Abstract
Although allelic diversity of genes has been shown to contribute to many phenotypic variations associated with different physiological processes in plants, information on allelic diversity of abiotic stress-responsive genes is limited. Here it is shown that the alleles OsWRKY45-1 and OsWRKY45-2 play different roles in abscisic acid (ABA) signalling and salt stress adaptation in rice. The two alleles had different transcriptional responses to ABA and salt stresses. OsWRKY45-1-overexpressing lines showed reduced ABA sensitivity, whereas OsWRKY45-1-knockout lines showed increased ABA sensitivity. OsWRKY45-1 transgenic plants showed no obvious difference from negative controls in response to salt stress. In contrast, OsWRKY45-2-overexpressing lines showed increased ABA sensitivity and reduced salt stress tolerance, and OsWRKY45-2-suppressing lines showed reduced ABA sensitivity and increased salt stress tolerance. OsWRKY45-1 and OsWRKY45-2 transgenic plants showed differential expression of a set of ABA- and abiotic stress-responsive genes, but they showed similar responses to cold and drought stresses. These results suggest that OsWRKY45-1 negatively and OsWRKY45-2 positively regulates ABA signalling and, in addition, OsWRKY45-2 but not OsWRKY45-1 negatively regulates rice response to salt stress. The different roles of the two alleles in ABA signalling and salt stress may be due to their transcriptional mediation of different signalling pathways.
Keywords: Abiotic stress, biotic stress, Oryza sativa, transcription factor
Introduction
In plants, naturally occurring intraspecific allelic diversity results in phenotypic variations. Different combinations of alleles generate numerous varieties in a species. Allelic diversity has been reported to contribute to variation in development, flower morphology, fruit and seed morphology and colour, grain weight and quality, plant architecture, disease resistance specificity, and pathogen-induced defence signalling (Jones, 2001; Alonso-Blanco et al., 2005; Fan et al., 2006; Bhave and Morris, 2008; Tao et al., 2009). Allelic diversity may also provide clues to understanding heterosis (Springer and Stupar, 2007).
Genomic analyses of the transcriptomes of plant genomes have revealed that many genes are involved in plant responses and adaptation to diverse abiotic stresses, such as drought, low temperature, and high salinity (Seki et al., 2002; Mahajan et al., 2005). Although relatively few genes have been characterized for their functions in plant tolerance to abiotic stresses, these genes function either positively or negatively in the pathways leading to stress adaptation. As regulatory proteins, transcription factors play important roles in almost every stress response. Much progress has been made in the characterization of rice transcription factors involved in response to abiotic stresses. At least four NAC-type transcription factors positively regulate rice tolerance to drought, salt, or cold stresses (Hu et al., 2006, 2008; Zheng et al., 2009; Jeong et al., 2010). Several bZIP-type transcription factors positively or negatively mediate rice adaptation to abiotic stresses (Xiang et al., 2008; Zou et al., 2008; Lu et al., 2009; Amir Hossain et al., 2010), and the transcription factors AP2/EREBP, MYB, and zinc-finger also play important roles in rice responses to various abiotic stresses (Xu et al., 2006; Chen et al., 2008; Huang et al., 2009; Ma et al., 2009).
The WRKY transcription factor family consists of 98–102 members in different rice varieties (Ross et al., 2007). The expression of ∼40% of the members was in response to abiotic stresses (Ramamoorthy et al., 2008), suggesting that WRKY-type regulators may play important roles in rice adaptation to abiotic stresses. However, only three rice WRKY genes have been functionally characterized for their roles in abiotic stress responses in rice. OsWRKY13 negatively regulates rice tolerance to salt and cold stresses, and its role in tolerance may be at least partly due to suppressing the expression of SNAC1, which encodes a transcription factor that regulates rice response to abiotic stresses (Hu et al., 2006; Qiu et al., 2008). Overexpression of OsWRKY89 enhanced tolerance to ultraviolet B irradiation (Wang et al., 2007). Overexpression of OsWRKY11 under the control of the heat shock protein 101 (HSP101) promoter enhanced heat and drought tolerance in rice (Wu et al., 2009). Although a number of genes have been shown to function in pathways leading to the adaptation of various abiotic stresses in plants, the allelic diversity of abiotic stress-responsive genes is poorly understood.
The phytohormone abscisic acid (ABA) is involved in abiotic stress responses (Verslues and Zhu, 2005), in addition functioning in various aspects of plant growth throughout development (Himmelbach et al., 2003; Finkelstein et al., 2008). ABA regulates stomatal opening and ion channel activity in plant guard cells when a plant adapts to water deficiency and salt stress (Verslues and Zhu, 2005). ABA is also involved in the transcriptional and post-transcriptional regulation of some stress-responsive genes (Chinnusamy et al., 2004). However, there is also ABA-independent signalling in plant responses to abiotic stresses (Chinnusamy et al., 2004; Yamaguchi-Shinozaki and Shinozaki, 2006). Accumulating evidence suggests that ABA is also involved in host–pathogen interactions. In most cases, ABA functions as a negative regulator in pathogen-induced defence signalling, although it sometimes acts as a positive regulator (Bari and Jones, 2009). The multiple roles of ABA in abiotic and biotic stresses suggest that it may be an important signalling molecule in the cross-talk of transduction pathways leading to abiotic stress tolerance and disease resistance. However, the molecular mechanisms of ABA in this cross-talk are poorly understood.
Asian cultivated rice (Oryza sativa L.) consists of two major groups, which are known by the subspecies names indica (O. sativa L. ssp. indica) and japonica (O. sativa L. ssp. japonica). A previous study revealed that OsWRKY45-1 from japonica rice and OsWRKY45-2 from indica rice are alleles located on rice chromosome 5 (Tao et al., 2009). The two alleles, which encode proteins with a difference of 10 amino acids, play opposite roles in rice resistance against bacterial pathogens. OsWRKY45-1 negatively and OsWRKY45-2 positively regulates rice resistance against Xanthomonas oryzae pv. oryzae (Xoo), which causes bacterial blight disease, and Xanthomonas oryzae pv. oryzicola, which causes bacterial streak disease. However, both alleles positively regulate rice resistance against Magnaporthe grisea, which causes fungal blast disease. The opposite roles of the two alleles in rice–bacterium interactions appear to be due to their mediation of different defence signalling pathways (Tao et al., 2009). In rice–pathogen interactions, the expression of OsWRKY45-1 and OsWRKY45-2 is regulated by another transcriptional regulator, OsWRKY13 (Qiu et al., 2007, 2009). Furthermore, both OsWRKY45-1 and OsWRKY45-2 appear to regulate OsWRKY13 expression in rice–Xoo interaction (Tao et al., 2009). OsWRKY13 positively regulates rice resistance against Xoo and M. grisea (Qiu et al., 2007) but negatively regulates rice tolerance to abiotic stresses (Qiu et al., 2008). These results suggest that OsWRKY45-1 and OsWRKY45-2 may also mediate the cross-talk between abiotic and biotic stresses, as does OsWRKY13. Here a series of experiments are reported whose results support this hypothesis. The two alleles are involved in rice response or adaptation to abiotic stresses. They play different roles, however, either negatively or positively regulating rice responses to ABA signalling and salt stress, but both negatively regulate rice responses to cold and drought stresses.
