic base within a G-tetrad. For example, guanine derivatives such as inosine (28–30) and 8-bromo-guanine (31,32) have been used as tools in structural studies to favor a single G-quadruplex in a sequence capable of forming multiple conformations.
Figure 1.
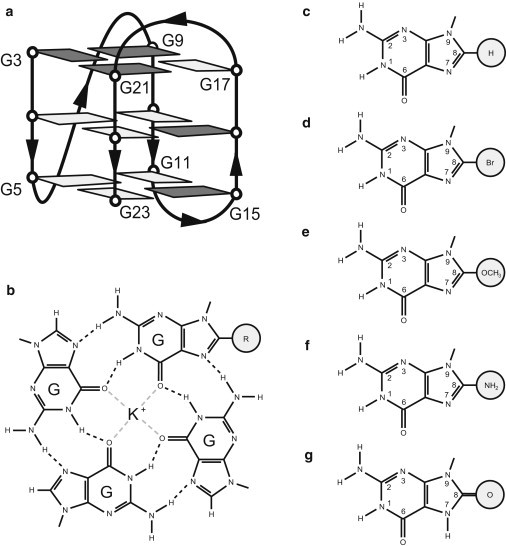
(a) Schematic structure of the (3 + 1) G-quadruplex platform used in this study. Anti and syn guanines are shown in light gray and dark gray, respectively. (b) Chemical schematic of a modified guanine tetrad with a centrally located K+ ion. The C8-modified guanine derivative is depicted by a modification group R. (c–g) The chemical schematics of guanine and the guanine derivatives explored in this work: (c) guanine, (d) 8-bromo-guanine, (e) 8-O-methyl-guanine, (f) 8-amino-guanine, and (g) 8-oxo-guanine.
The guanine derivative 8-bromo-guanine belongs to a specific class of modified bases involving substitutions of the C8 hydrogen with other modification groups (Fig. 1). C8 modifications can be made without disrupting Hoogsteen hydrogen-bonding within the tetrad. This strategy was successfully used to incorporate a fluorescent probe within a G-quadruplex structure (33). In contrast to base modifications that disrupt hydrogen bonding and destabilize G-quadruplexes (34,35), C8-modified derivatives 8-bromo-guanine (31,32) and 8-methyl-guanine (36) have shown a tendency to increase the thermal stability of a G-quadruplex when substituted with guanines that originally adopt syn glycosidic conformations in the G-tetrad core. In addition, C8-modified derivatives have been shown to influence the folding kinetics of G-quadruplex structures (34,37,38).
Past research investigating the effects of C8 modifications on G-quadruplexes has been carried out on both intramolecular (31–33,35,36) and intermolecular tetrameric structures (34,37–40). Intramolecular G-quadruplex scaffolds allow for single-position modifications to be implemented and to probe the effects of single modifications on syn/anti glycosidic conformations. In parallel-stranded tetrameric G-quadruplexes, all core guanines generally adopt anti conformations. In a tetrameric system, the substitution of a guanine by a C8-modified derivative is reflected about the entire tetrad due to the symmetry of the structure. C8-modified guanine substitutions in a tetrameric system have been reported to cause a change in the polarity of the modified tetrad (38–40). Some studies have noted position-dependent effects for the derivatives 8-bromo-guanine (34,39), 8-methyl-guanine (38,40), and 8-amino-guanine (37), including the ability of substitutions at the 5′ end to stabilize G-quadruplexes. Although studies involving tetrameric G-quadruplexes are valuable, conclusions arising from these studies could be fundamentally different in comparison to those arising from intramolecular scaffolds where only one guanine within a G-tetrad is substituted at a time. With evidence mounting for the in vivo roles of G-quadruplexes, the effects of C8-modified derivatives on these structures are of physiological importance, considering the numerous naturally occurring C8-modifications that are suggested to play roles in mutagenesis, including 8-oxo-guanine (41,42), 8-amino-guanine (43,44), and 8-methyl-guanine (45). From an engineering perspective, investigating the effects of various C8-modified derivatives on G-quadruplexes facilitates their effective application in engineered systems to achieve desired properties.
To understand how site-specific C8 modifications of guanines affect G-quadruplex stability and structure, this work explores the effects of substituting guanine derivatives 8-bromo-guanine, 8-O-methyl-guanine, 8-amino-guanine, and 8-oxo-guanine (Fig. 1, c–g) within a well-defined intramolecular (3 + 1) telomeric G-quadruplex structure (46) (Fig. 1 a) using a combination of ultraviolet (UV), circular dichroism (CD), and NMR spectroscopy and ab initio quantum mechanical calculations. The site-specific effects of substitutions to both syn and anti guanines were examined. Consistent folding topology was monitored, allowing for a meaningful thermal denaturing analysis within the context of a single conformer. The observation of an interesting pattern in NMR chemical shift change was reproduced using ab initio computational methods, providing insight into the structural effects of these modifications on G-tetrad geometry.
Methods
DNA sample preparation
The unlabeled and the site-specific low-enrichment (2% 15N-labeled) DNA oligonucleotides were chemically synthesized on a DNA synthesizer (model 394, Applied Biosystems, Foster City, CA) and purified as previously described (47). Phosphoramidites of guanine derivatives were purchased from Glen Research (Sterling, VA). 15N-labeled phosphoramidites were purchased from Cambridge Isotope (Andover, MA). Samples were dialyzed successively against KCl solution and against water.
UV spectroscopy
Experiments were performed on a Varian 300 Cary Bio UV-visible (UV-Vis) spectrophotometer. Samples contained 2–10 μM DNA in a solution of 20 mM potassium phosphate (pH 7). The thermal stability for each sequence was determined by recording the absorbance at 295 nm (48). Heating and cooling (15–88°C) was performed at a rate of 0.5°C/min. The quantitative melting temperature (Tm) data presented represent an average over a heating and cooling cycle. Hysteresis of <1.5°C was observed for all samples.
CD spectroscopy
Experiments were performed on a JASCO-815 spectropolarimeter (JASCO, Tokyo, Japan). Samples contained 2–10 μM DNA in a solution of 20 mM potassium phosphate (pH 7). Samples were heated to 90°C for 5 min and left to cool down to room temperature. CD spectra were monitored at 20°C from 220 to 320 nm and averaged over 10 accumulation cycles. Data were zero-corrected at 320 nm and finally normalized by strand concentration, determined by concurrent UV absorbance measurements at 260 nm.
NMR spectroscopy
Experiments were performed on 600 and 700 MHz Bruker (Billerica, MA) NMR spectrometers at 25°C using a jump-and-return-based water suppression pulse sequence (49,50). DNA concentration was typically 0.2–0.7 mM. Samples contained KCl (70 or 150 mM) and 20 mM potassium phosphate (pH 7). We also monitored several sequences in a low salt condition (20 mM potassium phosphate) and did not observe significant changes in NMR spectra. NMR chemical shifts were referenced with 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS). Resonances were assigned unambiguously for Gδ imino protons for most sequences using site-specific low-enrichment labeling (47).
Ab initio computation
Quantum mechanical calculations were performed using the GAUSSIAN 03 software (51). Molecular geometries were optimized at the Hartree-Fock level using four standard split-valance basis sets including 6-31G(d), 6-31G(d,p), 6-31++G(d,p), and 6-311++G(d,p) (52). Tight convergence criteria (51) were imposed for optimized geometries. NMR chemical shifts were calculated using the GIAO method (53) at the Hartree-Fock level using the above-mentioned standard split-valance basis sets as well as the 6-311+G(d) basis set (52).
