Abstract
Huntington disease (HD) is a progressive neurodegenerative disease that affects 30,000 individuals in North America. Treatments that slow its relentless course are not yet available, and biomarkers that can reliably measure disease activity and therapeutic response are urgently needed to facilitate their development. Here, we interrogated 119 human blood samples for transcripts associated with HD. We found that the dynamic regulator of chromatin plasticity H2A histone family, member Y (H2AFY) is specifically overexpressed in the blood and frontal cortex of patients with HD compared with controls. This association precedes the onset of clinical symptoms, was confirmed in two mouse models, and was independently replicated in cross-sectional and longitudinal clinical studies comprising 142 participants. A histone deacetylase inhibitor that suppresses neurodegeneration in animal models reduces H2AFY levels in a randomized phase II clinical trial. This study identifies the chromatin regulator H2AFY as a potential biomarker associated with disease activity and pharmacodynamic response that may become useful for enabling disease-modifying therapeutics for HD.
Keywords: macroH2A, transcriptional profiling, gene expression, sodium phenylbutyrate
Huntington disease (HD) is an autosomal dominant neurodegenerative disorder caused by an expansion of glutamine repeats at the N terminus of huntingtin protein (1). About 30,000 individuals in North America are affected by the disease, and no medication is available yet to slow the disease process. Rapidly advancing basic research is creating an expanding pipeline of candidate disease-modifying therapeutics that is entering clinical trials. Progress has been curtailed by the limited power of the clinical end points currently available to provide evidence for efficacy. In typical phase II clinical trials, obtaining preliminary evidence of disease modification based on clinical assessments alone is extremely challenging because the HD phenotype is highly variable and slowly progressive. As a result, much larger futility or phase III efficacy studies are required, involving hundreds of subjects and years of follow-up, to determine whether a treatment could be disease-modifying or is a likely failure. Simple and robust markers of disease activity that can serve as indicators of treatment response are critically needed to prioritize lead compounds for phase III clinical trials and will greatly accelerate the development of novel medications for patients with HD.
Although HD symptoms reflect preferential neuronal death in specific brain regions, huntingtin is expressed in almost all tissues and may cause detectable but clinically silent changes in gene expression and biochemistry in blood cells (2). Biochemical traits in blood cells of patients with HD encompass the biological processes of mitochondrial function (3), adenosine receptor signaling (4), and tryptophan metabolism (5). Although gene expression traits associated with HD have been reported in the brain (6), muscle (6), and blood (7, but note 8), a robust, generalizable, and potentially clinically useful blood test for HD has not yet been developed. A simple blood test of high accuracy and reproducibility would have many advantages over invasive cerebrospinal fluid tests or expensive imaging biomarkers. Likewise, genetic testing confirms the clinical impression of HD and identifies at-risk individuals but cannot dynamically track disease activity, progression of disease severity, and response to therapeutics or predict conversion from premanifest to clinical disease.
The US Food and Drug Administration-led Microarray Quality Control project has demonstrated that microarray results are generally repeatable within a laboratory, reproducible between laboratories, and comparable across different gene expression platforms (9). Indeed, gene expression microarray technology has matured to the point where some applications are deemed reliable enough for clinical and regulatory purposes (9). Transcriptome-wide scans can provide an unbiased global estimate of changes in gene expression and identify genes (10, 11) and pathways (12) causally, reactively, or independently associated with genetic, environmental, or complex disease etiologies (13, 14). Gene expression data can be used to rank individuals according to molecular characteristics (15) and to generate hypotheses about disease mechanisms (16), and such data may be particularly useful for identifying prototype biomarkers for quantitatively and longitudinally tracking dynamic disease traits that cannot readily be explained by static variation in DNA sequence.
Our data indicate that H2A histone family, member Y (H2AFY) expression in blood is robustly, specifically, and repeatedly associated with HD in four case–control studies of 472 samples and on two gene expression platforms and marks early stages of disease severity in human and mouse brains.
Results
H2AFY Is Specifically Overexpressed in Cellular Blood of Patients with HD.
