Abstract
The Human Proteome Organisation’s Proteomics Standards Initiative (HUPO-PSI) has developed the GelML data exchange format for representing gel electrophoresis experiments performed in proteomics investigations. The format closely follows the reporting guidelines for gel electrophoresis, which are part of the Minimum Information About a Proteomics Experiment (MIAPE) set of modules. GelML supports the capture of metadata (such as experimental protocols) and data (such as gel images) resulting from gel electrophoresis so that laboratories can be compliant with the MIAPE Gel Electrophoresis guidelines, while allowing such data sets to be exchanged or downloaded from public repositories. The format is sufficiently flexible to capture data from a broad range of experimental processes, and complements other PSI formats for mass spectrometry data and the results of protein and peptide identifications to capture entire gel-based proteome workflows. GelML has resulted from the open standardisation process of PSI consisting of both public consultation and anonymous review of the specifications.
Keywords: data standard, gel electrophoresis, database, ontology
Introduction
The technique of gel electrophoresis (GE) has been widely used for the large scale study of proteins for many years [1]. Gel electrophoresis is used, for example, to separate proteins in one dimension by their molecular weight, or in two dimensions (typically by pI and MW) to achieve separation of complex mixtures [2]. The proteins on the gel are usually stained or labelled to enable visualisation and the further assignment of qualitative or quantitative values, for instance by image analysis. It is widely acknowledged that specific stages of the process can affect measurements of protein abundance and the loss of the components of the proteome under investigation. Therefore, recording metadata about how the experiment was performed is a significant but important challenge.
Databases and data sharing
There are several public repositories for storing the final MS-based results of a proteomics study, including PRIDE [3], GPMDB [4], PeptideAtlas [5] and Tranche (which stores unprocessed data files https://proteomecommons.org/tranche/). Databases supporting gel-based proteomics include SWISS-2DPAGE [6], World-2DPAGE Repository [7], Gelbank [8], the ProteoRed MIAPE Generator Website [9] and many others smaller domain-specific websites (see World-2DPAGE List, http://www.expasy.org/ch2d/2d-index.html). Significantly, there is only limited support for the storage of detailed descriptions of all stages of a gel-based proteomics workflow, alongside MS data and identifications. Where such capabilities exist, information is mostly restricted to unstructured text paragraphs. One of the reasons for the lack of comprehensive workflow coverage in databases is the lack of widely accepted standards for representing gel data and the difficulties encountered modelling the range of workflows employed in different settings. The need for data sharing and common standards in proteomics has been clearly identified and requested by the proteomics community, funders and journal editors through numerous publications (e.g. [10-12]).
To allow systematic analysis of results, or the comparison of different experiments, the metadata and data must be captured in a format that can be interpreted by researchers within a laboratory from one day to the next, and potentially between collaborating researchers and laboratories. Each stage in the laboratory workflow could produce different data types or formats. Some of the data may be proprietary, requiring associated commercial software for interpretation or analysis. Other data may not be in a computationally amenable form, for example, in a paper-based laboratory notebook. The increasingly common way to ensure that data generated are persistent, interpretable and open to future computational analyses, is for the proteomics community to agree on a common representation for data.
Data standards
The Human Proteome Organisation (HUPO), a community of industry, academia and government groups (http://www.hupo.org/), has taken steps to devise community standards by creating the Proteomics Standards Initiative (PSI, http://www.psidev.info/). The PSI aims to develop global proteomics models to assist publication, data-storage, and integration by producing three specifications per designated technology: (i) minimum reporting requirements, (ii) a format for data exchange, and (iii) an ontology or controlled vocabulary (CV). In 2007, the PSI released the MIAPE (Minimum Information About a Proteomics Experiment) specification, which consists of a parent document [13] and a series of technology-specific modules which each contain a checklist of information that should be reported, for example when a data set is published. To date, there are seven published MIAPE modules (http://psidev.info/miape/), defining the minimum information required to report the use of Capillary Electrophoresis (MIAPE-CE [14]), Column Chromatography (-CC [15]), Gel Electrophoresis (-GE [16]), Gel Image Informatics (-GI [17]), Mass Spectrometry (-MS [18]), Mass Spectrometry Informatics (-MSI [19]) and molecular interactions (MIMIx [20]).
The PSI’s data exchange formats aim to facilitate the capture, storage and presentation of information prescribed within each MIAPE module. They are also designed to exchange data, such as between labs, for submission to a public database or for download of data produced by other groups. Providing an agreed format for both proprietary and non-proprietary applications should make exchange more straightforward and enable comparative analyses of data produced in different settings. The data formats typically have the ability to account for more information than is prescribed in the MIAPE modules, and therefore may be used as the basis of a LIMS.