Models consisted of four guanines arranged in a planar tetrad with two centrally located potassium ions placed at a distance of 1.7 Å above and below the tetrad plane. Initial guanines were arranged within the z=0 plane, allowing the tetrad to maintain planarity during optimization. The unmodified models underwent initial geometry optimization, resulting in a C4 symmetry with identical hydrogen-bond geometry. A single guanine was modified at the C8 position by adding a modification group (bromo-, amino-, or O-methyl-) within the plane of the base. In the case of OmetG, a cis conformation of the N7-C8-OR-CR bond was chosen. This was done to maintain planarity and was thought to be a more likely scenario than the trans conformation, which may clash with sugar-phosphate backbone in the G-quadruplex structure. Geometry optimization was again performed after substitution of a single guanine with a C8-modified guanine derivative. NMR chemical shift calculations were carried out on the unmodified and modified models.
A total of 16 individual molecular geometries (unmodified, bromo-, amino-, and O-methyl-containing models, each optimized with four different basis sets) were calculated. The geometries of modified tetrads were compared to the geometries of the unmodified tetrads (see Tables 5 and 6). A total of 80 individual chemical shifts (16 individual geometries evaluated with five different basis sets for NMR chemical-shift calculations) were computed (see Table 4 for a comparison of the chemical shifts of the modified and unmodified tetrads).
Table 5.
Computed effects of guanine substitutions on the N1-H1⋅⋅⋅O6 geometry of the tetrad
| Modification type | Optimization basis set | Computed change in H1⋅⋅⋅O6 length (pm) |
Computed change in N1–H1⋅⋅⋅O6 Angle (°) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Gα⋅Gβ | Gβ⋅Gγ | Gγ⋅Gδ | Gδ⋅Gα | Gα⋅Gβ | Gβ⋅Gγ | Gγ⋅Gδ | Gδ⋅Gα | ||
| 8-BromoG | 6-31G(d) | −0.97 | 0.99 | 1.57 | 5.02 | −1.12 | −0.12 | 0.00 | −1.51 |
| 6-31G(d,p) | −0.96 | 0.97 | 1.52 | 4.92 | −1.07 | −0.11 | 0.00 | −1.45 | |
| 6-31++G(d,p) | −1.05 | 0.44 | 0.66 | 2.96 | −0.60 | 0.00 | −0.01 | −0.69 | |
| 6-311++G(d,p) | −0.50 | 0.18 | 0.63 | 1.80 | −0.11 | 0.08 | −0.08 | −0.18 | |
| 8−OmethylG | 6-31G(d) | 1.08 | −1.97 | −2.11 | −10.14 | 3.80 | 0.46 | −0.22 | 3.35 |
| 6-31G(d,p) | 1.00 | −1.98 | −2.11 | −10.10 | 3.77 | 0.45 | −0.22 | 3.31 | |
| 6-31++G(d,p) | 1.13 | −2.10 | −2.11 | −10.26 | 3.89 | 0.47 | −0.21 | 3.41 | |
| 6-311++G(d,p) | 0.92 | −2.44 | −2.23 | −11.02 | 4.07 | 0.56 | −0.18 | 3.60 | |
| 8-AminoG | 6-31G(d) | −0.15 | −1.06 | −2.49 | −6.25 | 2.15 | 0.21 | 0.19 | 1.60 |
| 6-31G(d,p) | −0.14 | −1.08 | −2.50 | −6.30 | 2.12 | 0.21 | 0.18 | 1.60 | |
| 6-31++G(d,p) | −0.08 | −1.18 | −2.51 | −6.51 | 2.21 | 0.23 | 0.18 | 1.71 | |
| 6-311++G(d,p) | −0.35 | −1.36 | −2.70 | −6.83 | 2.30 | 0.29 | 0.24 | 1.79 | |
Geometry changes listed reflect deviations from the original values of bond length and angle as found in the unmodified optimized structures. Unmodified tetrad N1-H1⋅⋅⋅O6 hydrogen-bond geometries obtained are 6-31G(d), 214.33 pm and 157.04°; 6-31G(d,p), 214.07 pm and 157.18°; 6-31++G(d,p), 215.08 pm and 157.76°; and 6-311++G(d,p), 216.27 pm and 157.14°. Listed angles were measured toward the center of the tetrad. Variations of 2.20 pm and 0.72° were observed for hydrogen-bond length and angle, respectively, in the optimized geometries of unmodified tetrads across the basis sets explored.
Table 6.
Computed effects of guanine substitutions on the N2-H2⋅⋅⋅N7 geometry of the tetrad
| Modification type | Optimization basis set | Computed change in H2⋅⋅⋅N7 length (pm) |
Computed change in N2–H2⋅⋅⋅N7 Angle (°) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Gα⋅Gβ | Gβ⋅Gγ | Gγ⋅Gδ | Gδ⋅Gα | Gα⋅Gβ | Gβ⋅Gγ | Gγ⋅Gδ | Gδ⋅Gα | ||
| 8-BromoG | 6-31G(d) | 0.46 | 0.41 | 0.02 | 1.95 | −0.90 | −0.13 | −0.06 | −2.58 |
| 6-31G(d,p) | 0.41 | 0.38 | −0.01 | 2.03 | −0.86 | −0.12 | −0.05 | −2.48 | |
| 6-31++G(d,p) | −0.44 | 0.09 | −0.07 | 1.92 | −0.54 | −0.02 | −0.03 | −1.23 | |
| 6-311++G(d,p) | −0.58 | −0.17 | −0.13 | 2.76 | −0.17 | 0.05 | −0.08 | −0.49 | |
| 8−OmethylG | 6-31G(d) | −2.77 | −1.64 | −0.37 | 2.50 | 2.31 | 0.42 | −0.11 | 4.17 |
| 6-31G(d,p) | −2.88 | −1.67 | −0.36 | 2.50 | 2.30 | 0.42 | −0.11 | 4.12 | |
| 6-31++G(d,p) | −3.01 | −1.75 | −0.45 | 3.01 | 2.40 | 0.44 | −0.11 | 4.24 | |
| 6-311++G(d,p) | −3.25 | −1.89 | −0.52 | 2.97 | 2.52 | 0.52 | −0.08 | 4.33 | |
| 8-AminoG | 6-31G(d) | −0.84 | −0.76 | −0.28 | −2.78 | 1.02 | 0.20 | 0.22 | 2.53 |
| 6-31G(d,p) | −0.86 | −0.77 | −0.27 | −2.91 | 1.00 | 0.19 | 0.21 | 2.53 | |
| 6-31++G(d,p) | −0.98 | −0.82 | −0.30 | −2.56 | 1.07 | 0.21 | 0.21 | 2.65 | |
| 6-311++G(d,p) | −1.11 | −0.91 | −0.36 | −2.72 | 1.12 | 0.27 | 0.26 | 2.70 | |
Geometry changes listed reflect deviations from the original values of bond length and angle as found in the unmodified optimized structures. Unmodified tetrad N2-H2⋅⋅⋅N7 hydrogen-bond geometries obtained are 6-31G(d), 203.24 pm and 179.57°; 6-31G(d,p), 203.06 pm and 179.89°; 6-31++G(d,p), 204.72 pm and 180.28°; and 6-311++G(d,p), 204.60 pm and 180.08°. The angles listed were measured toward the center of the tetrad. Variations of 1.54 pm and 0.71° were observed for hydrogen-bond length and angle, respectively, in the optimized geometries of unmodified tetrads across the basis sets explored.
