Abstract
Known mechanisms of resistance to β-lactam antibiotics include β-lactamase expression, altered drug target, decreased bacterial permeability, and increased drug efflux. Here, we describe a unique mechanism of β-lactam resistance in the biothreat organism Burkholderia pseudomallei (the cause of melioidosis), associated with treatment failure during prolonged ceftazidime therapy of natural infection. Detailed comparisons of the initial ceftazidime-susceptible infecting isolate and subsequent ceftazidime-resistant variants from six patients led us to identify a common, large-scale genomic loss involving a minimum of 49 genes in all six resistant strains. Mutational analysis of wild-type B. pseudomallei demonstrated that ceftazidime resistance was due to deletion of a gene encoding a penicillin-binding protein 3 (BPSS1219) present within the region of genomic loss. The clinical ceftazidime-resistant variants failed to grow using commonly used laboratory culture media, including commercial blood cultures, rendering the variants almost undetectable in the diagnostic laboratory. Melioidosis is notoriously difficult to cure and clinical treatment failure is common in patients treated with ceftazidime, the drug of first choice across most of Southeast Asia where the majority of cases are reported. The mechanism described here represents an explanation for ceftazidime treatment failure, and may be a frequent but undetected resistance event.
The β-lactam antibiotics are widely used to treat community and healthcare-associated infections, and the emergence and dissemination of antimicrobial resistance to this family of drugs represents a significant threat to human health (1). Numerous resistance mechanisms have been described, including expression of drug-destroying enzymes such as β-lactamases (2, 3), altered drug targets such as conformational changes in penicillin-binding proteins (PBPs) (2), decreased bacterial permeability (2), and increased drug efflux (4). Defining the basis for resistance mechanisms is fundamental to surveillance, control of infection, and effective antimicrobial use.
Clinical treatment failure occurs in 11–17% of patients receiving ceftazidime for melioidosis (5), a severe Gram-negative infection caused by the biothreat agent Burkholderia pseudomallei (6). Ceftazidime represents the first-line therapy for melioidosis across much of Asia where most cases are reported. The basis for treatment failure is not known, but current evidence suggested that this did not result from antimicrobial resistance, because primary resistance to ceftazidime was reported to occur at a frequency of <0.2% (7), and secondary resistance was thought to be extremely rare and limited to isolated cases (8, 9). This, together with the observation that B. pseudomallei may become quiescent for many years in the human host between exposure and clinical manifestations of disease, has deflected the search for the basis of treatment failure away from drug resistance and toward other possibilities, such as dormancy and the presence of biofilm.
Here, we provide evidence that individuals with melioidosis caused by a ceftazidime-susceptible B. pseudomallei isolate who fail ceftazidime therapy may harbor a variant with high-level ceftazidime resistance. Of major clinical significance, we found that the phenotype rendered this almost undetectable in the diagnostic laboratory, because the variant had a marked growth defect and failed to grow using commonly used laboratory culture media, including commercial blood cultures. We present evidence to indicate that such variants arise in vivo. Detailed characterization of variants from six different patients combined with mutational analysis in wild-type B. pseudomallei led us to identify the mechanism of drug resistance, which involved the complete deletion of a penicillin-binding protein 3 gene associated with a large-scale genomic loss of at least 49 genes.
Results
Detection of Growth-Defective B. pseudomallei Associated with Antimicrobial Treatment Failure.
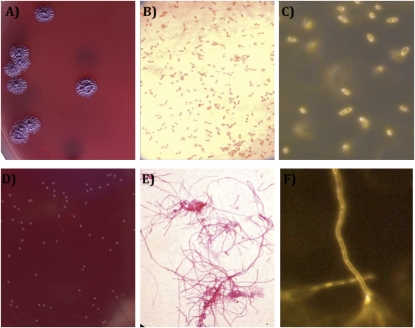
A patient presented to a hospital in northeast Thailand in 2006 with multiple abscesses in the spleen, an aspirate from which grew ceftazidime-susceptible B. pseudomallei. This isolate had an unremarkable Gram stain, colony morphology appearance, and growth characteristics on routine laboratory media (Fig. 1 A and B). Ceftazidime therapy was commenced on the day of admission. Despite spleen removal on day 19 and continuation of ceftazidime therapy, the patient remained febrile over the next 2 wk. A sample of drain fluid from the surgical wound taken on day 36 was culture negative on nonselective medium (blood agar), but a selective medium (Ashdown agar [ASH]) (10), used by a handful of laboratories worldwide for the isolation of B. pseudomallei from samples taken from the environment or colonizing body sites), supported the growth of a few pinpoint colonies after 48 h of incubation at 37 °C in air (Fig. 1D). This morphology is highly atypical for B. pseudomallei, which is described as “cornflower head” (purple and rugose) on ASH. Gram stain of the tiny colonies revealed long, Gram-negative filaments (Fig. 1E). Bacterial identification was not possible using biochemical tests or commercial kits because of the slow growth rate, but an agglutination test based on a monoclonal antibody that is highly specific to B. pseudomallei exopolysaccharide (11) was positive. The patient became afebrile after antimicrobial therapy was changed to standard oral therapy for melioidosis [trimethoprim-sulfamethoxazole (TMP-SMX) plus doxycycline], and was discharged home on day 53.
