Abstract
In the interleukin 3-dependent hematopoietic cell line Ba/F3, inhibition of mitogen-activated protein kinase, a member of the MAPK/c-Jun N-terminal kinase/stress-activated protein kinase kinase family that plays an important role in cell growth and death control, rapidly leads to severe apoptosis. However, most of the anti-apoptotic substrates of MAPK remain to be identified. Here we report that, upon interleukin-3 stimulation of Ba/F3 cells, the transcription factor GATA-1 is strongly phosphorylated at residue serine 26 by a MAPK-dependent pathway. Phosphorylation of GATA-1 increases GATA-1-mediated transcription of the E4bp4 survival gene without significantly changing the DNA-binding affinity of GATA-1. Further characterization of GATA-1 phosphorylation site mutants revealed that the anti-apoptotic function of GATA-1 is strongly dependent upon its phosphorylation at the Ser-26 position and is probably mediated through its up-regulation of Bcl-XL expression. Taken together, our data demonstrate that MAPK-dependent GATA-1 phosphorylation is important for its transactivation of the E4bp4 gene, Bcl-XL expression and cell survival. Therefore, GATA-1 may represent a novel MAPK substrate that plays an essential role in a cytokine-mediated antiapoptotic response.
GATA-1, the founding member of the GATA family of transcription factors, was initially identified as an erythroid-specific nuclear protein that binds to consensus GATA motifs, (A/T)GATA(A/G), present in various erythroid regulatory gene promoters, enhancers, and locus control regions (1, 2). GATA-1 is expressed in several tissues and participates in the transcriptional regulation of most erythroid and megakaryocytic expressed genes (3, 4). In addition to regulating differentiation, GATA-1 may also control cell survival. In GATA-1−/− erythroid precursors, cellular maturation is accompanied by apoptosis, suggesting a role of GATA-1 in cell survival (5, 6). Subsequent studies showed that the Bcl-x gene has two GATA consensus motifs in the 5′ promoter region (7), and ectopic expression of GATA-1 induces the expression of Bcl-XL in GATA-1−/− erythroid progenitor cells (8). GATA-1 also suppresses IL1-6-induced apoptosis and up-regulates the expression of Bcl-2 in M1 myeloid cells (9). Moreover, GATA elements are found in the promoters of other antiapoptotic genes such as nitric-oxide synthase (10, 11) and antioxidant enzymes (12). Therefore, GATA-1 may sustain the viability of terminally differentiated hematopoietic cells via a yet unidentified genetic pathway.
Phosphorylation plays a critical role in modulating the activity of transcription factors in vivo. This post-translational modification rapidly modulates protein activity in response to changes in environmental conditions, metabolic activity, or hormonal signaling (13). The effect on the activity of individual transcription factors is variable but includes a change in DNA binding, cellular localization, or the potential to interact with other transcription factors (13). Crossley and Orkin (14) have characterized the phosphorylation sites of GATA-1 in MEL cells before and after Me2SO-induced differentiation. GATA-1 possesses six constitutive phosphorylation sites at serines 26, 49, 72, 142, 178, and 187 and one inducible site at serine 310 that responds to Me2SO treatment (14). There is conflicting evidence regarding a role for phosphorylation of GATA-1 in influencing its DNA binding or transcriptional activity. In one study, no effect of phosphorylation of GATA-1 was found (14). However, in another study, the DNA binding of baculovirus-expressed human GATA-1 was reported to be sensitive to treatment by alkaline phosphatase (15). A more recent study showed that the level of GATA-1 phosphorylation increases after the induction of K562 cells with Me2SO and that phosphorylation increases the binding affinity of GATA-1 for a canonical binding site (16). These seemingly conflicting data may stem from different model systems utilized in each study.
In a previous study, we demonstrated that expression of the survival gene E4bp4 is transcriptionally regulated by IL-3 and that a GATA motif downstream of the major transcription initiation site is essential for E4bp4 expression in the IL-3 dependent Ba/F3 hematopoietic cell line (17). We also demonstrated that GATA-1 binds to the E4bp4 promoter in vivo and plays an important role in E4bp4 transcriptional activation. Furthermore, we showed that GATA-1 is involved in the anti-apoptotic signaling of IL-3 (17). Since GATA-1 appears to mediate IL-3-dependent survival responses, defining the mechanisms by which the GATA-1 gene and protein are regulated by IL-3-dependent signaling should provide important clues regarding antiapoptotic mechanisms in committed cells. In this study, we explore the mechanism by which IL-3 regulates the function of GATA-1. We show that phosphorylation of GATA-1 by ERK mediates its transcriptional activity induced by IL-3. Mutation of the GATA-1 phosphorylation site inhibits its anti-apoptotic function and induction of Bcl-XL. These data suggest that GATA-1 phosphorylation is essential for cell survival.
MATERIALS AND METHODS
Cell Cultures
Ba/F3 (murine IL-3-dependent pro-B cell line) cells and bone marrow-derived IL-3-dependent primary cells (18) were cultured as described previously (17). COS-1 cells were maintained in the standard culture medium (19), and Ba/F3 derivatives expressing GATA-1-ER (GER) (20), GATA-1S26E-ER (S26EER), and GATA-1S26A-ER (S26AER) were maintained with 5 μg/ml puromycin as described by Rao et al. (21). For experiments involving kinase inhibition, PD98059 (final concentration, 50 μM), U0126 (50 μM), wortmannin (0.1 μM), LY294002 (50 μM), JIP-I (1 μM), rapamycin (50 μM), SB203580 (5 μM), and SB202190 (5 μM) were all purchased from Calbiochem. Estrogen (β-estradiol, E2257) was purchased from Sigma and was used at a final concentration of 0.1 μM.
Western Blotting
Total cell lysates and nuclear extracts were prepared, fractionated, and visualized as described in the standard Western blot protocol described previously (17). The following rabbit polyclonal antibodies were used to detect the expression of specific antigens. Anti-GATA-1 (N6), anti-Bcl-XL (M-125), anti-Bcl-2 (ΔC21), and anti-Bax (N-20) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); anti-p-ERK1/2 and ERK1/2 were from Cell Signaling Technology, Inc.; anti-β-actin (AC-15) and FLAG (M2) were from Sigma. Anti-murine Mcl-1 antibody was kindly provided by Dr. H.-F. Yang-Yen (Academia Sinica). The polyclonal rabbit anti-p-GATA-1 was raised against the synthetic peptide SALVSpSPSDSTGFC (where pS represents phosphoserine) by Bethyl Laboratories Inc. (Montgomery, TX) with no ability to recognize the control peptide SALVSSPSDSTGFC (data not shown).
Phosphatase Treatment of Nuclear Extracts in Vitro
Nuclear extracts of Ba/F3 cells were prepared as described by Yu et al. (17) with Dignam extraction buffer. Fifteen μg of extracts were used for each reaction, and 1 unit of protein phosphatase 2A (Upstate Group, LLC) was added to the extracts with or without 1 μM of okadaic acid (Calbio-chem). The samples were then incubated for 1 h at 30 °C. Alternatively, calf intestinal alkaline phosphatase (New England Biolabs) was added to the nuclear extract. After 1 h at 37 °C, the reaction mixture was analyzed by immunoblotting with anti-GATA-1 antibody (N6; Santa Cruz Biotechnology).
Plasmid Construction
The pGEX-3X-based Escherichia coli expression constructs that would direct the synthesis of the GST-tagged various deletions or point mutations of GATA-1 are listed in Table I. All point mutants of GATA-1 were generated by PCR-based site-directed mutagenesis. For those constructs involving PCR or site-directed mutagenesis, the nucleotide sequences were confirmed by sequencing. W1 is a mammalian expression vector expressing FLAG-tagged GATA-1. W1M1, W1M2, W1M3, W1M4, and W1M5 are identical to W1 except that they encode a mutant GATA-1 with alanine or glutamic acid substitution at the following residues as indicated: W1M1, with an S26A mutation (Ser-26 mutated to Ala); W1M2, with S26A, T101A, S107A, and T108A mutations; W1M3, with S26A, S170A, S174A, and T176A mutations; W1M4, with S-26A, T101A, S107A, T108A, S170A, S174A, and T176A mutations; and W1M5, with an S26E mutation. W1, W1M1, and W1M5 were subcloned into the pCMXGal4-DBD vector (22) to generate W2, W2M1, and W2M5, respectively. Plasmid pEBB-GATA-1/ER was described previously (20). Expression plasmids encoding FLAG-tagged dominant negative MEK1, dominant negative ERK1, and dominant negative AKT, were provided by Dr. H.-F. Yang-Yen.
Table I.