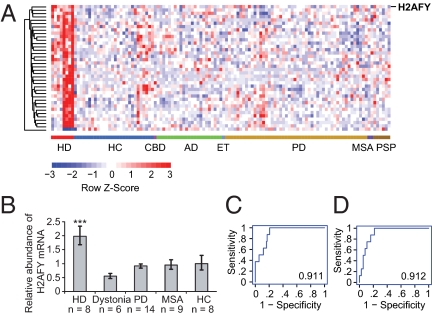
We performed a systematic transcriptome-wide analysis to interrogate the molecular processes perturbed in cellular blood of patients with HD for lead biomarkers. Genome-wide expression data of 119 transcriptomes from eight patients with HD and 111 controls, including 83 neurodegenerative disease controls, were analyzed. Stringent significance thresholds were set to control for false-positive results attributable to biological and technical noise and to correct for multiple testing. To correct for multiple testing, rigorous permutation analysis, significance analysis of microarrays (SAM) (17), was applied and genes with small effect sizes were excluded. Because technical variation is higher for genes with low average expression intensities on Affymetrix Human GeneChip U133A arrays, only genes with intensities ≥100 in at least one sample were considered for further analysis. This conservative statistical analysis keeps the number of false-positive results at a minimum, although the number of false-negative results is likely to remain high. Ninety-nine genes were significantly differentially expressed in patients with HD, with a fold change ≥1.5 or ≤0.66 and a false discovery rate (FDR) <0.00002 based on 50,000 permutations of the dataset (Table S1). These include the transcriptional modulator H2AFY. H2AFY was 1.6-fold overexpressed in cellular blood of patients with HD, with a FDR <0.00002. For visualization purposes, only the 38 genes with the lowest P values (P < 10−6 by a two-sample, two-sided t test) of all 99 significant genes are displayed in Fig. 1A (a complete list of all 99 genes associated with HD is provided in Table S1). H2AFY and its encoded histone variant, macroH2A1, modulate transcription factor binding (18), X-chromosome inactivation (19), and transcription repression (20). Interestingly, the highlighted genes (Table S1) include GAPDH, a “housekeeping” gene known to interact directly with mutant huntingtin (21), which mediates its nuclear translocation (22). High sensitivity and disease specificity are desirable (but not mandatory) for some clinically useful biomarkers. The area under the receiver-operating characteristics curve (AUC) for H2AFY was 0.911, indicating high sensitivity and specificity at various cutoffs (with an accuracy as high as 96% at the optimal cutoff; Fig. 1C). This 119-sample dataset included genome-wide expression data from 105 individuals with Parkinson disease (PD) and controls that we previously reported as part of our PD biomarker discovery efforts (15), as well as eight HD cases and six age- and sex-matched healthy controls (HCs) generally enrolled, processed, and assayed in parallel for the present study. If the analysis was restricted to this subset of eight patients with HD and six age- and sex-matched HCs without neurodegenerative diseases, H2AFY was still 1.5-fold overexpressed in the blood of patients with HD, with P = 0.01. Because transcriptional dysregulation is likely a pathogenic mechanism in HD (1), we prioritized H2AFY for further evaluation.
Fig. 1.
H2AFY is overexpressed in cellular blood of patients with HD in a discovery study comprising 119 individuals. (A) Genome-wide expression data of eight patients with HD and 111 controls, including 83 neurodegenerative disease controls, were analyzed. Expression levels for 38 genes most differentially expressed in patients with HD, including the transcriptional modulator H2AFY, are visualized (a complete list of all 99 genes significantly associated with HD is provided in Table S1). H2AFY is 1.6-fold overexpressed in cellular blood of patients with HD, with a FDR <0.00002 and P = 1.3 × 10−7 by two-sided t test. In the heat map, columns represent study subjects and rows represent genes. Expression higher than the mean is displayed as shades of red, and expression lower than the mean is displayed as shades of blue. AD, Alzheimer's disease; CBD, corticobasal degeneration; ET, essential tremor; PSP, progressive supranuclear palsy. (B) Overexpression of H2AFY in cellular blood of patients with HD was confirmed on the qPCR platform. H2AFY mRNA level are approximately twofold higher in blood of patients with HD [n = 8; mean age (y) ± SD: 51.8 ± 6.2] compared with that of HCs (n = 8; 50.9 ± 15.4; ***P = 0.0006) as well as patients with a hyperkinetic movement disorder other than HD (dystonia: n = 6; 56.3 ± 5.5; P < 0.0001) and patients with neurodegenerative movement disorders (PD: n = 14; 55.8 ± 4.8; P = 0.0004) or MSA (n = 9; 59.5 ± 6.8; P = 0.02). Error bars show SEM. The association between relative H2AFY mRNA abundance and HD is sensitive and specific with AUCs of 0.911 and 0.912 with accuracies of 96% and 87% at optimal cutoffs on the Affymetrix U133A (C) and qPCR (D) platforms, respectively.
To verify these results on an independent gene expression platform, we performed kinetic, real-time, quantitative PCR (qPCR) (Fig. 1B). We assayed relative H2AFY mRNA abundance in the 8 patients with HD and eight HCs, and we also included 29 new age- and sex-matched neurological disease controls (Fig. 1B). Six age- and sex-matched patients with a hyperkinetic movement disorder other than HD (dystonia) and treated with medications also commonly used for patients with HD (e.g., neuroleptics, anticholinergics) were enrolled to control for nonspecific medications and metabolic effects. Twenty-three age- and sex-matched patients with the neurodegenerative movement disorders, PD, or multiple system atrophy (MSA) were also enrolled to control for further nonspecific gene expression changes common to debilitating neurodegenerative diseases. To reduce bias from sample handling, the blood specimens were transported, processed, and stored in parallel in a uniform and standardized manner. HCs were recruited predominantly from nonaffected spouses of patients with HD, thus tightly matching the source population with similar environmental and lifestyle exposures. In this case–control study, H2AFY mRNA levels were approximately twofold higher in the blood of patients with HD compared with HCs (Fig. 1B; P = 0.0006 by general linear model regression analysis adjusted for age and sex with Tukey's studentized range test), neurologic disease controls with dystonia (P < 0.0001), patients with PD (P = 0.0004), and patients with MSA (P = 0.02). Relative H2AFY mRNA abundance was specifically associated with HD compared with clinically relevant movement disorders and HCs, with an accuracy of 87% and an AUC of 0.912 (Fig. 1D). The association between HD and elevated relative H2AFY mRNA abundance was repeatable and disease-specific on two gene expression platforms and in a substantial sample size (a total of 148 unique samples evaluated in this two-part discovery phase, as listed in Table S2).
Up-Regulation of H2AFY mRNA Levels in HD Is Independently Replicated in Two Validation Studies.