Table 4.
Computed change in imino proton (H1) NMR chemical shift of the Gδ residue
| Modification type | Optimization basis set | Computed change in Gδ H1 chemical shift (ppm) |
||||
|---|---|---|---|---|---|---|
| NMR basis set | ||||||
| 6-31G(d) | 6-31G(d,p) | 6-31++G(d,p) | 6-311+G(d) | 6-311++G(d,p) | ||
| 8-BromoG | 6-31G(d) | −0.289 | −0.311 | −0.304 | −0.300 | −0.307 |
| 6-31G(d,p) | −0.289 | −0.311 | −0.305 | −0.300 | −0.307 | |
| 6-31++G(d,p) | −0.235 | −0.249 | −0.243 | −0.245 | −0.242 | |
| 6-311++G(d,p) | −0.208 | −0.216 | −0.208 | −0.214 | −0.205 | |
| 8-OmethylG | 6-31G(d) | 0.398 | 0.461 | 0.442 | 0.347 | 0.453 |
| 6-31G(d,p) | 0.402 | 0.465 | 0.447 | 0.351 | 0.458 | |
| 6-31++G(d,p) | 0.398 | 0.461 | 0.443 | 0.350 | 0.455 | |
| 6-311++G(d,p) | 0.424 | 0.489 | 0.472 | 0.376 | 0.484 | |
| 8-AminoG | 6-31G(d) | 0.235 | 0.272 | 0.261 | 0.234 | 0.279 |
| 6-31G(d,p) | 0.241 | 0.279 | 0.267 | 0.239 | 0.285 | |
| 6-31++G(d,p) | 0.242 | 0.280 | 0.269 | 0.242 | 0.288 | |
| 6-311++G(d,p) | 0.248 | 0.286 | 0.275 | 0.247 | 0.294 | |
Values indicate the computed change in NMR imino proton chemical shift of the Gδ residue. Change in chemical shift is measured with respect to the computed spectra of the unmodified tetrad. NMR chemical-shift calculations were performed using multiple basis sets for each method of geometry optimization.
Results
Site-specific substitutions of a guanine with an 8-bromo-guanine, 8-O-methyl-guanine, 8-amino-guanine, or 8-oxo-guanine: G-quadruplex folding topology of modified sequences
In this study, we explored the effects of base modifications on an intramolecular (3 + 1) G-quadruplex structure (Fig. 1 a) formed by the human telomeric d[TT(GGGTTA)3GGGA] sequence (46), (henceforth referred to as the unmodified sequence). Individual guanines (G) (Fig. 1 c) were site-specifically substituted with four different guanine derivatives: 8-bromo-guanine (BrG) (Fig. 1 d), 8-O-methyl-guanine (OmetG) (Fig. 1 e), 8-amino-guanine (AmG) (Fig. 1 f), and 8-oxo-guanine (OxoG) (Fig. 1 g). Substitutions were made to guanines, which adopt syn (residues 3, 9, 15, 16, and 21) or anti (residue 22) glycosidic conformations in the original (3 + 1) G-quadruplex (Fig. 1 a). Each modified sequence used in this work contained only one or two substitutions (Table 1).
Table 1.
List of DNA sequences examined in this work
| Sequence name | Sequence |
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
19 |
20 |
21 |
22 |
23 |
24 |
|
| S | A | A | S | A | A | S | S | A | S | A | A | |||||||||||||
| Unmodified | T | T | G | G | G | T | T | A | G | G | G | T | T | A | G | G | G | T | T | A | G | G | G | A |
| Br-3 | T | T | B | G | G | T | T | A | G | G | G | T | T | A | G | G | G | T | T | A | G | G | G | A |
| Br-9 | T | T | G | G | G | T | T | A | B | G | G | T | T | A | G | G | G | T | T | A | G | G | G | A |
| Br-15 | T | T | G | G | G | T | T | A | G | G | G | T | T | A | B | G | G | T | T | A | G | G | G | A |
| Br-16 | T | T | G | G | G | T | T | A | G | G | G | T | T | A | G | B | G | T | T | A | G | G | G | A |
| Br-21 | T | T | G | G | G | T | T | A | G | G | G | T | T | A | G | G | G | T | T | A | B | G | G | A |
| Br-22 | T | T | G | G | G | T | T | A | G | G | G | T | T | A | G | G | G | T | T | A | G | B | G | A |
| Br-9-22 | T | T | G | G | G | T | T | A | B | G | G | T | T | A | G | G | G | T | T | A | G | B | G | A |
| Omet-3 | T | T | O | G | G | T | T | A | G | G | G | T | T | A | G | G | G | T | T | A | G | G | G | A |
| Omet-9 | T | T | G | G | G | T | T | A | O | G | G | T | T | A | G | G | G | T | T | A | G | G | G | A |
| Omet-15 | T | T | G | G | G | T | T | A | G | G | G | T | T | A | O | G | G | T | T | A | G | G | G | A |
| Omet-16 | T | T | G | G | G | T | T | A | G | G | G | T | T | A | G | O | G | T | T | A | G | G | G | A |
| Omet-21 | T | T | G | G | G | T | T | A | G | G | G | T | T | A | G | G | G | T | T | A | O | G | G | A |
| Omet-22 | T | T | G | G | G | T | T | A | G | G | G | T | T | A | G | G | G | T | T | A | G | O | G | A |
| Br-9-Omet-22 | T | T | G | G | G | T | T | A | B | G | G | T | T | A | G | G | G | T | T | A | G | O | G | A |
| Am-3 | T | T | M | G | G | T | T | A | G | G | G | T | T | A | G | G | G | T | T | A | G | G | G | A |
| Am-9 | T | T | G | G | G | T | T | A | M | G | G | T | T | A | G | G | G | T | T | A | G | G | G | A |
| Am-15 | T | T | G | G | G | T | T | A | G | G | G | T | T | A | M | G | G | T | T | A | G | G | G | A |
| Am-16 | T | T | G | G | G | T | T | A | G | G | G | T | T | A | G | M | G | T | T | A | G | G | G | A |
| Am-21 | T | T | G | G | G | T | T | A | G | G | G | T | T | A | G | G | G | T | T | A | M | G | G | A |
| Am-22 | T | T | G | G | G | T | T | A | G | G | G | T | T | A | G | G | G | T | T | A | G | M | G | A |
| Br-9-Am-22 | T | T | G | G | G | T | T | A | B | G | G | T | T | A | G | G | G | T | T | A | G | M | G | A |
| Oxo-3 | T | T | X | G | G | T | T | A | G | G | G | T | T | A | G | G | G | T | T | A | G | G | G | A |
| Oxo-9 | T | T | G | G | G | T | T | A | X | G | G | T | T | A | G | G | G | T | T | A | G | G | G | A |
| Oxo-15 | T | T | G | G | G | T | T | A | G | G | G | T | T | A | X | G | G | T | T | A | G | G | G | A |
Modified residues (bold letters) are 8-bromo-guanine (B), 8-O-methyl-guanine (O), 8-amino-guanine (M), and 8-oxo-guanine (X) in the syn (S) and anti (A) conformations. Numbers indicate residue positions (5′ to 3′).
CD profiles of modified sequences containing BrG, OmetG, and AmG modifications displayed a positive peak at 290 nm and a shoulder at 260 nm, similar to the CD profile of the unmodified sequence (Fig. 2 and Fig. S1), consistent with the formation of a (3 + 1) G-quadruplex as the major form in these sequences (54). Fig. 2 illustrates the CD spectra observed for sequences with single substitutions to syn position 9 and anti position 22, as well as double substitutions to both positions 9 and 22.