List of GST-GATA-1 fusion proteins
| Names | Position of mutated amino acid sequencea | Range of amino acid sequence in GATA-1 |
In vitro kinase reactionb |
|
|---|---|---|---|---|
| ERK1/2 immunoprecipitation complex (without/with IL-3) | Recombinant ERK1/2 | |||
| D1 | None | 2–193 | ++/++++ | ++++ |
| D1M1 | -VSS26P- → -VSA26P- | 2–193 | +/++ | ++ |
| D2 | None | 194–413 | −/− | − |
| D3 | None | 2–71 | −/++++ | ++++ |
| D3M1 | -VSS26P- → -VSA26P- | 2–71 | −/− | − |
| D3M2 | -STS49P- → -STA49P- | 2–71 | −/++++ | ++++ |
| D3M3 | -VSSP27- → -VSSA27- | 2–71 | −/− | − |
| D4 | None | 66–148 | ++/++ | − |
| D4M1 | -RHS72P- → -RHA72P- | 66–148 | ++/++ | − |
| D4M2 | -GGS91P- → -GGA91P- | 66–148 | ++/++ | − |
| D4M3 | -LDT135L- → -LDA135L- | 66–148 | ++/++ | − |
| D4M4 | -RLS142P- → -RLA142P- | 66–148 | ++/++ | − |
| D4M5 | -RHS72P–RLS142P- → -RHA72P–RLA142P- | 66–148 | ++/++ | − |
| D4M6 | -GKT101A–PAS107T108V- → -GKA101A–PAA107A108V- | 66–148 | −/− | − |
| D5 | None | 143–193 | −/+ | ++ |
| D5M1 | -FPS170P- → -FPA170P - | 143–193 | −/+ | + |
| D5M2 | -FFS174P- → -FFA174P - | 143–193 | −/+ | + |
| D5M3 | -SPT176G- → -SPA176G - | 143–193 | −/+ | + |
| D5M4 | -TGS178P- → -TGA178P- | 143–193 | −/+ | ++ |
| D5M5 | -YSS187P- → -YSA187P - | 143–193 | −/+ | ++ |
| D5M6 | -TGS178P–YSS187P- → -TGA178P–YSA187P- | 143–193 | −/+ | + |
| D5M7 | -FPS170P–TGS178P–YSS187P- → -FPA170P–TGA178P–YSA187P- | 143–193 | −/+ | + |
| D5M8 | -FPS170PFFS174PT176G- → -FPA170PFFA174PA176G- | 143–193 | −/− | − |
The putative phosphorylation sites are underlined.
++++, highly phosphorylated proteins; ++, medium phosphorylation; +, low phosphorylation; −, negative phosphorylation.
Reporter Gene Assay
Ba/F3 or COS-1 cells were transiently transfected with the indicated plasmids by electroporation or Lipofectamine transfection kit (Invitrogen), and the cell lysates were assayed for the luciferase activity as previously described (17).
Northern Blotting
Total RNA was isolated from Ba/F3 cells and bone marrow-derived IL-3-dependent primary cells of BALB/c mice with the TRIZOL reagent kit (Invitrogen). Approximately 30 μg of total RNA from each treatment were then subjected to the standard Northern blotting as previously described (17) using probes specific to the E4bp4 or G3pdh gene.
In Vitro Kinase Assay
Cell lysates were subjected to immunoprecipitation with ERK1/2 antibody, and the immunocomplexes were analyzed for kinase activity according to the vendor’s instruction (Cell Signaling Technology, Inc.). In some reactions, active recombinant ERK1 and ERK2 purchased from Upstate Biotechnology, Inc. (Lake Placid, NY; catalog number 14–439) were used to replace immunocomplexes in kinase reactions.
Quantitation of Apoptotic Cells and Mitochondrial Membrane Potential Analysis
Annexin V staining of the apoptotic cells (BioVision Research) was performed according to the vendor’s instructions and was described previously (17, 23). The viability of cells was determined by trypan blue exclusion. Loss of mitochondria membrane potential was monitored by rhodamine 123 staining (24).
RESULTS
IL-3 Induces Hyperphosphorylation of GATA-1
The genetic pathway of apoptosis is highly conserved throughout evolution from nematodes to humans (25). Within the regulatory pyramid of this pathway, E4BP4 acts as one of the initial responses to extracellular stimulation, suppresses the suicidal reaction, and prevents cells from disintegration. We previously demonstrated that GATA-1 is a key regulator of the E4bp4 gene and is also an essential component of the antiapoptotic network of IL-3 in an IL-3-dependent pro-B cell line (17). In this report, we sought to investigate how IL-3 regulates GATA-1, which in turn induces the expression of E4bp4.
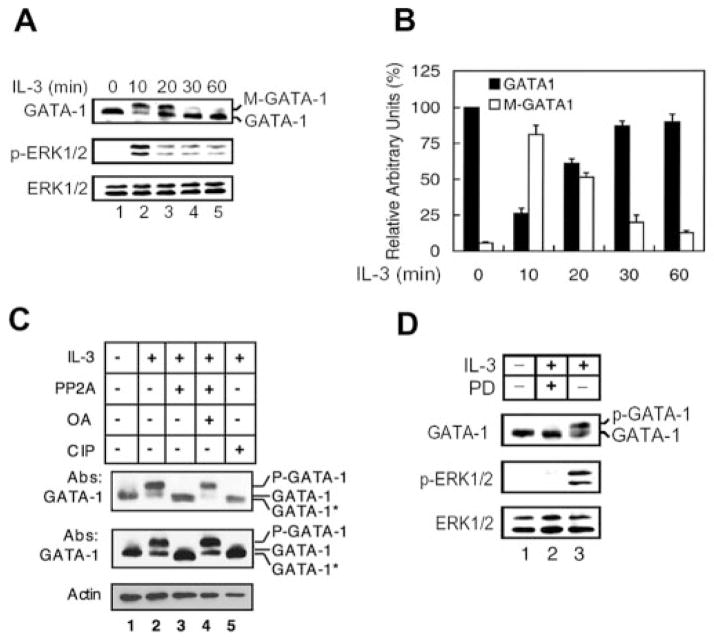
First we tested whether the protein level of GATA-1 is altered by IL-3. Western blot analyses of the total lysates of Ba/F3 cells with GATA-1-specific antibody revealed that the protein level of GATA-1 was rapidly decreased upon IL-3 stimulation at 10 min and then gradually returned to the base-line level (Fig. 1A, lanes 2–5). Interestingly, a slower migrating species of GATA-1 (M-GATA-1) is evident (Fig. 1A, top panel). Accumulation of this modified form of GATA-1 was maximal after 10–20 min of IL-3 induction (Fig. 1A, compare lanes 2 and 3 with lane 1) and declined thereafter to the basal level (Fig. 1A, lane 5). The phosphorylation status and total protein levels of ERK1/2 were monitored during IL-3 stimulation (Fig. 1A, middle and bottom panels). The result shows that both GATA-1 modification and ERK1/2 phosphorylation events follow similar kinetics. GATA-1 protein levels were quantified by densitometer (Fig. 1B), and the results suggested that the level of total GATA-1 protein, including M-GATA-1 and GATA-1, is largely unchanged during IL-3 treatment, whereas the ratio between slower and normal migrating species varied.
Fig. 1. IL-3 induces hyperphosphorylation of GATA-1.
A, cytokine-depleted Ba/F3 cells (lane 1) were treated with IL-3 as indicated (lanes 2–5), and total cell lysates were subjected to Western blotting using antibodies against GATA-1 (top panel), p-ERK1/2, and ERK1/2 (middle and bottom panels). The slower and normal migrating species of GATA-1 are indicated as M-GATA-1, and GATA-1, respectively. B, the protein levels of GATA-1 (black bars) or M-GATA-1 (open bars) in Western blots were determined by densitometry. The densitometric reading of GATA-1 at the zero time point was taken as 100%. Data are means ± S.D. from three independent experiments (one representative result is shown in A). C, nuclear extracts were prepared from Ba/F3 cells that were deprived of IL-3 for 12 h (lane 1) and restimulated with IL-3 for 10 min (lanes 2–5). The extracts were incubated in the presence of protein phosphatase 2A minus or plus okadaic acid (lanes 3 and 4) or in the presence of calf intestinal alkaline phosphatase (CIP) (lane 5) and then subjected to Western blotting with antibodies to GATA-1 (top and middle) and α-actin (bottom). Proteins loaded in the top panel are one-fifth the amount in the middle panel. p-GATA-1 and GATA-1* refer to the hyperphosphorylated and hypophosphorylated form of GATA-1, respectively. D, cytokine-depleted Ba/F3 cells (lane 1) were treated with IL-3 for 10 min (lanes 2 and 3) in the presence (lane 2) of the MEK inhibitor PD98059 or the vehicle control Me2SO (lane 3). Total cell lysates were subjected to Western blot analysis as described in the legend to A.