Fig. 1.
Comparison of the appearance of an initial ceftazidime-susceptible B. pseudomallei strain 415a and the ceftazidime-resistant variant strain 415e isolated from the same patient after prolonged ceftazidime therapy. Colony morphology (A and D), Gram stain and light microscopy (B and E), and unstained appearance by real-time microscopy (C and F) of initial (A–C) and variant strain (D–F). Colony morphology was observed after spread-plating on Ashdown agar and incubation for 4 d at 37 °C in air. Gram stain was observed through a 40× objective. Real-time microscopy was performed using an RTM-3 at 1,000× magnification.
Two similar cases were identified in 2007, which led us to examine historical laboratory records for further cases and to identify a further three cases based on a note by the technician of nutritional variants, suggesting that this is a recurring clinical event. These six patients had initial clinical manifestations that were entirely consistent with melioidosis, and had received prolonged ceftazidime therapy (median 26.5 d, range 18–36 d) before detection of the variant. Such protracted parenteral antibiotic therapy is not uncommon in our patient population with melioidosis. Two of the six patients died, which is comparable to the mortality rate from melioidosis of 43% in our hospital.
Bacteriological Analysis of Initial and Variant B. pseudomallei Isolates.
We recovered all six initial and variant B. pseudomallei isolates from frozen stocks, repeated bacterial identification tests, and determined their growth characteristics on commonly used laboratory media. All 12 strains were confirmed to be B. pseudomallei. The six initial isolates had typical growth characteristics and colonial appearances on tryptone soya agar, Columbia agar, B. cepacia agar, Mueller Hinton agar (MHA), blood agar, Luria–Bertani (LB) agar, and ASH after 48 h of incubation at 37 °C in air. These isolates also grew well in tryptone soya broth (TSB), Mueller Hinton broth (MHB), and commercial blood culture bottles (BacT/Alert FA). In contrast, the six variant B. pseudomallei strains failed to show signs of growth after subculture onto any of the solid media over a more protracted time course of 7 d incubation at 37 °C in air, with the exception of ASH for which pinpoint colonies comparable to those shown in Fig. 1D were visible by day 2 for all six strains. The six variants failed to grow in TSB or BacT/Alert FA blood culture bottles, and appeared to lyse as determined on days 1, 2, 3, and 7 of incubation by the absence of cloudy culture, lack of organisms on Gram stain, and lack of growth after subculture onto ASH.
Gram stain of the six initial B. pseudomallei isolates performed after 48 h of incubation was unremarkable after growth on all of the culture media tested. Gram stain of the six variant strains grown on ASH after 48 h of incubation revealed that all were Gram-negative filaments of comparable appearance. RTM-3 microscopy (which allows visualization of live bacteria in the absence of stains or fixatives) of the six initial strains demonstrated motile bacilli (Fig. 1C), whereas the six variant strains were nonmotile filaments with an appearance consistent with the presence of septa in the absence of cell division (Fig. 1F).
Variant B. pseudomallei Are Highly Resistant to Ceftazidime.
The clinical history of the six patients led us to suspect that the variants might be resistant to ceftazidime. We determined the minimum inhibitory concentration (MIC) of the first isolate from each of the six patients. For MHA-grown bacteria, the MICs ranged from 1.0 to 1.5 μg/mL (susceptible breakpoint ≤8 μg/mL) (12). The MIC of the parental isolates using a nonstandard ASH-based method described in Methods (used as it supported growth of the variant isolates) ranged between 4 and 6 μg/mL, whereas all six variant strains were highly resistant to ceftazidime with an MIC >256 μg/mL.
MICs were also determined using E-test methodology for other antimicrobial drugs in common use for the treatment of melioidosis. The six initial isolates were tested on MHA, and the six initial and six variant strains were tested using the modified ASH method. All six initial isolates were susceptible to imipenem, amoxicillin-clavulanic acid (AMC), doxycycline, and TMP-SMX. The MIC values of the variant strains to imipenem, AMC, doxycycline, and TMP-SMX on ASH were comparable to those for the initial strains on ASH.
Ceftazidime-Resistant B. pseudomallei Variants Arose in Vivo.
