Abstract
Over the years, researchers have developed several methods to deliver macromolecules into the cytosol and nucleus of living cells. However, there are limitations to all of these methods. The problems include (i) inefficient uptake, (ii) endosomal entrapment, (iii) delivery that is restricted to certain cell types, and (iv) damage to cells in the delivery process. Retroviral vectors are often used for gene delivery; however, integration of the genome of retroviral vector into the host genome can have serious consequences. Here we describe a safe alternative in which virus-like particles (VLPs), derived from an avian retrovirus, are used to deliver protein to cells. We show that these VLPs are a highly adaptable platform that can be used to deliver proteins either as part of Gag fusion proteins (intracellular delivery) or on the surface of VLPs. We generated VLPs that contain Gag-Cre recombinase, Gag-Fcy::Fur, and Gag-human caspase-8 as a proof-of-concept and demonstrated that the encapsidated proteins are active in recipient cells. In addition, we show that murine IFN-γ and human TNF-related apoptosis-inducing ligand can be displayed on the surface of VLPs, and that these modified VLPs can cause the appropriate response in cells, as evidenced by phosphorylation of STAT1 and induction of cell death, respectively.
Keywords: surface display, TRAIL
The goal of protein-based therapies is restoration of biological functions by supplementing the absent or defective form of an essential protein. Currently, a limited number of therapeutic proteins are used to treat affected individuals with genetic diseases (notably, diabetes and hemophilia). Originally, the therapeutic proteins were obtained by purification from human or animal sources; however, these proteins are now made using recombinant DNA techniques. Currently, successful protein-based therapies involve proteins, such as insulin, that are injected, and do not require delivery to a specific tissue or organ. The utility of protein-based therapy could be extended if there were a way to efficiently deliver therapeutic protein(s) into cells.
Retroviruses have a potential advantage for gene therapy. Because retroviruses enter cells through the interaction of the envelope protein on the surface of the virus and a specific host cell receptor, there is also the potential for developing retroviral vectors that can deliver their genes to specific tissues and cell types. Although this type of specific/targeted gene delivery has not yet been developed for gene therapy in humans, there are tissue-specific retroviral gene delivery systems that can be used in animal models (1, 2).
The insertion of the retroviral vector DNA into the host genome is a mixed blessing, however; retroviral vector DNAs can insert at a large number of sites in the host genome, and can insert near an oncogene, leading to the development of cancer. Although there has been progress in developing methods to control the sites at which retroviral DNA integrates, this technology is not yet suitable for human gene therapy (3, 4).
Nonviral methods, including microinjection and electroporation, can also be used to deliver proteins into cells (5–7). Both antibodies (8, 9) and mutant proteins (10) have been delivered into cells via microinjection. However, microinjection is limited to a relatively small number of target cells and needs specialized equipment. In terms of potential therapies, microinjection or electroporation are not appropriate methods to deliver proteins to cells or tissues in an intact animal or in patients.
Alternatively, proteins and peptides have been identified that are believed to cross biological membranes. The small protein transduction domains (PTDs) from these proteins (e.g., TAT, VP22, or Antp) can be fused to other peptides or proteins to cause them to be transported into cells (11). Transduction using these peptides fused to proteins has an advantage in that entry is rapid and concentration dependent, and the technology appears to work with different cell types (12). A major drawback is that PTD fusion proteins are initially taken up into endosomal vesicles, and the release of the fusion proteins from the vesicles into the cytosol is inefficient (13, 14). A number of studies have demonstrated that PTD fusion proteins are primarily localized in the perinuclear region, suggesting that they are sequestered within endosomal vesicles (13–19). These problems limit the use of PTD-mediated protein delivery.
In this report, we describe an efficient and tractable protein delivery system using virus-like particles (VLPs). The VLPs we generated are derived from a modified version of the viral genome; the VLPs do not contain the viral enzymes protease, reverse transcriptase, and integrase. During the past decade, VLPs have been mainly used as vaccines (20–24). In addition, there are a few reports where VLPs generated using nonstructural HIV proteins (e.g., Nef and Vpr) were used to deliver foreign proteins (25–27). Recently, Voelkel et al. (28) reported that they were able to deliver proteins (GFP and functional Flp recombinase) to cells using murine retroviral particles. However, in all these cases, Gag-Pol was used to generate the VLPs. The presence of reverse transcriptase and integrase in the particles is a potential concern, because the possibility exists for the conversion of RNAs in the virion into DNA, and the unwanted integration of that DNA into the host chromosome in a transduced cell. It has been previously shown that the Gag polyprotein is the only retroviral protein required for VLP formation (29–31).
Here we describe a safe and efficient system to deliver functional proteins into cells. This system is safe both because it is based on an avian retrovirus, which cannot replicate in human cells, and because the construct used to express the VLP proteins does not contain the Pol gene, and as a consequence, the VLPs do not contain either reverse transcriptase or integrase. The system is efficient because the VLPs are composed entirely of the Gag fusion protein, and a single VLP will deliver 2,000–5,000 copies of Gag fusion protein into a transduced cell (32). In addition to intracellular delivery, we created VLPs that display protein ligands on their surface. We used a truncated form of influenza neuraminidase (NA) or hemmagglutinin (HA) fused to murine IFN-γ and human TNF-related apoptosis-inducing ligand (TRAIL). The presence of the protein fusions on the surface of VLPs can activate the appropriate cellular receptors on cells.
Results and Discussion
Generation of Functional VLPs Containing Biologically Active Proteins.