To investigate whether this IL-3-induced modification of GATA-1 mobility is due to phosphorylation, we treated nuclear extracts with protein phosphatase 2A or calf intestinal phosphatase. Extracts from cells not treated with IL-3 were included as a control (Fig. 1C, lane 1), showing the migration of GATA-1 in unstimulated cells. Protein phosphatase 2A or calf intestinal alkaline phosphatase treatment of nuclear extracts from IL-3-stimulated cells resulted in the disappearance of the slower migrating form of GATA-1 (indicated as p-GATA-1) and the appearance of a band that ran slightly faster than the unstimulated GATA-1 (indicated as GATA-1*) (Fig. 1C, compare lanes 3 and 5 with lanes 2 and 4 in the middle panel). In the top panel, with fewer proteins loaded in the gel, the protein phosphatase 2A-treated product appeared to be a doublet, and the faster migrating GATA-1 (GATA-1*) presumably represents the hypophosphorylated form of GATA-1. Okadaic acid specifically inhibits protein phosphatase 2A and prevents the conversion of the hyperphosphorylated GATA-1 to the hypophosphorylated form (lane 4). When we treated the nuclear extracts with another potent protein phosphatase, λ-phage phosphatase, the same hypophosphorylated GATA-1 was observed (data not shown). These data suggested that GATA-1 is probably constitutively phosphorylated in the absence of IL-3 and rapidly becomes hyperphosphorylated upon IL-3 stimulation.
According to previous reports, IL-3 activates GATA-2 through the ERK pathway in hematopoietic cells (26). GATA-4 also is phosphorylated by ERK in cardiomyocytes (27). Therefore, we examined whether GATA-1 also is phosphorylated via the MEK/ERK pathway during IL-3 treatment. Pretreatment of cells with a specific inhibitor of MEK, PD98059, but not the vehicle control (Me2SO), effectively blocked induction of GATA-1 hyperphosphorylation (Fig. 1D, compare lane 2 with lane 3). Treatment with another MEK inhibitor, U0126, gave the same result (data not shown). However, unlike in vitro treatment with phosphatases (Fig. 1C), PD98059 treatment was not able to convert GATA-1 to the faster migrating form, suggesting that the MEK/ERK signaling pathway probably is involved in hyperphosphorylation but not in constitutive phosphorylation of GATA-1.
MEK/ERK Pathway-dependent Induction of E4bp4
To further investigate whether the MEK/ERK signaling pathway also mediates the induction of E4bp4 expression in response to IL-3, we examined the effect of various pharmacological agents on the activation of an E4bp4 promoter-reporter plasmid in Ba/F3 cells. In good agreement with the results in Fig. 1D showing that hyperphosphorylation of GATA-1 is completely blocked by the MEK inhibitors, we found that PD98059 and U0126 also completely blocked the IL-3-dependent induction of luciferase activity (Fig. 2A, compare lanes 3 and 4 with lane 2), whereas the phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 had only a partial inhibitory effect or no effect, respectively (Fig. 2A, lanes 5 and 6). Wortmannin has been shown to slightly inhibit MEK activity (28). Therefore, the partial inhibitory effect of wortmannin on the E4bp4 reporter may be due to inhibition of MEK instead of phosphatidylinositol 3-kinase. The c-Jun N-terminal kinase inhibitor JIP-I had no effect (Fig. 2A, lane 7). In contrast, the p70 S6K inhibitor rapamycin (Fig. 2A, lane 8) and the p38 MAPK inhibitors SB203580 and SB202190 (Fig. 2A, lanes 9 and 10) slightly, but reproducibly, potentiated the effect of IL-3 on the E4bp4 reporter activity, suggesting that p70 and p38 may inhibit E4bp4 transcription.
Fig. 2. MEK/ERK pathway-dependent induction of E4bp4.
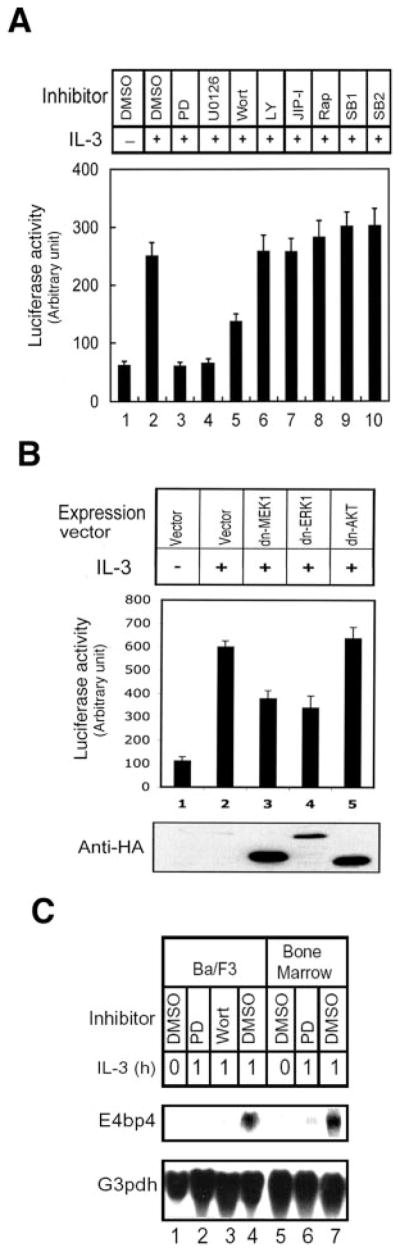
A, Ba/F3 cells were transfected with E4bp4-LUC and then incubated for 12 h in the absence (lane 1) or presence (lanes 2–10) of IL-3 and the indicated kinase inhibitors (lanes 3–10) or Me2SO (DMSO; lanes 1 and 2). Cell lysates were then assayed for luciferase activity. PD, PD98059; Wort, wortmannin; LY, LY294002; Rap, rapamycin; SB1, SB203580; SB2, SB202190. B, Ba/F3 cells were cotransfected with E4bp4-LUC and either expression vectors for the indicated dominant negative (dn) mutants (lanes 3–5) or the corresponding empty vector (lanes 1 and 2). Cells were incubated in the absence (lane 1) or presence (lanes 2–5) of IL-3, and cell lysates were assayed for luciferase activity. In A and B, data are means ± S.D. from three independent experiments performed in duplicate. C, Ba/F3 (lanes 1–4) and bone marrow-derived primary IL-3-dependent cells (lanes 5–7) were cultured in cytokine-free medium for 12 h (lanes 1 and 5) and restimulated with IL-3 for 1 h (lanes 2–4 and lanes 6 and 7). Cells were pretreated with Me2SO (DMSO; lanes 1, 4, 5, and 7), PD98059 (PD; lanes 2 and 6), or wortmannin (Wort; lane 3) 2 h before exposure to IL-3. Total RNA was prepared and subjected to Northern blotting with 32P-labeled probes specific to E4bp4 or G3pdh. The blot was visualized by autoradiography.
In order to further demonstrate the involvement of MEK/ERK activity in E4bp4 transcription, expression plasmids encoding dominant negative forms of several kinases were transfected along with the reporter construct into Ba/F3 cells. As shown in Fig. 2B, dominant negative mutants of MEK-1 (lane 3) and ERK1 (lane 4), but not that of AKT (lane 5), inhibited activation of the E4bp4 reporter gene by IL-3 (lanes 3–6). However, these suppressions were never greater than 50% even with much higher amounts of expression vectors used in the assay. The underlying mechanism of 50% suppression effect of dominant negative MEK1 and dominant negative ERK1 to the reporter gene activity is still unknown. We cannot rule out the possibility of technical limitation in exogenous gene expression. Alternatively, the involvement of kinases other than MEK1 and ERK1, like MEK5, in transducing IL-3 signal is another conceivable explanation. Nevertheless, our data suggest that the MEK1/ERK1 pathway is an important part of IL-3 induction of the E4bp4 gene.
We next investigated whether the MEK/ERK pathway mediates IL-3 induction of endogenous E4bp4. Ba/F3 cells and bone marrow-derived primary IL-3-dependent cells were treated with or without kinase inhibitors prior to IL-3 stimulation, and total RNAs were isolated from these cells and subjected to Northern blot analysis. Both PD98059 and wortmannin blocked the IL-3-induced expression of E4bp4 (Fig. 2C, lanes 2, 3, and 6). These data reveal that the MEK/ERK pathway plays an important role in mediating the IL-3-induced expression of E4bp4.