We hypothesized that variant strains arose in vivo from the infecting isolate, and compared the bacterial genotype of the initial isolate and the variant from each of the six patients using multilocus sequence typing (MLST) (13). Within-patient strain pairs shared the same sequence type (ST), suggesting that the variant arose from the infecting isolate during the course of infection. The possibility that the variant was present in the founder inoculum cannot be excluded, but this is a less plausible explanation because the variant had a marked growth defect on laboratory media and would be predicted to have a major fitness disadvantage in the environment where human infection is acquired.
We considered whether the variant arose in a single, possibly atypical lineage or whether this was more widespread in the B. pseudomallei population. MLST data indicated that each patient was infected with a different ST (ST 17, 174, 497, 498, 309, and 183 for patients 1–6, respectively). Each ST differed from the other five by at least three of the seven loci, with the exception of ST 17 and ST 309, which differed at a single locus. This finding indicates that the variant has arisen in multiple independent genetic lineages.
Known Mechanisms of Ceftazidime Resistance in B. pseudomallei Are Not Implicated.
Ceftazidime resistance in clinical B. pseudomallei isolates have been reported to be due to genetic alterations in penA (BPSS0946; Pro167Ser or Cys69Tyr according to Ambler's numbering system) (14) associated with ceftazidime MICs of 64 and 256 μg/mL, respectively (15, 16). A single-nucleotide change in penA (BPSS0946, strain K96243) leading to a Pro167Ser substitution was also the basis for ceftazidime resistance generated in the laboratory using B. pseudomallei strain LH-1 (17). The colony morphology and Gram stain appearance of these resistant strains were not reported to be unusual. We amplified and sequenced the penA gene for the initial and variant strains; this failed to reveal any de novo genetic alterations between paired strains (SI Text), suggesting a resistance mechanism in the variant strains that was independent of penA.
Exposure of B. pseudomallei strain KHW to sublethal concentrations of ceftazidime in vitro has been reported to induce bacterial filamentation that was reversible upon drug removal but resulted in a permanent increase in ceftazidime MIC from 1 to 4 μg/mL (18). We reproduced the development of filamentation following exposure to ceftazidime in vitro for all six initial strains, together with reversion to Gram-negative bacilli on removal of drug. We were unable, however, to replicate the postexposure increase in ceftazidime resistance following removal of the drug. Colony morphology on ASH was unchanged after drug exposure. We concluded that this experiment did not provide an adequate explanation for the high-level resistance in the six variant strains.
Ceftazidime Resistance Is Associated with a Large Genomic Deletion.
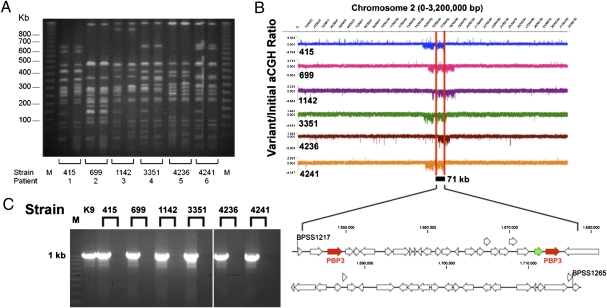
Serial passage (seven consecutive subcultures) of the six variant strains on ASH did not demonstrate reversion to wild-type colony or microscopic morphologies, suggesting the presence of an irreversible genetic alteration. To elucidate this, we analyzed the variant strains using two complementary genomic approaches: pulsed-field gel electrophoresis (PFGE) (19) and array-based comparative genomic hybridization (aCGH) (20). On PFGE, we observed distinct shifts in banding patterns between initial and variant strain pairs (Fig. 2A). In four of six strains, the loss of a 150-kb band was observed in the variant strains, suggesting the presence of a recurrent large-scale genomic deletion. This putative genomic deletion was investigated using aCGH in which the initial and variant strains were profiled on oligonucleotide arrays providing >97% tiling coverage of the entire nonrepetitive B. pseudomallei K96243 reference genome. A comparison of the initial and variant strain pairs revealed that all six variant strains exhibited genomic losses in a specific region of chromosome 2, the size of the loss ranging from 145 to 309 kb in individual variant strains (Fig. 2B). No genomic loss was observed for chromosome 1. By intersecting the regions of genomic loss observed in individual strains, we identified a minimal common region (MCR) of deletion comprising 49 genes (71 kb, BPSS1217–1265; Fig. 2B and Table S1). Other than this 71-kb region, no other recurrent regions of genomic loss were identified in the variant strains relative to parental controls.
Fig. 2.