Materials and methods
Rice materials and stress treatment
Independent homozygous T2 or T3 generations of the OsWRKY45-1-overexpressing (oe) lines (D113UM3, D113UM10, and D113UM11), OsWRKY45-2-oe lines (D114UM4, D114UM6, and D114UM11), the OsWRKY45-1-knockout (KO) line (2C-50229), and OsWRKY45-2-suppressing [RNA interference (RNAi)] lines (D115RMH1 and D115RMH6) were used. The OsWRKY45-1 gene from japonica rice variety Nipponbare (O. sativa L. ssp. japonica) and the OsWRKY45-2 gene from indica rice variety Minghui 63 (O. sativa L. ssp. indica) were previously used for generating overexpressing transgenic lines (Tao et al., 2009). The OsWRKY45-1-oe and OsWRKY45-2-oe plants were generated using the maize ubiquitin gene promoter to drive OsWRK45-1 or OsWRKY45-2, respectively, in the genetic background of rice variety Mudanjiang 8 (O. sativa L. ssp. japonica) that carried OsWRKY45-1 (Tao et al., 2009). The OsWRKY45-1-KO mutant, which had a T-DNA inserted into the promoter of OsWRKY45-1, had the background of variety Dongjin (O. sativa L. ssp. japonica) and was kindly provided by Professor Gynheung An (Jeong et al., 2006; Tao et al., 2009). The OsWRKY45-1 genes and their promoter regions in Mudanjiang 8, Dongjing, and Nipponbare have identical sequences (Tao et al., 2009). The OsWRKY45-2-RNAi plants were generated using the RNAi strategy in the genetic background of variety Minghui 63 (Tao et al., 2009). The negative siblings from corresponding transgenic segregating populations were used as controls. For experiments testing cold, drought, and salt stresses, plants were grown in a greenhouse with light strength maintained at 12 000–14 000 lux and with a 14 h light/10 h dark cycle at 25 °C until the 4- to 5-leaf stage. The humidity in the greenhouse was maintained at 50–60%. The stress treatments were performed as reported previously (Hu et al., 2008).
For analysing the influence of ABA on plant growth, plants germinated and grown on 1/2 strength MS medium at room temperature (25–28 °C) for 3 d were transferred to culture boxes containing 1/2 strength MS medium plus 3 μM ABA; each box contained two groups of plants, the transgenic and control plants. After 7–10 d of treatment (until the transgenic and control plants showed visible differences in growth rate), the phenotype was recorded.
For cold stress, plants were grown in soil in pots. Each pot contained two groups of plants, the transgenic and control plants. Seedlings were transferred to a growth chamber under a 14 h light /10 h dark cycle at 4 °C for 4–6 d (until almost all the leaves of one group in the pot became completely rolled and some leaves died); the plants were then recovered by maintaining them at room temperature for 3 d or 7 d and the phenotype was recorded.
For drought stress, water was withheld from seedlings of transgenic and control plants growing in the same pot filled with a mixture of sand and soil (1:1) for 3–5 d (until almost all the leaves of one group in the pot became completely rolled); water was then provided for 3–7 d and the survival rates were recorded.
For salt stress, seedlings of transgenic and control plants growing in the same pot filled with a mixture of sand and soil (1:1) were irrigated with a solution containing 200 mM NaCl for 4–6 d (until almost all the leaves of one group in the pot lost their green colour and some leaves died). During the treatment, phenotypes were recorded.
DNA sequencing and promoter analyses
The genomic fragment of OsWRKY45-1 was amplified from rice cultivar IRAT109 (O. sativa L. ssp. japonica) using primers w45F4 and w45R4, and sequenced using primers w45F4, w45R4, w45F6, and w45R6 (Supplementary Table S1 available at JXB online). To analyse the putative conserved cis-elements, which are strongly related to various abiotic and biotic stresses, in the promoters of OsWRKY45-1 and OsWRKY45-2, the ∼1500 nucleotide sequences upstream of the transcription initiation sites of the two genes were used to search the PLACE (Plant Cis-acting Regulatory DNA Elements, http://www.dna.affrc.go.jp/PLACE/index.html) database.
Gene expression analyses
To analyse the expression patterns of OsWRKY45-1 and OsWRKY45-2 under various abiotic stresses, rice cultivars IRAT109 and Zhenshan 97 (O. sativa L. ssp. indica) were grown in a greenhouse until the 4- to 5-leaf stage with a 14 h light/10 h dark cycle at 25 °C. For ABA treatments, seedlings were sprayed with a solution containing 0.1 mM ABA and 0.02% Tween-20. For cold stress, seedlings were transferred to a growth chamber under 14 h light/10 h darkness at 4 °C for 3–24 h. For drought stress, seedlings grown on sandy soil (Hu et al., 2008) were not watered for 3–6 d. For salt stress, seedlings were irrigated with 200 mM NaCl solution for 3–24 h. The quantitative reverse transcription-PCR (qRT-PCR) analysis was conducted as described previously (Qiu et al., 2007) using gene-specific primers (Supplementary Table S1 at JXB online). The expression level of the rice actin gene was used to standardize the RNA sample for each qRT-PCR. The expression level relative to the control was presented.
Statistical analysis
The significant differences between control and treatment of the samples or between control and transgenic plants were analysed by the pair-wise t-test installed in the Microsoft Office Excel program.