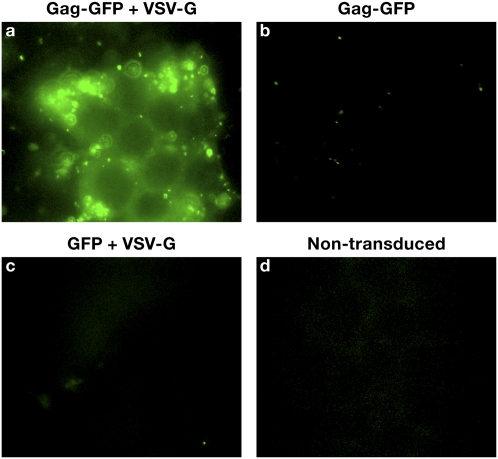
The VLPs were generated by transfection of 293T cells with two plasmids, one containing Gag fused with gene of interest (Gag fusion), and the other containing VSV-G as described in Materials and Methods. The Pol gene was not present in the expression plasmid used to express the Gag fusion and, as a consequence, neither reverse transcriptase nor integrase is present in the VLPs. A schematic representation of this system and mode of action of various fusion constructs we have generated are shown in Fig. 1 A and B. The coding sequence of Gag was cloned from the RCAS vector (33) and ligated into a modified mammalian expression vector, pDest472, as described in Materials and Methods and SI Materials and Methods. This vector is suitable for both recombinational cloning and restriction enzyme-mediated cloning of the cDNAs of proteins of interest to generate in-frame Gag fusion products. Cotransfection of an expression vector encoding the Gag protein of interest (e.g., GFP or Cre) with a vector expressing the VSV-G envelope protein resulted in generation of infectious, but replication-deficient, VLPs at a titer of ∼7 × 106 VLPs/mL. The VLPs titer was measured using a replication-competent avian sarcoma leukosis virus (RCAS)-GFP vector (with a titer of ∼1 × 107 infectious units/mL) stock prepared in and titered on the DF1 cell line. The amount of capsid protein (p27) in both samples (RCAS-GFP and Gag-GFP VLPs) was estimated by ELISA (Materials and Methods). The amount of capsid in the VLPs preparations was then used to estimate the titer of VLPs. Because the particle-to-infectivity ratio of retroviruses is low (∼1/100–1/1,000) (34), these titer calculations underestimate the number of VLPs by 100- to 1,000-fold. The size of VLPs was estimated to be between 80 and 130 nm, as determined by electron microscopy. The size range of the VLPs is similar to the size distribution of normal infectious retrovirus particles (35). To demonstrate the effectiveness of this approach, the 293T cell line was exposed to VLPs containing Gag-GFP fusion protein pseudotyped with VSV-G. The efficiency of transduction (as measured by counting GFP+ cells) was close to 100% (Fig. S1). Also, as shown in Fig. 2, the delivery of GFP was efficient when cells were exposed to VLPs containing Gag-GFP and VSV-G envelope. However, there was no detectable GFP fluorescence in the cells when they were exposed to VLPs that expressed Gag-GFP in the absence of VSV-G envelope, nor was there any fluorescence in cells that had been exposed to supernatants produced from cells transfected with VSV-G and GFP. This last control shows that transduction depends on the ability of Gag-GFP to form a VLP; VSV-G cannot transduce GFP containing cell debris/vesicles into cells. These results demonstrate the importance of having both the Gag fusion to form the VLP and a functional, fusogenic envelope on the surface of the VLP for efficient delivery of protein into cells.
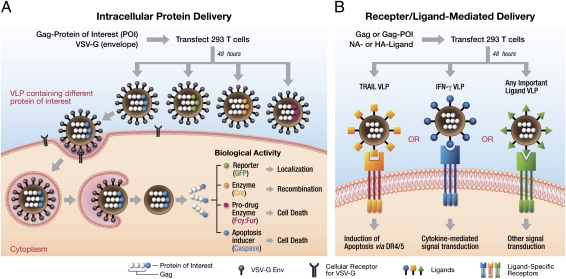
Fig. 1.
Generation of functional VLPs and mode of action. (A) A schematic representation of the assembly of VLPs and the biological functions of the proteins delivered intracellularly. Cotransfection of HEK 293T cells with pGag-POI and pCMV-VSV-G plasmids results in the formation and subsequent release of VLPs into the culture supernatant. Four different VLPs containing GFP, Cre, Fcy:Fur, and caspase 8 fused to Gag are shown. (B) A schematic representation of receptor/ligand-mediated protein delivery. VLPs were made as described above by cotransfection of HEK 293T cells with plasmids containing Gag (with or without a protein fusion) and the NA/HA ligand. Biological function mediated by signal transduction from the ligand is also shown.
Fig. 2.
VLP-mediated GFP delivery into cells. The HEK293T cell line was transduced with VLPs consisting of (A) Gag-GFP pseudotyped with VSV-G envelope, (B) Gag-GFP without VSV-G envelope, (C) GFP and VSV-G envelope, and (D) nontransduced cells.
Comparative Analysis of Protein Transduction Using Cell-Penetrating Peptides and Fusogenic VLPs.