Figure 2.
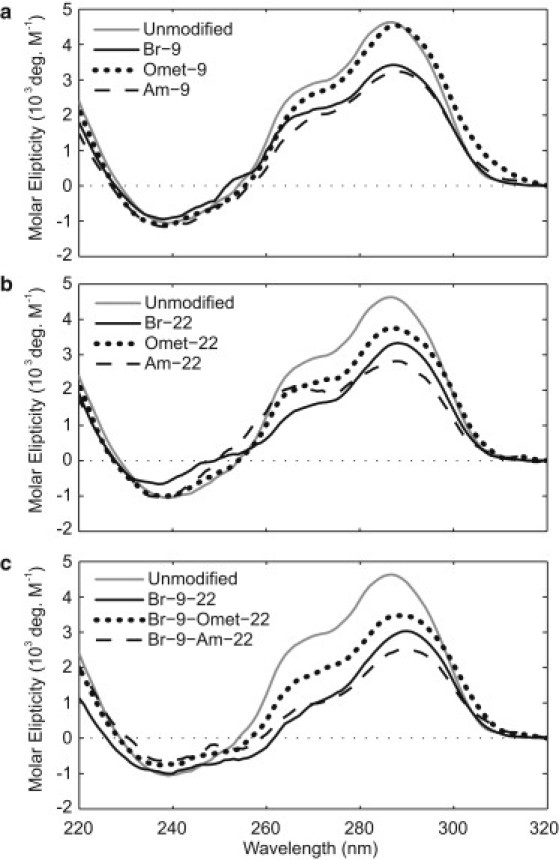
A comparison of CD spectra for sequences containing (a) single substitutions to syn residue 9, (b) single substitutions to anti residue 22, and (c) double substitutions to both syn residue 9 and anti residue 22. The CD spectrum of the unmodified sequence (gray line) is shown as a reference.
Imino proton NMR spectra were used to further assess the formation of G-quadruplexes. In all sequences containing BrG, OmetG, and AmG, the observation of ∼12 major imino protons at 10.5–12.5 ppm indicated the formation of a predominant G-quadruplex conformation (Fig. 3, a–d, Fig. S2, Fig. S3, Fig. S4, Fig. S5, Fig. S6, Fig. S7, Fig. S13, Fig. S14, and Fig. S15). The similar spectral pattern between these peaks and those of the unmodified sequence strongly suggest the formation of the same (3 + 1) G-quadruplex fold. Note that most changes in chemical shifts were observed only for protons from the G-tetrad containing the modified base (henceforth referred to as the modified tetrad). For example, in the spectra of sequences with a single BrG, OmetG, or AmG substitution at position 3 (Fig. 3), most changes were observed only for peaks from the modified G-tetrad G3⋅G21⋅G17⋅G9. This specific pattern in NMR chemical shift change is further investigated below.
Figure 3.
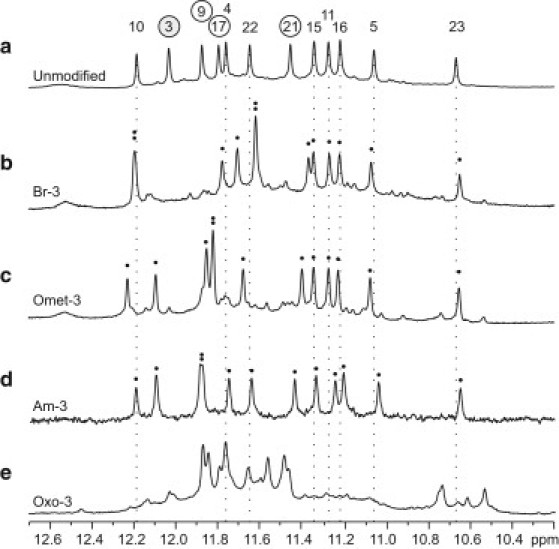
Imino proton NMR spectra for (a) the unmodified sequence, as well as modified sequences (b) Br-3, (c) Omet-3, (d) Am-3, and (e) Oxo-3 containing a single substitution to syn residue 3. The peaks in the unmodified spectrum are labeled with residue numbers, circled numbers representing guanines within the modified G3⋅G21⋅G17⋅G9 tetrad and the modified residue 3 shaded gray. Dotted lines identify the chemical shift of imino protons from the unmodified spectra for guanines located outside of the modified tetrad. Black dots indicate the presence of individual imino proton resonances.
The NMR spectra of sequences containing OxoG also gave imino proton peaks at 10.5–12.5 ppm (Fig. 3 e, Fig. S2, Fig. S3, and Fig. S16), indicating the formation of G-quadruplexes. However, these spectra were markedly different from that of the unmodified sequence, suggesting OxoG-containing sequences adopt different topologies.
Thermal stability of modified sequences
To characterize and compare the stability of G-quadruplexes formed by different modified sequences, UV melting experiments were performed by monitoring the absorption at 295 nm (48) (Fig. 4, Fig. S8, and Fig. S9). Reversible denaturing profiles were recorded for all sequences, and their melting temperatures (Tm) are summarized in Table 2.
Figure 4.
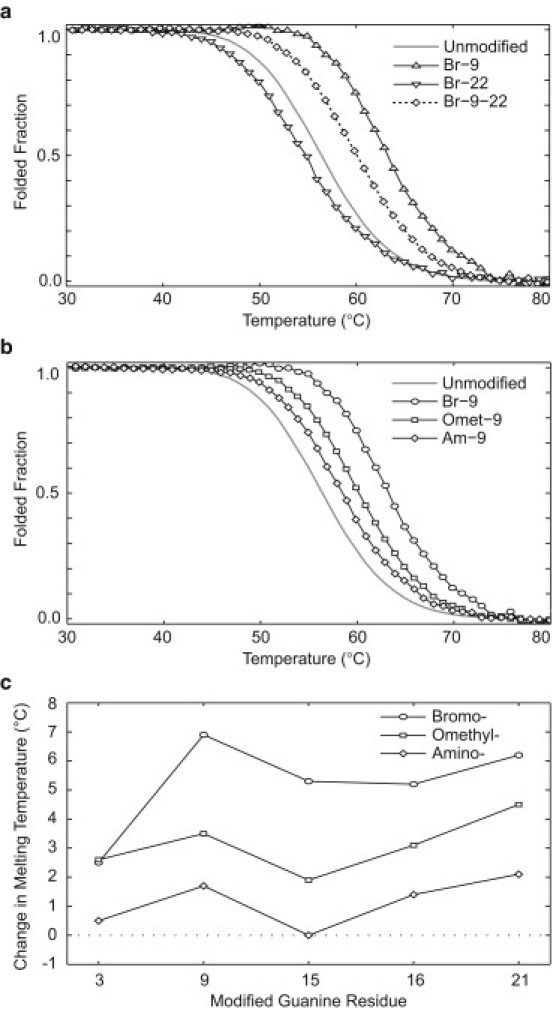
(a) Melting curves for sequences Br-9 (upward triangle solid line), containing a single syn substitution; Br-22 (downward triangle solid line), containing a single anti substitution; and Br-9-22 (diamonds with dotted line), containing both syn and anti substitutions. The curve for the unmodified sequence (gray solid line) serves as a reference. The additive effect of multiple substitutions on the Tm can be seen. (b) Modification-dependence stability (BrG > OmetG > AmG) was observed for substitutions to syn guanines, as illustrated by the melting curves of Br-9 (circles), Omet-9 (squares), and Am-9 (diamonds). The melting curve of the unmodified sequence is shown for reference. Melting curves presented are normalized UV cooling curves monitored at 295 nm. (c) Comparison of the change in Tm values for each guanine derivative over substitutions to various syn residues. Modifications are shown as circles (BrG), squares (OmetG), and diamonds (AmG).