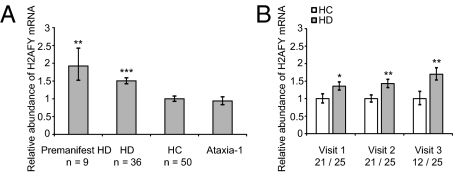
The high-throughput capability of gene expression technology comes at a price. Such multivariate analyses are susceptible to identifying false-positive associations between a transcript and a disease that are actually caused by chance, and are therefore not reproducible. To verify the observed association between elevated H2AFY mRNA levels and HD independently, we performed two validation studies. First, an independent, cross-sectional, case–control validation study of 96 individuals, including 36 patients with HD, 9 individuals with premanifest HD who carry the HD gene mutation without clinical phenotype, 50 HCs, and one subject with spinocerebellar ataxia-1 (Table S3), was performed. Validating the initial findings, H2AFY mRNA abundance was 1.5-fold elevated in patients with HD compared with HCs, with P = 0.0004 adjusting for age and sex (Fig. 2A). Consistent with a specific association with patients carrying an HD mutation (a CAG expansion in huntingtin), H2AFY mRNA abundance appeared unchanged in the two samples from the subject with spinocerebellar ataxia-1, who carries a CAG expansion in the ataxin-1 gene (Fig. 2A).
Fig. 2.
Association between relative H2AFY mRNA abundance and HD is independently replicated in two validation studies. (A) Statistically significantly elevated H2AFY mRNA levels are observed in blood of individuals with manifest HD [n = 36; mean age (y) ± SD: 49.4 ± 13.1 y] as well as premanifest HD (n = 9; 48.1 ± 7.5) compared with HCs (n = 50; 49.5 ± 8.2) with fold changes of 1.5, with P = 0.0004 (***) and 1.9 with P = 0.002 (**), respectively. No difference in H2AFY mRNA abundance is observed between HCs and two samples from a subject with spinocerebellar ataxia-1. (B) In a second, longitudinal study of 25 patients with HD and 21 tightly age- and sex-matched controls followed over 2–3 y, H2AFY is overexpressed in the HD group relative to controls at the baseline visit (1.4-fold; *P = 0.02) and remains overexpressed at the second (1.4-fold; **P = 0.005) and third (1.7-fold; **P = 0.008) annual visits. Numbers of HCs and patients with HD assayed at each visit are shown on the x axis (number of HCs/number of patients with HD). Error bars in A and B show SEM.
HD is an autosomal dominant disease with complete penetrance. Premanifest HD subjects are individuals at 100% risk for HD on the basis of documented positive genetic testing for the HD mutation but in whom clinical symptoms have not yet become manifest. The characterization of genes differentially expressed at the premanifest stage is of critical importance for therapeutic biomarker discovery because patients with premanifest and early-stage HD are most likely to benefit from disease-modifying interventions. There was a 1.9-fold increase in H2AFY mRNA levels in premanifest HD subjects compared with HCs (P = 0.002) indicating that H2AFY expression is already elevated early in the course of the disease.
Second, to further evaluate the stability of the association between increased H2AFY expression and HD over time, we performed an additional, longitudinal, case–control validation study (Fig. 2B) of 25 patients with HD and 21 tightly age- and sex-matched controls followed over 2–3 y (Table S4) with two (9 subjects) or three (37 subjects) annual follow-up visits. H2AFY was overexpressed in the HD group at the baseline visit (1.4-fold; P = 0.02 by general linear model regression analysis adjusted for age and sex) and remained overexpressed at the second (1.4-fold; P = 0.005) and third (1.7-fold; P = 0.008) annual visits (Fig. 2B).
Expression of the H2AFY-Encoded Protein MacroH2A1 Is Increased in Brains of Patients with Early-Grade HD, as Well as in Transgenic and Knock-In Mouse Models of HD.
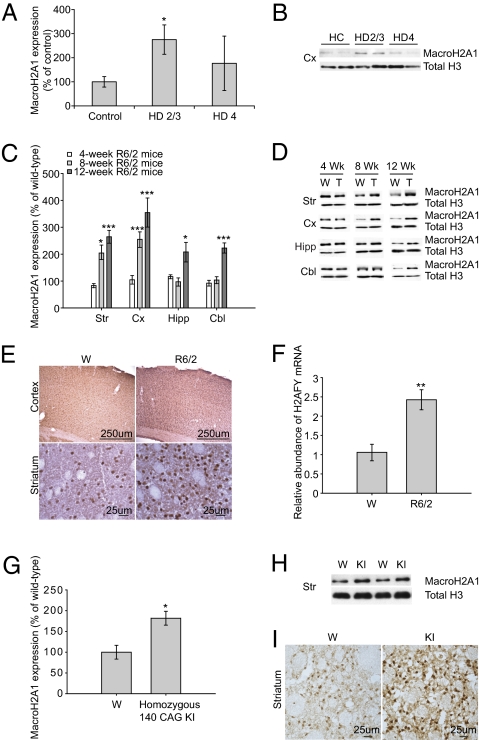
To determine whether macroH2A1 levels in the blood relate to disease progression in the brain, we extracted histones from the frontal cortex of 12 patients with HD (Table S5) and measured the expression of macroH2A1 by Western blotting. MacroH2A1 levels were elevated (P = 0.05) in grade 2/3 HD brain compared to controls but unaltered in grade 4 HD brain (Fig. 3 A and B) (23). Because H2AFY is so highly expressed in neurons, the profound neuron loss present in end-stage HD in humans (24) likely accounts for the late decline in H2AFY levels.
Fig. 3.