This article describes a data transfer format for gel electrophoresis, used in the context of proteomics, called the Gel electrophoresis Markup Language (GelML). GelML has been developed by extending the FuGE data model [21, 22]; FuGE is an object model describing the components of high-throughput experiments that are common across all types of technology, such as biological samples, protocols and multidimensional data. FuGE has been adopted in different ways by various standards bodies, including the PSI (in mzIdentML – the format for peptide and protein identifications [23]), the Flow Informatics and Computational Cytometry Society [24] and the Genetical Genomics consortium through an implementation in the MOLGENIS framework [25]. FuGE support has also been built into a draft proposal by the Metabolomics Standards Initiative (http://msiworkgroups.sourceforge.net/) which may be completed to a standard in due course. This effort towards structural similarity in data formats for different technologies should facilitate shared software development practices and data integration across life-science domains [26].
In tandem with the development of GelML, a controlled vocabulary has been created to standardise terms for protein and peptide separation, called sepCV (Sample Processing and Separation Techniques). A CV is required in this context to avoid different terms being used to represent the same concept. As a simple example, “2D”, “2DE”, “2D gel”, “2D-GE” and “two-dimensional gel electrophoresis” are different labels that refer to the same separation technique. Multiple terms or labels used to refer to the same concept (synonyms) can cause difficulties for users querying repositories or for automating data set comparisons. Similarly, confusion can arise when a single label can refer to distinct concepts in different contexts (homonym), for example the term “probe” has different specific meanings to particular technology practitioners. Therefore, it is essential to determine the intended semantics of the label in its particular context, especially if it is to be used in the systematic annotation of scientific data sets. The sepCV was first developed to satisfy the case-studies used to build the MIAPE GE/GI modules and GelML. After meeting the initial requirements, it has been registered in the Open Biomedical Ontologies (OBO) Foundry [27] and is available for term inclusion requests by the community (http://bioportal.bioontology.org/ontologies/39509).
Related data formats
There are several past formats developed for representing gel-based proteome data, which are summarised briefly here, including AGML [28], HUP-ML (from JHUPO http://www.jhupo.org/), and PEDRo [29]. Each of these formats contains models for representing gel data, and they were reviewed extensively during the development of GelML. The AGML data model clearly defines itself as a 2-DE centric representation, although it also incorporates a limited structure for associated mass-spectrometry components. AGML represents samples, equipment and protein detection procedures with free text elements, based on the a system using protocol templates previously stored in a database, whereas the PSI development approach aims to use controlled vocabulary terms extensively to facilitate automated comparison of data sets or future database searching. The AGML model contains some model components for representing intensities of features on gel images, which will be incorporated into future PSI models covering gel image informatics (see Discussion). HUP-ML was produced back in 2002, prior to the establishment of the HUPO-PSI, and was discussed as a starting point for gel modelling in the early PSI meetings. The model is also 2-DE centric and it allows the solutions and timings used in electrophoresis to be recorded. The process of acquiring an image of a gel, such as through the use of a scanner, is also accounted for but it does not possess a facility for representing different gel techniques, such as 1-DE or DIGE. PEDRo is a model which aims to represent the data-flow within a proteomics experiment [29]. Within PEDRo there exists the facility to capture limited information about other gel electrophoresis methods such as 1-DE, 2-DE and DIGE. However, PEDRo has a limited representation of the protocols for gel image acquisition and the resulting images. The ability to store electrophoresis conditions is also missing. It was felt by PSI that none of the existing formats had sufficient capabilities to support all the MIAPE GE guidelines in a structured format. However, the developers of PEDRo have been heavily involved in PSI efforts and GelML resulted from an evolution of this model. AGML developers have also joined the PSI working group to contribute to gel image informatics modelling. In the following sections, we briefly describe the development process of GelML, the main components of the data model, and how GelML relates to the MIAPE GE specification [16].
Methods
The GelML format was developed to meet the PSI data format requirements, aiming to support the following tasks: i) the discovery of relevant results, such as by querying public databases; ii) the sharing of best practice; iii) the evaluation of results and iv) the sharing of data sets. The primary focus of the model is to support long-term archiving and sharing of the results of gel electrophoresis experiments, rather than the representation of specific day-to-day laboratory management, although the model is designed to be extensible to support context-specific details where required.