Results
Both OsWRKY45-1 and OsWRKY45-2 a transcriptional response to ABA and abiotic stresses
The upland japonica rice variety IRAT109 (O. sativa L. ssp. japonica) is more drought resistant than the lowland indica rice variety Zhenshan 97 (O. sativa L. ssp. indica; Yue et al., 2006). Zhenshan 97 carries OsWRKY45-2 (Tao et al., 2009). Sequencing the DNA fragment (∼3.7 kb) harbouring the OsWRKY45 gene in IRAT109, including the 1.5 kb (upstream of the transcription initiation site) promoter region, coding region, and 5′- and 3′-untranslated regions, showed that it carried OsWRKY45-1, with its sequence identical to that in the japonica variety Mudanjiang 8 (Tao et al., 2009). To ascertain whether the two alleles play roles in rice responses to abiotic stresses, the expression patterns of the two genes were examined under drought, high salinity (250 mM NaCl), cold (4 °C), and ABA (100 μM) stresses by qRT-PCR.
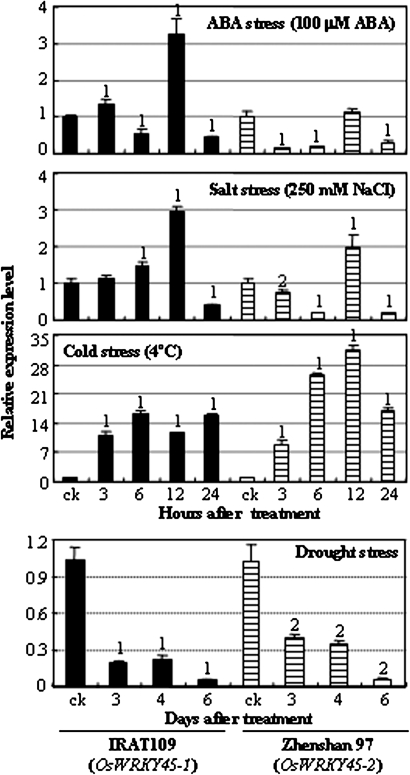
OsWRKY45-1 and OsWRKY45-2 showed a similar response to cold and drought stresses (Fig. 1). The expression of the two alleles was markedly induced after cold stress and suppressed after drought treatment. However, the two alleles showed different expression patterns after ABA and salt stresses (Fig. 1). In ABA treatment, OsWRKY45-1 expression was first slightly suppressed, then induced, and later suppressed again; OsWRKY45-2 expression was first rapidly suppressed, then recovered to a normal level, and later suppressed again. In salt stress, OsWRKY45-1 expression was first induced and then suppressed, whereas OsWRKY45-2 expression was first suppressed, then induced, and later suppressed again. These results suggest that the two alleles may be involved in rice responses to ABA signalling and abiotic stresses, but they may function differently in ABA and salt stresses.
Fig. 1.
Expression patterns of OsWRKY45-1 and OsWRKY45-2 in response to ABA and abiotic stresses in rice cultivars IRAT109 and Zhenshan 97. Bars represent the mean (three technical replicates) ±standard deviation. The ‘1’ or ‘2’ indicates that a significant difference was detected between treated plants and untreated control (ck) at P <0.01 or P <0.05, respectively.
The differential expression patterns of OsWRKY45-1 and OsWRKY45-2 in response to ABA and salt stresses prompted the examination of whether there are different ABA- and stress-responsive cis-acting elements in their promoters. Sequence analysis showed that both promoters contained ABRE, DRE, GCC, LTRE, MYB, and MYC cis-elements that have been reported to be responsive to ABA, cold, drought, or salt stresses (Supplementary Table S2 and Fig. S1 at JXB online). However, the OsWRKY45-2 promoter had two additional ABREs and one additional MYC element compared with the OsWRKY45-1 promoter. The different structures of the two promoters may explain the differential expression patterns of the two alleles in response to ABA and salt treatments.
OsWRKY45-1 and OsWRKY45-2 play different roles in response to ABA treatment
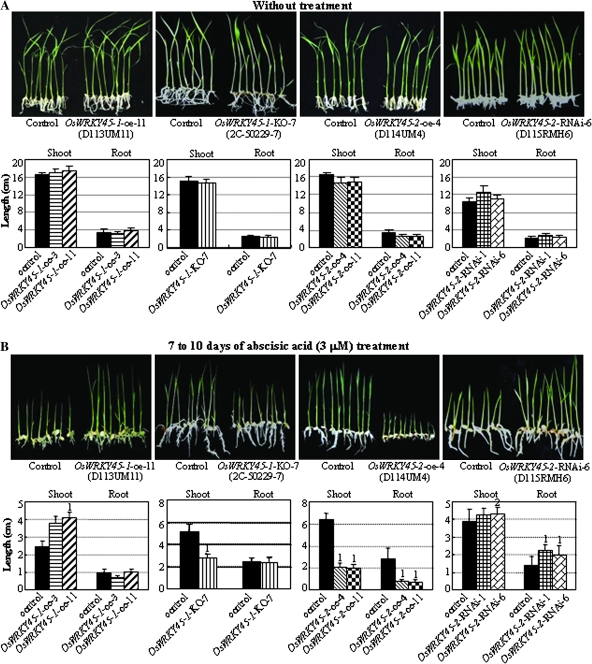
Xiang et al. (2008) reported that exogenous ABA suppresses the growth of rice seedlings. To ascertain whether this pair of alleles plays different roles in ABA signalling, the response of rice to ABA stress was analysed using different transgenic lines. The seedling growth of OsWRKY45-1-oe, OsWRKY45-1-KO, OsWRKY45-2-oe, and OsWRKY45-2-RNAi plants showed no obvious difference from that of corresponding control plants, when grown on medium without ABA (Fig. 2A; Supplementary Fig. S2A at JXB online). After treatment with 3 μM ABA, OsWRKY45-1-oe lines had significantly longer shoots than the control, whereas OsWRKY45-1-KO lines had significantly shorter shoots than the control (Fig. 2B; Supplementary Fig. S2B). Modulating OsWRKY45-1 expression appeared to have no obvious influence on the growth of root after ABA treatment compared with controls. In contrast, OsWRKY45-2-oe lines had significantly shorter roots and shoots than those of the control, whereas OsWRKY45-2-RNAi lines had significantly longer roots and somewhat longer shoots than those of the control. These results further suggest that the two alleles respond differently to ABA-mediated signalling, and OsWRKY45-1 negatively and OsWRKY45-2 positively regulates rice response to ABA signalling.
Fig. 2.
