Abstract
The non-canonical NF-κB pathway is an important arm of NF-κB signaling that predominantly targets activation of the p52/RelB NF-κB complex. This pathway depends on the inducible processing of p100, a molecule functioning as both the precursor of p52 and a RelB-specific inhibitor. A central signaling component of the non-canonical pathway is NF-κB-inducing kinase (NIK), which integrates signals from a subset of TNF receptor family members and activates a downstream kinase, IκB kinase-α (IKKα), for triggering p100 phosphorylation and processing. A unique mechanism of NIK regulation is through its fate control: the basal level of NIK is kept low by a TRAF-cIAP destruction complex and signal-induced non-canonical NF-κB signaling involves NIK stabilization. Tight control of the fate of NIK is important, since deregulated NIK accumulation is associated with lymphoid malignancies.
Keywords: NF-κB, non-canonical NF-κB, alternative NF-κB, p100, NIK, IKK, TRAF
Introduction
NF-κB forms a family of transcription factors that participates in various biological processes, including immune response, inflammation, cell growth and survival, and development 1, 2. Mammalian NF-κB family is composed of five members, including RelA (also named p65), RelB, c-Rel, NF-κB1 p50, and NF-κB2 p52, which form various dimeric complexes that transactivate numerous target genes via binding to the κB enhancer. The NF-κB proteins are normally sequestered in the cytoplasm by a family of inhibitors, including IκBα and other related ankyrin repeat-containing proteins. NF-κB1 and NF-κB2 are translated as precursor proteins, p105 and p100, which contain an IκB-like C-terminal portion and function as NF-κB inhibitors. Proteasome-mediated processing of p105 and p100 not only produces the mature NF-κB1 and NF-κB2 proteins (p50 and p52) but also results in disruption of the IκB-like function of these precursor proteins 3.
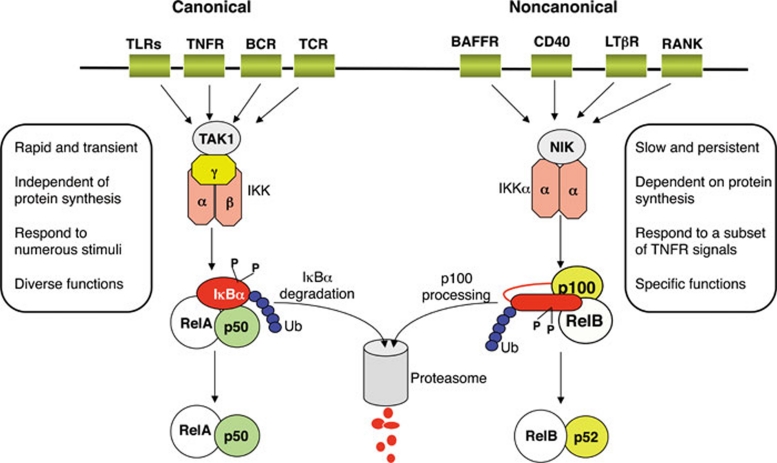
Canonical NF-κB pathway of NF-κB activation relies on inducible degradation of IκBs, particularly IκBα, leading to nuclear translocation of various NF-κB complexes, predominantly the p50/RelA dimer 1, 2 (Figure 1). The degradation of IκBα is mediated through its phosphorylation by the IκB kinase (IKK), a trimeric complex composed of two catalytic subunits, IKKα and IKKβ, and a regulatory subunit, IKKγ (also named NF-κB essential modulator or NEMO). In addition to this well-defined canonical pathway, other mechanisms exist to mediate activation of more specific NF-κB members 3. In particular, a non-canonical NF-κB pathway activates the RelB/p52 NF-κB complex using a mechanism that relies on the inducible processing of p100 instead of degradation of IκBα (Figure 1). Genetic evidence suggests that this NF-κB pathway regulates important biological functions, such as lymphoid organogenesis, B-cell survival and maturation, dendritic cell activation, and bone metabolism 4. Moreover, deregulated non-canonical NF-κB signaling is associated with lymphoid malignancies. Therefore, better understanding of the mechanism regulating non-canonical NF-κB activation has important therapeutic values. It is increasingly clear that this pathway of NF-κB activation differs significantly from the canonical NF-κB pathway in its signaling mechanisms. This review will focus on the molecular mechanisms by which non-canonical NF-κB signaling pathway is regulated under physiological and pathological conditions.
Figure 1.
Canonical and non-canonical NF-κB signaling pathways. Canonical pathway is triggered by numerous signals, including those mediated by innate and adaptive immune receptors. It involves activation of IKK complex by Tak1, IKK-mediated IκBα phosphorylation, and subsequent degradation, resulting in rapid and transient nuclear translocation of the prototypical NF-κB heterodimer RelA/p50. Non-canonical NF-κB pathway relies on phosphorylation-induced p100 processing, which is triggered by signaling from a subset of TNFR members. This pathway is dependent on NIK and IKKα, but not on the trimeric IKK complex, and mediates the persistent activation of RelB/p52 complex.
Inducible p100 processing: a central step of non-canonical NF-κB signaling
The discovery of non-canonical NF-κB signaling pathway came from the study of p100 processing 5. In addition to serving as the precursor of p52, p100 functions as an IκB-like molecule, preferentially inhibiting RelB nuclear translocation 6. Thus, the processing of p100 serves to both generate p52 and induce the nuclear translocation of the RelB/p52 heterodimer 7, 8. In contrast to the constitutive and cotranslational processing of p105 9, the processing of p100 is tightly regulated by both positive and negative domains 5. In most cell types, p100 is the predominant product of nfκb2, suggesting a lack of active processing of this precursor 10. Similarly, overexpressed p100 is barely converted to p52 in mammalian cells, as opposed to the constitutive production of p50 from p105 5, 9. However, p52 is actively generated in specific cell types, such as B cells, leading to the idea that p100 processing might be a signal-regulated event. Indeed, the NF-κB-inducing kinase (NIK) induces p100 processing in transfected cells and is required for in vivo p100 processing in splenocytes 5. Moreover, endogenous p100 processing can be induced by various receptor signals in a NIK-dependent manner 7, 11, 12, 13, 14.