The model was developed following a formal process, as defined by PSI [30]. Initially, efforts were put into collecting and collating opinion from a wide group of scientists and experts in gel electrophoresis, resulting in the MIAPE GE document. A set of use cases were then defined that should be supported by the format. The data format was initially developed by building an object model in the Unified Modeling Language, extending from FuGE, which was later mapped to XML (Extensible Markup Language), through the creation of an XML Schema (XSD: http://www.w3.org/XML/Schema) to control the allowed elements in a file for data exchange. The modelling processes was started at the PSI Autumn meeting in Geneva 2005, followed by conference calls which were open to all interested participants and development workshops at PSI meetings in Spring 2006 (San Francisco), Autumn 2006 (Washington) and Spring 2007 (Lyon). Two “milestone releases” of GelML were produced in June 2006 and March 2007. In June 2007, the GelML specifications were submitted to the PSI document process [30], which incorporates anonymous review of the specifications, similar to a journal article, and it is open to public comments. A version 1.0 release of the GelML specifications was made from the document process in late 2007. Since 2007, several implementations of GelML have been developed in different database systems, which have highlighted some minor bugs and issues (as is the case with the majority of software releases). The schema is currently fixed at a version 1.1 release (http://code.google.com/p/gelml/), which has been implemented in several systems. The specifications have thus received input from a large number of experts over several years in open and transparent process, since PSI meetings and conference calls have been open to any participants and the specifications have undergone public review.
Results
The following section presents a summary of the main components of the GelML model. For a comprehensive reference point, the technical detail is presented in the specification document (http://code.google.com/p/gelml/source/browse/#svn/trunk/SpecDoc).
GelML models the process of gel electrophoresis applied in the context of a proteomics experiment, after sample preparation and prior to image analysis or protein identification. The model supports the description of the protocols for electrophoresis, protein detection - either directly on the gel matrix or indirectly (e.g. Western blotting), and image acquisition from gel matrices. GelML is intended to be used in a modular way together with existing formats. It does not contain explicit models designed for sample processing or preparation, prior to applying a sample on a gel matrix, since such information can be captured in the core FuGE model, which is imported along with the GelML schema. GelML does not provide detailed support for describing the analysis of digitised imaged derived from gel matrices (see Discussion), although limited support is provided in GelML for capturing locations identified on gel images and related quantitative information. In addition, GelML does not describe the process of protein identification, for example by mass spectrometry, for which standards formats already exist (mzML [31] and mzIdentML as detailed on the PSI website, http://psidev.info/).
The GelML model can be broken down into various sub-sections. Each model represents a particular stage in a gel electrophoresis experiment, including: the gel materials and optionally the manufacture of the gel; one-dimensional gel electrophoresis; two dimensional gel electrophoresis; “non-standard”’ methods of gel electrophoresis that do not fit the traditional structure of 1-DE or 2-DE, such as 3-dimensional geometry gel electrophoresis; sample loading; electrophoresis; protein detection; image acquisition and the excision of locations on gels.
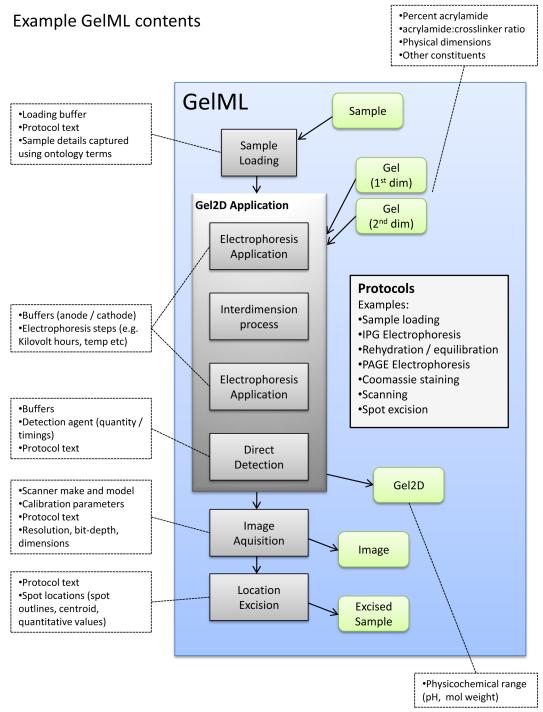
GelML makes uses of several structures of FuGE: models of protocols or procedures (Protocol), the running of the protocol and runtime parameters or readings (ProtocolApplication), all physical/biological materials (Material) and data files (Data). An overview of different parts of GelML is given in Figure 1 for a 2-DE example; similar workflows can also be constructed for a 1-DE or DIGE experiment. The backbone of a typical file is a series of ProtocolApplications (standard rectangles in Figure 1) that map inputs and outputs. The inputs and outputs to each ProtocolApplication can only be types of Material or Data (rounded rectangles). This structure allows some flexibility with regards to how workflows are constructed if non-standard procedures have been carried out. Each ProtocolApplication must reference a corresponding standard protocol, defined within the file. Each protocol consists of the main text of the protocol, parameters and equipment or software details. As such, if the same protocol is run many times, it only has to be recorded once in the file. Figure 1 is illustrated with several key details captured in each stage that are required by MIAPE GE.