Table 2.
Melting temperatures (Tm) determined by UV absorption at 295 nm
| Sequence name | Tm | ΔTm (°C) | Average ΔTm (°C)∗ |
|---|---|---|---|
| Unmodified | 57.0 | — | — |
| Br-3 | 59.5 | 2.5 | |
| Br-9 | 63.9 | 6.9 | |
| Br-15† | 62.3 | 5.3 | 5.2 ± 2.7 |
| Br-16 | 62.2 | 5.2 | |
| Br-21† | 63.2 | 6.2 | |
| Br-22 | 55.3 | −1.7 | — |
| Br-9-22 | 60.7 | 3.7 | — |
| Omet-3 | 59.6 | 2.6 | |
| Omet-9 | 60.5 | 3.5 | |
| Omet-15 | 58.9 | 1.9 | 3.1 ± 1.4 |
| Omet-16 | 60.1 | 3.1 | |
| Omet-21† | 61.5 | 4.5 | |
| Omet-22 | 59.0 | 2.0 | — |
| Br-9-Omet-22 | 64.6 | 7.6 | — |
| Am-3 | 57.5 | 0.5 | |
| Am-9 | 58.7 | 1.7 | |
| Am-15 | 57.0 | 0.0 | 1.1 ± 1.1 |
| Am-16 | 58.4 | 1.4 | |
| Am-21 | 59.1 | 2.1 | |
| Am-22 | 54.0 | −3.0 | — |
| Br-9-Am-22 | 59.3 | 2.3 | — |
| Oxo-3 | 47.2 | −9.8 | — |
| Oxo-9 | 38.7 | −18.3 | — |
| Oxo-15 | 38.5 | −18.5 | — |
Average Tm was calculated considering only the syn position substitutions for each modification type. Values are represented as the mean ± maximum deviation.
Samples with a small additional structural transition at low temperature in their denaturing profile.
We observed that single substitutions (BrG, OmetG, and AmG) at syn residues resulted in an increase in Tm (except for the AmG substitution at position 15, which showed no change), whereas single substitutions at anti position 22 showed different small Tm changes (Tm decreased for BrG and AmG and increased for OmetG) (Table 2 and Fig. S8). Among different syn guanines, the stabilization effect of these base substitutions was highly position-dependent, with somewhat similar trends for the three types of substitutions (Fig. 4 c). It can be seen that some residues, such as residues 3 and 15, showed low stability enhancement when substituted with C8-modified guanine derivatives, whereas others, such as residues 9 and 21, showed relatively high stability enhancement. In a comparison of the stability of sequences containing single-position syn substitutions, BrG generally had the highest stabilizing effect, followed by OmetG and AmG, respectively (Fig. 4, b and c). Alternatively, when substituting OxoG to syn positions 3, 9, and 15 a large destabilizing effect was observed. Due to the large differences in NMR spectra of OxoG-containing sequences compared to that of the unmodified sequence, we do not discuss the destabilizing effects of OxoG modifications in terms of the (3 + 1) G-quadruplex scaffold.
Sequences containing double substitutions showed a melting temperature reflective of the contributions of individual single substitution counterparts (Table 2). For example, the changes in Tm for sequences harboring BrG substitutions to syn residue 9 only, anti residue 22 only, and both residues 9 and 22, were +6.9, −1.7, and +3.7°C, respectively (Fig. 4 a and Table 2).
A single guanine substitution in a G-tetrad results in specific patterns of NMR chemical shift changes: experimental observations
Analysis of the imino proton spectra of various DNA sequences containing a single BrG, OmetG, or AmG substitution at a syn guanine (Table 1) revealed specific patterns of chemical-shift changes: 1), the most significant chemical-shift changes were observed for the modified G-tetrad; 2), the largest change was observed for the guanine that is hydrogen-bonded to the Hoogsteen edge of the modified base (note that these residues, although neighbors in the structure, are usually remote from each other in the sequence); and 3), the magnitude and direction of chemical-shift changes depended on the type of modification. Experimental data and details of these chemical-shift patterns are described below.
Considering a Gα⋅Gβ⋅Gγ⋅Gδ tetrad in which Gα is the modified base (Fig. 5 a), the largest change in imino proton chemical shift would be observed for Gδ, which is hydrogen-bonded to the Hoogsteen edge of Gα. For each sequence with a single BrG, OmetG, or AmG substitution at a syn guanine, the imino proton of Gδ was unambiguously assigned using a site-specific 15N-labeling approach (47) (see, e.g., Fig. 5 f, Fig. S10, Fig. S11, and Fig. S12). Table 3 lists the changes in chemical shifts for sequences containing single substitutions at syn guanines. For BrG substitutions, the Gδ imino proton peak was upfield-shifted; the chemical shift decreased by 0.26–0.39 ppm (Fig. 5 c and Table 3). Conversely, for OmetG and AmG substitutions, the Gδ imino proton peak was downfield-shifted; the chemical shift increased by 0.22–0.40 and 0.19–0.27 ppm, respectively (Fig. 5, d and e, and Table 3).
Figure 5.
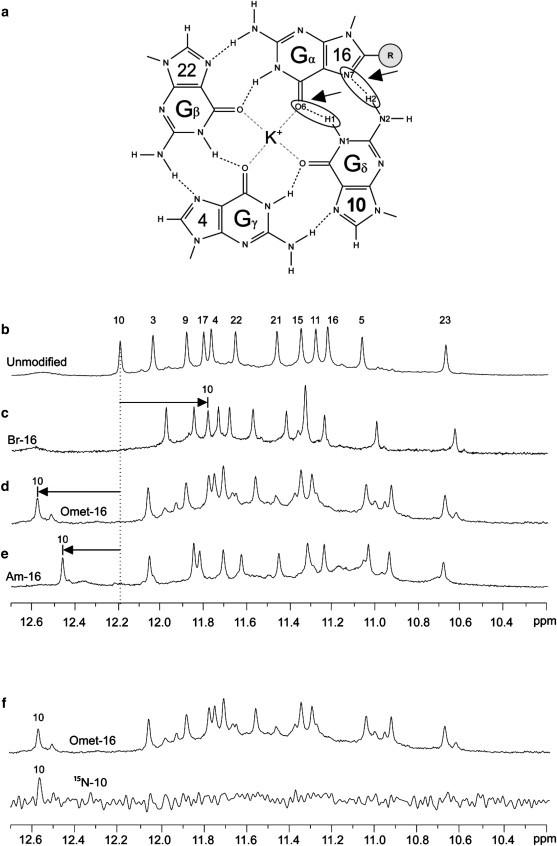
(a) Schematic of the modified G16⋅G22⋅G4⋅G10 tetrad is presented with the Gα⋅Gβ⋅Gγ⋅Gδ naming convention. Special attention is brought to the hydrogen bonds of the Gδ⋅Gα interface. (b–e) The NMR imino proton spectrum of (b) the unmodified sequence is presented along with the spectra of (c) Br-16, (d) Omet-16, and (e) Am-16. The chemical shift of the imino proton peak from the Gδ residue, G10, in the unmodified sequence is indicated by the dotted black line. Arrows indicate the positions of the Gδ imino proton peaks in the modified spectra. (f) Example of the unambiguous G10 imino proton assignment for Omet-16 using 15N-labeling.