Regulation by site-specific p100 phosphorylation
The C-terminal region of p100 (p100C) has a so-called NIK-responsive domain (Figure 2), since it is essential for NIK-induced p100 processing 5. This region of p100 contains two serine residues, S866 and S870, which resemble the phosphorylation site of IκBα 15. Mutation of one or both of these serines completely abolished the inducible processing of p100 5, 16. Initial in vitro kinase assays, using NIK immune complexes isolated from transfected HEK 293 cells, identified these two serines as potential phosphorylation sites of p100 5. This finding was later on confirmed by immunoblotting assays using phospho-specific anti-p100 antibodies 16. In both NIK-transfected 293 cells and signal-induced B cells and fibroblasts, the serines 860 and 870 of endogenous p100 are strongly phosphorylated. As seen with the induction of p100 processing 11, 17, the signal-induced p100 phosphorylation is dependent on de novo protein synthesis 16, and the potential underlying mechanism will be discussed in a following section.
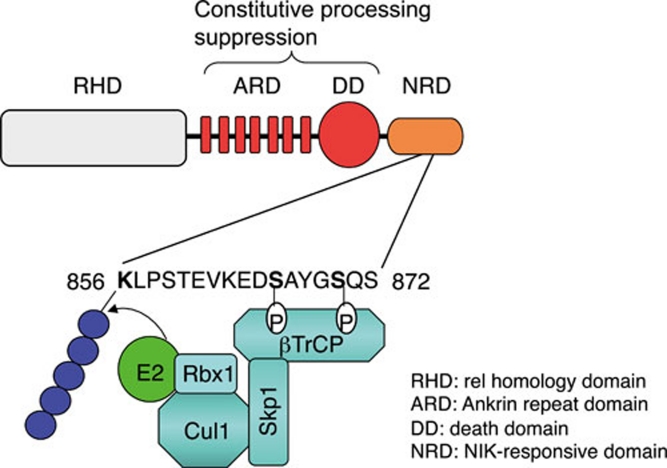
Figure 2.
Positive and negative domains regulating p100 processing. The tight control of p100 processing requires its DD as well as ARD, which serve as negative regulatory domains. The NRD, responsible for p100 inducible processing, contains a phospho-degron that is phosphorylated by IKKα and bound by βTrCP of the SCFβTrCP ubiquitin ligase complex.
Regulation by ubiquitination
NIK-induced p100 processing is associated with its ubiquitination 5. The amino-acid sequence of the p100 phoshorylation site resembles the binding sequence of βTrCP 15, substrate-binding subunit of the SCFβTrCP ubiquitin ligase 18. NIK induces the binding of βTrCP to p100, which is dependent on the two conserved serine phosphorylation residues, serines 866 and 870 (Figure 2). In vitro binding assays using phospho-peptides further confirmed that phosphorylation of the conserved serine residues within the phoshorylation site of p100 creates a binding site for βTrCP 16. Consistent with these findings, βTrCP knockdown by RNAi attenuates NIK-induced p100 ubiquitination and processing, thus establishing SCFβTrCP as a ubiquitin ligase mediating the inducible processing of p100 15. A lysine (K) residue, K856, located upstream of the phosphorylation site of p100 serves as the ubiquitin acceptor site 19 (Figure 2). This location, upstream of and adjacent to the phosphorylation residues of p100, is analogous to the ubiquitination site (K22) of IκBα 20. Mutation of K856 of p100 attenuates its inducible ubiquitination and processing 19.
The post-ubiquitination events involved in p100 processing are poorly understood. Proteasome recognition of ubiquitinated proteins is generally mediated by ubiquitin receptor proteins located in the base of the 19S regulatory particle 21. Interestingly, p100 also interacts with a protein, S9 (also known as PSMD11), located in the lid of the 19S regulatory particle 22. The binding of S9 to p100 is greatly promoted by NIK and dependent on p100 ubiquitination. However, S9 does not seem to recognize the ubiquitin chains but rather binds to the death domain (DD) of p100. It is possible that ubiquitination of p100 may cause a conformational change, thereby facilitating the binding of S9 to the DD. The p100/S9 interaction is important for NIK-induced p100 processing 22, although it is unclear whether this interaction contributes to the proteasome recruitment of p100 or the translocation of p100 to the catalytic chamber of the 20S proteasome particle.
Regulation by sumoylation
Sumoylation is a posttranslational mechanism of protein modification that regulates diverse biological processes, such as protein-protein interaction, protein ubiquitination, protein phosphorylation, and gene transcription 23. A recent study suggests the involvement of p100 sumoylation in the regulation of its ubiquitination and processing 24. In cell lines and MEFs (murine embryonic fibroblasts), a proportion of p100 is constitutively conjugated with SUMO1. Mutation of the putative sumoylation sites of p100 or RNA interference-mediated knockdown of the SUMO-conjugating enzyme Ubc9 attenuates the inducible processing of p100. It appears that the basal sumoylation of p100 is required for its phosphorylation both in vivo and in vitro, although the underlying mechanism is unknown 24. Since protein sumoylation may create binding sites for protein-protein interactions, it is intriguing to examine whether p100 sumoylation facilitates its binding by IKKα or NIK.
Suppression of constitutive processing
As mentioned above, p100 barely undergoes constitutive processing. This tightly regulated nature of p100 is due to the processing-suppressive function of its C-terminal portion 5, 25. This portion contains a processing inhibitory domain (PID) as well as the ankyrin repeat domain (ARD). The PID is notable for covering the sequence that forms a DD (Figure 2). Disruption of either the DD or the ARD leads to constitutive processing of p100 26. As will be discussed later, the presence of a negative regulatory mechanism in p100 processing may be critical for maintaining the normal biological function of this NF-κB precursor. Since p100 functions as an IκB-like molecule, uncontrolled p100 processing would result in both overproduction of p52 and disruption of its IκB function.
Although how the DD and ARD inhibit p100 processing is incompletely understood, the ARD is known to interact with the Rel-homology domain (RHD) that forms the N-terminal portion of p100. It is thus likely that the ARD, possibly with the help of DD, interacts with the N-terminal RHD of p100, thereby forming a three-dimensional structure that prevents constitutive processing of p100. Regarding the underlying mechanism, formation of such a three-dimensional structure would mask the nuclear localization signal of p100, which in turn appears to serve as a critical mechanism that prevents constitutive processing of p100 26. Strong biochemical evidence suggests that nuclear translocation of p100ΔC mutants is required for their constitutive processing 26.