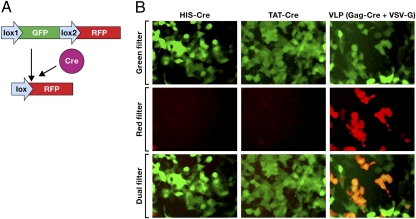
Cell-penetrating peptides or PTDs (e.g., HIV TAT peptide fused to proteins of interest) have been used to facilitate transduction of proteins across the plasma membrane into the cytoplasm. To evaluate the effectiveness of PTD-mediated protein transduction, we performed a comparative study using the HIV TAT peptide fused to Cre recombinase (HIV-TAT-Cre) vs. VLPs consisting of a Gag-Cre recombinase fusion protein pseudotyped with the fusogenic VSV-G envelope glycoprotein. For the protein transduction experiments, His-Cre (used as a negative control) and HIV-TAT-His-Cre recombinase (TAT-Cre) proteins were purified from bacteria. The 293T cells that contained a stably integrated reporter cassette (Lox1-GFP-Lox2-RFP) were used to measure the effectiveness of the protein transduction (36). Normally, these cells are green; however, when active Cre is present, it will excise the Lox1-GFP-lox2 cassette, leading to the expression of red fluorescent protein (RFP; Fig. 3A). As demonstrated in Fig. 3B, Middle Left and Middle Center, treatment of the 293T reporter cell line containing a stably integrated expression cassette in the chromosome with His- Cre or TAT-Cre did not result in any evidence of expression of RFP. To make sure that His-Cre and TAT-Cre had every opportunity to give positive result, we used far more of these control proteins compared with Gag-Cre in the VLPs (∼1.5 × 1015 molecules TAT-Cre or His-Cre compared with ∼4 × 1012 molecules of Gag-Cre. However, exposing the same cell line to VLPs comprised of Gag-Cre and VSV-G envelope resulted in efficient expression of RFP, thus demonstrating efficient delivery of active Cre in this cell line (Fig. 3B, Middle Right). In the short-term experiment shown in Fig. 3B, Top Right, the cells that have the target gene converted from expressing GFP to RFP still fluoresce both green and red because the GFP turns over (decays) slowly (37). The fact that even in the presence of high levels of TAT-Cre and high levels of His-Cre in the culture, media were unable to act on the loxP sites in an integrated target gene may be due to an inability of this fusion protein to escape endosomal entrapment and translocate into the nucleus. These results support an earlier finding that TAT-mediated protein transduction does not lead to the efficient delivery of proteins to cellular compartments beyond the endosomes (14).
Fig. 3.
Cre recombinase delivery using a cell-penetrating peptide fusion (HIV TAT) and VLPs. (A) Activation of RFP expression following the action of Cre in a reporter cell line containing a stably integrated expression cassette (Lox1-GFP-Lox2-RFP). (B) A reporter 293T cell line containing a stably integrated expression cassette (Lox1-GFP-Lox2-RFP) was exposed to His-Cre, TAT-Cre, or VLP consisting of Gag-Cre pseudotyped with VSV-G. Expression of RFP shows that active Cre recombinase was able convert the genomic copy of the GFP/RFP expression cassette.
Gag-Cre Expression and Protein Processing.
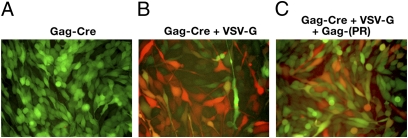
Although the experiments performed with Gag-Cre show that Cre is active when it is part of a Gag fusion protein, other proteins may not be functional if they are part of a similar Gag fusion. Retroviruses contain a protease that processes both the Gag and the Gag-Pol polyprotein. In most retroviruses, protease is part of Gag-Pol. One of the advantages of using the avian sarcoma leukosis viruses (ASLVs) to produce the VLPs is that, in contrast to most retroviruses, in the ASLVs, protease is normally part of Gag. To produce VLPs that contained protease, we mixed an expression construct that expresses the Gag-Cre fusion with one that expresses a full-length Gag that does encode protease. It is not possible to use this cotransfection system to produce VLPs that contain the amount of protease that is normally found in an ASLV virion (1:1 with Gag). We did experiments with various ratios of Gag-Cre and Gag-protease and found that a 10:1 ratio was optimal for formation of the chimeric VLPs. To determine whether viral protease can process the Gag-fusion proteins to generate the native (unfused) protein, the proteins in the VLPs were fractionated on PAGE and a Western blot was performed and probed with anti-capsid (CA) antibody or anti-Cre antibody. The Cre protein was cleaved from the Gag fusion by the viral protease during virion maturation (Fig. S2 A and B). The processed capsid (p27) and Cre recombinase are present only when VLPs also contain intact Gag. Presumably, processing is inefficient, because compared with ASLV (RCAS) virions where one protease is present for every Gag, in these experiments the ratio of Gag-protease to Gag-Cre is 1:10 (based on the amount of DNA used in the transfection). Because the Gag-Cre fusion is active, we cannot conclusively prove that the cleaved Cre is active (Fig. 4); however, this experiment does show that the viral protease can be used to free the fusion protein from Gag, if that is needed for the protein to be active.
Fig. 4.
Activity and processing of Gag-Cre. Incorporation of a functional retroviral protease into VLPs caused the release of the Cre protein from the Gag-Cre fusion. (A) VLP without VSV-G envelope (control); (B) Gag-Cre VLPs pseudotyped with VSV-G envelope; and (C) Gag-Cre and Gag-protease VLPs pseudotyped with VSV-G.
VLPs Containing Gag-Cytosine Deaminase and Uracil Phosphorybosyltransferase (Gag-Fcy::Fur).