Table 3.
Experimentally observed changes in NMR chemical shift of the Gδ imino proton (H1)
| Modification type | Change in Gδ H1 chemical shift (ppm) |
||||
|---|---|---|---|---|---|
| Neighboring residue (Gδ)⋅Modified residue (Gα) | |||||
| 9⋅3 | 17⋅9 | 11⋅15 | 10⋅16 | 3⋅21 | |
| 8-BromoG | −0.26 | −0.32 | −0.28 | −0.39 | −0.28 |
| 8-OmethylG | 0.22 | 0.24 | 0.40 | 0.39 | 0.30 |
| 8-AminoG | 0.21 | 0.26 | 0.26 | 0.27 | 0.19 |
Values indicate the observed change in imino proton chemical shift of the Gδ residue as measured for single-position substitutions to syn residues. Identified by unambiguous 15N-labeling experiments, change in chemical shift is measured with respect to the spectra of the unmodified sequence.
Tentative spectral assignments also suggested a specific pattern of chemical-shift changes for the imino proton of the modified guanine Gα, which was in the opposite direction of the Gδ imino proton and in agreement with computational studies (data not shown). However, as these assignments were not unambiguously confirmed, we do not discuss their patterns here.
A single guanine substitution in a G-tetrad results in specific patterns of NMR chemical-shift changes: computational studies
To better understand the properties of a G-tetrad, we attempted to reproduce the experimentally observed patterns of chemical-shift changes due to base modifications using a series of ab initio computational studies. Computations were performed at the Hartree-Fock level (52) using a variety of basis sets, as described in the Methods section. NMR chemical-shift calculations were performed on optimized molecular geometries for modified and unmodified tetrads. The changes in NMR chemical shifts between the unmodified and modified systems are shown in Table 4. All calculation protocols employed were able to reproduce the experimentally observed trends in Gδ imino proton chemical-shift changes, with a decrease of 0.21–0.31 ppm calculated for the BrG-containing model and increases of 0.35–0.49 and 0.23–0.29 ppm calculated for the OmetG- and AmG-containing models, respectively. Good agreement was found between the computed and experimentally observed values for chemical-shift changes (Fig. 6).
Figure 6.
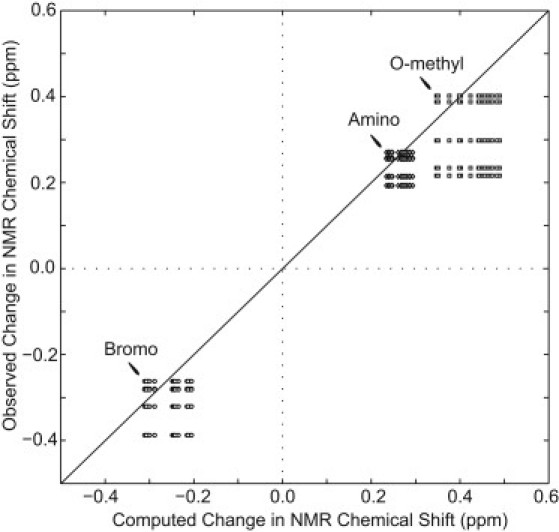
Comparison of experimental and computed NMR chemical-shift changes. Experimental data are observed chemical-shift changes of the Gδ imino protons upon substitutions to syn residues 3, 9, 15, 16, and 21 by guanine derivatives BrG (circles), OmetG (squares), and AmG (diamonds). Computational data are the results of different NMR chemical-shift calculations, as shown in Table 4. The solid black line represents exact agreement between computational and experimental results. Data points are shown for every combination of experimentally observed and computed changes in chemical shift for each type of guanine derivative.
Structural effects of modifications on G-tetrad geometry: an ab initio computational study
As our computational studies correctly reproduced the experimentally observed chemical-shift patterns, we used these computational methods to analyze the structural effects of base modifications on the G-tetrad geometry. Generally, the most significant changes in geometry occurred at the Gδ⋅Gα and Gα⋅Gβ interfaces (refer to Fig. 5 a). Calculated changes to imino and amino hydrogen-bond geometries at the Gδ⋅Gα interface are presented in Tables 5 and 6, respectively. Comparison between the changes in Gδ imino proton chemical shifts (Table 3) and the changes in H1δ⋅⋅⋅O6α bond length (Table 5) shows an increase in imino proton chemical shifts with a decrease in hydrogen-bond lengths, and vice versa. We also observed the largest changes in hydrogen-bond geometry at the Gδ⋅Gα interface regarding N1δ-H1δ⋅⋅⋅O6α angle (Table 5), H2δ⋅⋅⋅N7α bond length (Table 6), and N2δ-H2δ⋅⋅⋅N7α angle (Table 6). However, it is difficult to relate these changes to the Gδ imino proton chemical shifts. In addition, partial atomic charges were derived from the electrostatic potentials of tetrads with optimized geometries. We were unable to observe a relationship between partial atomic charges and NMR chemical-shift changes, as the calculated partial charge highly depended on the basis set used for geometry optimization.
Discussion
This study examines the effects of site-specific base modifications of four C8-substituted guanine derivatives (BrG, OmetG, AmG, and OxoG) in various positions of a G-quadruplex structure. The chosen DNA sequence formed an intramolacular (3 + 1) G-quadruplex with well-defined spectroscopic characteristics, and the folding topology of the G-quadruplex was controlled and monitored by CD and NMR spectra, allowing a quantitative comparison of the effects of base modifications in different positions of the same G-quadruplex fold for BrG, OmetG, and AmG modifications. Although minor conformations were present for some modified sequences (Fig. S13, Fig. S14, Fig. S15, and Fig. S16), their relatively small populations should not affect the interpretations regarding the stability of the major fold.
This work reports that single-position BrG, OmetG, and AmG substitutions to the five syn guanines of a (3 + 1) G-quadruplex scaffold generally lead to an increase in thermal stability. These results agree with past findings that substitutions of syn guanines by C8-modified guanine derivatives 8-bromo-guanine (31,32) and 8-methyl-guanine (36) stabilize G-quadruplexes and extend this conclusion for 8-O-methyl-guanine and 8-amino-guanine substitutions. Among the C8-modifications explored, the most stabilizing was generally found to be BrG, followed by OmetG and AmG (Fig. 4 b and Table 2). Thermal denaturing results indicated a position-dependent stabilization effect of single C8-modified derivatives among the various syn residues (Fig. 4 c). This position dependence may be explained by the different local structural environments among the various syn positions, such as neighboring loops, grooves, and tetrad layers. A position-dependence effect has also been demonstrated in other scaffolds containing C8-modifications (31,36).