Expression of histone protein macroH2A1 is increased in the brains of patients with early-grade HD as well as in R6/2 and knock-in mouse models. (A and B) Western blots show levels of macroH2A1 in histone extracts from the frontal cortex (Cx) of patients with HD and controls. MacroH2A1 expression is increased in HD grade 2/3 frontal cortex (*P = 0.05). A representative Western blot of frontal cortex from two controls, two individuals with early-grade, and two individuals with advanced HD is shown. The level of macroH2A1 is normalized to total histone H3 level. (C and D) Western blots show levels of macroH2A1 in histone extracts from the striatum (Str), cortex (Cx), hippocampus (Hipp) and cerebellum (Cbl) of mice. Representative Western blots comparing wild-type (W) and transgenic R6/2 (T) mice at 4, 8, and 12 weeks of age, respectively, are shown. At 4 wk, there is no significant difference in macroH2A1 levels in any of the brain regions. Levels of macroH2A1 are significantly increased in the striatum and cortex at 8 and 12 wk of age (n = 8). The level of macroH2A1 is normalized to the total histone H3 level. Striatum, *P = 0.02 (8 wk), ***P < 0.0001 (12 wk); cortex, ***P = 0.0001 (8 wk), ***P = 0.001 (12 wk); hippocampus, *P = 0.03 (12 wk); cerebellum, ***P < 0.0001 (12 wk). (E) Immunohistochemical staining for macroH2A1 is markedly increased in the cortex and striatum of R6/2 mice compared with wild-type mice. (F) qPCR confirmed an increase in relative H2AFY mRNA abundance in the cortex of R6/2 mice compared with wild-type mice, with P = 0.004 (**). Error bars in A, C, and F show SEM. (G and H) Western blots show levels of macroH2A1 in histone extracts from the striatum (Str) of 12-mo-old wild-type and homozygous 140-CAG knock-in mice. A representative Western blot is shown. Levels of macroH2A1 are increased in the striatum of knock-in mice (n = 4). MacroH2A1 is normalized to the total histone H3 level. *P = 0.014. KI, knock-in; W, wild type. (I) Immunostaining for macroH2A1 is also increased in the striatum of homozygous 140-CAG knock-in mice compared with wild-type mice.
We further investigated whether macroH2A1 expression correlates with disease progression in mouse models of HD. The R6/2 mouse model expresses an exon 1 fragment of the human huntingtin gene and recapitulates many behavioral and neuropathological features observed during early disease stages in patients with HD (25). These mice exhibit progressive cognitive and motor deficits starting from 5 wk; neuropathological abnormalities, including a decline in brain weight and neostriatal volume by 8 wk; reduction in striatal neuronal number by 12 wk; and death by 13–15 wk of age. Levels of macroH2A1 were quantified in the striatum, cortex, hippocampus, and cerebellum of R6/2 HD mice (CAG repeat length of 135–140) at 4, 8, and 12 wk of age using Western blotting. We found a progressive increase in the levels of macroH2A1 in R6/2 brain with disease progression. Levels of macroH2A1 were unaltered at 4 wk of age and dramatically increased in the cortex and striatum (regions mostly affected in HD) at 8 and 12 wk of age (Fig. 3 C and D), whereas the hippocampus and cerebellum (less affected in HD) showed elevated macroH2A1 levels only at 12 wk of age, indicating that macroH2A1 correlates with disease progression and severity. Consistent with the increase in protein expression, immunohistochemistry of 12-wk-old R6/2 brains revealed a striking increase in macroH2A1 immunoreactivity in the striatum (Fig. 3E), especially in neuronal nuclei, where it plays an active role in transcriptional silencing. In addition to macroH2A1 protein, the relative abundance of H2AFY mRNA is increased 2.5-fold (P = 0.004) in the cortex of R6/2 mice as quantified by qPCR (Fig. 3F).
By contrast to florid mouse models carrying an N-terminal huntingtin fragment (e.g., R6/2 mice), mice carrying a chimeric mouse/human exon 1 containing 140 CAG repeats inserted in the murine huntingtin gene more faithfully model the disease genetics and have normal life spans (26, 27), and they develop a subtle pathologic phenotype that is most analogous to presymptomatic and early symptomatic stages of the human disease (26). Despite a mild phenotype, knock-in mice have abnormal motor behavior that can be quantified by formal behavioral testing and aged mice show characteristic striatal nuclear huntingtin aggregates (26, 27) as well as ∼20% underexpression of select striatal enriched transcripts [e.g., D2 receptor and dopamine- and cAMP-regulated neuronal phosphoprotein (DARPP-32)] (27) in the absence of neuronal cell loss. To validate our findings in this full-length huntingtin knock-in model, we assayed macroH2A1 levels in histone extracts from the striatum of homozygous 140-CAG knock-in mice compared with age- and sex-matched wild-type mice. Consistent with our observation in patients with early disease stages, striatal levels of macroH2A1 were increased in homozygous knock-in mice (Fig. 3 G–I). Western blot results revealed 1.8-fold increased levels of macroH2A1 in histone extract from the striatum of homozygous 140-CAG knock-in mice compared to 12-mo-old wild-type mice (P = 0.014; n = 4). Consistent with the Western blotting results, immunohistochemical staining for macroH2A1 was also remarkably increased in the striatum of homozygous 140-CAG knock-in mice compared with wild-type mice (Fig. 3I).
To confirm the results from our in vivo analyses further in the 140-CAG, full-length, knock-in mouse model as well as in the transgenic R6/2 mouse model of HD, we analyzed in silico genome-wide gene expression datasets (reported in 28) from a related knock-in mouse model of HD, the CHL2 mouse (29) as well as two distinct datasets from R6/2 mice (Fig. S1). Consistent with our in vivo studies, relative H2AFY abundance was 1.43-fold increased in the striatum of CHL2 knock-in mice (P = 0.0003). In the two R6/2 mouse datasets, relative H2AFY abundance was 1.18-fold and 1.27-fold increased, respectively (P = 0.03 and 0.01, respectively).