Why do p100ΔC mutants have to be in the nucleus for their constitutive processing? One possibility is that some critical factors mediating constitutive p100 processing are located in the nucleus. For example, a number of ubiquitin ligases, including SCFβTrCP, are predominantly localized in the nucleus 27, 28, 29, 30, 31. Although SCFβTrCP is dispensable for constitutive processing of p100 15, the possibility for the involvement of another nuclear ubiquitin ligase(s) in this molecular event cannot be excluded. It is also likely that the proteasome targeting or processing of p100ΔC mutants requires a nuclear factor. In this regard, a recent study suggests that the constitutive processing of p100ΔC mutants requires their binding to promoter DNA via κB sites 32. This study also reveals that the constitutive p100 processing is initiated by a proteasome-mediated endoproteolytic cleavage at a specific residue (aspartic acid 415) of p100ΔCs. Thus, formation of a stable complex with DNA may promote the exposure of the endoproteolytic site and/or facilitate the insertion of p100 C-terminal portion to the catalytic chamber of the proteasome 32. This idea is also supported by the finding that proteolysis of RelA is triggered by its association with κB-specific promoter DNAs 33.
NIK and IKKα as key non-canonical NF-κB signaling components
The intracellular signaling components mediating non-canonical NF-κB activation differ significantly from those involved in canonical NF-κB activation, which is why most of the canonical NF-κB inducers are incapable of stimulating p100 processing. As mentioned above, the first component of the non-canonical NF-κB signaling pathway to be identified is NIK 5, a MAP kinase kinase kinase (MAP3K) member that was originally implicated in NF-κB activation by the TNF receptor (TNFR) pathway 34. NIK gene mutation in the alymphoplasia (aly) or NIK knockout mice, however, has no obvious effect on TNFα-stimulated NF-κB activation, but completely blocks the processing of p100 5, 35, 36. Consistently, NIK stimulates the phosphorylation, ubiquitination, and processing of p100 in transfected cells 5, 16. To date, all of the non-canonical NF-κB inducers identified so far are known to signal through NIK 7, 11, 12, 13, 14. These findings establish NIK as a signal integrator and, thus, a central component of the non-canonical NF-κB pathway.
Although NIK stimulates p100 phosphorylation in vivo, recombinant NIK is unable to directly phosphorylate p100 in vitro 16, 37. NIK functions through activation of a downstream kinase, IKKα 37. Whereas IKKβ and IKKγ are essential components of the canonical NF-κB pathway, IKKα, but not IKKβ or IKKγ, is required for non-canonical NF-κB signaling 11, 12, 16, 37. Thus, inactivation of IKKα in mice leads to phenotypes that are similar, although not identical, to those seen in the aly mice or NIK knockout mice, including impaired B-cell maturation and lymphoid organogenesis 37, 38. However, how precisely IKKα regulates non-canonical NF-κB signaling is incompletely understood. Compared with NIK, IKKα is much less effective in inducing p100 processing 5. It appears that NIK not only activates IKKα but also promotes the binding of IKKα to its substrate p100 39. It also remains possible that NIK induces additional signaling factor(s) that act(s) cooperatively with IKKα in the induction of effective p100 processing.
In vitro kinase assays reveal that serines 866 and 870 of p100 are surprisingly dispensable for p100C phsophorylation by recombinant IKKα 39. Instead, mutation of serine 872 abolishes IKKα-mediated in vitro phsophorylation of p100C. Although these in vitro studies suggest serine 872 as the phosphorylation site of IKKα, in vivo phosphorylation analyses of p100 using phospho-specific antibodies reveal serines 866 and 870 as the major phosphorylation sites of p100 in both IKKα- and NIK-expressing cells 16. These two serines are also phosphorylated in cells stimulated with the non-canonical NF-κB inducers. It is currently unclear whether IKKα has different specificities under in vitro and in vivo conditions or IKKα stimulates another kinase that phosphorylates serines 866 and 870 in vivo. In any case, since mutation of serine 872 only weakly affects the ubiquitination and processing of p100 16, 39, this in vitro IKKα target site may not be the major functional phosphorylation site of p100. Clearly, precisely how IKKα and NIK induce p100 processing needs to be clarified by additional studies.
Receptors eliciting non-canonical NF-κB signaling
In contrast to the canonical NF-κB signaling pathway, which responds to signals elicited by diverse receptors, the non-canonical NF-κB pathway is targeted by a specific set of receptors 40. To date, the best-characterized non-canonical NF-κB receptors are a subset of TNFR superfamily members, including B-cell-activating factor belonging to TNF family receptor (BAFFR) 11, 13, CD40 7, lymphotoxin β-receptor (LTβR) 12, receptor activator for nuclear factor κB (RANK) 14, TNFR2 41, 42, Fn14 43, 44, etc. These receptors each mediate specific biological roles of the non-canonical NF-κB. A common feature of the non-canonical NF-κB-stimulating receptors is the possession of a TRAF-binding motif, which recruits different TRAF members, particularly TRAF2 and TRAF3, to the receptor complex during ligand ligation 45. The receptor recruitment of these TRAF members is important for triggering their degradation, a critical step leading to the activation of NIK and induction of p100 processing 46.
LTβR
LTβR is a TNFR superfamily member that is expressed in lymphoid stromal and epithelial cells and binds to two different ligands: lymphotoxin and LIGHT (homologous to lymphotoxin, exhibits inducible expression and competes with HSV glycoprotein D for herpes virus entry mediator, a receptor expressed by T lymphocytes) both being primarily expressed in lymphocytes 47. A major function of LTβR is to mediate the development and maintenance of peripheral lymphoid organs. The lack of lymph nodes and Peyer's patches in aly mice, which harbor NIK gene mutation 35, suggests a connection between LTβR and NIK activation. The initial evidence for the involvement of LTβR in NIK activation was provided by a transient transfection study, in which overexpressed LTβR strongly promotes NIK-mediated p100 processing 5. This finding was then confirmed by a separate study demonstrating the induction of NIK- and IKKα-dependent p100 processing in embryonic fibroblasts stimulated through LTβR ligation 12. The cytoplasmic domain of LTβR contains motifs that associate with several TRAF members, including TRAF2, 3, and 5, thereby stimulating both the canonical and non-canonical NF-κB pathways 47. The LTβR-mediated non-canonical NF-κB signaling pathway is required for induction of a number of chemokines, such as SLC, ELC, BLC, and SDF-1a, which are required for organization of lymphoid organs and lymphocyte homing to peripheral lymphoid tissues 12.