Ser-26 of GATA-1 Is the Major Inducible Phosphorylation Site
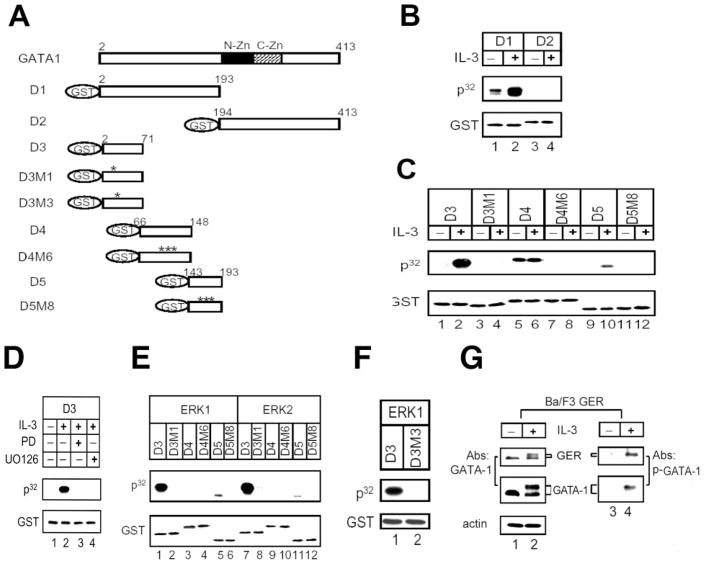
To investigate the role of phosphorylation in regulating the transcriptional function of GATA-1, we identified and then mutated the IL-3-inducible and constitutive phosphorylation sites in GATA-1. The coding sequence of GATA-1 was split into the N-and C-terminal fragments (plasmids D1 and D2 in Table I), which were each expressed as recombinant GST fusion proteins (Fig. 3A, D1 and D2). The recombinant proteins were subjected to an in vitro kinase assay with complexes immunoprecipitated by MAPK antibody from Ba/F3 cell lysates. As shown in Fig. 3B, the D1 fragment of GATA-1 was initially slightly phosphorylated in the absence of IL-3 and became highly phosphorylated upon IL-3 treatment (compare lane 1 with lane 2). In contrast, the D2 fragment was not phosphorylated in the presence or absence of IL-3 (lanes 4 and 3, respectively). These data suggested that both the constitutive and IL-3-inducible sites are probably located in the N-terminal domain of GATA-1.
Fig. 3. Ser-26 of GATA-1 is the major inducible phosphorylation site of ERK1/2 in vitro and in vivo.
A, schematic representation of various mutants of GST-GATA-1 fusion proteins as listed in Table I. The asterisk indicates the amino acids that were replaced with alanine. N-Zn, N-zinc finger in GATA-1; C-Zn, C-zinc finger. B, in vitro kinase assays were performed using D1 or D2 GST-GATA-1 fusion proteins with an ERK1/2 immunocomplex precipitated from lysates of Ba/F3 cells treated with (lanes 2 and 4) or without (lanes 1 and 3) IL-3. GST, the immunoblot probed with anti-glutathione S-transferase antibody, which serves as the internal control. P32, the autoradiograph. C–F, various mutants of GST-GATA-1 fusion proteins as indicated in each panel were subjected to in vitro kinase assays as described in B, except that cells were grown in the presence of the vehicle control Me2SO (lane 2), the MEK inhibitor PD98059 (PD, lane 3), or U0126 (lane 4) during IL-3 restimulation (D) or using active recombinant ERK1 (lanes 1–6 of E; lanes 1 and 2 of F) or ERK2 (lanes 7–12 of E). G, in vivo Ser-26 phosphorylation detected by anti-p-GATA-1 antibody. The Ba/F3 cells expressing GATA-1-ER fusion protein (GER) were stimulated with IL-3, and cell lysates were subjected to Western blot analysis with either anti-GATA-1 antibody (lanes 1 and 2) or anti-p-GATA-1 (Ser-26) antibody (see “Materials and Methods”) (lanes 3 and 4).
To further narrow down the location of phosphorylation site(s), the D1 peptide was split into three smaller peptides (see Table I, D3, D4, and D5) that were expressed as GST fusion proteins (Fig. 3A, D3, D4, and D5) and then subjected to the in vitro kinase assay as shown in Fig. 3C. The D3 peptide was not phosphorylated with lysates prepared from IL-3-depleted cells (Fig. 3C, lane 1), but it was highly phosphorylated when cells were stimulated with IL-3 (Fig. 3C, lane 2), indicating that D3 peptide contains a strong inducible site(s) but not the constitutive site. Analysis of D4 and D5 recombinant proteins suggested that the D4 peptide contains a weak constitutive site(s) but not an inducible site (Fig. 3C, lanes 5 and 6), and the D5 peptide contains a minor inducible site(s) (Fig. 3C, lanes 9 and 10). To identify the phosphorylated amino acid residues in these peptides, alanine substitutions were introduced into these three peptides at putative MAPK substrate sites (29, 30) or at phosphorylated residues identified by tandem mass spectrum analysis of phosphorylated and nonphosphorylated D4 and D5 peptides (LCQ DECA XPPlus; ThermoFinnigan, San Jose, CA). Recombinant D3, D4, and D5 proteins containing single or combined point mutations were produced and subjected to the in vitro kinase reaction (see Table I). In the D3 peptide, alanine substitution at Ser-26 (Fig. 3A, D3M1) completely blocked the IL-3-inducible phosphorylation by MAPK immunocomplexes (Fig. 3C, compare lane 2 with lane 4). A combination of three single-point mutations at residues 101, 107, and 108 in the D4 peptide (Fig. 3A, D4M6) dramatically eliminated constitutive phosphorylation (Fig. 3C, lanes 5–8). Another combination of mutations at residues 170, 174, and 176 in the D5 peptide (Fig. 3A, D5M8) abolished the minor phosphorylation by IL-3-inducible MAPK immunocomplexes (Fig. 3C, compare lane 10 with lane 12). Taken together, these results suggest that under the in vitro kinase assay condition described above, candidate constitutive phosphorylation sites include Thr-101, Ser-107, and Thr-108; candidate minor IL-3-inducible sites include Ser-170, Ser-174, and Thr-176, and a major IL-3-inducible site resides at Ser-26. As shown in Fig. 3D, phosphorylation of the D3 peptide was completely inhibited in extracts of cells that were pretreated with PD98059 or U0126 (compare lane 2 with lanes 3 and 4), suggesting that the major inducible site in the D3 peptide is most likely the substrate of kinases in the MEK/ERK pathway.
ERK1/2 Directly Phosphorylates GATA-1 in Vitro
We next examined whether active, recombinant ERK can directly phosphorylate any of these sites mentioned above. In vitro kinase reactions of GATA-1 fragments were carried out using active recombinant ERK1 and ERK2 (see “Materials and Methods”). As shown in Fig. 3E, the D3 and D5 peptides were directly phosphorylated by both kinases (lanes 1, 5, 7, and 11), whereas the D3M1 and D5M8 peptides were resistant to these kinases (lanes 2, 6, 8, and 12), consistent with our observations using MAPK immunocomplexes (Fig. 3C). These data suggest that the major site at Ser-26 and minor sites at Ser-170, Ser-174, and Thr-176 are all direct phosphorylation targets for MAPK in vitro. Interestingly, the D4 peptide was not phosphorylated by either ERK1 or ERK2 (Fig. 3E, lanes 3 and 9). Therefore, the kinase(s) that constitutively phosphorylates D4 peptide in the absence of IL-3 and is insensitive to the presence of PD98059 is unlikely to be a component in the MEK1/ERK1 signaling pathway, although it was co-immunoprecipitated with MAPK from cell lysates.
Inspection of the peptide sequence around the major inducible MAPK site shows that only the serine-proline dipeptide sequence of the consensus MAPK substrate peptide sequence that is deduced from synthetic peptide libraries (29, 30) is conserved. To investigate whether this serine-proline dipeptide is essential to be recognized by MAPK, we mutated Pro-27 to alanine (Fig. 3A, D3M3) and subjected it to the ERK1 kinase reaction. As shown in Fig. 3F, the D3M3 peptide is highly resistant to phosphorylation by ERK1 (compare lane 1 with lane 2), suggesting that Pro-27 of GATA-1 is indeed part of the MAPK substrate recognition site and required for Ser-26 phosphorylation.
To further demonstrate that the phosphorylation of GATA-1 at Ser-26 occurs in vivo upon IL-3 induction, we raised a rabbit polyclonal peptide antibody specifically against a phospho-Ser-26-containing peptide of GATA-1 (residues 21–34). This antibody does not recognize nonphosphorylated control peptide (data not shown). In the Ba/F3 cells expressing GER (see below), anti-p-GATA-1 antibody only recognized single bands for GATA-1 and GER that co-migrated with the slower migrating hyperphosphorylated GATA-1 and GER, respectively, indicating that both endogenous GATA-1 and exogenous GER are phosphorylated at residue Ser-26 (Fig. 3G, lane 2). Therefore, our data suggest that the serine 26 phosphorylation occurs both in vitro and in vivo.