Figure 1.
A graphical representation of example components from a GelML file, and certain key details that should be captured in each section. Standard rectangles indicate ProtocolApplications, rounded rectangles indicate Materials or Data.
In the rest of this section, a brief summary is given of several components of the model from the point of view of a “standard” 2-DE experiment, illustrating how these components could represent a MIAPE GE compliant data set.
Gel model
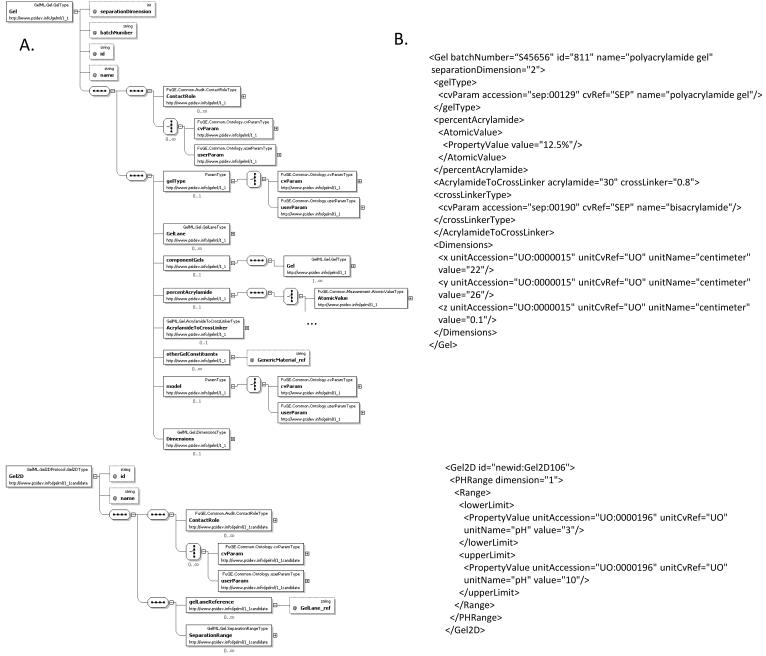
The MIAPE GE document requires that users report a description of the gel matrix, the physical dimensions, the concentration of acrylamide and the crosslinking agent. GelML has model to support these details, as outlined in Figure 2A as a representative example of GelML (detailed diagrams of other key model components can be found in the supplementary figures). There is an additional model (not shown) which allows the protocol for the gel manufacture to be recorded if the gel was not purchased pre-cast, which is also required by MIAPE GE. The Gel element has attributes for specifying the separation dimension and the batch number. Associations to other elements can be used to capture the dimensions of the gel(s), the ratio of acrylamide to a crosslinker (such as bisacrylamide), the overall percentage of acrylamide, the model number and identifiers for any lanes within the gel. All of these characteristics can affect the quality of the resulting protein separation and estimates of protein quantities, so it is important that such details are stored in a structured format. There is a separate element representing a 1-D or 2-D gel after electrophoresis has been performed (Gel1D, Gel2D) which can be used to specify the range of physicochemical separation performed, such as molecular weight or pH. In Figure 2B, example instances of the XML format are shown.
Figure 2.
A. The model in XSD of the gel material prior to (Gel) and following electrophoresis (ElectrophoresedGel and the sub-elements: Gel2D, Gel1D, OtherGel). B. Examples in XML of one instance of Gel and Gel2D, the relationships between Gel and Gel2D are captured by the application of protocols that reference these elements as inputs/outputs (not shown).
Electrophoresis
MIAPE GE requests that users report the electrophoresis protocols employed, allowing, for example, database users to apply protocols in their own labs. The protocol, as represented in GelML, consists of the main protocol text and references to buffer details and equipment, such as gel tanks (Supplementary Figure 1). Earlier iterations of GelML modelled electrophoresis protocols by breaking down each step of the protocol into individual parameters, with values and units (rather than plain text). However, there are currently no software packages able to export these protocols directly from electrophoresis control software, and our experience testing implementations has shown that users are generally not willing to complete complex forms manually with such high granularity information.