BAFFR
The second TNFR family member found to stimulate p100 processing is BAFFR 11, 13, which is predominantly expressed in B cells, and plays an important role in mediating the survival and maturation of peripheral B cells 48. BAFFR differs from many other TNFR superfamily members in that it predominantly activates the non-canonical NF-κB signaling pathway with only weak activity in the induction of canonical NF-κB pathway 11, 49. This unique feature of BAFFR is primarily due to its possession of an atypical TRAF-binding sequence, which interacts with TRAF3 but not with TRAF2 49. Thus, BAFFR crosslinking triggers degradation of TRAF3, but not TRAF2, as opposed to the degradation of both TRAFs by many other TNFRs. As will be discussed in a following section, degradation of TRAF3 is sufficient for triggering non-canonical NF-κB signaling. However, recruitment of TRAF2 is required for activation of the canonical NF-κB pathway. Indeed, a two-amino-acid substitution that converts this atypical TRAF-binding motif of BAFFR to a typical TRAF-binding motif renders BAFFR competent in binding TRAF2 and inducing canonical NF-κB activation 49. BAFFR-mediated induction of p100 processing contributes to the survival of transitional and mature B cells, probably through induction of anti-apoptotic genes like bcl-2 and bcl-x 11.
CD40
CD40 is a TNFR member that is expressed on various cell types, including B cells, dendritic cells, monocytes, endothelial epithelial cells, and neurons 50. The ligand of CD40, CD40L (also known as CD154), is primarily expressed by activated T cells. In the immune system, a major function of CD40 signaling is to regulate B-cell activation and differentiation events, including proliferation and survival of activated B cells, germinal center formation, and antibody isotype switching. Another major function of CD40 is to mediate dendritic cell maturation and antigen presentation. Unlike BAFFR, CD40 elicits strong signals that target both the canonical and non-canonical NF-κB pathways. Upon ligation by CD40L, CD40 interacts via two different TRAF-binding motifs with several TRAF members, including TRAF1, 2, 3, 5, and 6, and this leads to proteolysis of both TRAF2 and TRAF3 51, 52. As mentioned above, the degradation of TRAF2 and TRAF3 represents an important step in the activation of the non-canonical NF-κB signaling pathway 46.
RANK
RANK is best known for its role in osteoclastogenesis, but it also regulates important immune functions, such as dendritic cell survival and lymphoid organogenesis 53. RANK is expressed on osteoclast precursors, dendritic cells, and activated B cells, and in general, RANK signaling promotes cell survival and differentiation. As seen with CD40, the cytoplasmic domain of RANK binds TRAF1, 2, 3, 5, and 6 and mediates activation of both canonical and non-canonical NF-κB signaling pathways. Genetic evidence suggests an essential role for RANK-stimulated activation of non-canonical NF-κB activation in osteoclastogenesis and bone metabolism 14, 54.
Fn14
Fn14 serves as the receptor of TNF-like weak inducer of apoptosis (TWEAK) 55, which is known to activate canonical and non-canonical NF-κB pathways 43, 56. The cytoplasmic domain of Fn14 contains a TRAF-binding motif capable of associating with TRAF1, 2, 3, and 5 55, 56. Like the other non-canonical NF-κB-stimulating receptors, TWEAK/Fn14 binding induces NIK activation through targeting the degradation of TRAF members, particularly TRAF2 57. It has also been suggested that TWEAK may induce the degradation of cIAP1 and cIAP2, major components of a NIK ubiquitin ligase complex 58 (discussed in the following section).
Other receptors
Several other TNFR family members have been shown to induce p100 processing; these include TNFR2 41, 42, CD30 59, 60, and CD27 61. In addition, a toll-like receptor (TLR) member, TLR4, may have a role in non-canonical NF-κB signaling, since its ligand LPS induces p100 processing in a B-cell line 17. However, it is important to note that compared with the typical non-canonical NF-κB stimuli, LPS is a weaker inducer of p100 processing. It is unclear whether this function of LPS is executed through activation of NIK and IKKα or indirectly mediated by inducing the synthesis of a NIK/IKKα activator. Nevertheless, the former possibility is supported by a recent finding that LPS stimulates NIK phoshorylation 62.
Controlling NIK stability as a central point of non-canonical NF-κB signaling
Signal transduction is usually regulated by catalytic activation/inactivation of central signaling components. However, a unique feature of the non-canonical NF-κB signaling pathway is its dependence on the steady level of NIK expression. The level of NIK is controlled largely through its ubiquitin-dependent degradation.
Negative regulation of NIK by TRAF3
Under normal conditions, the steady level of NIK protein is extremely low, which is apparently due to its constant degradation targeted by a ubiquitination-dependent mechanism 63. A major player of this negative regulatory mechanism is TRAF3, which was identified as a NIK-binding protein in a yeast two-hybrid screening 63. TRAF3 interacts with an N-terminal domain of NIK containing a novel sequence motif, ISIIAQA. Through this molecular interaction, TRAF3 targets NIK for constant ubiquitination and proteasomal degradation (Figure 3A). This negative regulatory mechanism may ensure that the steady level of NIK is kept at an extremely low level, thus preventing signal-independent processing of p100. TRAF3 knockdown by RNAi or TRAF3 knockout by gene targeting is sufficient for triggering NIK accumulation and constitutive p100 processing 63, 64. This finding suggests that the signaling function of NIK is primarily regulated through the steady level of this protein, which is in turn subject to control by the negative regulator TRAF3.
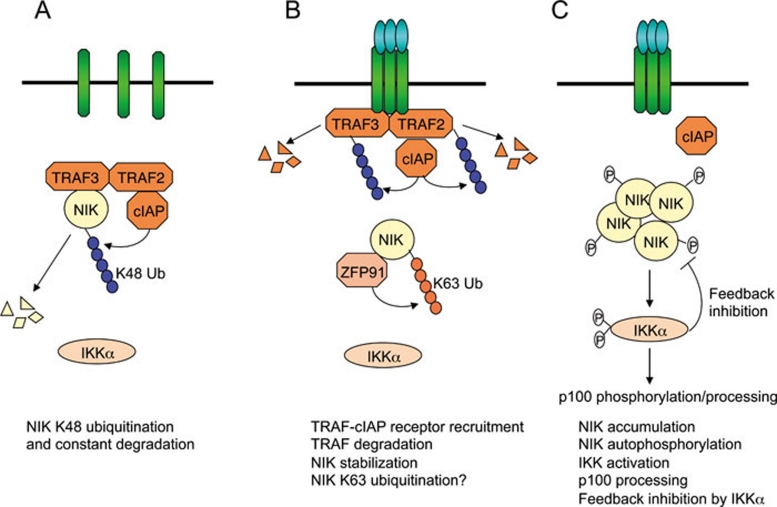
Figure 3.