Thus, expression of the H2AFY-encoded protein macroH2A1 is increased in the frontal cortex of patients with early-grade HD, as well as in transgenic and knock-in mice thought to model premanifest and early/mid-disease stages.
H2AFY Expression Marks Clinical Response to Treatment with Sodium Phenylbutyrate, Which Suppresses Mutant Huntingtin-Induced Neurodegeneration in Mice.
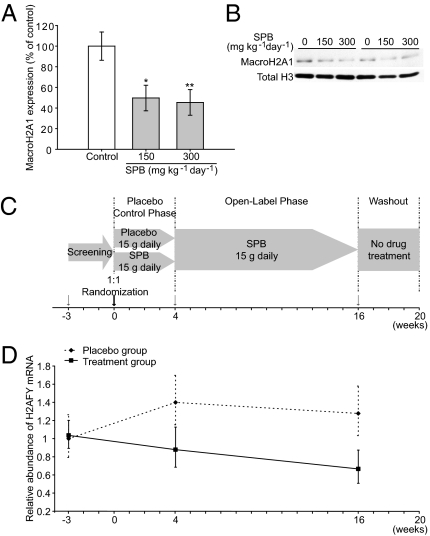
There is evidence that mutant huntingtin leads to reduced histone acetylation and altered transcription (30). Consequently, drug-like compounds that inhibit histone deacetylases (HDACs) are emerging as a leading class of potentially disease-modifying therapeutics for HD. Clinically, HDAC inhibitors, such as sodium phenylbutyrate (SPB), are already advancing through phase I and II drug trials. Administration of SPB or sodium butyrate increases brain histone acetylation (31), corrects neuronal mRNA dysregulation (31), significantly extends survival, and attenuates both gross brain and neuronal atrophy in R6/2 (32) as well as N171-82Q transgenic mouse models of HD (33). In a Drosophila model, HDAC inhibitors suppress neurodegeneration (34). To test whether macroH2A1 levels can be used to monitor a pharmacodynamic response to treatment with HDAC inhibitors, we treated R6/2 mice with SPB. Mice were treated with either 150 or 300 mg⋅kg−1⋅d−1 of SPB for 2 wk, and levels of macroH2A1 were determined in the striatum 2 h after the last injection. MacroH2A1 levels were dramatically reduced after SPB treatment (Fig. 4 A and B), thus linking the biomarker response to SPB and the known neuroprotective effects of SPB (32, 33).
Fig. 4.
H2AFY levels mark the pharmacodynamic response to treatment with the HDAC inhibitor SPB, a drug known to suppress huntingtin-induced neurodegeneration in mice. (A and B) R6/2 8-wk-old mice were treated with 150 or 300 mg⋅kg−1⋅d−1 of SPB for 2 wk, and macroH2A1 levels were determined in striatal histone extracts by semiquantitative Western blotting. R6/2 mice treated with 150 or 300 mg⋅kg−1⋅d−1 of SPB show a marked reduction in macroH2A1 levels compared with PBS-treated control mice with P = 0.02 (*) and P = 0.01 (**), respectively. (C and D) H2AFY levels in blood of patients with HD enrolled in a randomized, double-blind, placebo-controlled, phase II clinical trial (Phenylbutyrate Development for Huntington's Disease study). The relative abundance of H2AFY mRNA in SPB-treated and placebo groups was examined at baseline (week −3/visit 1), the end of the placebo-control phase (week 4/visit 4), and the end of the open-label phase (week 16/visit 7) as indicated by arrows. H2AFY expression is significantly down-regulated over time with SPB treatment. Error bars in A and D show SEM. By contrast, time alone (weeks) in the study (adjusting for drug treatment) is associated with a trend toward increasing H2AFY abundance [note subtle increase in H2AFY mRNA abundance in patients with HD (dotted line from week −3 to week 4) treated with placebo only].
To test whether H2AFY levels can be used to monitor a response to treatment with SPB in patients with HD as well, we analyzed frozen blood samples from participants enrolled in the Phenylbutyrate Development for Huntington's Disease (PHEND-HD) trial, a randomized, double-blind, placebo-controlled, phase II clinical trial. High-quality RNA (mean RNA integrity number ± SD, 8.1 ± 0.7) was extracted from blood samples collected and frozen in tubes containing an RNA-quality preserving reagent. H2AFY mRNA levels were assayed by qPCR in blood samples collected from 28 trial participants at the baseline visit (week −3/visit 1); at the end of the 4-wk randomized, double-blind, placebo-controlled phase (week 4/visit 4); and at the end of the 12-wk open-label phase (week 16/visit 7) (Fig. 4C). In this standard trial design, patients are first randomized to either study drug (SPB) or placebo during the 4-wk-long placebo-controlled phase; then, after completing the placebo-controlled phase, all participants go on to receive the study drug for 12 wk (both the “placebo” and “study drug” groups). Because HDAC inhibitors are known to exert their effects through both acute (35) and delayed time-dependent pharmacodynamic effects (36–38), we asked whether H2AFY mRNA abundance measured in blood can be used to monitor a pharmacodynamic response to treatment with the potentially disease-modifying drug SPB over time. To test for the effects of time on SPB on H2AFY abundance, we used a mixed random and fixed coefficient regression model, wherein weeks on SPB was a fixed predictor (irrespective of weeks in the study, which was a separate simultaneous fixed predictor; the intercept was random, whereas slopes for weeks on SPB and weeks in the study did not show significant variance across subjects to warrant them being analyzed as random effects). Thus, given the study design (Fig. 4C), the SPB group was 16 wk on SPB (sampled at study weeks 4 and 16) and the placebo group was 12 wk on SPB (sampled at study week 16). We found a significant linear effect of both weeks on SPB and time (weeks in the study) on relative H2AFY mRNA abundance detectable in blood (Fig. 4D). The number of weeks a participant was treated with SPB was associated with decreasing relative H2AFY mRNA abundance, with P = 0.0275. By contrast, time alone (weeks) in the study (adjusting for drug treatment) was associated with increasing H2AFY abundance, with P = 0.0577, canceling each other out to some extent when they occurred at the same time.