Protein Detection
Proteins are detected or visualised on a gel by either a direct method, such as staining, or an indirect method in which they are transferred to another medium such as a Western blot. Choosing the appropriate detection agent, such as silver, Coomassie blue or fluorescent stains (for example used in DIGE), is based on the concentration and abundance of the sample. The choice of detection agent is also influenced by the information required in the post gel processing steps, such as mass spectrometry. The overall details of the procedure are captured as plain text in GelML. The protocol references a controlled vocabulary term for the name of the detection agent (which would allow a database to be queried for this property) and the quantity of the agent as a volume, mass or concentration (Supplementary Figure 2). The model can also capture indirect detection procedures, such as Western blots in which proteins are first transferred to a new medium (e.g. a nitrocellulose membrane).
Gel image acquisition
The protocol for acquiring a digitised image can be captured as plain text in GelML with a set of parameters including a specification of how scanner calibration was performed (Supplementary Figure 3). The model also captures the make and model of the scanner. The application of the protocol has an input of the gel on which proteins were detected by a direct process or the medium on which indirect detection was performed (not shown). The output of the ProtocolApplication is the image itself, with attributes for capturing the image dimensions, the bit depth, resolution and file format (information required by MIAPE GE). In a DIGE experiment, several instances of the ProtocolApplication are created, each producing one Image, to capture the procedure of scanning at several different wavelengths.
Spot or band excision
In a typical 1-DE or 2-DE experiment, following protein detection (and image acquisition), individual spots or bands are excised and progress to mass spectrometry for protein identification. The PSI format for mass spectrometry data, mzML [31] can specify a reference to an input sample. In a gel-based experiment, the ProtocolApplication for excision produces a series of samples (ExcisedSample) with unique identifiers which could be referenced within mzML. This link would allow a mass spectrum to be linked back to a complete trace of the gel, and associated protocols, from which it was extracted.
GelML contains a model for linking the samples back to the corresponding locations on images, and for capturing a protocol describing how excision is performed (Supplementary Figure 4). Locations on a gel, such as spots or bands, can be captured in several different ways depending on how the images have been analysed, such as pairs of X/Y coordinates, circular or rectangular locations. If spot locations have a complex shape, as produced by image analysis software, the location can be specified by a set of X/Y boundary points by an ordered chain of boundary points (see specification document for more detail). Gel locations can be annotated with additional measurements, which could be used to store quantitative values derived from image analysis, such as spot density or volume.
Controlled vocabulary
The PSI-Gel workgroup has developed the controlled vocabulary sepCV, which contains terms specific to the methods and techniques of protein separation using gel electrophoresis. It covers gel manufacture and preparation, running conditions, protein detection techniques as well as imaging methods. Several key parts of GelML require CV terms to be sourced from sepCV, such as the protein detection agent and the type of crosslinker in the gel. The description of the starting sample requires the use of CV terms to capture its important characteristics, as defined by the investigators, for example sourced from an organism-specific ontology within the OBO Foundry [27]. The Unit Ontology should be used with GelML to standardise the naming of units, which is also part of the OBO Foundry. The use of CV terms is controlled by a mapping file that specifies exactly which CV terms are allowed within each part of the schema. The usage can then be checked using the PSI’s semantic validation technology [32] for which a test implementation has been created by the OpenMS developers (details at http://www.psidev.info/validator/).
Implementations of GelML
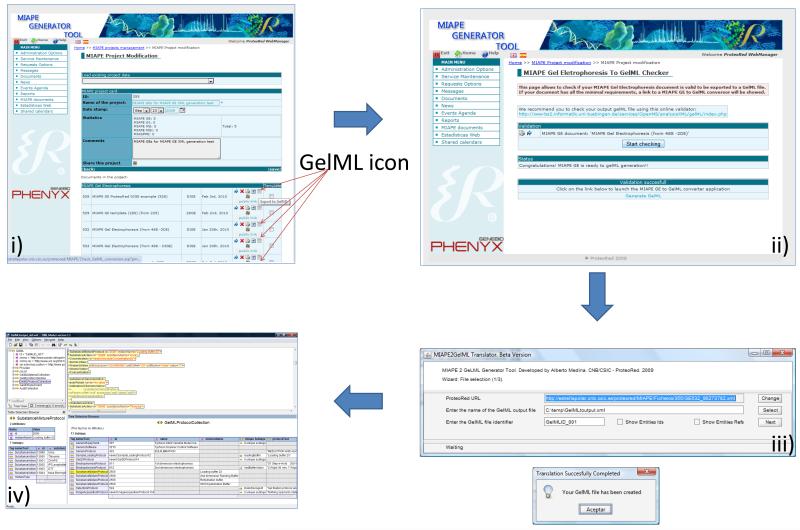
The first implementations of GelML within database systems have recently been developed. The ProteoRed consortium has developed the MIAPE generator tool that automates the process of collecting methods and data sets for proteomics, compliant with the MIAPE guidelines [9]. The tool guides users through each stage of an experimental process, capturing key details as specified in each MIAPE module. The sepCV and unitCV vocabularies have been implemented to ensure that consistent method descriptions and units are provided throughout. At the end of the process, the user can verify that their submission is MIAPE GE compliant. Other users can browse the MIAPE database, and have the opportunity to download descriptions of methods in “Report format” (as pdf). A tool has been developed for mapping the internal ProteoRed format to GelML, using a Java Webstart application (Figure 3). The ProteoRed database covers protein separation and electrophoresis protocols in much greater detail than the EBI PRIDE database format, while PRIDE provides a central repository for protein identifications based on mass spectrometry. Thus, data from gel-based proteomics workflows can be accommodated by a dual submission of methodology description, gel images and image features to ProteoRed with protein identifications stored in PRIDE. A mechanism has been created for linking the two submissions by unique identifiers. Users of the system can therefore also download linked files in PRIDE XML and GelML format for local analysis.