Modulating OsWRKY45-1 and OsWRKY45-2 expression influenced rice response to ABA treatment. Control, negative siblings from corresponding transgenic segregating populations. (A) Rice seedlings before ABA treatment. Plants grown on MS medium for 10 d. (B) Phenotypes of transgenic plants after ABA treatment. Rice seedlings were grown on MS medium containing ABA for 7–10 d (until the transgenic and control plants showed marked differences in growth rate) and then phenotypes were recorded. Bars represent the mean (8–10 plants) ±standard deviation. The ‘1’ or ‘2’ indicates that a significant difference was detected between a positive transgenic line and its negative control at P <0.01 or P <0.05, respectively. Similar results were obtained in two biological repeats and only data from one repeat are presented. Because of the varying durations of ABA treatment (7–10 d) in different groups of plants, the control plants with the same genetic background in the OsWRKY45-1-oe and OsWRKY45-2-oe groups exhibited different lengths of shoot and root.
OsWRKY45-1 and OsWRKY45-2 function differently in regulating the salt stress response
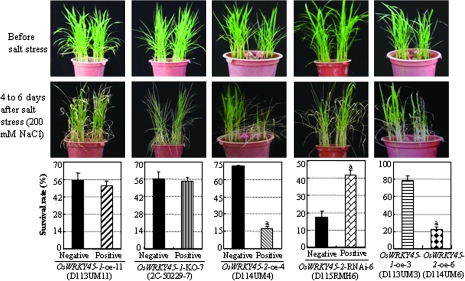
Transgenic lines with modified expression of either OsWRKY45-1 or OsWRKY45-2 showed no obvious difference from corresponding controls before salt stress. After treating the plants with 200 mM NaCl for 5 d, the survival rates of OsWRKY45-1-oe and OsWRKY45-1-KO lines were similar to those of corresponding controls, whereas the survival rates of OsWRKY45-2-oe and OsWRKY45-2-RNAi lines were significantly lower and higher than corresponding controls, respectively (Fig. 3; Supplementary Fig. S3 at JXB online). When OsWRKY45-1-oe and OsWRKY45-2-oe plants, which had the same genetic background, were grown in the same pot, the former was significantly more tolerant to salt stress than the latter; the survival rate of the OsWRKY45-1-oe plants was 3.6-fold higher than that of the OsWRKY45-2-oe plants (Fig. 3). These results suggest that OsWRKY45-2 negatively regulates the salt stress response and OsWRKY45-1 is insensitive to salt stress, although it was transcriptionally induced by salt stress (Fig. 1).
Fig. 3.
Modulating the expression of OsWRKY45-2 but not OsWRKY45-1 influenced rice response to salt stress. Rice seedlings were irrigated with a solution containing 200 mM NaCl at the 4- to 5-leaf stage. After 4–6 d of stress (until almost all the leaves of one group in the pot lost their green colour and some leaves died), the survival rates were recorded. Negative, negative siblings from corresponding transgenic segregating populations; positive, positive transgenic plants. Bars represent the mean (three technical replicates with each replicate containing 16–20 plants) ±standard deviation. The ‘1’ indicates that a significant difference was detected between a positive transgenic line and its negative control at P <0.01. Similar results were obtained in three biological repeats and only data from one repeat are presented. Because of the varying durations of salt stress (4–6 d) in different groups of plants, the control plants with the same genetic background in the OsWRKY45-1-oe and OsWRKY45-2-oe groups exhibited different survival rates.
OsWRKY45-1 and OsWRKY45-2 play similar roles in regulating the cold stress response
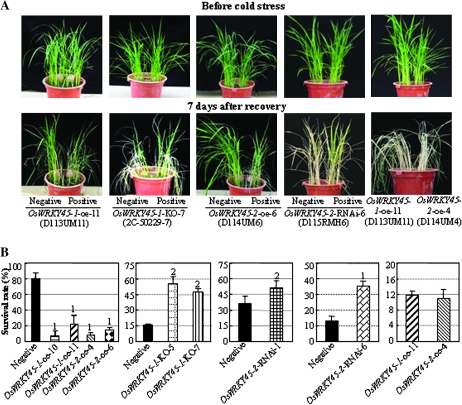
Both OsWRKY45-1-oe and OsWRKY45-2-oe plants showed a similar level of sensitivity to cold. The leaves of OsWRKY45-oe plants rolled extensively in a cold environment. After recovery, these plants died quickly compared with the controls (Fig. 4A; Supplementary Fig. S4 at JXB online). After 7 d of recovery, the average survival rate of different OsWRKY45-oe lines was 6–18% compared with 76% for the control (Fig. 4A). In contrast, OsWRKY45-1-KO and OsWRKY45-2-RNAi plants were more tolerant to cold than the control (Fig. 4A; Supplementary Fig. S4). After 7 d of recovery, the average survival rate of OsWRKY45-1-KO plants was 48–55% compared with 16% for the control; the average survival rates of the two OsWRKY45-2-RNAi lines were 36% and 51% compared with 13% and 36% for corresponding negative controls, respectively (Fig. 4B). When OsWRKY45-1-oe and OsWRKY45-2-oe lines were grown in the same pot, the two lines showed a similar level of sensitivity to cold stress (Fig. 4). The marked difference of the survival rates among different control plants could be due to the difference in genetic background or due to the varying durations of cold stress (4–6 d) for different groups of plants. These results suggest that both OsWRKY45-1 and OsWRKY45-2 negatively regulate rice response to cold stress.
Fig. 4.
Modulating OsWRKY45-1 and OsWRKY45-2 expression influenced rice response to cold stress. Rice seedlings were kept at 4 °C for 4–6 d (until almost all the leaves of one group in the pot became completely rolled and some leaves died) and then transferred to room temperature (25–28 °C) for recovery. Negative, negative siblings from corresponding transgenic segregating populations; positive, positive transgenic plants. (A) Phenotypes of transgenic plants before and after cold treatment. (B) Survival rate of cold-treated plants. Bars represent the mean (two or three replicates with each replicate containing 16–20 plants) ±standard deviation. The ‘1’ or ‘2’ indicates that a significant difference was detected between a positive transgenic line and its negative control at P <0.01 or P <0.05, respectively. Similar results were obtained in three biological repeats and only data from one repeat are presented.