Thus, H2AFY levels mark the pharmacodynamic response to treatment with SPB, a drug known to suppress mutant huntingtin-induced neurodegeneration in mouse models of HD (33), in the brain of the widely used R6/2 mouse model as well as in the blood of patients with HD enrolled in a randomized, double-blind, placebo-controlled, phase II clinical trial.
Collectively, our data indicate that H2AFY expression is robustly, specifically, and repeatedly associated with HD in four case–control studies of 472 samples and on two gene expression platforms (Table S2) and marks the early stages of disease severity in human and mouse brains. Linking H2AFY levels to disease activity, reduction of H2AFY levels in a mouse model of HD correlates with known disease modification. Potentially important for accelerating drug development, although current clinical parameters are not powered to detect treatment effects in small trials (14), in a phase II clinical trial, this simple H2AFY blood assay detected a molecular response to a drug known to ameliorate huntingtin-induced neurodegeneration in model systems.
Discussion
The biomarker validation process involves several validation studies, each adding information and confidence to a biomarker. This process replaces the idea of absolute “validation” with a scientific understanding of degrees of certainty in various dimensions (39). In principle, the scientific assessment of a biomarker progresses through different phases of assay development and study design, advancing from discovery to small- to medium-scale cross-sectional studies [which we termed phase I biomarker studies (14)], to large-scale prospective studies (phase II biomarker studies), to clinical phase III trials of biomarker and disease-modifying drug combinations (combined phase III clinical and biomarker trial). This involves a careful and step-wise process of biomarker development similar to the phases of drug development.
Our study should be interpreted keeping several limitations in mind. First, although we assessed a substantial number of cases and controls across four case–control studies and a randomized, placebo-controlled, phase II trial, additional well-powered patient populations will need to be evaluated to confirm the generalizability of these observations (14). It will be particularly important to confirm whether H2AFY robustly tracks HD activity over time in a much larger study population.
Second, lurking bias is a key threat to all biomarker studies (14, 40), and a panel of 12 RNA biomarkers for HD highlighted in a previous small study (7) was not replicated (8). We have attempted to minimize bias by adhering to rigorous proposed “rules of evidence” in study design, conduct, and interpretation (40). We established quality control measures for sample collection, processing, and storage, and we monitored for potentially confounding demographic and sample handling variables (41) and included disease controls. Samples were predominantly collected and processed in parallel for cases and controls using one predefined standard operating procedure for both cases and controls. Case and control samples were assayed in parallel. Controls were recruited from the source population (largely unaffected spouses of patients with HD or spouses of patients with other neurodegenerative diseases receiving care at Partners HealthCare). Despite these efforts, cross-sectional studies may remain vulnerable to bias from confounders that we did not recognize. The randomized, placebo-controlled, phase II trial also included in this study, however, follows a study design thought to be least susceptible to bias (40). Third, substantial sex-related (8) and diurnal rhythm-related changes in blood gene expression (42) have been observed. In our analysis, we thus used multivariate analyses adjusted for sex and age, and blood draws of cases and controls were performed predominantly in the afternoon to guard against diurnal bias in gene expression. Fourth, genome-wide expression studies like those we conducted in the discovery phase are vulnerable to false-positive results as a result of overfitting. We have made several provisions to guard against overfitting. We used a simple analytical procedure that adjusts for multiple testing utilizing a permutation-based FDR, and such simple analytics may be less vulnerable to overfitting. Most importantly, we specifically reproduced the association of elevated relative H2AFY mRNA abundance with HD in three case–control studies on the qPCR platform, thus eliminating the possibility of a false-positive result attributable to computational overfitting. Fifth, in contrast to assays that quantify absolute analyte levels, both the microarray and qPCR technology estimate relative mRNA abundance by comparing a target gene's abundance with a reference measure but do not directly inform on absolute changes in mRNA levels.
It has become clear that chromatin structure and function are regulated not only through posttranslational modification of histones, such as acetylation, but through specialized replacement histones (43). Such alterations contribute to a dynamic composition of the nucleosome and allow specific regions of the chromatin fiber to have specialized functions (43). Excitingly, the H2AFY-encoded atypical histone variant macroH2A1 that is pervasively overexpressed in HD is emerging as a hallmark of discrete heterochromatic foci and as a dynamic regulator of chromatin plasticity and gene repression (44). Although mutant huntingtin has long been thought to induce aberrant posttranslational histone modifications, the robust and specific association between H2AFY and HD seen in situ in human blood and brain and in vivo in a variety of HD models hints at a potentially powerful new link between the specialized histone H2AFY and the dysfunctional chromatin architecture of HD. This view and the precise mechanistic role of H2AFY in HD can now be thoroughly investigated.
If prospectively and mechanistically confirmed, H2AFY may become a biomarker of disease activity and therapeutic response useful for prioritizing lead compounds for phase III clinical trials and for bridging the “valley of death” between preclinical drug discovery and clinical drug trials.