Figure 3.
Screenshots from the ProteoRed MIAPE Generator Website, showing the pipeline for generating GelML files from an existing MIAPE experiment: i) The user is guided through the data input process and is provided with the option to export to GelML; ii) a validator is run to check the GelML document against the MIAPE guidelines; iii) a Java Web Start application opens on the users desktop to convert the internal XML representation to GelML; iv) a valid XML file is produced on the user’s machine.
A second beta implementation is under development at the Swiss Institute of Bioinformatics, in which MIAPE-compliant submissions to the World-2DPAGE Repository can be created using the MIAPEGelDB interface [33]. An example file can be viewed at http://miapegeldb.expasy.org/experiment/2/gel/102/as_xml/. Since the GelML model is based on FuGE, FuGE-based software can be adapted relatively simply to provide implementations for GelML. A toolkit has been developed that provides one such mechanism, comprising a software application to facilitate the collection, storage and the browsing of FuGE compliant models and FuGE extensions such as GelML [34]. A mapping has also been created from GelML as part of the ISA-TAB project, allowing the XML to be rendered in a tab-based format for simpler visualisation (http://isatab.sourceforge.net/examples.html). The ISA-TAB mechanism is also used in the Bioinvestigation Index (http://www.ebi.ac.uk/bioinvindex) project to submit data to ‘omics databases hosted at the European Bioinformatics Institute.
The PSI Protein Separation (PSI-PS) work group has an active team of developers working on software implementations. The group is committed to providing on-going documentation and help guides for GelML, and will provide support for other groups implementing GelML through the group’s mailing list (see the workgroup home page http://www.psidev.info/index.php?q=node/83).
Discussion
The HUPO PSI defines community standards for data representation in proteomics to facilitate data comparison, exchange and verification. The Gel working group is developing standards for describing the use of gel electrophoresis and the informatics analysis on the derived gel images [35]. These standards currently consist of MIAPE GE, MIAPE GI reporting guidelines, GelML and sepCV. Access to structured data sets will then allow the data to be re-used or re-assessed to gain greater knowledge about a proteome or to be re-analysed with more powerful computational methods as and when they become available. We anticipate that the standard representation of data will enable elucidation of previously unobserved details and subtle trends and permit integration with other data sets such as gene expression or metabolomics data, forming data sets that may prove useful in systems biology investigations.
GelML can represent 1-DE, 2-DE and DIGE, and attempts to anticipate non-standard forms of gel electrophoresis. Furthermore, it allows information to be stored in significantly higher detail than MIAPE GE if required, for use in internal pipelines and databases. The implementation within ProteoRed’s MIAPE generator provides the ability for researchers to mine the documents for specific information, for example, images acquired by a particular scanner, or gels visualised with a particular stain. This mechanism thus allows researchers to assemble or integrate large collections of gel-based data for local analyses.