Variant ceftazidime-resistant B. pseudomallei strains exhibit a recurrent genomic deletion in chromosome 2. (A) PFGE banding patterns of paired B. pseudomallei strains from six patients in whom the infecting isolate developed secondary ceftazidime resistance during therapy. M represents lambda ladder. (B) Variant B. pseudomallei strains exhibited a common region of genomic loss in chromosome 2. (Upper) Chromosome 2 aCGH (variant/initial) signals for all six B. pseudomallei pairs. A region of deletion was identified as a drop in the aCGH signal. Each variant strain exhibited a regional deletion of chromosome 2 compared with the initial strain. The 71-kb minimal common region (MCR) of deletion is demarcated by red lines. (Lower) Genomic organization of the 49 genes in the 71-kb MCR region. The two PBP 3 homologs (BPSS1219 and BPSS1240) are red, and BPSS1239 (carboxypeptidase) is green. (C) PCR validation of aCGH results. PCR primers designed to amplify a 1-Kb region in the MCR were used to amplify the cognate region in initial and variant strains. M, molecular marker; the 1-kb size range is indicated. Strains are ordered as follows: K9 (BpK96243 reference strain) and the six pairs (initial followed by variant).
To validate the microarray data, we used PCR primers to amplify a 1-kb region predicted to lie within the MCR in the six variant strains (BPSS1245–1246). As shown in Fig. 2C, PCR amplification products were successfully obtained for all six initial strains, but not the variant strains, indicating the presence of this region in the former but not the latter. These results provide further evidence for the recurrent genomic loss in chromosome 2 associated with ceftazidime-resistant variant strains.
The genomic losses observed in the variant strains may represent a true deletion or, alternatively, this region may have been replaced in the variant strains by an unrelated region of novel genetic material (20). To distinguish between these possibilities, we used PCR primers targeting regions flanking the predicted regions of deletion in each strain and attempted to amplify products across the deleted region. No amplification products were obtained when these primers were applied to the initial strains, most likely because of the large size of the intervening region (145–309 kb). In contrast, amplification products of the predicted size were obtained for all six variants, and subsequent DNA sequencing analysis confirmed that the deleted regions were devoid of novel genetic material (Fig. S1). Analysis of the flanking sequences did not identify any distinctive motifs, such as sequence repeats, associated with the breakpoints. These observations suggest that the most likely mechanism for the generation of the deletions is random recombination, selected for by the presence of ceftazidime.
Ceftazidime-Resistant B. pseudomallei Have Lost Three Penicillin-Binding Protein Genes.
We surmised that loss of one or more of the genes in the deleted region were associated with ceftazidime resistance. Bioinformatic curation of the 49 genes present in the MCR identified three potential candidate genes: BPSS1219, BPSS1239, and BPSS1240. Both BPSS1219 and BPSS1240 encode penicillin-binding proteins (PBPs) of the PBP 3 family. These PBPs are specifically involved in septal peptidoglycan synthesis. The K96243 genome encodes three PBP 3 proteins, with the additional homolog, BPSL3031, present on the large chromosome (21). The three homologs, BPSS1219, BPSS1240, and BPSL3031, are 35.6%, 34.8%, and 37.7% identical to the E. coli PBP 3 protein (22). The two PBP 3 proteins on the small chromosome are 46.7% identical to each other, and 47.5% (BPSS1219) and 39.3% (BPSS1240) identical to the large chromosome PBP 3 protein. The deleted region also contained a gene (BPSS1239) that encodes a putative d-alanyl-d-alanine carboxypeptidase that belongs to the penicillin-binding-protein 5/6 family. The K96243 genome contains another homolog of this family of penicillin-binding proteins (BPSL0408) that is 33.5% identical at the amino acid level to BPSS1239.
Comparative genomic analysis with other Burkholderia genomes showed that BPSS1219 had orthologs in other members of the pseudomallei group (B. mallei and B. thailandensis), Burkholderia cepacia complex (B. multivorans and B. cenocepacia), and more distantly related Burkholderia species (B. vietnamiensis and B. ambifaria, B. glumae, B. xenovorans, B. phymatum, and B. phytofirmans), whereas BPSS1239 and BPSS1240 only had orthologs with other members of the pseudomallei group. We hypothesized that BPSS1219 encoded a core function, whereas BPSS1239 and BPSS1240 may represent accessory PBPs.
Loss of PBP 3 Encoded by BPSS1219 Is Responsible for Ceftazidime Resistance and the Growth-Impaired Phenotype.
A genetic approach in a wild-type background was used to demonstrate a direct link between ceftazidime resistance, a growth-impaired phenotype, and loss of function of one or more of the deleted PBP 3-encoding genes (BPSS1219, BPSS1239, and BPSS1240). Single-deletion mutants defective in BPSS1239 (Bp276) or BPSS1240 (Bp307) and a ΔBPSS1239 ΔBPSS1240 double mutant (Bp308) were readily created in B. pseudomallei strain 1026b (a ceftazidime-susceptible clinical strain originating from Thailand). These three mutants did not show an altered growth phenotype or Gram stain appearance, and remained susceptible to ceftazidime with no change in MIC value. In contrast, BPSS1219 could not be mutated despite numerous attempts and different selection parameters, indicating that this gene was essential under the experimental conditions used. A strategy was developed to determine the essentiality of this gene (Fig. 3A). A copy of BPSS1219 was cloned into a mini-Tn7 vector where it was under control of the B. thailandensis ribosomal s12 gene promoter and flanked by loxP sites. Strain Bp483 was obtained by integration of this Tn7 construct into chromosome 1 of strain 1026b. The resident copy of BPSS1219 on chromosome 2 could then be readily deleted from Bp483, resulting in the BPSS1219 haploid strain Bp561. The rescue copy of BPSS1219 was then deleted from Bp561 by Cre recombinase-mediated excision to create the BPSS1219 null mutant Bp560.
