Abstract
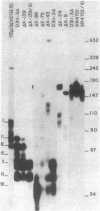
The 5' termini of yeast CYC1 RNA molecules have been mapped, by nuclease S1 digestion of mRNA . DNA duplexes, to seven locations from 29 to 93 base pairs upstream from the initiating ATG codon. When the CYC1 gene is introduced into yeast in plasmid YEp13, substantially the same transcripts are made. Using this system to study in vivo gene expression, we measured the capacity of enzymatically produced DNA deletions to form the normal set of RNAs. Four regions of 5'-flanking DNA were identified as functional. Sequences within the region -242 to -139 are required for maximal CYC1 transcript formation; their deletion reduces transcription by a factor of 15 but does not change the pattern of 5' ends observed. Deletion of the sequence between -242 and -99 does not further change the overall transcript level but does affect the specificity of CYC1 mRNA starting. A deletion that extends from -242 to -75 causes both an additional shift in the pattern of 5' ends observed and a further large decrease (factor of 10--20) in CYC1 RNA level. Deletions that extend from -242 to -43, and particularly two deletions that extend still closer to the initiating ATG, cause the appearance of an abundant transcript which starts upstream of position -1078 and of minor transcripts starting in the region -325 to -245.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenmeier E. H., Brown D. D., Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978 Nov;15(3):1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Claisse M. L., Péré-Aubert G. A., Clavilier L. P., Slonimski P. P. Méthode d'estimation de la concentration des cytochromes dans les cellules entières de levure. Eur J Biochem. 1970 Nov;16(3):430–438. doi: 10.1111/j.1432-1033.1970.tb01098.x. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Fowlkes D. M., Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980 Nov;22(2 Pt 2):405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski R. A., Clarkson S. G., Kurjan J., Hall B. D., Smith M. Mutations of the yeast SUP4 tRNATyr locus: transcription of the mutant genes in vitro. Cell. 1980 Nov;22(2 Pt 2):415–425. doi: 10.1016/0092-8674(80)90352-9. [DOI] [PubMed] [Google Scholar]
- Kressmann A., Clarkson S. G., Pirrotta V., Birnstiel M. L. Transcription of cloned tRNA gene fragments and subfragments injected into the oocyte nucleus of Xenopus laevis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1176–1180. doi: 10.1073/pnas.75.3.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressmann A., Hofstetter H., Di Capua E., Grosschedl R., Birnstiel M. L. A tRNA gene of Xenopus laevis contains at least two sites promoting transcription. Nucleic Acids Res. 1979 Dec 11;7(7):1749–1763. doi: 10.1093/nar/7.7.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Hall B. D., Gillam S., Smith M. Identification and isolation of the yeast cytochrome c gene. Cell. 1978 Jul;14(3):673–680. doi: 10.1016/0092-8674(78)90250-7. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K., Hall B. D., Astell C., Smith M. A position effect in the control of transcription at yeast mating type loci. Nature. 1981 Jan 22;289(5795):244–250. doi: 10.1038/289244a0. [DOI] [PubMed] [Google Scholar]
- Petrochilo E., Verdiere J. A glycine to serine substitution identifies the CYP 3 locus as the structural gene of iso 2 cytochrome c in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1977 Nov 21;79(2):364–371. doi: 10.1016/0006-291x(77)90166-8. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalit P., Loughney K., Olson M. V., Hall B. D. Physical analysis of the CYC1-sup4 interval in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Mar;1(3):228–236. doi: 10.1128/mcb.1.3.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Sherman F. Deletions of the iso-1-cytochrome c and adjacent genes of yeast: discovery of the OSM1 gene controlling osmotic sensitivity. Genetics. 1978 Aug;89(4):653–665. doi: 10.1093/genetics/89.4.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M., Leung D. W., Gillam S., Astell C. R., Montgomery D. L., Hall B. D. Sequence of the gene for iso-1-cytochrome c in Saccharomyces cerevisiae. Cell. 1979 Apr;16(4):753–761. doi: 10.1016/0092-8674(79)90091-6. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Kédinger C., Corden J., Brison O., Chambon P. Specific in vitro initiation of transcription on conalbumin and ovalbumin genes and comparison with adenovirus-2 early and late genes. Nature. 1980 Jun 5;285(5764):367–373. doi: 10.1038/285367a0. [DOI] [PubMed] [Google Scholar]