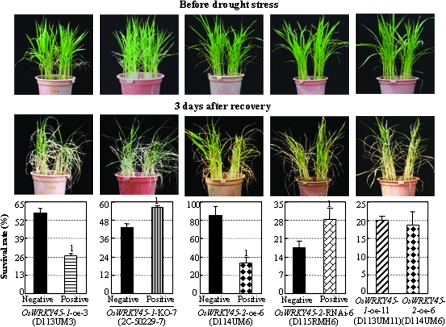
OsWRKY45-1 and OsWRKY45-2 play similar roles in regulating the drought stress response
Both OsWRKY45-1-oe and OsWRKY45-2-oe plants showed a similar level of sensitivity to drought stress. After drought stress, these plants have a significantly lower survival rate than corresponding controls (Fig. 5; Supplementary Fig. S5 at JXB online). After 3 d of recovery, the survive rates of the two OsWRKY45-1-oe lines were 27% and 22% compared with 57% and 47% for the controls, respectively; the survival rates of the two OsWRKY45-2-oe lines were 33% and 31% compared with 86% and 72% for the controls, respectively. When the OsWRKY45-1-oe and OsWRKY45-2-oe plants were grown in the same pot, they showed no obvious difference in the sensitivity to drought stress (Fig. 5). In contrast, OsWRKY45-1-KO and OsWRKY45-2-RNAi plants were significantly more tolerant to drought stress than the controls (Fig. 5; Supplementary Fig. S5). After 3 d of recovery, the survival rate of OsWRKY45-1-KO line was 56% compared with 43% for the control; the survival rates of the two OsWRKY45-2-RNAi lines were 28% and 28% compared with 20% and 18% for their corresponding controls, respectively (Fig. 5; Supplementary Fig. S5). The marked difference in the survival rates among different control plants could be due to the difference in genetic background or due to the varying durations of drought stress (3–5 d) for the different groups of plants. These results suggest that both OsWRKY45-1 and OsWRKY45-2 negatively regulate rice response to drought stress.
Fig. 5.
Modulating OsWRKY45-1 and OsWRKY45-2 expression influenced rice response to drought stress. Water was withheld from rice seedlings at the 4- to 5-leaf stage for 3–5 d (until almost all the leaves of one group in the pot became completely rolled). After 3 d of recovery, the survival rates were recorded. Negative, negative siblings from corresponding transgenic segregating populations; positive, positive transgenic plants. Bars represent the mean (two or three replicates with each replicate containing 16–20 plants) ±standard deviation. The ‘1’ indicates that a significant difference was detected between a positive transgenic line and its negative control at P <0.01. Similar results were obtained in the three biological repeats and only data from one repeat are presented. Because of the varying durations of drought stress (3–5 d) in different groups of plants, the control plants with the same genetic background in the OsWRKY45-1-oe and OsWRKY45-2-oe groups exhibited different survival rates.
Modulating OsWRKY45-1 and OsWRKY45-2 expression influences expression of ABA- and abiotic stress-responsive genes
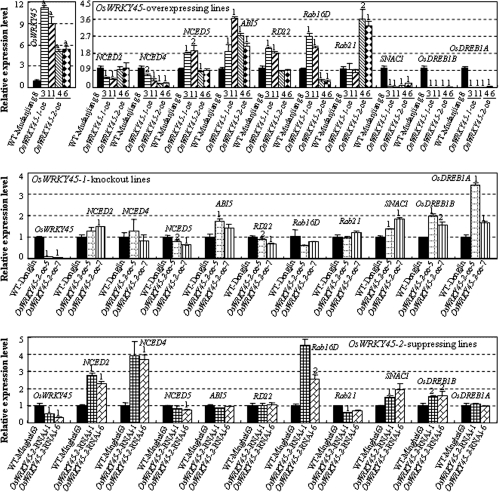
To investigate differences in the function of OsWRKY45-1 and OsWRKY45-2 in the regulation of ABA signalling and abiotic stress responses, the expression of 10 ABA- and putative stress-related genes in plant leaves was analysed. These genes included three ABA biosynthesis genes, NCED2 (AK120176), NCED4 (AK119780), and NCED5 (AK107649) (Zhu et al., 2009); four ABA- or abiotic stress-responsive genes, ABI5 (AK070998), RD22 (AK065358), Rab16D (AK109096), and Rab21 (AK121952) (Buchanan et al., 2004; Zou et al., 2008; Hou et al., 2009); and three known abiotic stress-resistant genes, SNAC1 (AK067690), OsDREB1A (AY345233), and OsDREB1B (AY166833) (Dubouzet et al., 2003; Hu et al., 2006; Ito et al., 2006).
All the genes showed differential expression in at least one type of transgenic plant as compared with their expression in corresponding wild-type plants (Fig. 6). Nine of the genes (NCED2, NCED4, NCED5, RD22, Rab16D, Rab21, SNAC1, OsDREB1B, and OsDREB1A) showed opposite expression patterns in the overexpressing and knockout/suppressing plants for at least one OsWRKY45 allele. Among the nine genes, SNAC1 and OsDREB1B showed similar expression patterns in both the OsWRKY45-1 and OsWRKY45-2 transgenic plants; the two genes were significantly suppressed in OsWRKY45-oe plants and significantly induced in OsWRKY45-KO and OsWRKY45-RNAi plants. Rab16D showed opposite expression patterns in OsWRKY45-1 and OsWRKY45-2 transgenic plants; this gene was significantly induced in OsWRKY45-1-oe lines and appeared to be suppressed in OsWRKY45-1-KO lines, but it was significantly suppressed in OsWRKY45-2-oe lines and significantly induced in OsWRKY45-2-RNAi lines. NCED2, OsDREB1A, NCED5, and RD22 only showed opposite expression patterns in OsWRKY45-1-oe and OsWRKY45-1-KO plants (Fig. 6). NCED2 and OsDREB1A were significantly suppressed in OsWRKY45-1-oe lines and significantly induced in OsWRKY45-1-KO lines. NCED5 and RD22 were significantly induced in OsWRKY45-1-oe lines and significantly suppressed in OsWRKY45-1-KO lines. NCED4 only showed opposite expression patterns in OsWRKY45-2-oe and OsWRKY45-2-RNAi plants (Fig. 6); it was significantly suppressed in OsWRKY45-2-oe lines and significantly induced in OsWRKY45-2-RNAi lines. These results suggest that OsWRKY45 directly or indirectly regulates the expression of at least nine of the 10 genes. The differential expression of most of the genes examined in OsWRKY45-1 and OsWRKY45-2 transgenic plants also suggests that this pair of alleles may regulate ABA signalling and abiotic stress responses through different pathways.
Fig. 6.
Modulating OsWRKY45-1 and OsWRKY45-2 expression influenced the expression of ABA- and abiotic stress-related genes. Bars represent the mean (three technical replicates) ±standard deviation. The ‘1’ or ‘2’ indicates that a significant difference was detected between a transgenic line and its corresponding wild-type (WT) plant at P <0.01 or P <0.05, respectively.