Although the expected stabilization of G-quadruplexes by C8-modified guanine substitutions BrG, OmetG, and AmG to syn positions was confirmed, the observation of a stabilizing effect from an OmetG substitution to anti guanine 22 was unanticipated, as previous studies exploring C8-modified guanine substitutions to anti positions in intramolecular scaffolds have reported destabilizing effects (31,32,55). To our knowledge, this is the first reported observation of a site-specific C8-modified substitution (8-O-methyl-guanine) stabilizing an intramolecular G-quadruplex system when substituted into an anti position. Similarities between the CD and imino proton NMR spectra of the sequence containing an OmetG substitution at position 22 and those of the unmodified sequence suggest that the same folding topology is maintained and the O-methyl-guanine adopts an anti conformation in this modified sequence. It is speculated that OmetG may constitute a unique type of modification due to its ability to rotate about the N7-C8-OR-CR bond. The ability for anti positions to tolerate the inclusion of C8-modified guanine derivatives may be dependent on specific structural features. However, the generalization of such dependence would require more systematic study and is outside the scope of this work. Substitutions were also made to anti residue 23, with destabilizing effects observed for BrG, AmG, and OmetG (data not shown). The effects of modifications to residue 23 are not discussed here, as the folding topology could not be confirmed due to significant changes in CD and NMR spectra.
The observation of specific NMR chemical-shift patterns due to base modifications could be a tool in structural elucidation or verification, as the observed cross-hydrogen-bond effect occurs at a remote site in the sequence. For example, in the (3 + 1) human telomeric G-quadruplex structure considered above, a modification at position 3 would cause a predictable change in the imino proton chemical shift of residue 9, whereas a modification at position 9 would cause a change in position 17, and so forth. In this manner, site-specific modifications could be used in NMR studies to identify Hoogsteen hydrogen-bound guanines within a G-quadruplex structure.
Quantum mechanical calculations of NMR chemical shifts provide a valuable link between experimentally observable chemical shifts and calculated molecular geometries (56,57). Previously, NMR was used to probe the effects of ion type and temperature on hydrogen-bond length within G-tetrads (58). The changes in NMR observables, namely N7 chemical shifts and J-couplings, were interpreted in terms of hydrogen-bond length and substantiated by quantum mechanical calculations. A similar approach was also used to study hydrogen-bond length and strength in a triple helix (57).
In this manner, observed chemical-shift patterns were used to investigate the effects of modifications on hydrogen-bond geometry. In this study, ab initio NMR chemical shift calculations were found to reproduce the observed changes in Gδ imino proton chemical shifts to a high degree (Fig. 6 and Tables 3 and 4). The difference between modification-driven chemical-shift changes calculated by different methods was in the vicinity of 0.11 ppm, 0.14 ppm, and 0.06 ppm for the BrG-, OmetG- and AmG-containing models, respectively. These data are in agreement with studies by Kumar and McAllister (56), indicating that relative NMR chemical shift calculations are quite accurate.
The computed effects of modifications on tetrad geometries revealed that the largest change in hydrogen-bond geometry was at the Gδ⋅Gα interface (Tables 5 and 6). Calculations revealed a relationship between decreasing H1δ⋅⋅⋅O6α hydrogen-bond length and downfield shifting (increased ppm) Gδ of imino proton chemical shifts and vice versa. This trend was obtained across all choices of geometry optimization basis sets explored in this study. These results are in agreement with those of previous studies (56,57) that reported a similar relationship between hydrogen-bond length and proton chemical shift. It is important to note that NMR chemical-shift calculations performed on modified tetrads before geometry reoptimization (data not shown) did not fully reproduce experimental data, especially for the OmetG- and AmG-containing models. This indicates that a structural change in G-tetrad geometry is an important feature of modification-driven changes in NMR chemical shift.
A modification-driven change in hydrogen-bond geometry may not be the only factor contributing to the observed changes in imino proton chemical shift. Changes in base-stacking geometry caused by C8-substitutions may lead to changes in the ring-current-driven magnetic shielding experienced by imino protons. However, a similar pattern in NMR chemical-shift change was observed in experiments for guanines from all three G-tetrad layers in the structure. This indicates that for the modifications explored here, the effect of base-stacking changes may be small.
The effects of C8 modifications on thermal stability, as well as the hydrogen-bond geometry of a G-tetrad are important fundamentals of G-quadruplex structures. Understanding these fundamental properties should be valuable in the engineering of new G-quadruplexes, as well as the design of molecules targeting these structures. Computational models explored here, as supported by the agreement between computational and experimental NMR data, show great potential for investigation of the effects of different guanine derivatives on tetrad structure.
Conclusion
We examined the effects of site-specific substitution of a guanine by guanine derivatives 8-bromo-guanine, 8-O-methyl-guanine, 8-amino-guanine, and 8-oxo-guanine, using a well-defined (3 + 1) human telomeric G-quadruplex as the platform. It was observed that 8-bromo-guanine has the highest stabilizing effect when substituted with a single syn guanine, followed by 8-O-methyl-guanine and 8-amino-guanine, respectively. Substitutions of 8-oxo-guanine lead to drastic changes in NMR imino proton spectra and a large decrease in stability. The effect of substitution on the stability of the G-quadruplex is highly position-dependent among different guanines in the structure. A C8 modification of a guanine results in a specific pattern of chemical-shift changes: the imino proton of the guanine that is hydrogen-bonded to the modified base experiences the largest chemical shift change, which depends on the type of modification. This specific chemical-shift pattern was successfully reproduced by ab initio quantum mechanical computations. The computational study performed here suggests the effects of various C8-modified guanine substitutions on the Hoogsteen hydrogen-bond geometry within a G-tetrad.
Acknowledgments
We thank Prof. Kuo Jer-Lai and Prof. Shen Zexiang for the use of computational resources.
This research was supported by Singapore Biomedical Research Council grant 07/1/22/19/542 and Singapore Ministry of Education grant ARC30/07 to A.T.P.