Materials and Methods
Genome-Wide Expression Analysis.
Microarray experiments were carried out as we have previously described (15). Briefly, 4 μg of total RNA was used for cDNA synthesis and transcribed in vitro to cRNA labeled with biotin-11-UTP (PerkinElmer), followed by cRNA purification using an RNeasy Mini kit (Qiagen). Purified cRNA was hybridized to Human Genome U133A arrays (Affymetrix). The arrays were washed, stained, and scanned on an HP Gene Array scanner (Affymetrix).
Real-Time qPCR.
Real-time qPCR in human tissues was performed similar to what we have previously described (15). The TaqMan Assay-on-demand (Applied Biosystems) probe for H2AFY was designed according to the manufacturer's “rules” (primer/probe information is available on request). The ribosomal gene RPL13 was used to control for input RNA. qPCR using an Applied Biosystems 7900HT Fast Real-Time PCR System was performed according to the manufacturer's protocols. Samples were loaded in duplicate, and no-template, no-reverse transcriptase, and plate-to-plate controls were included. The comparative threshold cycle method was used for analysis. Equal amplification efficiencies were confirmed for the gene of interest and reference gene in the range of input amounts observed in this study. qPCR studies in mice were done using D-LUX fluorogenic primers (Invitrogen) and a Bio-Rad iCycler system according to the manufacturers’ protocols. β-Actin was used to control for input RNA.
A detailed description of clinical studies, human biospecimens, animal studies, RNA/histone extraction, Western blotting, immunohistochemistry, and biostatistics can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Marian DiFiglia and Antonio Valencia for 140-CAG knock-in mouse brain tissues. This study was funded by National Institutes of Health Grants P01NS058793 (to C.R.S., S.M.H., H.D.R., and R.J.F.), R01NS064155 (to C.R.S.), R21NS060227 (to C.R.S.), P01NS045242 (to S.M.H. and H.D.R.), and R01NS042861 (to H.D.R.); the Maximillian E. & Marion O. Hoffman Foundation (to C.R.S.); the RJG Foundation (to C.R.S.); the Huntington's Disease Society of America (HDSA) Coalition for the Cure (to S.M.H. and V.C.); and the HDSA New England Center for Excellence for Huntington Disease (to S.M.H. and H.D.R.).
Footnotes
Conflict of interest statement: C.R.S. has served as a consultant to Link Medicine Corporation and was a scientific collaborator of DiaGenic in a study funded by the Michael J. Fox Foundation. He is listed as coinventor on US patent/applications held by the Brigham and Women's Hospital relating to diagnostics in neurodegenerative diseases. S.M.H. is a scientific collaborator with the Novartis Institutes for Biomedical Research. He is listed as coinventor on US patent/applications held by the Massachusetts General Hospital relating to therapeutics and diagnostics in neurodegenerative diseases.
Data deposition: Gene expression data for the eight patients with Huntington disease and the six age- and sex-matched healthy controls in this paper have been submitted to the Gene Expression Omnibus, www.ncbi.nlm.nih.gov/geo (accession no. GSE24250). Gene expression data for the 105 individuals with Parkinson disease and controls were previously reported as part of our Parkinson disease biomarker discovery efforts and are available under accession no. GSE6613.
*This Direct Submission article had a prearranged editor.
See Commentary on page 16867.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104409108/-/DCSupplemental.
References
- 1.Sugars KL, Rubinsztein DC. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003;19:233–238. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- 2.van der Burg JMM, Björkqvist M, Brundin P. Beyond the brain: Widespread pathology in Huntington's disease. Lancet Neurol. 2009;8:765–774. doi: 10.1016/S1474-4422(09)70178-4. [DOI] [PubMed] [Google Scholar]
- 3.Panov AV, et al. Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 4.Maglione V, et al. The platelet maximum number of A2A-receptor binding sites (Bmax) linearly correlates with age at onset and CAG repeat expansion in Huntington's disease patients with predominant chorea. Neurosci Lett. 2006;393:27–30. doi: 10.1016/j.neulet.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 5.Stoy N, et al. Tryptophan metabolism and oxidative stress in patients with Huntington's disease. J Neurochem. 2005;93:611–623. doi: 10.1111/j.1471-4159.2005.03070.x. [DOI] [PubMed] [Google Scholar]
- 6.Luthi-Carter R, et al. Dysregulation of gene expression in the R6/2 model of polyglutamine disease: Parallel changes in muscle and brain. Hum Mol Genet. 2002;11:1911–1926. doi: 10.1093/hmg/11.17.1911. [DOI] [PubMed] [Google Scholar]
- 7.Borovecki F, et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proc Natl Acad Sci USA. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Runne H, et al. Analysis of potential transcriptomic biomarkers for Huntington's disease in peripheral blood. Proc Natl Acad Sci USA. 2007;104:14424–14429. doi: 10.1073/pnas.0703652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi LM, et al. MAQC Consortium The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scherzer CR, Jensen RV, Gullans SR, Feany MB. Gene expression changes presage neurodegeneration in a Drosophila model of Parkinson's disease. Hum Mol Genet. 2003;12:2457–2466. doi: 10.1093/hmg/ddg265. [DOI] [PubMed] [Google Scholar]
- 11.Scherzer CR, et al. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch Neurol. 2004;61:1200–1205. doi: 10.1001/archneur.61.8.1200. [DOI] [PubMed] [Google Scholar]