Phosphorylation of GATA-1 Enhances E4bp4 Transcription Activity
We showed previously that GATA-1 is able to trans-activate the survival gene E4bp4 (17). We therefore investigated whether phosphorylation of GATA-1 at Ser-26 is important for transactivation of the E4bp4 gene. To address this issue, the FLAG-tagged wild type (W1) or various alanine (W1M1, W1M2, W1M3, W1M4) or glutamic acid substitution mutants (W1M5) of GATA-1 (Fig. 4A) were transiently cotransfected into Ba/F3 cells together with the E4bp4 promoter-luciferase reporter (Fig. 4B). As shown in the inset of Fig. 4B, in this transient assay, all mutant proteins were expressed at a level that was very similar to that of the wild-type GATA-1 protein. Without IL-3 stimulation, overexpression of wild-type GATA-1 alone transactivated the E4bp4 reporter gene ~10-fold (compare lane 2 with lane 1). Under the same experimental conditions, GATA-1 with a single S24A mutation (W1M1) manifested a slightly reduced transactivation activity (i.e. ~70% of that of the wild-type protein) (compare lanes 2 and 3 of Fig. 4B). Additional alanine substitution mutations at the minor inducible and/or constitutive sites as in W1M2, W1M3, and W1M4 did not further reduce the transactivation activity of W1M1 on the E4bp4 reporter gene (compare lanes 4–6 with lane 3 of Fig. 4B). In contrast, the S26E mutant (W1M5) exerted a much higher transactivation activity (~2-fold increase) than that of the wild-type protein (compare lanes 2 and 7 of Fig. 4B). We next examined whether IL-3 stimulation would have a differential effect on the transactivation activity of these proteins. We noticed first that, in the absence of a co-transfected GATA-1 expression vector (i.e. replaced with an empty control vector, V), the E4bp4 reporter was slightly stimulated by IL-3 (compare lanes 1 and 8 of Fig. 4B), a result probably due to the presence of the endogenous GATA-1 in the Ba/F3 cells (hereafter referred to as the “basal” stimulation activity). Upon IL-3 stimulation, after the subtraction of the “basal” stimulation activity, the transactivation activity of the wild-type GATA-1 protein was further increased to a level that was ~160% of that observed under conditions without IL-3 stimulation (compare lanes 2 and 9 of Fig. 4B and lane 1 of Fig. 4C). Using the same normalization method, the transactivation activities of all alanine (W1M1, W1M2, W1M3, and W1M4) and glutamic acid (W1M5) substitution mutants were not further enhanced by IL-3 treatment of cells (compare lanes 3–7 with lanes 10–14 of Fig. 4B and lanes 2 and 3 of Fig. 4C). These data suggest that the alanine substitution of serine at residue 26 (S26A) abolishes phosphorylation and keeps GATA-1 in a transcriptionally less active state, whereas the glutamic acid substitution (S26E) mimics phosphorylation and maintains the function of GATA-1 in a more active state. To investigate whether this phosphorylation-dependent increment of GATA-1 transactivation ability is specific to hematopoietic cells, the same reporter gene assay was carried out in a nonhematopoietic cell line COS-1. Results obtained in COS-1 cells (Fig. 4D) were very similar to that obtained in Ba/F3 cells under conditions without IL-3 stimulation (Fig. 4B); i.e. the transactivation activity of wild-type GATA-1 (W1, lane 2) was only one-third that of S26E (W1M5, lane 4), and the S26A mutant (W1M1, lane 3) manifested a transaction activity slightly reduced compared with that of the wild-type protein. Taken together, these data suggest that GATA-1 phosphorylation may be essential for full activation of the E4bp4 transcription activity and that the transcriptional machinery that can detect the phosphorylation status at Ser-26 of GATA-1 may be present in both hematopoietic and nonhematopoietic cell types.
Fig. 4. Phosphorylation of Ser-26 enhances GATA-1 transactivation of the E4bp4 promoter.
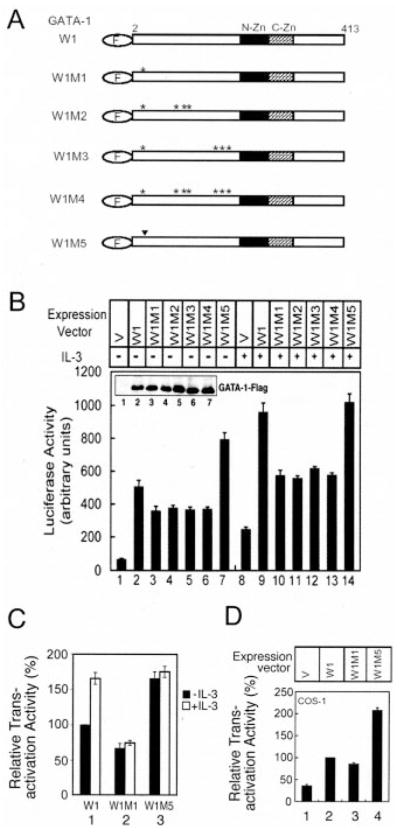
A, schematic representation of FLAG-tagged, wild-type (W1) or mutant (W1M1–W1M5) GATA-1 proteins. F in the oval indicates FLAG tag; the asterisk and inverted triangle indicate the amino acids that were replaced with alanine and glutamic acid, respectively. N-Zn, N-zinc finger in GATA-1; C-Zn, C-zinc finger. B, the E4bp4-LUC plasmid was transfected into Ba/F3 cells along with an empty expression vector (V; lanes 1 and 8) or expression vectors encoding W1 (lanes 2 and 9) or the mutants of W1M1–W1M5 (lanes 3–7 and 10–14). Transfected cells were incubated in medium without (lanes 1–7) or with IL-3 (lanes 8–14) for 12 h, and cell lysates were prepared and analyzed for luciferase activity or for expression of FLAG-tagged GATA-1 proteins (see inset in B). C, normalized results of those shown in B to illustrate the relative transactivation activity of wild-type or mutant GATA-1 with or without IL-3 stimulation. For this normalization method, the basal activities caused by the vector control alone (with or without IL-3 stimulation) (lanes 1 and 8) were each subtracted from those activities shown in lanes 2–7 and lanes 9–14, respectively. After subtraction, the activity of wild-type GATA-1 (W1) in the absence of IL-3 (lane 2) was assigned a value of 100, and the activities shown in all other lanes were normalized to this activity. The normalized results of W1M2, W1M3, and W1M4 are very similar to that of W1M1 (data not shown). D, the expression vector (V; lane 1) or plasmids encoding W1 (lane 2) or the mutants W1M1 or W1M5 (lanes 3 and 4) were transfected into COS-1 cells and were analyzed as described in B. In B–D, data are means ± S.D. from five independent experiments performed in duplicate.
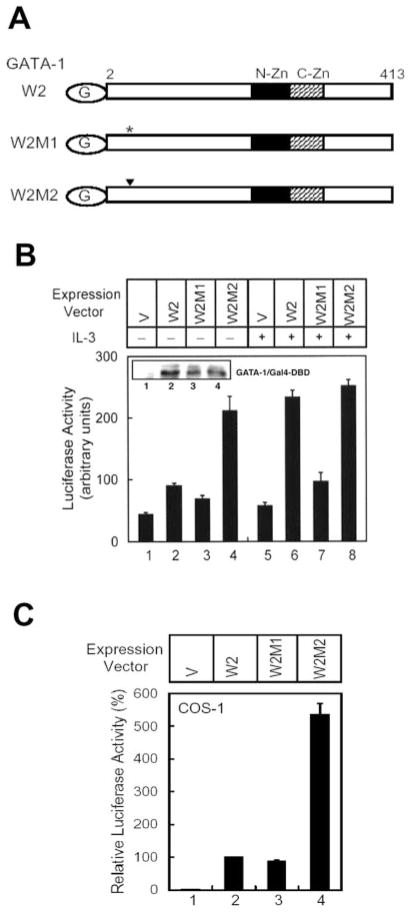
Ser-26 Is Part of the Phosphorylation-dependent Transactivation Domain
Phosphorylation might facilitate translocation of GATA-1 from the cytoplasm to the nucleus. However, immunostaining of ectopically expressed GATA-1 and the mutant S26A suggested that both proteins are present inside the nucleus (data not shown). Alternatively, phosphorylation might increase the binding affinity of GATA-1 for its DNA recognition element. However, we observed that λ-protein phosphatase treatment only reduces 10–20% of the GATA-1 binding activity in IL-3-stimulated nuclear extracts (data not shown) and that the in vitro translated S26A and S26E mutants of GATA-1 manifested a DNA binding activity that is very similar to that of the wild-type protein (data not shown). These two results suggest that phosphorylation of GATA-1 may not significantly alter its DNA-binding activity. On the other hand, once GATA-1 binds to DNA, phosphorylation may facilitate transcriptional activation. To explore this possibility, wild-type GATA-1, S26A, and S26E mutants were each fused to the Gal4 DNA binding domain to generate the W2, W2M1, and W2M2 mutants, respectively (Fig. 5A). Plasmids encoding these proteins were cotransfected into Ba/F3 cells along with the Gal4-LUC reporter plasmid. Western blot analysis indicated that all three Gal4 fusion proteins were expressed at similar levels (inset in Fig. 5B). In the presence of IL-3, the W2 protein activated the reporter gene equally well as the W2M2 (Fig. 5B,lanes 6 and 8). However, in the absence of IL-3, the W2 protein’s activity was much weaker than that of W2M2 and was similar to that of W2M1 (Fig. 5B, lanes 2–4). Similar results were observed when the experiments were carried out in nonhematopoietic COS-1 cells (Fig. 5C). Therefore, phosphorylation at residue 26 is essential for the full transactivation activity of the Gal4-GATA-1 fusion protein, and the results suggest that serine 26 is a part of the phosphorylation-dependent transactivation domain.