The combination of inactive prodrugs and enzymes that convert the prodrugs to an active form has been widely used in chemotherapeutic approaches for the treatment of cancer. 5-Fluorocytosine (5FC) has been introduced as a prodrug, which in combination with the appropriate cytosine deaminase, can cause cell death. The protein Fcy converts 5-FC into 5-fluorouracil (5-FU), a highly cytotoxic compound routinely used in cancer chemotherapy. The protein Fur converts 5-FU to 5-F UMP, an irreversible inhibitor of thymidylate synthase, which blocks DNA synthesis. However, this approach often relies on the establishment of cell lines that express the enzymes required to convert the prodrug into the cytotoxic component. We have approached this problem by generating VLPs that contain a Gag-Fcy::Fur fusion and VSV-G envelope to assess the efficiency of this system for the delivery of enzymes that could be used for prodrug-to-active drug conversion.
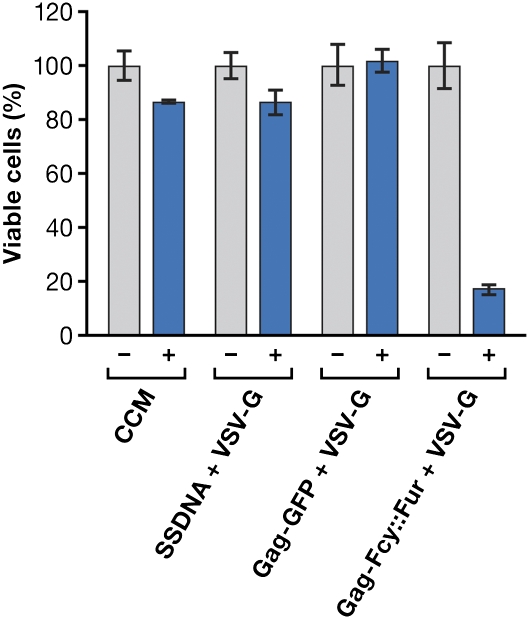
The cytotoxic effect of 5-FC, which is a substrate for Fcy::Fur, was determined using the prostate cancer cell line PC3. Cells were incubated with 0–250 μg/mL of 5-FC (Fig. S3) to determine an appropriate concentration for subsequent experiments (a concentration that would cause minimal cell death when the prodrug was administered by itself). Based on the results of these experiments, a 5-FC concentration of 60 μg/mL was selected to test the ability of the VLPs to deliver Gag-Fcy::Fur into these cells. As shown in Fig. 5, there was minimal reduction in the viability of cells treated either with 5-FC alone or with supernatants produced by transducing the packaging cell line with salmon sperm DNA alone or with VSV-G expressing DNA alone, or when the cells were treated with VLPs containing Gag-GFP pseudotyped with VSV-G. These results indicate that there was no appreciable conversion of the prodrug to the cytotoxic form under these conditions. However, treatment of cells with VLPs consisting of Gag-Fcy::Fur pseudotyped with VSV-G in the presence of 5-FC caused up to an 80% reduction in cell viability. These findings show that VLPs can be used to deliver enzymes that can mediate prodrug conversion to a more cytotoxic form to a majority of the cells in an experiment, and suggest that the VLPs have the potential to be useful in some types of in vivo chemotherapy.
Fig. 5.
Cytotoxic effects of 5FC in the presence of Gag-Fcy::Fur on the PC3 cell line. PC3 (a human prostate cancer line) cells were treated with complete culture medium (CCM) from untransfected cells; CCM from transfected cells with salmon sperm DNA (ssDNA) + VSV-G envelope; VLPs that contain Gag-GFP and VSV-G envelope or VLPs consisting of Gag-Fcy::Fur and VSV-G in the presence (+) or absence (−) of 60 μg/mL of 5FC.
Application of VLPs for Delivery of Proapoptotic Enzymes.
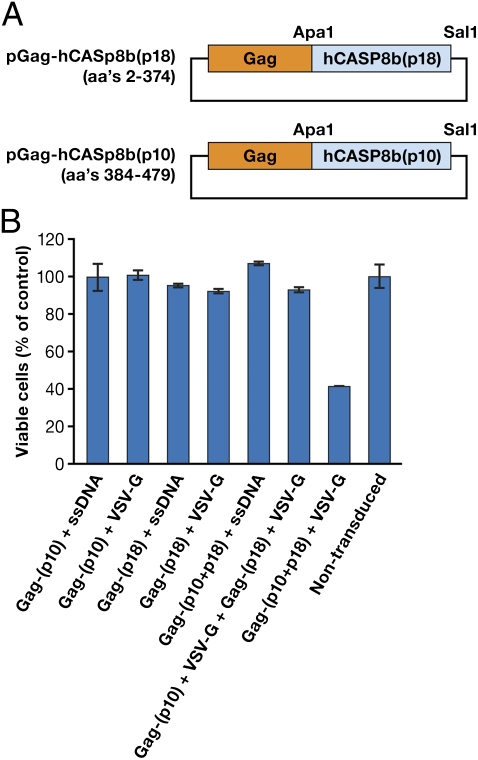
One of the disadvantages of the prodrug approach is that once the compounds are activated to the toxic state, they can leak out of cells, and may have unwanted toxic effects on neighboring cells. A better approach might be to use an enzyme, like human caspase 8b (hCasp8B), which cannot leak out of, or enter, cells and is an efficient inducer of apoptosis. We have incorporated Gag-hCaspase8 into VLPs to determine the effectiveness of this approach. The initial transfection with plasmid containing full-length human caspase 8 linked to Gag (Gag-hCaspase8) caused the death of the packaging cells (293T), indicating that expression of the intact and active caspase was incompatible with cell viability. To reduce this toxicity, we generated “split” caspase Gag fusion vectors. The coding sequence of hCaspase8 was divided into two separate domains: a small (p10) and a large (p18) domain, and these domains were individually cloned in frame with Gag (Fig. 6A). The packaging cell line was viable when it was cotransfected with the expression plasmids for Gag fused to each of the two domains and the VSV-G envelope. The VLPs were collected and exposed to the PC3 cell line. As shown in Fig. 6B, no cell death was observed with VLP containing only one of the two caspase components. However, VLPs that contained both Gag-p10 and Gag-p18 in the same VLPs caused a 60% reduction in cell viability. Interestingly, no cell death was observed after exposure of PC3 cells with two separate VLPs containing p10 and p18, respectively. For cell death to occur, the two caspase components, p10 and p18, must combine to form an active caspase; this happens efficiently when the two caspase components are linked to Gag and coassemble into a single particle, a process that is mediated by Gag–Gag interactions. However, if two VLPs, each of which contains one of the caspase fragments transduce the same cell, the concentration of the caspase fragments may be too low to form functional caspase. These results demonstrate the ability of VLP-mediated protein delivery to cause direct cell cytotoxicity using caspase.