Discussion
To compare the functions of OsWRKY45 alleles, they were separately overexpressed in the same genetic background. Although the OsWRKY45-2-oe plants expressed OsWRKY45-2 at a high level and background OsWRKY45-1, the observed phenotypes should be contributed by the constitutively overexpressed OsWRKY45-2. This inference is supported by the following evidence. First, the OsWRKY45-2-oe and OsWRKY45-2-RNAi plants showed opposite phenotypes in response to ABA and abiotic stresses. Secondly, OsWRKY45-1-oe and OsWRKY45-2-oe plants showed different responses to ABA and salt stresses. The present findings indicate that OsWRKY45 alleles play opposite roles in ABA signalling and salt stress responses in rice, although they have similar functions in responses to cold and drought stresses. The opposite phenotypes of OsWRKY45-oe and OsWRKY45-KO and OsWRKY45-RNAi plants suggest that OsWRKY45-1 negatively and OsWRKY45-2 positively regulates ABA signalling. Furthermore, OsWRKY45-2 negatively regulates rice response to salt stress. OsWRKY45-1 does not influence the response to salt stress, although its transcriptional expression was changed by this stress. However, both OsWRKY45-1 and OsWRKY45-2 negatively regulate rice responses to cold and drought stresses. These observations not only remind us to consider the roles of intraspecific allelic variation in plant tolerance and adaptation to abiotic stresses, but they also provide a novel molecular-based example of how a pair of alleles functions distinctly in abiotic stress adaptation.
Alteration of the protein and promoter sequences may result in different functions of OsWRKY45 alleles
Intraspecific allelic diversity can be caused by nucleotide sequence differences in coding regions, differential expression, and epigenetic changes (Guo et al., 2004; Shiba and Takayama, 2007; Springer and Stupar, 2007). The first type of allelic diversity results in alteration of protein function, whereas the last two types cause variation in the quantity of the same protein. The difference in 10 amino acids between OsWRKY45-1 and OsWRKY45-2 may be a factor that results in the phenotypic variation of transgenic plants, as supported by the following evidence. First, the two alleles in OsWRKY45-1-oe and OsWRKY45-2-oe plants were regulated by the same promoter. In addition, the OsWRKY45-1-oe and OsWRKY45-2-oe plants had the same genetic background, which can eliminate the putative differential transcriptional and epigenetic regulations in the present experimental conditions.
The promoter regions (∼1.5 kb upstream of the transcription initiation sites) of OsWRKY45-1 and OsWRKY45-2 differ in 19 nucleotides (Tao et al., 2009). The OsWRKY45-2 promoter has two additional ABRE-type cis-acting elements and one additional MYC-type cis-element (Supplementary Table S1 and Fig. S1 at JXB online). The ABRE for binding basic leucine zipper-type proteins and the MYC element for binding helix–loop–helix and leucine zipper-type proteins are important regulating elements in ABA-responsive gene expression in two ABA-dependent signalling pathways involved in drought and salt stresses (Agarwal et al., 2006; Valliyodan and Nguyen, 2006; Yamaguchi-Shinozaki and Shinozaki, 2006). Thus, the two alleles may function differently in ABA signalling and salt tolerance due to differentially regulating their expression via trans-acting factors under physiological conditions. This hypothesis is supported by evidence that the two alleles showed different expression patterns in response to ABA and salt stresses (Fig. 1).
The different roles of the alleles in ABA signalling and salt stress tolerance may be due to differential regulation of ABA- and abiotic stress-responsive genes
OsWRKY45-1 functions as a transcriptional regulator (Shimono et al., 2007). It is argued that OsWRKY45-2 is also a transcription factor. ABA is an important signalling molecule when a plant faces abiotic stress (Verslues and Zhu, 2005). The two alleles differentially regulate the expression of ABA synthesis-related NCED genes, which encode 9-cis-epoxycarotenoid dioxygenases and function in the ABA biosynthetic pathway (Nambara and Marion-Poll, 2005). The opposite expression patterns of NCED2 and NCED5 in OsWRKY45-1-oe and OsWRKY45-1-KO plants suggest that the two genes may be more closely associated with the negative role of OsWRKY45-1 in ABA signalling. In contrast, the opposite expression patterns of NCED4 in OsWRKY45-2-oe and OsWRKY45-2-RNAi plants suggest that this gene may be more closely associated with the positive role of OsWRKY45-2 in ABA signalling. Thus, OsWRKY45 transcriptionally regulates ABA biosynthesis, which in turn may influence the expression of ABA-responsive genes and abiotic stress-responsive genes involved in salt stress response.
This hypothesis is supported by the different expression patterns of RD22, Rab16D, and Rab21 in OsWRKY45-1 and OsWRKY45-2 transgenic plants. The Arabidopsis homologues of rice RD22, Rab16D, and Rab21 are well-documented ABA- and abiotic stress-responsive genes (Yamaguchi-Shinozaki and Shinozaki, 1993; Nylander et al., 2001). Rice Rab16D, encoding a dehydrin-like protein, was rapidly induced by ABA application and abiotic stresses (Rabbani et al., 2003; Buchanan et al., 2004). The opposite expression patterns of Rab16D in OsWRKY45-1 and OsWRKY45-2 transgenic plants suggest that OsWRKY45-1 is a transcriptional activator and OsWRKY45-2 is a transcriptional repressor of Rab16D. Although Rab21, which also encodes a dehydrin-like protein, was transcriptionally responsive to abiotic stress, its expression level was reduced in transgenic plants showing enhanced drought tolerance (Hou et al., 2009). The result is consistent with this previous report. Transcriptional modulation of OsWRKY45-2 but not OsWRKY45-1 influenced Rab21 expression; the suppression of OsWRKY45-2 accompanied by enhanced abiotic stress tolerance was associated with suppressed Rab21 expression (Fig. 6). The induction of RD22, encoding a BURP domain-containing protein, was associated with enhanced drought tolerance in transgenic plants (Hou et al., 2009). The present result is also consistent with this report. Transcriptional modulation of OsWRKY45-1 but not OsWRKY45-2 influenced RD22 expression; activation of OsWRKY45-1 accompanied by enhanced ABA stress tolerance and non-sensitivity to salt stress was associated with induced RD22 expression (Fig. 6). Although the definite functions of the three genes in adaptation to abiotic stresses remain to be elucidated in rice, the present results suggest that these genes may be involved in the different roles of OsWRKY45-1 and OsWRKY45-2 in ABA signalling and the response to salt stress.