Fig. 5. Ser-26 is part of the phosphorylation-dependent trans-activation domain.
A, schematic representation of wild-type (W2), S26A (W2M1), and S26E (W2M2) GATA-1 proteins fused to the Gal4 DNA binding domain. G in the oval indicates the Gal4 tag; the asterisk and the inverted triangle indicates the amino acid that was replaced with alanine and glutamic acid, respectively. N-Zn, N-zinc finger in GATA-1; C-Zn, C-zinc finger. B, the Gal4-LUC plasmid was transfected into Ba/F3 cells along with an empty expression vector (V; lanes 1 and 5) or expression vectors encoding W2 (lanes 2 and 6), W2M1 (lanes 3 and 7), or W2M2 (lanes 4 and 8). Transfected cells were incubated in medium without (lanes 1–4) or with IL-3 (lanes 5–8) for 12 h, and cell lysates were analyzed for luciferase activity or for expression of Gal4-GATA-1 fusion protein (see inset in B). C, the experiment described in B was carried out in COS-1 cells again. Data are means ± S.D. from three independent experiments performed in duplicate.
Phosphorylation of Ser-26 Is Essential for the Cell Survival Activity of GATA-1
We previously demonstrated that overexpression of GATA-1 sustained the viability of Ba/F3 cells in the absence of IL-3 (17). Here, we seek to investigate whether phosphorylation of GATA-1 Ser-26 is required for its activity in the survival response. Several Ba/F3 derivative cell lines stably expressing the FLAG-tagged W1, W1M1, or W1M5 mutant protein were established. Fig. 6A shows similar levels of the protein expression of one representative clone of these lines. We then studied the kinetics of cell apoptosis by the annexin V staining method (Fig. 6B) during the first 24 h after IL-3 deprivation. As shown in Fig. 6B, the percentages of apoptosis of a S26E-GATA-1-expressing cell line (W1M5, solid circles) and of a mixed population of many stable S26E clones (data not shown) were significantly lower than that of cells expressing wild-type GATA-1 (W1, triangle) 12 and 24 h after IL-3 withdrawal. On the other hand, under the same conditions, the apoptosis percentages of S26A-GATA-1-expressing cells (W1M1, solid square) and the mixed population of many stable S26A cells (data not shown) were slightly, but reproducibly, higher than cells expressing vector (diamond) or the wild-type GATA-1 protein (W1) (Fig. 6B). These data are consistent with our previous observations (17) and demonstrate that phosphorylation of GATA-1 is crucial for its survival activity.
Fig. 6. Phosphorylation of Ser-26 is essential for the survival activity of GATA-1.
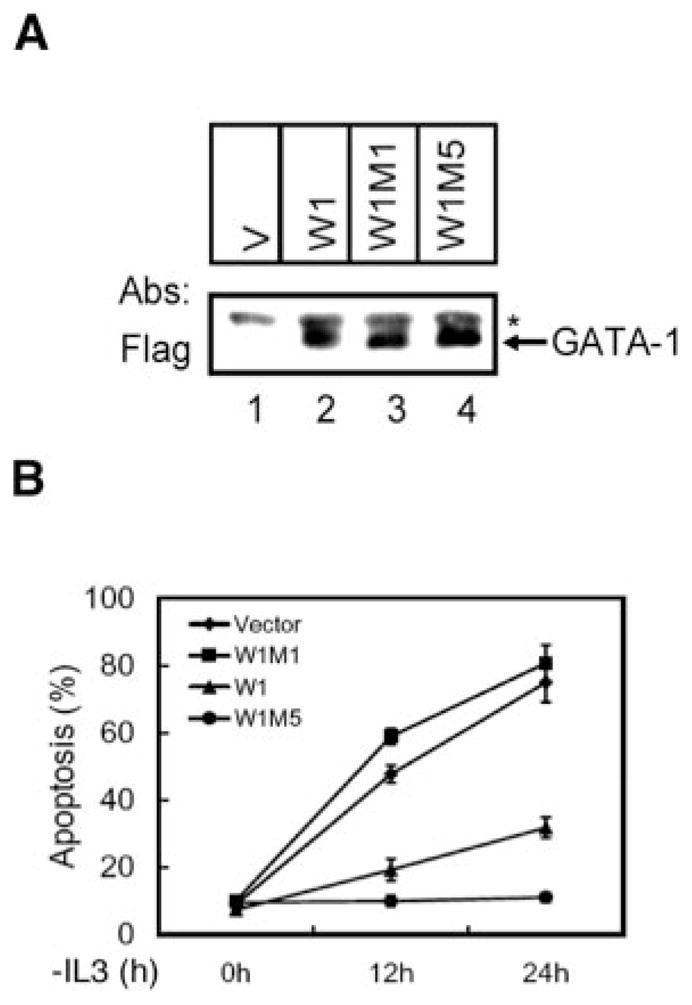
A, lysates were prepared from Ba/F3 stable lines expressing an empty expression vector (V; lane 1) or expression plasmids encoding W1 (lane 2), W1M1 (lane 3), or W1M5 (lane 4) and then analyzed by Western blotting using an antibody specific to FLAG. The arrow points to the ectopically expressed GATA-1 or its derivatives. The asterisk indicates the nonspecific loading control. B, Ba/F3 stable transfected cell lines as indicated were cultured in IL-3-free medium for various times as indicated before the extent of cells undergoing apoptosis was quantified by the annexin V staining method (B). The values are means ± S.D. of three independent experiments.
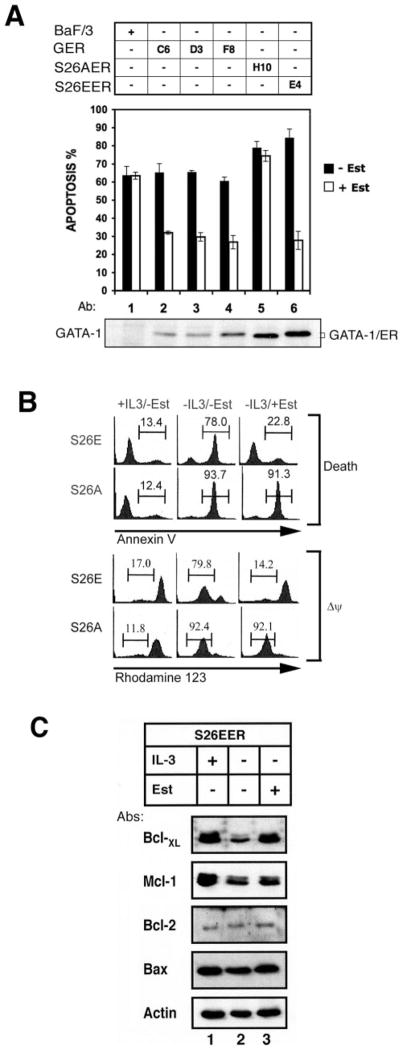
We next determined whether GATA-1 could protect Ba/F3 cells from IL-3 withdrawal-induced apoptosis in an inducible system. To do this, we established Ba/F3 cell lines expressing estrogen receptor fusion proteins of wild-type or mutant GATA-1. Upon estrogen stimulation, these fusion proteins translocate into the nucleus and activate the GATA-1 target genes (21). To be certain that fusion with ER does not interfere with MAPK-mediated GATA-1 phosphorylation, GER was expressed in Ba/F3 cells, and its phosphorylation status was examined before and after treatment with IL-3. As shown in Fig. 3G, in the presence of IL-3, a slower migrating band of GER appeared concomitantly with the appearance of the endogenous hyperphosphorylated GATA-1 (Fig. 3G, lanes 1 and 2). This slower migrating GER was also detected by anti-p-GATA-1 antibody (Fig. 3G, lane 4), suggesting that GER was susceptible to IL-3 signaling and phosphorylated at Ser-26 similar to wild-type GATA-1 in an appropriate intra-cellular context.
As shown in Fig. 7A, estrogen has no antiapoptotic activity in the parental Ba/F3 cells nor in cells expressing the S26AER fusion protein (lanes 1 and 5), albeit the S26AER protein was expressed at a higher level than GER (lanes 2 and 5). However, apoptosis could be effectively protected in cells expressing either GER (about 50%, lanes 2–4) or S26E ER fusion proteins (about 66%, lane 6). The protection efficiency of each cell line is independent of GATA-1 protein expression levels (Fig. 7A, bottom panel).
Fig. 7. The S26E GATA-1 mutant protects mitochondria integrity and induces Bcl-XL expression.