Fig. 6.
Biological activity of VLPs containing split human caspase 8b. (A) The coding sequence of human capsase 8b was split into two separate components (p10 and p18) that were cloned in-frame into the Gag coding sequence. (B) Biological activity of split caspase 8b delivered via VLPs. Target cells (PC3) were either left untreated or incubated for 48 h with control VLPs consisting of Gag-(p10) + ssDNA (salmon sperm DNA), Gag-(p10) + VSV-G, Gag-(p18) + ssDNA, Gag-(p18) + VSV-G, Gag-(p10 + p18) + ssDNA, Gag-(p10) + VSV-G, and Gag-(p18) + VSV-G or with VLPs consisting of Gag-(p10 + p18) + VSV-G within the same particles. As shown, only the cells transduced with VLPs that contained both p10 and p18 and also contained VSV-G caused a significant reduction in cell viability.
VLP Surface Display and Delivery of Functional TRAIL Ligand.
TRAIL has been shown to exhibit potent and specific apoptotic activity against tumor cells (38). TRAIL induces apoptosis through death receptors (DR4 and DR5) present on the surface of the cells. Although DR4 and DR5 are known to be present in both normal and cancer cells, TRAIL has been shown to act only on cancer cells, sparing normal cells (39, 40). This property made TRAIL an attractive molecule for cancer therapy. Currently, recombinant soluble TRAIL is being used against cancer cells.
We sought to develop a delivery method based on using modified VLPs to deliver human TRAIL to cells in culture. Both influenza neuraminidase (NA) and hemmagglutinin (HA) are readily incorporated into VLPs, and truncated forms of these proteins can be fused to cell-specific ligands (41). These truncated forms of both HA and NA lacked multimerization domains (42). We used truncated influenza NA (membrane protein class 2) to display protein ligands fused at the C terminus of NA, or hemmagglutinin (membrane protein class 1) to display protein ligands fused at the N terminus of HA (Fig. S4A). We cloned the TRAIL ligand containing a histidine tag (His-6) into the expression plasmid that encodes the truncated NA called pCMV-NA. VLPs were generated after transfection of 293T cells with plasmids containing Gag-GFP and NA-TRAIL. The VLPs were analyzed by Western blot using an anti-capsid antibody (anti-p27) that can detect Gag. A control virus (RCAS) was also included as positive control for the ability of the antibody to detect processed p27. Fig. S4B shows that the VLPs have been made, because the anti-p27 antibody recognized the Gag-GFP fusion. The NA-TRAIL ligand contains an internal His tag. When the same VLPs were tested, by Western blot, with anti-His antibody, the NA-TRAIL ligand was also shown to be present on the VLPs.
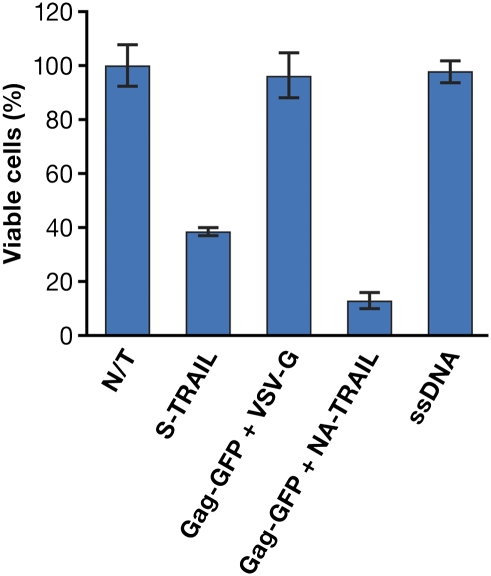
These VLPs were tested for biological activity (i.e., their ability to cause a loss of viability of PC3 cells; Fig. 7). The commercially available soluble human TRAIL ligand (100 ng/mL) was used as a positive control. As shown, the treatment of PC3 cells with soluble TRAIL resulted in significant cytotoxicity (60% cell death). When cells were treated with culture medium collected from the packaging cell line transfected with carrier salmon sperm DNA or with VLPs consisting of Gag-GFP pseudotyped with VSV-G (negative controls), no cytotoxicity was observed. However, treatment of PC3 cells with VLPs consisting of Gag-GFP and surface-displayed NA-TRAIL resulted in an 80–90% reduction in cell viability. To rule out an effect of the free form of the fusion protein, which might have been released from the transfected cells, the culture supernatant was passed through a 300-KD cutoff filter. Treatment of the PC3 cells with the eluate made from Gag-GFP + NA-TRAIL VLPs resulted in 70–80% cell viability (Fig. S5), which suggests that, though there is a modest amount of toxicity from the free form of the TRAIL fusion protein, which would come through in the filtrate, the majority of the cell death is caused by VLPs containing NA-TRAIL.