OsWRKY45-1 and OsWRKY45-2 may regulate abiotic stress responses via different pathways
Transgenic rice plants overexpressing SNAC1, which encodes a transcription factor, showed improved drought and salt tolerance but increased ABA sensitivity (Hu et al., 2006). Overexpressing OsDREB1B, which encodes a transcription factor, enhanced rice tolerance to drought, salt, and cold stresses (Ito et al., 2006). OsDREB1B is under the regulation of SNAC1, and the activation of SNAC1 transcriptionally induced OsDREB1B, suggesting that SNAC1 and OsDREB1B may function in the same pathway in rice responses to abiotic stresses (Qiu et al., 2008). This hypothesis is further supported by the similar expression patterns of the two genes in OsWRKY45 transgenic plants (Fig. 6). Although both OsWRKY45-1 and OsWRKY45-2 suppressed the expression of SNAC1 and OsDREB1B, the phenotypes of transgenic plants suggest that OsWRKY45-2 negatively regulates rice responses to cold, drought, and salt stresses, and OsWRKY45-1 negatively regulates rice responses to cold and drought stresses but it does not obviously influence the response to salt stress. These results suggest that both OsWRKY45-1 and OsWRKY45-2 negatively regulate rice responses to cold, drought, and salt stresses through the pathway involving SNAC1 and OsDREB1B. In addition, OsWRKY45-1 may positively regulate a salt tolerance pathway that could compensate for the suppression of the pathway involving SNAC1 and OsDREB1B in salt stress response. This hypothesis is supported by evidence that OsWRKY45-1 and OsWRKY45-2 differentially regulate the expression of RD22, Rab16D, and Rab21, whose transcript levels are associated with rice adaptation to abiotic stresses (Rabbani et al., 2003; Buchanan et al., 2004; Hou et al., 2009). However, further research is required to examine this hypothesis.
OsWRKY45-1 and OsWRKY45-2 differentially mediate the cross-talk between abiotic and biotic stress defence pathways
Plants are constantly exposed to diverse abiotic and biotic stresses and have evolved efficient ways to reallocate metabolic resources rapidly among different physiological signalling pathways in order to adapt to a changing environment. Convergence points of the cross-talk between abiotic and biotic stress signalling are emerging (Fujita et al., 2006; Park et al., 2007; Qiu et al., 2008), but our understanding of the molecular mechanisms underlying this cross-talk in rice remains preliminary. Different proteins, including transcription factors, kinases, and other enzymes, have been reported to play roles in this cross-talk (Xiong and Yang, 2003; Anderson et al., 2004; Park et al., 2007; Qiu et al., 2008). Abiotic and biotic stress signalling pathways frequently interact antagonistically (Fujita et al., 2006) because the proteins functioning in the convergence points of the cross-talk have two functions. For example, the proteins may function as positive regulators in abiotic stress tolerance but as negative regulators in disease resistance, such as rice OsMPK5 kinase (Xiong and Yang, 2003) and Arabidopsis AtMYC2 transcription factor (Anderson et al., 2004), or they may function as negative regulators in abiotic stress tolerance but as positive regulators in disease resistance, such as rice OsWRKY13 transcription regulator (Qiu et al., 2007, 2008). However, the Arabidopsis protein WES1, a putative GH3-type auxin-conjugating enzyme, functions as a positive regulator in both abiotic stress tolerance and disease resistance (Park et al., 2007).
Adding to the short list of proteins known to regulate cross-talk, here it is shown that OsWRKY45-1 and OsWRKY45-2 are also players in the cross-talk between abiotic and biotic stress signalling. Previously, it was reported that the two alleles had opposite roles in rice–bacterium interactions and had the same role in rice–fungus interactions (Tao et al., 2009). Based on the present and previous findings (Tao et al., 2009), OsWRKY45-1 negatively regulates bacterial pathogen resistance, cold and drought tolerance, and ABA-mediated signalling, and positively regulates fungal pathogen resistance. In contrast, OsWRKY45-2 positively regulates bacterial and fungal pathogen resistance and ABA-mediated signalling, and negatively regulates cold, drought, and salt tolerance. The different roles of the OsWRKY45 alleles in the cross-talk may result at least partly from their differential regulation of ABA-mediated signalling. ABA is extensively involved in plant responses to abiotic stresses and is also implicated in plant–pathogen interactions (Verslues and Zhu, 2005; Bari and Jones, 2009).
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Alignment of the promoter regions of OsWRKY45-1 and OsWRKY45-2.
Figure S2. Modulating OsWRKY45-1 and OsWRKY45-2 expression influenced rice response to abscisic acid (ABA) treatment.
Figure S3. Modulating the expression of OsWRKY45-2 but not OsWRKY45-1 markedly influenced rice response to salt stress.
Figure S4. Modulating OsWRKY45-1 and OsWRKY45-2 expression influenced rice response to cold stress.
Figure S5. Modulating OsWRKY45-1 and OsWRKY45-2 expression influenced rice response to drought stress.
Table S1. Primers used for PCR amplification
Table S2. The numbers of known abiotic stress-responsive cis-acting elements in the promoters of OsWRKY45-1 and OsWRKY45-2.
Acknowledgments
We thank Mr Jun You and Professor Lizhong Xiong of Huazhong Agricultural University for providing ABA- and abiotic stress-treated RNA samples. This work was supported by grants from the National Natural Science Foundation of China (30930063, 30921091).