A key strategy throughout the development work was the engagement of the proteomics community, both for their opinions in the direction of the research and for the testing of the artefacts at each stage in the development process. A notable omission from GelML is the ability to represent the protocols employed in analysis of gel images to detect quantitative differences in proteins, through the use of gel image informatics software. The creation of a gel informatics data exchange standard would necessarily require significant efforts from vendors of gel informatics software to agree on a common representation. A first step has been made through the creation of a reporting guidelines document (MIAPE Gel informatics [17]), which was written and reviewed by a number of software vendors. The PSI is also actively developing a format for proteome quantification by mass spectrometry, called mzQuantML. The early draft of mzQuantML captures features on 2-D plots (for example retention time versus mass/charge), matches between features on different plots (for example for label free quantification), the combination of data across replicates and links to peptide and protein identification evidence, for example represented in the PSI mzIdentML standard. We believe that mzQuantML can be easily adapted to capture gel image informatics data, since quantification by label free MS has similar metadata requirements. The PSI would like to encourage further input and opinion on the community requirements for gel image informatics standards, through the mailing list or via attendance at a PSI meeting.
Supplementary Material
Acknowledgments
ARJ gratefully acknowledges funding from BBSRC [BB/G010781/1]. Work in Manchester by ARJ on GelML was funded by the BBSRC (grant to Prof Steve Oliver and NWP), whose support we gratefully acknowledge. JSA and RS acknowledge NIH/CTSA award no. 1UL1RR024148. We would like to thank the steering committee of the PSI and the individuals who provided anonymous reviews on the GelML specifications. We also thank Andreas Bertsch from the University of Tuebingen for creating the GelML validator.
Abbreviations
- MIAPE
Minimum Information About a Proteomics Experiment
- GE
Gel electrophoresis
- CV
Controlled Vocabulary
References
- [1].Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- [2].O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- [3].Jones P, Cote RG, Martens L, Quinn AF, et al. PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucl. Acids Res. 2006;34:D659–663. doi: 10.1093/nar/gkj138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Craig R, Cortens JP, Beavis RC. Open Source System for Analyzing, Validating, and Storing Protein Identification Data. Journal of Proteome Research. 2004;3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- [5].Desiere F, Deutsch EW, King NL, Nesvizhskii AI, et al. The PeptideAtlas project. Nucl. Acids Res. 2006;34:D655–658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hoogland C, Mostaguir K, Sanchez J-C, Hochstrasser DF, Appel RD. SWISS-2DPAGE, ten years later. PROTEOMICS. 2004;4:2352–2356. doi: 10.1002/pmic.200300830. [DOI] [PubMed] [Google Scholar]
- [7].Hoogland C, Mostaguir K, Appel RD, Lisacek F. The World-2DPAGE Constellation to promote and publish gel-based proteomics data through the ExPASy server. Journal of Proteomics. 2008;71:245–248. doi: 10.1016/j.jprot.2008.02.005. [DOI] [PubMed] [Google Scholar]
- [8].Babnigg G, Giometti CS. GELBANK: a database of annotated two-dimensional gel electrophoresis patterns of biological systems with completed genomes. Nucleic acids research. 2004;32 doi: 10.1093/nar/gkh089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Martínez-Bartolomé S, Medina-Aunon JA, Jones AR, Albar JP. Semi-automatic tool to describe, store and compare proteomics experiments based on MIAPE compliant reports. PROTEOMICS. 2010;10:1256–1260. doi: 10.1002/pmic.200900367. [DOI] [PubMed] [Google Scholar]
- [10].Time for leadership. Nat Biotech. 2007;25:821–821. doi: 10.1038/nbt0807-821. [DOI] [PubMed] [Google Scholar]
- [11].Democratizing proteomics data. Nat Biotech. 2007;25:262–262. doi: 10.1038/nbt0307-262b. [DOI] [PubMed] [Google Scholar]
- [12].Data’s shameful neglect. Nature. 2009;461:145–145. doi: 10.1038/461145a. [DOI] [PubMed] [Google Scholar]
- [13].Taylor C, Paton N, Lilley K, Binz P-A, et al. The minimum information about a proteomics experiment (MIAPE) Nature Biotechnology. 2007;25:887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- [14].Domann PJ, Akashi S, Barbas C, Huang L, et al. Guidelines for reporting the use of capillary electrophoresis in proteomics. Nature Biotechnology. 2010 doi: 10.1038/nbt0710-654b. in press. [DOI] [PubMed] [Google Scholar]