Fig. 7.
TRAIL-mediated cell death. The PC3 cell line was treated with supernatants consisting of complete culture medium (N/T) or culture medium from a packaging cell line transfected with carrier ssDNA or culture medium supplemented with soluble human TRAIL or supernatants containing VLPs as Gag-GFP + VSV-G, Gag-GFP + NA-TRAIL. As shown, cell death is observed when cells were treated either with s-TRAIL or with VLPs consisting of Gag-GFP and NA-TRAIL. GFP was introduced into the VLPs to make it simple to monitor VLP formation.
VLPs Displaying Mouse IFN-γ on Their Surface.
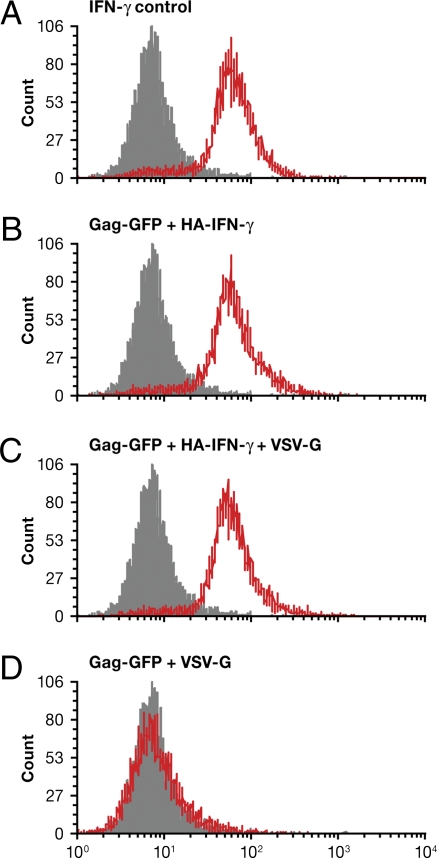
We next sought to determine if VLPs could deliver cytokines to appropriate target cells. The same truncated version of influenza virus HA and NA containing the cytoplasmic and the transmembrane domains were used to express these ligands on the surface of VLPs. We chose IFN-γ because it is known to be active as a homodimer, and this homodimer interacts with a cell surface receptor. The mouse IFN-γ was displayed on VLP as either an HA fusion (HA–IFN-γ) or a NA fusion (NA–IFN-γ), as described in SI Materials and Methods. A mouse macrophage cell line developed from a C57BL/6 mouse that expresses IFN-γ receptor was used. Activation of the receptor was monitored by the phosphorylation of STAT1, a target of the activated receptor, as shown using control IFN-γ (Fig. 8A). As shown, VLPs containing HA–IFN-γ caused STAT1 phosphorylation at levels comparable to soluble IFN-γ in the absence (Fig. 8B) or presence of VSV-G (Fig. 8C). However, VLPs containing NA–IFN-γ induced slightly lower levels of phosphorylation of STAT1, irrespective of the absence (Fig. S6A) or the presence (Fig. S6B) of VSV-G. This difference in activity could be due to orientation of the IFN-γ fusion with respect to NA and HA. There was no detectable STAT1 phosphorylation in the absence of IFN-γ (Fig. 8D). Thus, VLPs are capable of delivering a cytokine signal to an appropriate target cell.
Fig. 8.
Murine macrophage cells respond to VLPs that carry mouse IFN-γ. The mouse macrophage cell line was treated with VLPs containing HA–IFN-γ constructs. Cells were analyzed for the presence of phospho-STAT1 by flow cytometry. (A) Positive control (soluble IFN-γ). (B) VLPs containing HA–IFN-γ. (C) Gag-GFP + HA–IFN-γ + VSV-G. (D) Gag-GFP + VSV-G (negative control).
Conclusions
We have developed a safer and robust protein delivery using VLPs that can deliver proteins either to the surface or the interior of cells. We have demonstrated the effectiveness of this technology by delivering a number of biologically active proteins, including GFP, Cre recombinase, cytotoxic enzymes Fcy::Fur (cytosine deaminase and uracil phosphoribosyl-transferase fusion protein), and human Caspase 8, to the inside of cells. In addition to intracellular delivery of proteins, we have shown that protein ligands, such as TRAIL and IFN-γ, can be displayed on the surface of VLPs and can cause appropriate signaling in cells. We have shown that VLPs that display TRAIL are active and can induce cell death. In addition, we have also shown that IFN-γ is active toward the target cells, as evidenced in STAT1 phosphorylation.
Materials and Methods
Generation of the VLPs.