Supporting Material
References
- 1.Davis J.T. G-quartets 40 years later: From 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 2.Patel D.J., Phan A.T., Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Zahler A.M., Williamson J.R., Prescott D.M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 5.Paeschke K., Simonsson T., Lipps H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 6.Oganesian L., Moon I.K., Jarstfer M.B. Extension of G-quadruplex DNA by ciliate telomerase. EMBO J. 2006;25:1148–1159. doi: 10.1038/sj.emboj.7601006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 8.Lipps H.J., Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Siddiqui-Jain A., Grand C.L., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumari S., Bugaut A., Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun D.Y., Thompson B., Hurley L.H. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 12.Mergny J.L., Hélène C. G-quadruplex DNA: a target for drug design. Nat. Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 13.Balasubramanian S., Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bock L.C., Griffin L.C., Toole J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 15.Schultze P., Macaya R.F., Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 16.Martino L., Virno A., Mayol L. A new modified thrombin binding aptamer containing a 5′-5′ inversion of polarity site. Nucleic Acids Res. 2006;34:6653–6662. doi: 10.1093/nar/gkl915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saneyoshi H., Mazzini S., Eritja R. Conformationally rigid nucleoside probes help understand the role of sugar pucker and nucleobase orientation in the thrombin-binding aptamer. Nucleic Acids Res. 2009;37:5589–5601. doi: 10.1093/nar/gkp598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyatt J.R., Vickers T.A., Ecker D.J. Combinatorially selected guanosine-quartet structure is a potent inhibitor of human immunodeficiency virus envelope-mediated cell fusion. Proc. Natl. Acad. Sci. USA. 1994;91:1356–1360. doi: 10.1073/pnas.91.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jing N., Marchand C., Pommier Y. Mechanism of inhibition of HIV-1 integrase by G-tetrad-forming oligonucleotides in vitro. J. Biol. Chem. 2000;275:21460–21467. doi: 10.1074/jbc.M001436200. [DOI] [PubMed] [Google Scholar]
- 20.de Soultrait V.R., Lozach P.Y., Andréola M.L. DNA aptamers derived from HIV-1 RNase H inhibitors are strong anti-integrase agents. J. Mol. Biol. 2002;324:195–203. doi: 10.1016/s0022-2836(02)01064-1. [DOI] [PubMed] [Google Scholar]
- 21.Phan A.T., Kuryavyi V., Patel D.J. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc. Natl. Acad. Sci. USA. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bates P.J., Kahlon J.B., Miller D.M. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J. Biol. Chem. 1999;274:26369–26377. doi: 10.1074/jbc.274.37.26369. [DOI] [PubMed] [Google Scholar]
- 23.Simonsson T., Henriksson M. c-myc Suppression in Burkitt's lymphoma cells. Biochem. Biophys. Res. Commun. 2002;290:11–15. doi: 10.1006/bbrc.2001.6096. [DOI] [PubMed] [Google Scholar]
- 24.Jing N., Li Y., Tweardy D.J. G-quartet oligonucleotides: a new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004;64:6603–6609. doi: 10.1158/0008-5472.CAN-03-4041. [DOI] [PubMed] [Google Scholar]
- 25.Qi H.Y., Lin C.P., Liu L.F. G-quadruplexes induce apoptosis in tumor cells. Cancer Res. 2006;66:11808–11816. doi: 10.1158/0008-5472.CAN-06-1225. [DOI] [PubMed] [Google Scholar]
- 26.Gellert M., Lipsett M.N., Davies D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phan A.T. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 28.Phan A.T., Kuryavyi V., Patel D.J. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu L.Y., Lim K.W., Phan A.T. Giardia telomeric sequence d(TAGGG)4 forms two intramolecular G-quadruplexes in K+ solution: effect of loop length and sequence on the folding topology. J. Am. Chem. Soc. 2009;131:16824–16831. doi: 10.1021/ja905611c. [DOI] [PubMed] [Google Scholar]
- 30.Lim K.W., Lacroix L., Phan A.T. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 2010;132:12331–12342. doi: 10.1021/ja101252n. [DOI] [PubMed] [Google Scholar]
- 31.Dias E., Battiste J.L., Williamson J.R. Chemical probe for glycosidic conformation in telomeric DNAs. J. Am. Chem. Soc. 1994;116:4479–4480. [Google Scholar]
- 32.Xu Y., Noguchi Y., Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 33.Dumas A., Luedtke N.W. Cation-mediated energy transfer in G-quadruplexes revealed by an internal fluorescent probe. J. Am. Chem. Soc. 2010;132:18004–18007. doi: 10.1021/ja1079578. [DOI] [PubMed] [Google Scholar]
- 34.Gros J., Rosu F., Mergny J.L. Guanines are a quartet's best friend: impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Res. 2007;35:3064–3075. doi: 10.1093/nar/gkm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mekmaysy C.S., Petraccone L., Chaires J.B. Effect of O6-methylguanine on the stability of G-quadruplex DNA. J. Am. Chem. Soc. 2008;130:6710–6711. doi: 10.1021/ja801976h. [DOI] [PubMed] [Google Scholar]
- 36.Xu Y., Sugiyama H. Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb) Nucleic Acids Res. 2006;34:949–954. doi: 10.1093/nar/gkj485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gros J., Aviñó A., Mergny J.L. 8-Amino guanine accelerates tetramolecular G-quadruplex formation. Chem. Commun. (Camb.) 2008;25:2926–2928. doi: 10.1039/b801221k. [DOI] [PubMed] [Google Scholar]
- 38.Tran P.L., Virgilio A., Galeone A. Effects of 8-methylguanine on structure, stability and kinetics of formation of tetramolecular quadruplexes. Biochimie. 2011;93:399–408. doi: 10.1016/j.biochi.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 39.Esposito V., Randazzo A., Mayol L. Effects of an 8-bromodeoxyguanosine incorporation on the parallel quadruplex structure [d(TGGGT)]4. Org. Biomol. Chem. 2004;2:313–318. doi: 10.1039/b314672c. [DOI] [PubMed] [Google Scholar]
- 40.Virgilio A., Esposito V., Galeone A. 8-methyl-2′-deoxyguanosine incorporation into parallel DNA quadruplex structures. Nucleic Acids Res. 2005;33:6188–6195. doi: 10.1093/nar/gki924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grollman A.P., Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 42.David S.S., O'Shea V.L., Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan X.Z., Suzuki N., Shibutani S. Mutagenic properties of the 8-amino-2′-deoxyguanosine DNA adduct in mammalian cells. Nucleic Acids Res. 1999;27:2310–2314. doi: 10.1093/nar/27.11.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venkatarangan L., Sivaprasad A., Basu A.K. Site-specifically located 8-amino-2′-deoxyguanosine: thermodynamic stability and mutagenic properties in Escherichia coli. Nucleic Acids Res. 2001;29:1458–1463. doi: 10.1093/nar/29.7.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shrivastav N., Li D.Y., Essigmann J.M. Chemical biology of mutagenesis and DNA repair: cellular responses to DNA alkylation. Carcinogenesis. 2010;31:59–70. doi: 10.1093/carcin/bgp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luu K.N., Phan A.T., Patel D.J. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phan A.T., Patel D.J. A site-specific low-enrichment 15N,13C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002;124:1160–1161. doi: 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]
- 48.Mergny J.L., Phan A.T., Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 49.Plateau P., Gueron M. Exchangeable proton NMR without base-line distortion, using new strong-pulse sequences. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- 50.Phan A.T., Guéron M., Leroy J.L. Investigation of unusual DNA motifs. Methods Enzymol. 2001;338:341–371. doi: 10.1016/s0076-6879(02)38228-4. [DOI] [PubMed] [Google Scholar]
- 51.Frisch, M. J., G. W. Trucks, …, S. S. Iyengar. 2004. Gaussian 03, Rev. e.01. Gaussian, Wallingford, CT.
- 52.Hehre W.J., Radom L., Pople J.A. Wiley-Interscience; New York: 1986. Ab Initio Molecular Orbital Theory. [Google Scholar]
- 53.Ditchfield R. Self-consistent perturbation theory of diamagnetism. 1. Gauge-invariant LCAO method for NMR chemical shifts. Mol. Phys. 1974;27:789–807. [Google Scholar]
- 54.Gray D.M., Wen J.D., Fleischhauer J. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality. 2008;20:431–440. doi: 10.1002/chir.20455. [DOI] [PubMed] [Google Scholar]
- 55.López de la Osa J., González C., Eritja R. Destabilization of quadruplex DNA by 8-aminoguanine. Chem. Bio. Chem. 2006;7:46–48. doi: 10.1002/cbic.200500281. [DOI] [PubMed] [Google Scholar]
- 56.Kumar G.A., McAllister M.A. Theoretical investigation of the relationship between proton NMR chemical shift and hydrogen bond strength. J. Org. Chem. 1998;63:6968–6972. doi: 10.1021/jo980759h. [DOI] [PubMed] [Google Scholar]
- 57.Barfield M., Dingley A.J., Grzesiek S. A DFT study of the interresidue dependencies of scalar J-coupling and magnetic shielding in the hydrogen-bonding regions of a DNA triplex. J. Am. Chem. Soc. 2001;123:4014–4022. doi: 10.1021/ja003781c. [DOI] [PubMed] [Google Scholar]
- 58.Dingley A.J., Peterson R.D., Feigon J. Characterization of the cation and temperature dependence of DNA quadruplex hydrogen bond properties using high-resolution NMR. J. Am. Chem. Soc. 2005;127:14466–14472. doi: 10.1021/ja0540369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