References
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Reports. 2006;25:1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Mendez-Vigo B, Koornneef M. From phenotypic to molecular polymorphisms involved in naturally occurring variation of plant development. International Journal of Developmental Biology. 2005;49:717–732. doi: 10.1387/ijdb.051994ca. [DOI] [PubMed] [Google Scholar]
- Amir Hossain M, Lee Y, Cho JI, Ahn CH, Lee SK, Jeon JS, Kang H, Lee CH, An G, Park PB. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Molecular Biology. 2010;72:557–566. doi: 10.1007/s11103-009-9592-9. [DOI] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K. Antagonistic interaction between abscisic acid and jasmonates–ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. The Plant Cell. 2004;16:3460–3479. doi: 10.1105/tpc.104.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant Molecular Biology. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- Bhave M, Morris CF. Molecular genetics of puroindolines and related genes: allelic diversity in wheat and other grasses. Plant Molecular Biology. 2008;66:205–219. doi: 10.1007/s11103-007-9263-7. [DOI] [PubMed] [Google Scholar]
- Buchanan CD, Klein PE, Mullet JE. Phylogenetic analysis of 5'-noncoding regions from the ABA-responsive rab16/17 gene family of sorghum, maize and rice provides insight into the composition, organization and function of cis-regulatory modules. Genetics. 2004;168:1639–1654. doi: 10.1534/genetics.104.030346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JQ, Meng XP, Zhang Y, Xia M, Wang XP. Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnology Letters. 2008;30:2191–2198. doi: 10.1007/s10529-008-9811-5. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Schumaker K, Zhu JK. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. Journal of Experimental Botany. 2004;55:225–236. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuqa M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaquchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. The Plant Journal. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theoretical and Applied Genetics. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annual Review of Plant Biology. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Current Opinion of Plant Biology. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Zinselmeier C, Habben J, Bowen BA, Smith OS. Allelic variation of gene expression in maize hybrids. The Plant Cell. 2004;16:1707–1716. doi: 10.1105/tpc.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E. Relay and control of abscisic acid signaling. Current Opinion in Plant Biology. 2003;6:470–479. doi: 10.1016/s1369-5266(03)00090-6. [DOI] [PubMed] [Google Scholar]
- Hou X, Xie K, Yao J, Qi Z, Xiong L. A homolog of human ski-interacting protein in rice positively regulates cell viability and stress tolerance. Proceedings of the National Academy of Sciences, USA. 2009;106:6410–6415. doi: 10.1073/pnas.0901940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences, USA. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Molecular Biology. 2008;67:169–181. doi: 10.1007/s11103-008-9309-5. [DOI] [PubMed] [Google Scholar]
- Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes and Development. 2009;23:1805–1817. doi: 10.1101/gad.1812409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant and Cell Physiology. 2006;47:141–153. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Park S, et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. The Plant Journal. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiology. 2010;153:185–197. doi: 10.1104/pp.110.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD. Putting knowledge of plant disease resistance genes to work. Current Opinion in Plant Biology. 2001;4:281–287. doi: 10.1016/s1369-5266(00)00174-6. [DOI] [PubMed] [Google Scholar]
- Lu G, Gao C, Zheng X, Han B. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta. 2009;229:605–615. doi: 10.1007/s00425-008-0857-3. [DOI] [PubMed] [Google Scholar]
- Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X, Chong K. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiology. 2009;150:244–256. doi: 10.1104/pp.108.133454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Archives of Biochemistry and Biophysics. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV. Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Molecular Biology. 2001;45:263–279. doi: 10.1023/a:1006469128280. [DOI] [PubMed] [Google Scholar]
- Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. Journal of Biological Chemistry. 2007;282:10036–10046. doi: 10.1074/jbc.M610524200. [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Molecular Plant-Microbe Interactions. 2007;20:492–499. doi: 10.1094/MPMI-20-5-0492. [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Cheng H, Li X, Wang S. Exploring transcriptional signaling mediated by OsWRKY13, a potential regulator of multiple physiological processes in rice. BMC Plant Biology. 2009;9:74. doi: 10.1186/1471-2229-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Liu H, Li X, Xiong L, Wang S. Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Molecular Plant. 2008;1:538–551. doi: 10.1093/mp/ssn012. [DOI] [PubMed] [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiology. 2003;133:1755–1767. doi: 10.1104/pp.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy R, Jiang SY, Kumar N, Venkatesh PN, Ramachandran S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant and Cell Physiology. 2008;49:865–879. doi: 10.1093/pcp/pcn061. [DOI] [PubMed] [Google Scholar]
- Ross CA, Liu Y, Shen QJ. The WRKY gene family in rice (Oryza sativa) Journal of Integrative Plant Biology. 2007;49:827–842. [Google Scholar]
- Seki M, Ishida J, Narusaka M, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Shiba H, Takayama S. RNA silencing systems and their relevance to allele-specific DNA methylation in plants. Bioscience, Biotechnology, and Biochemistry. 2007;71:2632–2646. doi: 10.1271/bbb.70339. [DOI] [PubMed] [Google Scholar]
- Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, Takatsuji H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. The Plant Cell. 2007;19:2064–2076. doi: 10.1105/tpc.106.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Stupar RM. Allelic variation and heterosis in maize: how do two halves make more than a whole? Genome Research. 2007;17:264–275. doi: 10.1101/gr.5347007. [DOI] [PubMed] [Google Scholar]
- Tao Z, Liu H, Qiu D, Zhou Y, Li X, Xu C, Wang S. A pair of allelic WRKY genes play opposite roles in rice–bacteria interactions. Plant Physiology. 2009;151:936–948. doi: 10.1104/pp.109.145623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valliyodan B, Nguyen HT. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Current Opinion in Plant Biology. 2006;9:189–195. doi: 10.1016/j.pbi.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Verslues PE, Zhu JK. Before and beyond ABA: upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochemical Society Transactions. 2005;33:375–379. doi: 10.1042/BST0330375. [DOI] [PubMed] [Google Scholar]
- Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, Peng Y, Guo Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Molecular Biology. 2007;65:799–815. doi: 10.1007/s11103-007-9244-x. [DOI] [PubMed] [Google Scholar]
- Wu X, Shiroto Y, Kishitani S, Ito Y, Toriyama K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Reports. 2009;28:21–30. doi: 10.1007/s00299-008-0614-x. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiology. 2008;148:1938–1952. doi: 10.1104/pp.108.128199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Yang Y. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. The Plant Cell. 2003;15:745–759. [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Molecular and General Genetics. 1993;238:17–25. doi: 10.1007/BF00279525. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yue B, Xue W, Xiong L, Yu X, Luo L, Cui K, Jin D, Xing Y, Zhang Q. Genetic basis of drought resistance at reproductive stage in rice: separation of drought tolerance from drought avoidance. Genetics. 2006;172:1213–1228. doi: 10.1534/genetics.105.045062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Chen B, Lu G, Han B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochemical and Biophysical Research Communications. 2009;379:985–989. doi: 10.1016/j.bbrc.2008.12.163. [DOI] [PubMed] [Google Scholar]
- Zhu G, Ye N, Zhang J. Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant and Cell Physiology. 2009;50:644–651. doi: 10.1093/pcp/pcp022. [DOI] [PubMed] [Google Scholar]
- Zou M, Guan Y, Ren H, Zhang F, Chen F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Molecular Biology. 2008;66:675–683. doi: 10.1007/s11103-008-9298-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.