The 293T cell line was used to generate the VLPs. The 3.3 × 106 cells were plated in a 10-cm dish (10 mL of DMEM containing 10% FBS was added 24 h before transfection). On the day of transfection, the culture medium was replaced with 10 mL of fresh medium. Cells were transfected 2 h later with a solution of DNA and FuGene 6 (Roche), used according to the manufacturer's instructions. Each dish was transfected with a total of 15 μg of DNA. The Gag-Cre transfection used 9 μg of pDest472-Gag-Cre plasmid and 6 μg of salmon sperm DNA (ssDNA), and the Gag-Cre + VSV-G transfection used 9 μg of pDest472-Gag-Cre plasmid, 5 μg of pCMV-VSV-G plasmid, and 1 μg of ssDNA. The Gag-Cre + VSV-G + Gag-Protease (a gift from J. W. Wills, Pennsylvania State University, University Park, PA) transfection used 9 μg of pDest472-Gag-Cre plasmid, 5 μg of pCMV-VSV-G plasmid, and 1 μg of pCMV-Gag-Protease plasmid. The transfection reaction for VLPs consisting of Gag-Fcy::Fur used 10 μg of pDest472-Gag-Fcy::Fur and 5 μg of pCMV-SVG or 5 μg of ssDNA, whereas Gag-GFP + NA-TRAIL used 8 μg of pDest472-Gag-GFP + 4 μg of pNA-TRAIL and 4 μg of pCMV-VSV-G or 4 μg of ssDNA. Following transfection, supernatants were collected 48–72 h later and clarified by centrifugation at 2200 × g for 10 min at 4 °C. Clarified supernatants were treated with DNase1 (10 units/mL) for 30 min at room temperature and then filtered through a 0.45-μm filter. The VLPs were used for immediate transduction or frozen on dry ice and stored at −80 °C.
Supplementary Material
Acknowledgments
We thank Drs. James Hartley and Timothy Harris for support and encouragement throughout the work. We also thank Michael Sanford for the flow cytometry experiment, Marritta Grau for editing the manuscript, and Jiro Wada for preparation of the figures. Funding was received from National Cancer Institute, National Institutes of Health Grant HHSN 261 2008 00001E.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101874108/-/DCSupplemental.
References
- 1.Federspiel MJ, Bates P, Young JA, Varmus HE, Hughes SH. A system for tissue-specific gene targeting: Transgenic mice susceptible to subgroup A avian leukosis virus-based retroviral vectors. Proc Natl Acad Sci USA. 1994;91:11241–11245. doi: 10.1073/pnas.91.23.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bockamp E, et al. Conditional transgenic mouse models: From the basics to genome-wide sets of knockouts and current studies of tissue regeneration. Regen Med. 2008;3:217–235. doi: 10.2217/17460751.3.2.217. [DOI] [PubMed] [Google Scholar]
- 3.Hare S, et al. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 2009;5:e1000259. doi: 10.1371/journal.ppat.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang S, Kawabe Y, Ito A, Kamihira M. Cre recombinase-mediated site-specific modification of a cellular genome using an integrase-defective retroviral vector. Biotechnol Bioeng. 2010;107:717–729. doi: 10.1002/bit.22863. [DOI] [PubMed] [Google Scholar]
- 5.Capecchi MR. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell. 1980;22:479–488. doi: 10.1016/0092-8674(80)90358-x. [DOI] [PubMed] [Google Scholar]
- 6.Bar-Sagi D, Feramisco JR. Microinjection of the ras oncogene protein into PC12 cells induces morphological differentiation. Cell. 1985;42:841–848. doi: 10.1016/0092-8674(85)90280-6. [DOI] [PubMed] [Google Scholar]
- 7.Campbell PL, McCluskey J, Yeo JP, Toh BH. Electroporation of antibodies into mammalian cells. Methods Mol Biol. 1995;48:83–92. doi: 10.1385/0-89603-304-X:83. [DOI] [PubMed] [Google Scholar]
- 8.Abarzúa P, LoSardo JE, Gubler ML, Neri A. Microinjection of monoclonal antibody PAb421 into human SW480 colorectal carcinoma cells restores the transcription activation function to mutant p53. Cancer Res. 1995;55:3490–3494. [PubMed] [Google Scholar]
- 9.Theiss C, Meller K. Microinjected anti-actin antibodies decrease gap junctional intercellular commmunication in cultured astrocytes. Exp Cell Res. 2002;281:197–204. doi: 10.1006/excr.2002.5652. [DOI] [PubMed] [Google Scholar]
- 10.Narayanan A, et al. Nuclear RanGTP is not required for targeting small nucleolar RNAs to the nucleolus. J Cell Sci. 2003;116:177–186. doi: 10.1242/jcs.00176. [DOI] [PubMed] [Google Scholar]
- 11.Schwarze SR, Hruska KA, Dowdy SF. Protein transduction: Unrestricted delivery into all cells? Trends Cell Biol. 2000;10:290–295. doi: 10.1016/s0962-8924(00)01771-2. [DOI] [PubMed] [Google Scholar]
- 12.Fenton M, Bone N, Sinclair AJ. The efficient and rapid import of a peptide into primary B and T lymphocytes and a lymphoblastoid cell line. J Immunol Methods. 1998;212:41–48. doi: 10.1016/s0022-1759(97)00208-1. [DOI] [PubMed] [Google Scholar]
- 13.Murriel CL, Dowdy SF. Influence of protein transduction domains on intracellular delivery of macromolecules. Expert Opin Drug Deliv. 2006;3:739–746. doi: 10.1517/17425247.3.6.739. [DOI] [PubMed] [Google Scholar]
- 14.Richard JP, et al. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J Biol Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 15.Schwarze SR, Dowdy SF. In vivo protein transduction: Intracellular delivery of biologically active proteins, compounds and DNA. Trends Pharmacol Sci. 2000;21:45–48. doi: 10.1016/s0165-6147(99)01429-7. [DOI] [PubMed] [Google Scholar]