A, the Ba/F3 parental cell line (lane 1) and its derivative lines stably expressing GER (clones C6, D3, and F8; lanes 2–4), S26AER (clone H10; lane 5), or S26EER (clone E4; lane 6) were cultured in IL-3-free medium without (■) or with (□) estradiol (Est) for 16 h and were analyzed for cells undergoing apoptosis by the annexin V staining method. The expression level of each ER fusion protein is shown in the bottom panel of A. Data are means ± S.D. from three independent experiments performed in duplicate. B, S26EER (S26E) and S26AER (S26A) cells cultivated under three different conditions for 16 h as indicated at the top of each panel were subjected to flow cytometric quantification of cells undergoing apoptosis (death) or losing mitochondrial membrane potential (ΔΨ). The percentage of cells undergoing apoptosis (annexin V-positive) (upper panels) or of cells losing mitochondrial membrane potential (rhodamine 123-negative) (lower panels) is indicated above each bracket. C, Bcl-XL expression was induced by estradiol in S26EER cells. S26EER cells were cultivated in medium as indicated, and cell lysates were analyzed for the expression of various Bcl-2 family proteins as indicated. Data shown in B and C are one representative result from three independent experiments.
The ability of the ER fusion protein to protect cells from apoptosis seemed to correlate well with its ability to protect mitochondrial membrane integrity and maintain membrane potential. In the presence or absence of IL-3 (Fig. 7B, columns 1 and 2), the percentage of cells losing membrane potential (Δψ) was closely associated with the extent of cells undergoing apoptosis (Death). Moreover, in the presence of estrogen, S26EER, but not S26AER, preserved membrane potential and maintained cell viability very well (Fig. 7B, column 3). Therefore, activation of the GATA transcription factor probably leads to synthesis of certain antiapoptotic proteins that stabilize mitochondrial membrane integrity. There are several prosurvival Bcl-2 family members, including Mcl-1, Bcl-XL, and Bcl-2 (31), that can protect mitochondrial membrane integrity. In order to determine whether Blc-2 family members are involved in the antiapoptotic activity of GATA-1, total cell lysates prepared from S26EER cells cultured in the presence or absence of estrogen were subjected to Western blot analysis for the expression of Bcl-2 family proteins (Fig. 7C). In the presence of IL-3, both Bcl-XL and Mcl-1 were induced, but not Bcl-2 and Bax (Fig. 7C, lanes 1 and 2). Interestingly, in the absence of IL-3, GATA-1 activation by estrogen only stimulated the expression of Bcl-XL but not that of Mcl-1 (lanes 2 and 3). These results suggest that induction of Bcl-XL, instead of Mcl-1, might account for the antiapoptotic activity of the estrogen-activated S26EER protein (Fig. 7C, lanes 2 and 3). Since IL-3-induced Mcl-1 expression is mainly mediated via the phosphatidylinositol 3-kinase/Akt and p38, but not by the MEK/MAPK pathways (32, 33), it is consistent with our observation that Mcl-1 could not be induced by GATA-1 that is mainly regulated by the MEK/MAPK signaling pathway. Taken together, these data suggest that phosphorylation at Ser-26 of GATA-1 plays a significant role in preventing apoptosis and that Bcl-XL expression may mediate the antiapoptotic response of GATA-1.
DISCUSSION
Crossley and Orkin (14) have carefully characterized the phosphorylation sites of GATA-1 in MEL cells. GATA-1 possesses six constitutive phosphorylation sites at serines 26, 49, 72, 142, 178, and 187 and one inducible site at serine 310 that responds to Me2SO treatment. However, in this report, our in vitro kinase reaction data suggested that in the murine pro-B cell line Ba/F3 the profile of phosphorylation sites in mouse GATA-1 might be very different. Our data suggest that in the absence of IL-3 GATA-1 is phosphorylated at Thr-101, Ser-107, and/or Thr-108 by an unidentified kinase that could be co-immunoprecipitated with ERK1 and ERK2. In response to IL-3 signaling, GATA-1 is further phosphorylated by kinases downstream of MEK1, most likely ERK1 and ERK2, mainly at Ser-26 and to a much lesser extent at Ser-170, Ser-174, and/or Thr-176. Intriguingly, a mutant without any of these phosphorylation sites (W1M4 in Fig. 4A) still retains significant transcriptional activity in the reporter gene assay (Fig. 4B) and is capable of induction of erythroid differentiation.2 Both PD98059 and U0126 could completely suppress the IL-3-activated E4bp4 reporter activity (Fig. 2A). However, neither the MEK nor the ERK dominant negative mutants could do so (only ~50% inhibition) (Fig. 2B). Taken together, our results suggest a more complicated signaling scenario for the involvement of MEK/ERK in regulating GATA-1 function. PD98059 and U0126 may inhibit the function of proteins involved in modifying GATA-1 by means other than phosphorylation. Alternatively, since the MEK5/ERK5 pathway is also sensitive to the treatment of PD98059 and U0126 (34), it is possible that this pathway is also involved in mediating the modulatory effect of IL-3 on GATA-1. However, since the Ba/F3 cells do not contain detectable levels of ERK5,3 this latter possibility, if it exists, would contribute minimally to the regulation of the GATA-1 functions. More experiments are certainly required to investigate all of these possibilities.
Of particular note, the IL-3-inducible MAPK-mediated phosphorylation sites we described, including both major and minor sites, do not fulfill perfectly the consensus sequence of the MAPK substrate site (PX(S/T)P) (29, 30) (see Table I). Nevertheless, our data indicate that Pro-27 is essential for MAPK-mediated Ser-26 phosphorylation, since the P27A mutation completely abolished this event (Fig. 3F). This suggests that the reaction at this major site still follows the serine-proline dipeptide rule for MAPK. Furthermore, since the D3 peptide (amino acids 2–71) was strongly phosphorylated in vitro by ERK1/2, it suggests that ERK1/2 may dock and recognize the substrate site very efficiently. There is a tentative FF motif (Phe-33 and Phe-34) near the major site Ser-26 that may serve as an alternative docking sequence for MAPKs, like the one described in GATA-2 (35).
Phosphorylation is a common post-translational modification for various nuclear transcriptional factors to regulate protein function. GATA-4 is phosphorylated in response to cardiac hypertrophic stimuli and hepatocyte growth factor in cardiomyocytes (27, 36). This phosphorylation event appears to be functionally critical in the transcriptional activation and DNA binding activity of GATA-4 (27) and is required for cellular hypertrophy (27, 36). Similarly, GATA-3 may be involved in regulation of IL-5 gene expression in T helper 2 cell differentiation, probably via a p38 MAPK-dependent phosphorylation pathway (37). In hematopoietic cells, both GATA-1 and GATA-2 are phosphorylated upon stimulation by Me2SO (14), epidermal growth factor, and cytokines (38), probably through the ERK pathway (26, 38). However, a role for these phosphorylation events in controlling either the DNA binding or the transactivation activity by GATA-1 and GATA-2 was not directly demonstrated.
In this report, we provide the first evidence that phosphorylation of GATA-1 at Ser-26 is critical for its transcriptional activation of the E4bp4 gene and for its biological activity in promoting a cellular survival response. Despite the fact that ectopic GATA-1 expression is a very potent signal for cell survival, we know very little about its antiapoptotic mechanism, except for induction of the survival genes E4bp4 and Bcl-XL. E4bp4 could act downstream of GATA-1 and upstream of Bcl-XL; alternatively, GATA-1 might activate the Bcl-XL and E4bp4 antiapoptotic genes in a parallel manner. If the latter scenario is true, it implies that transactivation of the Bcl-XL gene will also depend upon GATA-1 Ser-26 phosphorylation. Furthermore, the promoter architecture of the Bcl-XL gene would be similar to that of E4bp4. Intriguingly, the promoter region of the Bcl-XL gene in K562 and FL5.12 hematopoietic cell lines contains no canonical TATA box. Instead, there is a consensus YYAN(T/A)YY initiator element (30) at positions −149 to −142 near a GATA element (7). It will be of great interest to determine whether GATA-1 phosphorylation affects this promoter. Although Mcl-1 has been shown to be one survival gene activated by IL-3 in Ba/F3 cells (33), our data indicated that activation of GATA-1 did not lead to synthesis of the Mcl-1 protein. Therefore, the IL-3 survival signal probably induces the expression of Mcl-1 and Bcl-XL via two separate signaling pathways (32, 33) that converge at mitochondria to protect membrane integrity and sustain viability.