Fig. 3.
Deletion of BPSS1219 from B. pseudomallei strain 1026b. (A) Deletion of the BPSS1219 gene from chromosome 2 (Chr 2) was achieved in several steps. In step 1, a mini-Tn7 element containing the BPSS1219 gene flanked by loxP sites and expressed from a B. thailandensis s12 promoter (Ps12) was inserted at the glmS2 attTn7 site on chromosome 1 (Chr 1), resulting in Bp483 (1026b::mini-Tn7-Ps12-BPSS1219+ cBPSS1219+). In step 2, the resident chromosomal BPSS1219 gene (cBPSS1219) was deleted from Chr 2 using a gene replacement method yielding Bp561(1026b::mini-Tn7-Ps12-BPSS1219+ ΔcBPSS1219). In step 3, the BPSS1219 rescue copy was deleted using Cre recombinase-mediated excision, resulting in Bp560 (1026b::mini-Tn7-ΔBPSS1219 ΔcBPSS1219). (B) Growth curves of 1026b (red), Bp561 (magenta), Bp483 (green), and Bp560 (blue) of bacteria in LB medium.
Growth curves of Bp560 in LB medium revealed complete growth attenuation (Fig. 3B), and the cells were filamentous on Gram stain. Filamentation and lack of growth were not due to expression of BPSS1219 from the constitutively expressed Tn7 copy, because Bp483 and Bp561 cells grew well in LB medium (Fig. 3B) and looked normal in a Gram stain. Whereas 1026b was susceptible to ceftazidime (MIC = 3 μg/mL), Bp560 was highly ceftazidime resistant (MIC >256 μg/mL). Ceftazidime susceptibility (MIC = 3 μg/mL) was the same in strains 1026b and Bp562, the BPSS1219 null mutant Bp560 expressing BPSS1219 from a complementing mini-Tn7 element integrated at glmS3. The MICs for other drugs, including AMC, TMP-SMX, and doxycycline, were not substantially affected by deletion of and/or complementation with BPSS1219. A notable exception was the imipenem MIC, which was not affected by constitutive BPSS1219 expression in 1026b (0.75 μg/mL), but dropped >10-fold in Bp560 (0.064 μg/mL) and was restored in Bp562 after complementation with BPSS1219 (MIC = 0.5 μg/mL).
Despite different degrees of fitness and clumping of the ceftazidime-resistant (MIC >256 μg/mL) clinical variants 415e, 699d, and 1142b isolates in LB medium, expression of BPSS1219 from a chromosomally integrated mini-Tn7 element strain 1026b in these growth-impaired strains restored growth rates that were comparable to those observed in the corresponding primary isolates 415a, 699c, and 1142b. This near-normal growth phenotype was accompanied by restoration of ceftazidime susceptibility (MIC = 1.5–3 μg/mL on MHA).
From these data, we conclude that deletion of BPSS1219 was responsible for the severe growth defect and the ceftazidime-resistance phenotype of the variant strains.
Discussion
We have described a unique mechanism of antimicrobial resistance involving the entire deletion of a PBP 3 gene in B. pseudomallei. Defining the candidate gene responsible for the resistance phenotype was made more challenging by the finding that ceftazidime resistance was associated with a minimum region of deletion of 49 genes. Using a mutagenesis approach we were able to unequivocally demonstrate, however, that the loss of PBP 3 encoded by BPSS1219 was responsible. The filamentous appearance on Gram stain and poor growth of the BPSS1219 defective mutant was consistent with the characteristics of the clinical ceftazidime-resistant variants and with E. coli following inactivation of PBP 3, which has been reported to result in inhibition of cell division and growth into long filaments (23, 24). Also compatible with our findings is that in other Gram-negative bacilli, including E. coli and P. aeruginosa, ceftazidime owes its antibacterial activity to a high affinity for PBP 3 (25). We consider it unlikely that these strains will find their way back into the environment and spread through the bacterial population because of the growth defect in laboratory media, and the resulting likely fitness disadvantage.