NIK stabilization as a mechanism of non-canonical NF-κB signaling. (A) Under normal conditions, NIK is bound by TRAF3 and recruited to the cIAP1/2 ubiquitin ligase via TRAF3 dimerization with TRAF2. The T3-T2-cIAP E3 complex mediates constant ubiquitination and proteasomal degradation of NIK, thus preventing non-canonical NF-κB activation. (B) In response to receptor crosslinking, TRAFs and cIAP1/2 are recruited to the receptor, where cIAP1/2 ubiquitinates TRAF2 and TRAF3 and stimulates their degradation. ZFP91 mediates K63 ubiquitination of NIK, which may promote stability and catalytic activity of NIK. (C) Accumulated NIK activates IKKα, which in turn phosphorylates p100, leading to p100 processing. IKKα also phosphorylates NIK to promote NIK degradation, a feedback mechanism that may control the magnitude of NIK activation.
Binding of TRAF3 to the N-terminal domain of NIK is essential for controlling NIK function, since deletion of the core sequence (ISIIAQA) of the TRAF3-binding domain generates a NIK mutant (NIKΔ78-84) that is completely insensitive to TRAF3-mediated negative regulation. This NIK mutant is stably expressed even in the presence of TRAF3, thus functioning as a constitutively active NIK 63. Transgenic mice expressing the TRAF3-uncoupled NIK mutant in B cells display maximal p100 processing, leading to B-cell hyperplasia even in the absence of the BAFFR signal 65.
TRAF3 as a component of a multi-subunit NIK ubiquitin ligase
Although TRAF3 induces NIK ubiquitination and degradation in vivo, TRAF3 has no intrinsic function to catalyze the formation of K48-linked ubiquitin chains, and it was thus speculated that TRAF3 might function as a critical component of an ubiquitin ligase mediating NIK ubiquitination 63. Indeed, several recent studies demonstrate that the NIK ubiquitination involves a multi-subunit ubiquitin ligase complex composed of TRAF3, TRAF2, and cIAP1 (or cIAP2, hereafter named cIAP1/2) 58, 66, 67, 68. Within this NIK ubiquitin ligase complex (hereafter named T3-T2-cIAP E3 complex), TRAF2, but not TRAF3, directly interacts with cIAP1/2. TRAF3, connected to cIAP1/2 via dimerization with TRAF2, serves as an adaptor to recruit this multi-subunit E3 complex to NIK (Figure 3A). Thus, negative control of NIK function requires all three molecular components of the T3-T2-cIAP complex. Notably, genetic deficiencies in TRAF2 or TRAF3, or degradation of cIAP1/2 by specific antagonists, lead to accumulation of NIK and aberrant p100 processing 64, 69, 70. The importance of NIK negative regulation is underscored by the recent finding that genetic deficiencies in the T3-T2-cIAP E3 components or NIK gene amplification are associated with aberrant non-canonical NF-κB activation and B-cell malignancies, particularly multiple myeloma 71, 72.
Receptor-stimulated NIK stabilization
A hallmark of non-canonical NF-κB signaling is the requirement of de novo protein synthesis 11, 16. Although the precise mechanism by which protein synthesis regulates p100 processing remains incompletely understood, emerging evidence suggests the requirement of NIK synthesis and accumulation during the induction of non-canonical NF-κB signaling. An initial study demonstrates that induction of p100 processing in B cells involves persistent degradation of TRAF3, which is coupled with marked increase in NIK steady level 63. The NIK accumulation is obviously a result of both its stabilization and de novo synthesis. Since the level of NIK is extremely low in unstimulated cells, the signal-induced NIK accumulation is likely an essential step for triggering the downstream signaling events in non-canonical NF-κB pathway. In further support of this idea, protein synthesis is also essential for signal-induced p100 phosphorylation 16.
Strong evidence suggests that disruption of the T3-T2-cIAP E3 complex is sufficient for triggering non-canonical NF-κB signaling. Germ line inactivation of either TRAF2 or TRAF3 leads to NIK accumulation and constitutive p100 processing 64, 69, 70. Similar effect is seen in cells treated with the cIAP1/2 antagonists, which induce the degradation of cIAP1 and cIAP2 58, 67. Thus, signal-induced disruption of the T3-T2-cIAP complex is a critical mechanism mediating non-canonical NF-κB activation. This idea is consistent with the finding that receptor-mediated non-canonical NF-κB activation is associated with degradation of TRAF3 and/or TRAF2.
Recent studies have provided important insights into the mechanism by which receptor signals induce the degradation of TRAF2 and TRAF3. A study using BAFFR and CD40 suggests that recruitment of these TRAF members to the receptor complex is essential for their degradation 49. CD40 contains a typical TRAF-binding motif capable of recruiting both TRAF2 and TRAF3, whereas BAFFR contains an atypical TRAF-binding motif that selectively recruits TRAF3. Consequently, ligation of CD40 leads to degradation of both TRAF2 and TRAF3, whereas ligation of BAFFR leads to degradation of only TRAF3. A two-amino-acid substitution to convert the atypical TRAF-binding motif of BAFFR to a typical TRAF-binding motif renders the modified BAFFR capable of recruiting and degrading both TRAF2 and TRAF3 49. More recent studies reveal that the inducible degradation of TRAF2 and TRAF3 is mediated through their ubiquitination by cIAP1 and cIAP2 66. Thus, cIAP1/2 targets NIK for ubiquitination and degradation under unstimulated conditions but redirects its destructive action toward TRAF2 and TRAF3 in response to receptor signals (Figure 3B). A crucial trigger for this substrate switch seems to be the receptor recruitment of cIAP1/2 along with the TRAFs. Within the receptor complex, TRAF2 stimulates the K48-ubiquitin ligase function of cIAP1/2 by mediating conjugation of K63-linked ubiquitin chains to cIAP1 and cIAP2 66. This finding explains why signal-induced degradation of TRAF3 requires TRAF2 73.