- [15].Jones AR, Carroll K, Knight D, MacLellan K, et al. Guidelines for reporting the use of column chromatography in proteomics. Nat Biotech. 2010 doi: 10.1038/nbt0710-654a. in press. [DOI] [PubMed] [Google Scholar]
- [16].Gibson F, Anderson L, Babnigg G, Baker M, et al. Guidelines for reporting the use of gel electrophoresis in proteomics. Nat Biotech. 2008;26:863–864. doi: 10.1038/nbt0808-863. [DOI] [PubMed] [Google Scholar]
- [17].Hoogland C, O’Gorman M, Bogard P, Gibson F, et al. Guidelines for reporting the use of gel image informatics in proteomics. Nat Biotech. 2010 doi: 10.1038/nbt0710-655. in press. [DOI] [PubMed] [Google Scholar]
- [18].Taylor CF, Binz P-A, Aebersold R, Affolter M, et al. Guidelines for reporting the use of mass spectrometry in proteomics. Nat Biotech. 2008;26:860–861. doi: 10.1038/nbt0808-860. [DOI] [PubMed] [Google Scholar]
- [19].Binz P-A, Barkovich R, Beavis RC, Creasy D, et al. Guidelines for reporting the use of mass spectrometry informatics in proteomics. Nat Biotech. 2008;26:862–862. doi: 10.1038/nbt0808-862. [DOI] [PubMed] [Google Scholar]
- [20].Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat Biotech. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- [21].Jones A, Miller M, Aebersold R, Apweiler R, et al. The Functional Genomics Experiment model (FuGE): an extensible framework for standards in functional genomics. Nat Biotech. 2007;25:1127–1133. doi: 10.1038/nbt1347. [DOI] [PubMed] [Google Scholar]
- [22].Jones AR, Pizarro A, Spellman P, Miller M. FuGE: Functional Genomics Experiment Object Model. OMICS: A Journal of Integrative Biology. 2006;10:179–184. doi: 10.1089/omi.2006.10.179. [DOI] [PubMed] [Google Scholar]
- [23].mzIdentML: exchange format for peptides and proteins identified from mass spectra. http://www.psidev.info/files/mzIdentML1.0.0.pdf.
- [24].Qian Y, Tchuvatkina O, Spidlen J, Wilkinson P, et al. FuGEFlow: data model and markup language for flow cytometry. BMC Bioinformatics. 2009;10:184. doi: 10.1186/1471-2105-10-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Swertz M, Velde KJ, Tesson B, Scheltema R, et al. XGAP: a uniform and extensible data model and software platform for genotype and phenotype experiments. Genome Biology. 2010;11:R27. doi: 10.1186/gb-2010-11-3-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jones AR, Lister AL, Hermida L, Wilkinson P, et al. Modeling and Managing Experimental Data Using FuGE. OMICS: A Journal of Integrative Biology. 2009;13:239–251. doi: 10.1089/omi.2008.0080. [DOI] [PubMed] [Google Scholar]
- [27].Smith B, Ashburner M, Rosse C, Bard J, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotech. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Stanislaus R, Arthur J, Rajagopalan B, Moerschell R, et al. An open-source representation for 2-DE-centric proteomics and support infrastructure for data storage and analysis. BMC Bioinformatics. 2008;9:4. doi: 10.1186/1471-2105-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Taylor CF, Paton NW, Garwood KL, Kirby PD, et al. A systematic approach to modeling, capturing, and disseminating proteomics experimental data. Nat Biotech. 2003;21:247–254. doi: 10.1038/nbt0303-247. [DOI] [PubMed] [Google Scholar]
- [30].Vizcaíno JA, Martens L, Hermjakob H, Julian RK, Paton NW. The PSI formal document process and its implementation on the PSI website. PROTEOMICS. 2007;7:2355–2357. doi: 10.1002/pmic.200700064. [DOI] [PubMed] [Google Scholar]
- [31].Deutsch E. mzML: A single, unifying data format for mass spectrometer output. PROTEOMICS. 2008;8:2776–2777. doi: 10.1002/pmic.200890049. [DOI] [PubMed] [Google Scholar]
- [32].Montecchi-Palazzi L, Kerrien S, Reisinger F, Aranda B, et al. The PSI semantic validator: A framework to check MIAPE compliance of proteomics data. PROTEOMICS. 2009;9:5112–5119. doi: 10.1002/pmic.200900189. [DOI] [PubMed] [Google Scholar]
- [33].Robin X, Hoogland C, Appel RD, Lisacek F. MIAPEGelDB, a web-based submission tool and public repository for MIAPE gel electrophoresis documents. Journal of Proteomics. 2008;71:249–251. doi: 10.1016/j.jprot.2008.06.005. [DOI] [PubMed] [Google Scholar]
- [34].Belhajjame K, Jones AR, Paton NW. A toolkit for capturing and sharing FuGE experiments. Bioinformatics. 2008;24:2647–2649. doi: 10.1093/bioinformatics/btn496. [DOI] [PubMed] [Google Scholar]
- [35].Jones AR, Gibson F. An Update on Data Standards for Gel Electrophoresis. Proteomics. 2007;7:35–40. doi: 10.1002/pmic.200700171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.