Small-colony variants of B. pseudomallei with high-level resistance to ceftazidime have been recovered previously from experimental animals and in vitro cultures exposed to antibiotics (26). Several features suggest a different mechanism for resistance to the one described here, including an increase in MIC to various unrelated classes of antibiotics, no change in PFGE banding pattern, and reversion to wild type. Small-colony variants have also been described in other organisms associated with chronic disease, including those caused by P. aeruginosa (predominantly in people with cystic fibrosis) and Staphylococcus aureus (27, 28), but the mechanism described here appears to be unique.
We postulate that the ability of the ceftazidime-resistant variants to grow (albeit slowly) on ASH but not on other routinely used agar or in liquid broth may be related to osmotic effects and bacterial lysis, with growth on ASH being supported by the presence of 4% glycerol. Evidence for this belief comes from the observation that variants demonstrated no growth in TSB or LB, but slow growth in TSB or LB supplemented with 4% glycerol. No other phenotypic trait was observed in association with the absence of other genes in the large genomic deletion in the variant strains, although it is highly likely that this impacts numerous bacterial functions. For example, the minimum region of deletion included numerous genes involved in metabolic pathways, as well as several transporters and regulators. It is not clear why such a large genomic region is deleted from these variants as opposed to a more limited genetic change, such as target mutation via a smaller deletion or point mutation associated with an equivalent resistant phenotype, and it is possible that there is an additional biological advantage associated with loss of one or more other genes in this region. The finding that two of the six patients described here died indicates that untreated infection with a residual population of ceftazidime-resistant variants may still be lethal, and the clear history of treatment failure and lack of clinical improvement until antimicrobial therapy was changed in all six cases is consistent with the bacteria's ability to persist in the human host despite an apparent lack of fitness in vitro.
We were unable to identify a common mechanism associated with deletion in the six variant strains studied, but propose that this was due to random recombination. Natural, large-scale deletion of genomic material in B. pseudomallei has been reported once in a rare, gentamicin-susceptible strain (a phenotype which occurs in 1 in 1,000 clinical isolates) that was found to have undergone a deletion of a region of >130 kb, including the amrAB-oprA operon, which encodes an efflux pump (29). This finding suggests that gene deletion may be a mechanism associated with adaptation and evolution of B. pseudomallei as a result of selective pressure. The six resistant strains in our study belonged to several phylogenetic lineages, suggesting that this genetic event is a generic property of the species rather than an obscurity in a rare genetic background.
An important but unanswered question is whether the process of PBP 3 deletion and development of secondary resistance to ceftazidime is a more frequent cause of antimicrobial failure in patients with melioidosis than the number of cases detected in our study would suggest. Clinical treatment failure occurs in up to one-sixth of cases (5), and we suggest that the six cases reported here represent the tip of the iceberg. Samples from normally sterile sites from patients who are recultured in response to antimicrobial therapy are not routinely plated onto ASH, and variants in these cases would be missed. This report explains failure of therapy for melioidosis, and though it remains to be seen whether this is the only bacterial mechanism, we postulate that it may be a major cause.
Methods
Bacterial Strains and Culture Media.
B. pseudomallei strains used in this study other than the clinical isolates described elsewhere in the text are listed in Table S2. Culture media used for genetic constructs is described in SI Text.
Culture and Microscopy.
Identification of the six primary B. pseudomallei isolates from admission cultures was performed using standard methodology (30) (details are presented in SI Text). The variants were identified using a highly specific latex agglutination test based on a monoclonal antibody to exopolysaccharide (11), because the growth defect precluded the use of standard methodology. Growth of the six strain pairs was tested on tryptone soya agar, Columbia agar, B. cepacia agar, Mueller Hinton agar, blood agar (all from Oxoid, Ltd.), and in Luria–Bertani (LB) agar (Becton Dickinson), tryptone soya broth, Mueller Hinton broth, and BacT/Alert FA blood culture bottles (AB bioMèrieux). A Gram stain was performed on aliquots of each liquid culture at 24, 48, and 72 h, and day 7, and an aliquot of each liquid culture was streak inoculated onto ASH at these time points, incubated at 37 °C in air, and examined daily for 7 d. Real-time microscopy (RTM-3; Richardson Technologies, Inc.) was performed on colonies picked from ASH after incubating at 37 °C in air for 48 h and observed at 1,000× magnification. Bacterial growth curves were performed using microtiter plates and a Synergy HT Multi-Mode Microplate Reader (BioTek). Plates were incubated at 37 °C with constant shaking at 200 rpm, and the optical density at 600 nm was read every 30 min for 48 h.
Antimicrobial Susceptibility Testing.