Other potential mechanisms of NIK activation
NIK belongs to the family of MAP3Ks that are known to be activated through T-loop phosphorylation. One major remaining question is whether NIK activation is also triggered by its phosphorylation, as seen with other MAP3Ks. This possibility is suggested by the finding that T559 phoshorylation of NIK is required for its activity 74. However, the T559 phosphorylation of NIK is likely mediated by autophosphorylation, which could be triggered through NIK protein accumulation by the mechanism discussed above. On the other hand, emerging evidence indicates that NIK may also be activated by mechanisms independent of its protein accumulation. For example, induction of p100 processing by CD27 involves recruitment of NIK to the receptor complex 61. The receptor recruitment of NIK would increase its local concentration and thus may trigger its autophosphorylation without the need of increased NIK expression. It is possible that such a mechanism of NIK activation occurs during the early phase of non-canonical NF-κB signaling, whereas NIK accumulation may contribute to the sustained non-canonical NF-κB signaling. However, it is also possible that NIK activation by certain receptor signals, or within specific cell types, solely relies on the early phase mechanism. In this regard, a recent study suggests that LPS stimulates NIK phoshorylation without altering its expression level 62. It is unknown whether NIK is recruited to TLR4 or the MyD88 signaling complex. Rapid NIK phosphorylation is also stimulated by the T-cell receptor (TCR)/CD28 signals or T-cell mitogens through a PKCθ-dependent mechanism 75. Since the TCR/CD28 signals and T-cell mitogens do not stimulate an appreciable level of p100 processing 75, 76, it is currently unclear whether TCR-stimulated NIK phosphorylation is linked to weak non-canonical NF-κB signaling or some other functions. An example regarding the latter possibility is that NIK promotes TCR-stimulated STAT3 phosphorylation, thereby regulating the differentiation of the Th17 subset of CD4 T cells 77.
Feedback regulation of NIK by IKKα
A hallmark of the non-canonical NF-κB signaling pathway is its persistent kinetics, which differs from the transient nature of the canonical NF-κB pathway. However, the magnitude of the non-canonical NF-κB signaling appears to be subject to feedback regulation. It is notable that the signal-induced non-canonical NF-κB signaling involves persistent degradation of the T3-T2-cIAP E3 components (TRAF3 and/or TRAF2) 63, 78, although the accumulation of NIK is stopped early on and maintained at a steady level during the non-canonical NF-κB signaling 78. This is largely due to a feedback mechanism of NIK regulation mediated by its downstream kinase, IKKα 78. Upon activation by NIK, IKKα phosphorylates NIK, triggering its proteolysis 78 (Figure 3C). Thus, in IKKα-deficient cells, NIK is continuously accumulated in response to non-canonical NF-κB inducers. The IKKα-mediated feedback regulation of NIK appears to control the magnitude of NIK activation but cannot replace the T3-T2-cIAP E3 complex for controlling the basal level of NIK. This is why disruption of the T3-T2-cIAP E3 complex leads to NIK accumulation and constitutive p100 processing, even in the presence of wild-type IKKα. Thus, NIK stability, and thus non-canonical NF-κB signaling, are controlled by both basal and feedback mechanisms 46.
Novel factors implicated in the regulation of non-canonical NF-κB signaling
To date, the best-characterized components of the non-canonical NF-κB signaling pathway include NIK and IKKα, as well as the T3-T2-cIAP1/2 NIK ubiquitin ligase complex. However, several additional factors have recently been implicated in the regulation of non-canonical NF-κB activation. These novel factors are mostly involved in the regulation of NIK or IKKα.
NIK regulators
One potential negative regulator of NIK is TRAF- and NIK-associated protein (TNAP), which was identified by yeast two-hybrid screen of an adult human brain cDNA library using NIK as bait 79. The isolated TNAP clone encodes the C-terminal 140 amino acids of a truncated protein. TNAP interacts with NIK, TRAF2, and TRAF3, but it does not bind IKKα or IKKβ. Transfected TNAP inhibits the kinase activity of NIK and suppresses the induction of both p100 processing and canonical NF-κB activation. Whether TNAP has a physiological role in regulating canonical or non-canonical NF-κB signaling and how TNAP possibly regulates NIK activity remain unknown. Notably, Blast search reveals significant sequence homology between TNAP and the dynein axonemal heavy chain protein (data not shown). Clearly, additional work is required to further assess the role of this novel protein in NF-κB signaling.
Another negative regulator of NIK is Monarch-1 (also named NLRP12), a member of the NLR/CATERPILLER family of proteins characterized by the possession of nucleotide binding domain and leucine-rich repeats 80. Monarch-1 suppresses CD40-stimulated p100 processing in the human monocytic cell line THP-1 81. Monarch-1 interacts with NIK in a CD40-inducible manner and induces NIK degradation through a proteasome-dependent pathway. Whether the NIK degradation by Monarch-1 involves the T3-T2-cIAP ubiquitin ligase complex is unknown. Furthermore, the physiological role of Monarch-1 in non-canonical NF-κB signaling is yet to be assessed, since the current finding is based on experiments using Monarch-1-transfected THP-1 cells 81.
A positive regulator of NIK, zinc finger protein 91 (Zfp91), has recently been identified through microarray analysis of NF-κB-regulated genes 82. Zfp91 physically associates with NIK, causes stabilization and activation of NIK, and induces p100 processing under overexpression conditions 82. Zfp91 induces conjugation of K63-linked ubiquitin chains to NIK, which is associated with NIK phosphorylation at threonine 559 (T559), an activation loop phosphorylation site of NIK required for its catalytic activation 74 (Figure 3B). Although Zfp91 lacks a typical E3 domain, it appears to have intrinsic ubiquitin ligase function, since purified Zfp91 catalyzes NIK ubiquitination in vitro 82. It is currently unclear whether Zfp91-induced NIK ubiquitination directly triggers its kinase activity or promotes NIK activation indirectly through inhibition of NIK degradation. How Zfp91 regulates non-canonical NF-κB signaling function in vivo also awaits gene-targeting studies, although an essential role of Zfp91 in CD40-mediated NIK activation and p100 processing has been revealed by Zfp91 RNA interference experiments 82.
Another potential non-canonical NF-κB signaling component is MALT1, a para-caspase initially identified as a component of antigen receptor-stimulated canonical NF-κB pathway 83, 84. Unlike its essential role in TCR signaling, MALT1 is largely dispensable for canonical NF-κB activation by BCR 84. Interestingly, MALT1 has recently been shown to regulate BAFFR-mediated p100 processing in B cells 85. MALT1 physically interacts with TRAF3 and appears to induce degradation of this negative regulator of NIK. It appears that MALT1 functions as a scaffold that promotes cIAP1/2-mediated TRAF3 degradation upon BAFFR stimulation.