- 16.Säälik P, et al. Protein cargo delivery properties of cell-penetrating peptides. A comparative study. Bioconjug Chem. 2004;15:1246–1253. doi: 10.1021/bc049938y. [DOI] [PubMed] [Google Scholar]
- 17.Drin G, Cottin S, Blanc E, Rees AR, Temsamani J. Studies on the internalization mechanism of cationic cell-penetrating peptides. J Biol Chem. 2003;278:31192–31201. doi: 10.1074/jbc.M303938200. [DOI] [PubMed] [Google Scholar]
- 18.Fischer R, Köhler K, Fotin-Mleczek M, Brock R. A stepwise dissection of the intracellular fate of cationic cell-penetrating peptides. J Biol Chem. 2004;279:12625–12635. doi: 10.1074/jbc.M311461200. [DOI] [PubMed] [Google Scholar]
- 19.Wagstaff KM, Jans DA. Protein transduction: Cell penetrating peptides and their therapeutic applications. Curr Med Chem. 2006;13:1371–1387. doi: 10.2174/092986706776872871. [DOI] [PubMed] [Google Scholar]
- 20.Ramqvist T, Andreasson K, Dalianis T. Vaccination, immune and gene therapy based on virus-like particles against viral infections and cancer. Expert Opin Biol Ther. 2007;7:997–1007. doi: 10.1517/14712598.7.7.997. [DOI] [PubMed] [Google Scholar]
- 21.Grgacic EV, Anderson DA. Virus-like particles: Passport to immune recognition. Methods. 2006;40:60–65. doi: 10.1016/j.ymeth.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcea RL, Gissmann L. Virus-like particles as vaccines and vessels for the delivery of small molecules. Curr Opin Biotechnol. 2004;15:513–517. doi: 10.1016/j.copbio.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Ludwig C, Wagner R. Virus-like particles-universal molecular toolboxes. Curr Opin Biotechnol. 2007;18:537–545. doi: 10.1016/j.copbio.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jennings GT, Bachmann MF. The coming of age of virus-like particle vaccines. Biol Chem. 2008;389:521–536. doi: 10.1515/bc.2008.064. [DOI] [PubMed] [Google Scholar]
- 25.Peretti S, Schiavoni I, Pugliese K, Federico M. Cell death induced by the herpes simplex virus-1 thymidine kinase delivered by human immunodeficiency virus-1-based virus-like particles. Mol Ther. 2005;12:1185–1196. doi: 10.1016/j.ymthe.2005.06.474. [DOI] [PubMed] [Google Scholar]
- 26.Peretti S, Schiavoni I, Pugliese K, Federico M. Selective elimination of HIV-1-infected cells by Env-directed, HIV-1-based virus-like particles. Virology. 2006;345:115–126. doi: 10.1016/j.virol.2005.09.054. [DOI] [PubMed] [Google Scholar]
- 27.Muratori C, Bona R, Federico M. Lentivirus-based virus-like particles as a new protein delivery tool. Methods Mol Biol. 2010;614:111–124. doi: 10.1007/978-1-60761-533-0_7. [DOI] [PubMed] [Google Scholar]
- 28.Voelkel C, et al. Protein transduction from retroviral Gag precursors. Proc Natl Acad Sci USA. 2010;107:7805–7810. doi: 10.1073/pnas.0914517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voynow SL, Coffin JMJ. Truncated gag-related proteins are produced by large deletion mutants of Rous sarcoma virus and form virus particles. J Virol. 1985;55:79–85. doi: 10.1128/jvi.55.1.79-85.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishna NK, Campbell S, Vogt VM, Wills JW. Genetic determinants of Rous sarcoma virus particles size. J Virol. 1998;72:564–577. doi: 10.1128/jvi.72.1.564-577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weldon RA, Jr, Erdie CR, Oliver MG, Wills JW. Incorporation of chimeric gag protein into retroviral particles. J Virol. 1990;64:4169–4179. doi: 10.1128/jvi.64.9.4169-4179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogt VM, Simon MN. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J Virol. 1999;73:7050–7055. doi: 10.1128/jvi.73.8.7050-7055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes SH, Greenhouse JJ, Petropoulos CJ, Sutrave P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1997. [PubMed] [Google Scholar]
- 35.Lindberg LG Virus-like particles in Rous rat sarcoma. Acta Pathol Microbiol Scand. 1968;74:189–198. doi: 10.1111/j.1699-0463.1968.tb03470.x. [DOI] [PubMed] [Google Scholar]
- 36.Kaczmarczyk SJ, Green JE. A single vector containing modified cre recombinase and LOX recombination sequences for inducible tissue-specific amplification of gene expression. Nucleic Acids Res. 2001;29:e56. doi: 10.1093/nar/29.12.e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12:1035–1040. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- 38.LeBlanc HN, Ashkenazi A. Apo2L/TRAIL and its death and decoy receptors. Cell Death Differ. 2003;10:66–75. doi: 10.1038/sj.cdd.4401187. [DOI] [PubMed] [Google Scholar]
- 39.Falschlehner C, Emmerich CH, Gerlach B, Walczak H. TRAIL signalling: Decisions between life and death. Int J Biochem Cell Biol. 2007;39:1462–1475. doi: 10.1016/j.biocel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Walczak H, et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y, Leggat D, Herbert A, Roberts PC, Sundick RS. A novel method to incorporate bioactive cytokines as adjuvants on the surface of virus particles. J Interferon Cytokine Res. 2009;29:9–22. doi: 10.1089/jir.2008.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.