MICs for the six initial B. pseudomallei isolates were determined for ceftazidime, imipenem, AMC, TMP-SMX, and doxycycline using the E-test method (AB bioMérieux) according to the manufacturer's instructions. The six variant strains did not grow on MHA (the medium used for the E test), and a nonstandard method was developed on ASH. B. pseudomallei was subcultured on ASH and incubated for 2 d at 37 °C in air. Several colonies were picked and suspended in normal saline and adjusted based on OD to an approximate bacterial concentration of 1 × 108 cfu/mL; this was spread-plated onto the agar surface using a sterile cotton swab. An E-test strip was applied and the plate incubated at 37 °C in air for 18 h before reading MIC values.
In Vitro Induction and Reversion of Bacterial Filamentation.
The six primary ceftazidime-susceptible B. pseudomallei isolates from study patients (415a, 699c, 1142a, 3351c, 4236a, and 4241a) were tested for in vitro induction of filamentation following exposure to ceftazidime following a previously described method detailed in S1 Text.
Bacterial Genotyping.
MLST of B. pseudomallei was performed as described previously (13). The alleles at each of the seven loci were assigned and STs defined using the B. pseudomallei MLST Web site (http://www.mlst.net/). PFGE of B. pseudomallei was performed using SpeI and the banding patterns analyzed using BioNumerics software version 2.5 (Applied Maths) as described previously (19).
High-Resolution Comparative Genomic Hybridization Array (aCGH).
aCGH experiments were performed using oligonucleotide microarrays comprising >390,000 50-mer probes providing tiling coverage of the Bp K96243 reference genome (15-bp overlap) (NimbleGen) (20). Arrays were probed with labeled genomic DNA fragments from parental and variant B. pseudomallei strains as described in SI Text. For each parental vs. variant comparison, reciprocal dye-swap experiments were performed and the resulting signals averaged. Normalized log2 ratios of aCGH signals for parental over variant strains were visualized in SignalMap. The microarray data has been deposited into the Gene Expression Omnibus (accession no. GSE25832). Bioinformatic analysis is described in SI Text.
PCR, Sequencing, and Genetic Constructs.
PCR primer sequences for DNA amplification and sequencing are listed in Table S2. The methods used for construction and integration of a BPSS1219 expression construct and deletion of BPSS1239 and BPSS1240 are described in SI Text.
Supplementary Material
Acknowledgments
Funding for this work was provided by the Wellcome Trust (N.C., V.W., D.L., P.A., A.T., N.P.D., and S.J.P.). N.C. is the recipient of the Wellcome Trust Career Development in Public Health and Tropical Medicine Grant 087769/Z/08/Z. Funding was also provided by an institutional grant from the Genome Institute of Singapore, an institute of the Agency for Science, Technology and Research (B.S., H.H.C., W.F.O. and P.T.), National Institutes of Health National Institute of Allergy and Infectious Diseases Grant AI065357 (to H.P.S. and D.A.R.), and the National Institute for Health Research Cambridge Biomedical Research Center (S.J.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE25832).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111020108/-/DCSupplemental.
References
- 1.Woodford N, Livermore DM. Infections caused by Gram-positive bacteria: A review of the global challenge. J Infect. 2009;59(Suppl 1):S4–S16. doi: 10.1016/S0163-4453(09)60003-7. [DOI] [PubMed] [Google Scholar]
- 2.Livermore DM. Mechanisms of resistance to cephalosporin antibiotics. Drugs. 1987;34(Suppl 2):64–88. doi: 10.2165/00003495-198700342-00007. [DOI] [PubMed] [Google Scholar]
- 3.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: A clinical update. Clin Microbiol Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chierakul W, et al. Two randomized controlled trials of ceftazidime alone versus ceftazidime in combination with trimethoprim-sulfamethoxazole for the treatment of severe melioidosis. Clin Infect Dis. 2005;41:1105–1113. doi: 10.1086/444456. [DOI] [PubMed] [Google Scholar]
- 6.Wiersinga WJ, van der Poll T, White NJ, Day NP, Peacock SJ. Melioidosis: Insights into the pathogenicity of Burkholderia pseudomallei. Nat Rev Microbiol. 2006;4:272–282. doi: 10.1038/nrmicro1385. [DOI] [PubMed] [Google Scholar]
- 7.Wuthiekanun V, Peacock SJ. Management of melioidosis. Expert Rev Anti Infect Ther. 2006;4:445–455. doi: 10.1586/14787210.4.3.445. [DOI] [PubMed] [Google Scholar]
- 8.Dance DA, Wuthiekanun V, Chaowagul W, White NJ. The antimicrobial susceptibility of Pseudomonas pseudomallei. Emergence of resistance in vitro and during treatment. J Antimicrob Chemother. 1989;24:295–309. doi: 10.1093/jac/24.3.295. [DOI] [PubMed] [Google Scholar]