Bcl10, a partner protein of MALT1, has also been found to regulate non-canonical NF-κB signaling in LPS-stimulated human colonic epithelial cells based on studies using Bcl10 RNA interference 62. In contrast to other regulators, Bcl10 does not regulate the level of NIK but rather promotes NIK phosphorylation. Furthermore, mice expressing an Emu-driven Bcl10 transgene exhibit constitutive non-canonical, as well as canonical, NF-κB activity in B cells 86. However, the activation of non-canonical NF-κB, in this case, appears to be due to overproduction of the BAFFR ligand, BAFF, which in turn induces the activation of non-canonical NF-κB. Thus, Bcl10 may not be an intrinsic regulator of the non-canonical NF-κB pathway.
IKKα regulators
Based On overexpression experiments in cancer cell lines, a STAT family member, STAT3, was shown to induce p100 processing through activation of IKKα 87. This function of STAT3 requires its acetylation by the CBP/p300 acetyltransferase. Whereas this finding is interesting, it is currently unclear whether endogenous STAT3 plays a role in mediating receptor-stimulated p100 processing or the constitutive p100 processing occurring in cancer cells. It is also unknown whether STAT3 directly activates IKKα or acts indirectly through inducing the expression of a non-canonical NF-κB-stimulating factor.
A recent study identified specific microRNAs, miR-223, miR-15a, and miR-16, as negative regulators of IKKα 88. During the differentiation of human monocytes to macrophages, the expression level of these microRNAs is considerably decreased, leading to heightened IKKα expression and p52 generation from p100. In addition to IKKα stimulation, the monocyte differentiation also causes stabilization of NIK, although the underlying mechanism is unclear.
Nuclear regulators of p52/RelB
Strong evidence suggests that non-canonical NF-κB contributes to the induction of specific genes, such as several chemokines (SLC, BLC, ELC, SDF1) that are involved in lymphoid organogenesis 12. The molecular mechanism governing the specificity of gene transcription by non-canonical NF-κB is elusive. It has been suggested that the promoter region of the non-canonical NF-κB target genes contains κB sites that are preferentially recognized by the non-canonical NF-κB dimer RelB/p52 89. However, this idea was later on challenged by an in vitro study, suggesting similar DNA-binding specificity of canonical and non-canonical NF-κB members 90. Another possibility is the involvement of chromatin remodeling factors in the specific control of non-canonical NF-κB. In this regard, a recent study suggests that RelB/p52 is linked to the SWI/SNF chromatin remodeling complex via an adaptor protein, requiem 91. In response to lymphotoxin stimulation, requiem forms a large complex with the SWI/SNF catalytic subunit, Brm, and RelB/p52, which is recruited to the promoter of the BLC gene. Consistently, both requiem and Brm are required for lymphotoxin-stimulated BLC gene expression. This finding suggests that the gene-specific function of RelB/p52 may involve recruitment of SWI/SNF by specific adaptors, such as requiem.
Deregulation of non-canonical NF-κB signaling
The processing of p100 regulates important physiological functions, including the survival and maturation of B cells, development of peripheral lymphoid organs, thymic deletion of autoimmune T cells, and bone metabolism. Thus, deregulated activation of the non-canonical NF-κB is associated with severe disorders, such as autoimmunity, inflammation, and osteoporosis. More importantly, aberrant non-canonical NF-κB signaling contributes to the development of lymphoid malignancies. The deregulation of non-canonical NF-κB signaling can be due to genetic alterations of the nfκb2 gene, which lead to structural changes in p100, or modulation of the non-canonical signaling components by pathogens.
Nfκb2 chromosomal translocations and loss of p100 PID
The importance of p100 in preventing abnormal lymphocyte growth was first suggested by the finding that nfκb2 gene is involved in chromosomal translocations in some lymphomas 92. A common feature of the rearranged nfκb2 gene products is the lack of C-terminal PID of p100. In some cases, the C-terminal region is replaced with a heterologus gene product. As predicted from the p100 truncation studies, these rearranged nfκb2 gene products undergo constitutive processing 5. It is likely that the deregulated processing of p100, along with additional mechanisms, contribute to the development of lymphoma. The pathological consequence of p100 disruption has also been revealed by a study using nfκb2 knockin mice that express p52 in the absence of p100 93. These mice display marked gastric and lymphoid hyperplasia and early postnatal lethality.
Deregulated NIK stabilization and expression
As discussed already, the steady level of NIK is normally low due to its negative regulation by the T3-T2-cIAP ubiquitin ligase complex; thus, mutation of any of the negative regulatory components of NIK can lead to its deregulated accumulation. Recent studies reveal aberrant NIK steady levels in a large proportion of multiple myeloma tumors and cell lines 71, 72, 94. This in turn is due to mutations in TRAF2, TRAF3, cIAP1/2, as well as gain-of-function mutations in CD40 and NIK. Notably, in line with a study using TRAF3 knockout cells 95, the NIK accumulation in multiple myeloma cells leads to activation of both non-canonical and canonical pathways 94. These findings establish NIK as an attractive candidate target for drug therapies in the treatment of multiple myeloma and possibly other cancers.
Deregulated NIK gene transcription may also be a mechanism of its deregulation. It has been shown that the level of NIK mRNA is upregulated in adult T-cell leukemia (ATL) and Hodgkin Reed-Sternberg cells 96. ATL is an acute T-cell malignancy caused by infection of the human T-cell leukemia virus type 1 (HTLV1). As will be discussed below, HTLV Tax protein activates non-canonical NF-κB via a NIK-independent mechanism. However, the deregulation of NIK may be important for constitutive non-canonical NF-κB activation in ATL cells with low or undetectable levels of Tax 96.
Persistent activation by oncogenic viruses and bacteria
The involvement of non-canonical NF-κB in virus-induced tumorigenesis was first suggested by the finding that this pathway is targeted by the HTLV1-encoded oncoprotein Tax 76. As indicated above, HTLV1 is the etiological agent of ATL, which occurs in a small percentage of HTLV1-infected individuals following many years of clinical latency 97. Tax stimulates the activation of both canonical and non-canonical NF-κB pathways, which serves as an essential mechanism for HTLV1-mediated T-cell transformation 98. The activation of non-canonical NF-κB by Tax is particularly unique for HTLV1-infected T cells, since normal T cells usually elicit only the canonical NF-κB pathway upon stimulation by the TCR signal 76. In contrast to the cellular non-canonical NF-κB pathway, Tax induction of p100 processing does not seem to require NIK, since it is insensitive to a dominant-negative NIK mutant. Furthermore, the Tax-specific pathway requires the IKK regulatory subunit IKKγ in addition to IKKα 76. This is because IKKγ functions as an adaptor of Tax and IKKα and is involved in the formation of a Tax-IKKα signaling complex. Formation of this Tax-specific signaling complex also requires physical interaction between Tax and p100. Thus, Tax-stimulated constitutive p100 processing may involve IKKα activation via a physical crosslinking mechanism. Consistently, Tax induction of p100 processing requires the phosphorylation sites, serines 866 and 870, of p100 76.