On the other hand, phosphorylation of Ser-26 appears to be dispensable for erythyroid-specific gene expression (e.g. α-globin) and for erythrocyte differentiation.2 The discrepancy between the E4bp4 and globin gene expression could be due to different promoter architecture. Particularly, the E4bp4 promoter, as we pointed out previously, possesses an initiator element (17) that is essential for its transcriptional activation.2 Perhaps the transcription initiation complex binding to the E4bp4 initiator is different from that binding to the TATA-containing core promoter of α-globin (40) and that different protein components in the core promoter complexes determine the requirement of the phosphorylation status of GATA-1 at Ser-26. ERK-mediated phosphorylation of GATA-1 also leads to a pronounced shift in electrophoretic mobility in SDS-polyacrylamide gels, suggestive of a conformational change. Phosphorylation at Ser-26 might modulate the interaction of the GATA-1-inducible activation domain with a coactivator or some other component of the transcriptional machinery. In support of this possibility, GATA-1 has been reported recently to interact with a variety of coactivators and components of the transcription complexes, including p300/CREB-binding protein, E2A, SCL, LMO2, and Ldb-1 (40). Mutations at Ser-26 and Ser-178 significantly interfered with interaction of GATA-1 and the transcriptional factor LIM-only protein LMO2 (38). Therefore, it is likely that the transcription potential and expression of a GATA target gene can be determined by the phosphorylation status of GATA-1 in addition to specific cis-acting elements, trans-acting factors, and extracellular signals.
Supplementary Material
Acknowledgments
We thank Dr. Hsin-Fang Yang-Yen for the plasmids expressing dominant negative MEK, dominant negative ERK1, and dominant negative AKT and for careful reading of the manuscript, and we thank Dr. Hsiu-Ming Shih for the plasmids Gal4-LUC and pCMXGal4-DBD.
Footnotes
This work was supported in part by an intramural fund from Academia Sinica and by National Science Council of Taiwan Grants NSC-91-2320-B-001-046 and NSC92-2320-B-001-039 (to J. J. Y. Y.).
The on-line version of this article (available at http://www.jbc.org) contains an additional four figures and one table.
The abbreviations used are: IL, interleukin; MAPK, mitogen-activated protein kinase; GST, glutathione S-transferase; ER, estrogen receptor; GER, GATA-1-ER; S26EER, GATA-1S26E-ER; S26AER, GATA-1S26A-ER; ERK, extracelluar signal-regulated kinase; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; CREB, cAMP-response element-binding protein.
Y.-L. Yu, Y.-J. Chiang, Y.-C. Chen, M. Papetti, C.-G. Juo, A. I. Skoultchi, and J. J. Y. Yen, unpublished data.
Y.-L. Yu, Y.-J. Chiang, Y.-C. Chen, M. Papetti, C.-G. Juo, A. I. Skoultchi, and J. J. Y. Yen, unpublished result.
References
- 1.Evans T, Felsenfeld G. Cell. 1989;58:877–885. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- 2.Tsai SF, Martin DI, Zon LI, D’Andrea AD, Wong GG, Orkin SH. Nature. 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 3.Orkin SH. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 4.Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss MJ, Orkin SH. Proc Natl Acad Sci U S A. 1995;92:9623–9627. doi: 10.1073/pnas.92.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pevny L, Lin CS, D’Agati V, Simon MC, Orkin SH, Costantini F. Development. 1995;121:163–172. doi: 10.1242/dev.121.1.163. [DOI] [PubMed] [Google Scholar]
- 7.Grillot DA, Gonzalez-Garcia M, Ekhterae D, Duan L, Inohara N, Ohta S, Seldin MF, Nunez G. J Immunol. 1997;158:4750–4757. [PubMed] [Google Scholar]
- 8.Gregory T, Yu C, Ma A, Orkin SH, Blobel GA, Weiss MJ. Blood. 1999;94:87–96. [PubMed] [Google Scholar]
- 9.Tanaka H, Matsumura I, Nakajima K, Daino H, Sonoyama J, Yoshida H, Oritani K, Machii T, Yamamoto M, Hirano T, Kanakura Y. Blood. 2000;95:1264–1273. [PubMed] [Google Scholar]
- 10.Keinanen R, Vartiainen N, Koistinaho J. Gene (Amst) 1999;234:297–305. doi: 10.1016/s0378-1119(99)00196-1. [DOI] [PubMed] [Google Scholar]
- 11.Zhang R, Min W, Sessa WC. J Biol Chem. 1995;270:15320–15326. doi: 10.1074/jbc.270.25.15320. [DOI] [PubMed] [Google Scholar]
- 12.O’Prey J, Ramsay S, Chambers I, Harrison PR. Mol Cell Biol. 1993;13:6290–6303. doi: 10.1128/mcb.13.10.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karin M. Curr Opin Cell Biol. 1994;6:415–424. doi: 10.1016/0955-0674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 14.Crossley M, Orkin SH. J Biol Chem. 1994;269:16589–16596. [PubMed] [Google Scholar]
- 15.Taxman DJ, Sonsteby SK, Wojchowski DM. Protein Expression Purif. 1994;5:587–594. doi: 10.1006/prep.1994.1081. [DOI] [PubMed] [Google Scholar]
- 16.Partington GA, Patient RK. Nucleic Acids Res. 1999;27:1168–1175. doi: 10.1093/nar/27.4.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu YL, Chiang YJ, Yen JJ. J Biol Chem. 2002;277:27144–27153. doi: 10.1074/jbc.M200924200. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Tarduchy G, Collins M, Lopez-Rivas A. EMBO J. 1990;9:2997–3002. doi: 10.1002/j.1460-2075.1990.tb07492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rekhtman N, Choe KS, Matushansky I, Murray S, Stopka T, Skoultchi AI. Mol Cell Biol. 2003;23:7460–7474. doi: 10.1128/MCB.23.21.7460-7474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choe KS, Radparvar F, Matushansky I, Rekhtman N, Han X, Skoultchi AI. Cancer Res. 2003;63:6363–6369. [PubMed] [Google Scholar]
- 21.Rao G, Rekhtman N, Cheng G, Krasikov T, Skoultchi AI. Oncogene. 1997;14:123–131. doi: 10.1038/sj.onc.1200807. [DOI] [PubMed] [Google Scholar]
- 22.Lin DY, Shih HM. J Biol Chem. 2002;277:25446–25456. doi: 10.1074/jbc.M200633200. [DOI] [PubMed] [Google Scholar]
- 23.Huang HM, Li JC, Hsieh YC, Yang-Yen HF, Yen JJ. Blood. 1999;93:2569–2577. [PubMed] [Google Scholar]
- 24.Lee SF, Huang HM, Chao JR, Lin S, Yang-Yen HF, Yen JJ. Mol Cell Biol. 1999;19:7399–7409. doi: 10.1128/mcb.19.11.7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis RE, Yuan JY, Horvitz HR. Annu Rev Cell Biol. 1991;7:663–698. doi: 10.1146/annurev.cb.07.110191.003311. [DOI] [PubMed] [Google Scholar]
- 26.Towatari M, May GE, Marais R, Perkins GR, Marshall CJ, Cowley S, Enver T. J Biol Chem. 1995;270:4101–4107. doi: 10.1074/jbc.270.8.4101. [DOI] [PubMed] [Google Scholar]
- 27.Liang Q, Wiese RJ, Bueno OF, Dai YS, Markham BE, Molkentin JD. Mol Cell Biol. 2001;21:7460–7469. doi: 10.1128/MCB.21.21.7460-7469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferby IM, Waga I, Hoshino M, Kume K, Shimizu T. J Biol Chem. 1996;271:11684–11688. doi: 10.1074/jbc.271.20.11684. [DOI] [PubMed] [Google Scholar]
- 29.Clark-Lewis I, Sanghera JS, Pelech SL. J Biol Chem. 1991;266:15180–15184. [PubMed] [Google Scholar]
- 30.Gonzalez FA, Raden DL, Davis RJ. J Biol Chem. 1991;266:22159–22163. [PubMed] [Google Scholar]
- 31.Adams JM, Cory S. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 32.Wang JM, Chao JR, Chen W, Kuo ML, Yen JJ, Yang-Yen HF. Mol Cell Biol. 1999;19:6195–6206. doi: 10.1128/mcb.19.9.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang JM, Lai MZ, Yang-Yen HF. Mol Cell Biol. 2003;23:1896–1909. doi: 10.1128/MCB.23.6.1896-1909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamakura S, Moriguchi T, Nishida E. J Biol Chem. 1999;274:26563–26571. doi: 10.1074/jbc.274.37.26563. [DOI] [PubMed] [Google Scholar]
- 35.Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- 36.Kitta K, Day RM, Kim Y, Torregroza I, Evans T, Suzuki YJ. J Biol Chem. 2003;278:4705–4712. doi: 10.1074/jbc.M211616200. [DOI] [PubMed] [Google Scholar]
- 37.Chen CH, Zhang DH, LaPorte JM, Ray A. J Immunol. 2000;165:5597–5605. doi: 10.4049/jimmunol.165.10.5597. [DOI] [PubMed] [Google Scholar]
- 38.Towatari M, Ciro M, Ottolenghi S, Tsuzuki S, Enver T. Hematol J. 2004;5:262–272. doi: 10.1038/sj.thj.6200345. [DOI] [PubMed] [Google Scholar]
- 39.Smale ST. Genes Dev. 2001;15:2503–2508. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- 40.Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR. EMBO J. 2004;23:2841–2852. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.