- 9.Jenney AW, Lum G, Fisher DA, Currie BJ. Antibiotic susceptibility of Burkholderia pseudomallei from tropical northern Australia and implications for therapy of melioidosis. Int J Antimicrob Agents. 2001;17:109–113. doi: 10.1016/s0924-8579(00)00334-4. [DOI] [PubMed] [Google Scholar]
- 10.Ashdown LR. An improved screening technique for isolation of Pseudomonas pseudomallei from clinical specimens. Pathology. 1979;11:293–297. doi: 10.3109/00313027909061954. [DOI] [PubMed] [Google Scholar]
- 11.Wuthiekanun V, Anuntagool N, White NJ, Sirisinha S. Short report: A rapid method for the differentiation of Burkholderia pseudomallei and Burkholderia thailandensis. Am J Trop Med Hyg. 2002;66:759–761. doi: 10.4269/ajtmh.2002.66.759. [DOI] [PubMed] [Google Scholar]
- 12.Cockerill FR., III . Performance Standards for Antimicrobial Susceptibility Testing, Twentieth Informational Supplement, M100-S20. Vol 30. Wayne, PA: Clinical and Laboratory Standards Inst; 2010. p. No 1. [Google Scholar]
- 13.Godoy D, et al. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol. 2003;41:2068–2079. doi: 10.1128/JCM.41.5.2068-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambler RP, et al. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tribuddharat C, Moore RA, Baker P, Woods DE. Burkholderia pseudomallei class a beta-lactamase mutations that confer selective resistance against ceftazidime or clavulanic acid inhibition. Antimicrob Agents Chemother. 2003;47:2082–2087. doi: 10.1128/AAC.47.7.2082-2087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sam IC, See KH, Puthucheary SD. Variations in ceftazidime and amoxicillin-clavulanate susceptibilities within a clonal infection of Burkholderia pseudomallei. J Clin Microbiol. 2009;47:1556–1558. doi: 10.1128/JCM.01657-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho PL, Cheung TK, Yam WC, Yuen KY. Characterization of a laboratory-generated variant of BPS beta-lactamase from Burkholderia pseudomallei that hydrolyses ceftazidime. J Antimicrob Chemother. 2002;50:723–726. doi: 10.1093/jac/dkf208. [DOI] [PubMed] [Google Scholar]
- 18.Chen K, Sun GW, Chua KL, Gan YH. Modified virulence of antibiotic-induced Burkholderia pseudomallei filaments. Antimicrob Agents Chemother. 2005;49:1002–1009. doi: 10.1128/AAC.49.3.1002-1009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maharjan B, et al. Recurrent melioidosis in patients in northeast Thailand is frequently due to reinfection rather than relapse. J Clin Microbiol. 2005;43:6032–6034. doi: 10.1128/JCM.43.12.6032-6034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sim BM, et al. Genomic acquisition of a capsular polysaccharide virulence cluster by non-pathogenic Burkholderia isolates. Genome Biol. 2010;11:R89. doi: 10.1186/gb-2010-11-8-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holden MT, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura M, et al. On the process of cellular division in Escherichia coli: Nucleotide sequence of the gene for penicillin-binding protein 3. Mol Gen Genet. 1983;191:1–9. doi: 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- 23.Spratt BG. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci USA. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spratt BG. Temperature-sensitive cell division mutants of Escherichia coli with thermolabile penicillin-binding proteins. J Bacteriol. 1977;131:293–305. doi: 10.1128/jb.131.1.293-305.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayes MV, Orr DC. Mode of action of ceftazidime: Affinity for the penicillin-binding proteins of Escherichia coli K12, Pseudomonas aeruginosa and Staphylococcus aureus. J Antimicrob Chemother. 1983;12:119–126. doi: 10.1093/jac/12.2.119. [DOI] [PubMed] [Google Scholar]
- 26.Häussler S, Rohde M, Steinmetz I. Highly resistant Burkholderia pseudomallei small colony variants isolated in vitro and in experimental melioidosis. Med Microbiol Immunol (Berl) 1999;188:91–97. doi: 10.1007/s004300050110. [DOI] [PubMed] [Google Scholar]
- 27.Proctor RA, et al. Small colony variants: A pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol. 2006;4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 28.Häussler S, Tümmler B, Weissbrodt H, Rohde M, Steinmetz I. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin Infect Dis. 1999;29:621–625. doi: 10.1086/598644. [DOI] [PubMed] [Google Scholar]
- 29.Trunck LA, et al. Molecular basis of rare aminoglycoside susceptibility and pathogenesis of Burkholderia pseudomallei clinical isolates from Thailand. PLoS Negl Trop Dis. 2009;3:e519. doi: 10.1371/journal.pntd.0000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dance DA, Wuthiekanun V, Naigowit P, White NJ. Identification of Pseudomonas pseudomallei in clinical practice: Use of simple screening tests and API 20NE. J Clin Pathol. 1989;42:645–648. doi: 10.1136/jcp.42.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.