- 12.Zheng B, et al. PGC-1α, a potential therapeutic target for early intervention in Parkinson's disease. Sci Transl Med. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schadt EE, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scherzer CR. Chipping away at diagnostics for neurodegenerative diseases. Neurobiol Dis. 2009;35:148–156. doi: 10.1016/j.nbd.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scherzer CR, et al. Molecular markers of early Parkinson's disease based on gene expression in blood. Proc Natl Acad Sci USA. 2007;104:955–960. doi: 10.1073/pnas.0610204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scherzer CR, et al. GATA transcription factors directly regulate the Parkinson's disease-linked gene alpha-synuclein. Proc Natl Acad Sci USA. 2008;105:10907–10912. doi: 10.1073/pnas.0802437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Angelov D, et al. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell. 2003;11:1033–1041. doi: 10.1016/s1097-2765(03)00100-x. [DOI] [PubMed] [Google Scholar]
- 19.Mermoud JE, Costanzi C, Pehrson JR, Brockdorff N. Histone macroH2A1.2 relocates to the inactive X chromosome after initiation and propagation of X-inactivation. J Cell Biol. 1999;147:1399–1408. doi: 10.1083/jcb.147.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyen CM, et al. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol Cell Biol. 2006;26:1156–1164. doi: 10.1128/MCB.26.3.1156-1164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burke JR, et al. Huntingtin and DRPLA proteins selectively interact with the enzyme GAPDH. Nat Med. 1996;2:347–350. doi: 10.1038/nm0396-347. [DOI] [PubMed] [Google Scholar]
- 22.Bae B-I, et al. Mutant huntingtin: Nuclear translocation and cytotoxicity mediated by GAPDH. Proc Natl Acad Sci USA. 2006;103:3405–3409. doi: 10.1073/pnas.0511316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vonsattel JP, et al. Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Selemon LD, Rajkowska G, Goldman-Rakic PS. Evidence for progression in frontal cortical pathology in late-stage Huntington's disease. J Comp Neurol. 2004;468:190–204. doi: 10.1002/cne.10938. [DOI] [PubMed] [Google Scholar]
- 25.Beal MF, Ferrante RJ. Experimental therapeutics in transgenic mouse models of Huntington's disease. Nat Rev Neurosci. 2004;5:373–384. doi: 10.1038/nrn1386. [DOI] [PubMed] [Google Scholar]
- 26.Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington's disease with 140 CAG repeats. J Comp Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 27.Rising AC, et al. Longitudinal behavioral, cross-sectional transcriptional and histopathological characterization of a knock-in mouse model of Huntington's disease with 140 CAG repeats. Exp Neurol. 2011;228:173–182. doi: 10.1016/j.expneurol.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhn A, et al. Mutant huntingtin's effects on striatal gene expression in mice recapitulate changes observed in human Huntington's disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet. 2007;16:1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- 29.Lin CH, et al. Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum Mol Genet. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 30.Sadri-Vakili G, Cha JHJ. Mechanisms of disease: Histone modifications in Huntington's disease. Nat Clin Pract Neurol. 2006;2:330–338. doi: 10.1038/ncpneuro0199. [DOI] [PubMed] [Google Scholar]
- 31.Sadri-Vakili G, et al. Histones associated with downregulated genes are hypo-acetylated in Huntington's disease models. Hum Mol Genet. 2007;16:1293–1306. doi: 10.1093/hmg/ddm078. [DOI] [PubMed] [Google Scholar]
- 32.Ferrante RJ, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington's disease mice. J Neurosci. 2003;23:9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardian G, et al. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington's disease. J Biol Chem. 2005;280:556–563. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- 34.Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 35.Li XN, et al. Phenylbutyrate and phenylacetate induce differentiation and inhibit proliferation of human medulloblastoma cells. Clin Cancer Res. 2004;10:1150–1159. doi: 10.1158/1078-0432.ccr-0747-3. [DOI] [PubMed] [Google Scholar]
- 36.Ryu H, et al. Sodium phenylbutyrate prolongs survival and regulates expression of anti-apoptotic genes in transgenic amyotrophic lateral sclerosis mice. J Neurochem. 2005;93:1087–1098. doi: 10.1111/j.1471-4159.2005.03077.x. [DOI] [PubMed] [Google Scholar]
- 37.Collins AF, et al. Oral sodium phenylbutyrate therapy in homozygous beta thalassemia: A clinical trial. Blood. 1995;85:43–49. [PubMed] [Google Scholar]
- 38.Mercuri E, et al. Randomized, double-blind, placebo-controlled trial of phenylbutyrate in spinal muscular atrophy. Neurology. 2007;68:51–55. doi: 10.1212/01.wnl.0000249142.82285.d6. [DOI] [PubMed] [Google Scholar]
- 39.Woodcock J. A framework for biomarker and surrogate endpoint use in drug development, US Food and Drug Administration. 2004. Available at www.fda.gov/ohrms/dockets/ac/04/.../2004-4079S2_03_Woodcock.ppt. Accessed on November 4, 2010.
- 40.Ransohoff DF. Rules of evidence for cancer molecular-marker discovery and validation. Nat Rev Cancer. 2004;4:309–314. doi: 10.1038/nrc1322. [DOI] [PubMed] [Google Scholar]
- 41.Hennecke G, Scherzer CR. RNA biomarkers of Parkinson's disease: Developing tools for novel therapies. Biomarkers in Medicine. 2008;2:41–53. doi: 10.2217/17520363.2.1.41. [DOI] [PubMed] [Google Scholar]
- 42.Whitney AR, et al. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci USA. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 44.Timinszky G, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.