In addition to HTLV1, several other human oncogenic viruses activate NF-κB; these include the Kaposi's sarcoma-associated herpes virus (KSHV) and the Epstein Bar virus (EBV). KSHV is tightly associated with Kaposi's sarcoma, a cancer commonly seen in individuals infected with the human immunodeficiency virus 99. In addition, KSHV is also found in several lymphoproliferative disorders, such as primary effusion lymphoma. KSHV encodes a viral homolog of the cellular FLICE inhibitor protein (FLIP), termed vFLIP, which functions as an anti-apoptotic protein. Like HTLV1 Tax, vFLIP stimulates both the canonical and non-canonical NF-κB pathway via a mechanism that involves interaction with IKKγ 99. Binding of vFLIP to IKKγ induces the conversion of IKKγ from a helical bundle conformation to an open conformation, which is thought to contribute to the activation of IKK catalytic subunits 100. Thus, like Tax, vFLIP activates the non-canonical NF-κB pathway via a NIK-independent and IKKγ- and IKKα-dependent mechanism.
EBV is known to persistently infect most healthy adults without normally causing overt diseases. However, accumulating evidence suggests that EBV may be associated with certain forms of lymphomas under immunodeficient conditions 99. EBV-encoded LMP1 protein is a major viral activator of NF-κB that targets both the canonical and non-canonical pathways. Unlike the intracellular Tax and vFLIP proteins, LMP1 is a six membrane-spanning molecule that mimics a constitutively activated TNFR and thus persistently activates the NF-κB signaling pathways through TRAF proteins 101. LMP1 contains two C-terminal activation regions (CTAR1 and CTAR2); whereas CTAR2 recruits TRAF6 and activates canonical NF-κB, CTAR1 recruits TRAF 1, 2, 3, and 5 and activates non-canonical NF-κB 101. Similar to cellular receptors, LMP1 stimulates p100 processing via a NIK- and IKKα-dependent mechanism.
Herpesvirus ateles, a monkey virus inducing T-cell transformation, activates both canonical and non-canonical NF-κBs 102. This virus encodes a transmembrane oncoprotein, Tio, which interacts with TRAF6 for canonical NF-κB activation. Tio-mediated non-canonical NF-κB activation is independent of TRAF6 or the canonical NF-κB signaling components IKKγ and IKKβ. Interestingly, Tio induces the stabilization of NIK, although the underlying mechanism is unknown.
Non-canonical NF-κB pathway is also known as a target of certain bacterial pathogens. One example is Helicobacter pylori, a Gram-negative bacterium that infects human gastric mucosa 103. The H. pylori infection causes chronic gastric inflammation, which in turn promotes the development of gastric cancer 104. H. pylori is known to induce canonical NF-κB activation in gastric epithelial cells via its virulence factor CagA, which activates IKK through stimulating K63 ubiquitination of the IKK-activating kinase Tak1 105. On the other hand, H. pylori induces the non-canonical NF-κB pathway in B cells, which is independent of CagA but dependent on LPS 103. Precisely how the H. pylori LPS stimulates non-canonical NF-κB signaling is unknown, although this action requires NIK and IKKα 103. Another bacterial pathogen known to stimulate both the canonical and non-canonical NF-κB pathways is Legionella pneumophila, a pathogen that infects alveolar lung macrophages and causes Legionnaire's disease 106. In contrast to the other NF-κB-inducing microbes, L. pneumophila activates NF-κB independently of IKKs and other cellular signaling components. Instead, it encodes a kinase, LegK1, which phosphorylates IκBα and p100, leading to the activation of canonical and non-canonical NF-κB pathways 106.
Concluding remarks
Since its discovery about 10 years ago, the non-canonical NF-κB pathway has become a hot area of research. It is now clear that this pathway differs substantially from the canonical NF-κB pathway in not only the composition of IKK complex but also, more importantly, the signaling mechanisms. This knowledge has opened new opportunities for drug therapies in the treatment of NF-κB-associated diseases, such as cancers. However, a number of outstanding questions are yet to be addressed for better understanding of the mechanism of non-canonical NF-κB signaling and for exploiting this pathway in therapeutic approaches.
One major question is how NIK and IKKα stimulate p100 processing. It is remarkable that the currently known non-canonical NF-κB inducers are all dependent on NIK. Is IKKα activation solely mediated by NIK or is active IKKα insufficient for triggering p100 processing without NIK? In other words, does NIK induce any other signaling factors that synergize with IKKα? Another question is about the mechanism of NIK activation. Is receptor-induced TRAF2/3 degradation and NIK accumulation the primary mechanism of NIK activation? Does NIK have to be recruited to receptors for its activation? Missing links also exist regarding the functions of non-canonical NF-κB. The currently known functions of this pathway are limited to a few cell types. Generation of conditional knockout mice is critical for systemically assessing the functions of this pathway in different cell types. Another future challenge is to understand the mechanism that mediates the specificity of non-canonical NF-κB in gene induction. Better understanding of these problems will help design more specific and efficient NF-κB-based therapies.
References
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- Sun SC, Ley SC. New insights into NF-kappaB regulation and function. Trends Immunol. 2008;29:469–478. doi: 10.1016/j.it.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin E. The alternative NF-kappaB pathway from biochemistry to biology: pitfalls and promises for future drug development. Biochem Pharmacol. 2006;72:1161–1179. doi: 10.1016/j.bcp.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Xiao G, Harhaj EW, Sun SC. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- Solan NJ, Miyoshi H, Carmona EM, Bren GD, Paya CV. RelB cellular regulation and transcriptional activity are regulated by p100. J Biol Chem. 2002;277:1405–1418. doi: 10.1074/jbc.M109619200. [DOI] [PubMed] [Google Scholar]
- Coope HJ, Atkinson PG, Huhse B, et al. CD40 regulates the processing of NF-kappaB2 p100 to p52. EMBO J. 2002;15:5375–5385. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derudder E, Dejardin E, Pritchard LL, et al. RelB/p50 dimers are differentially regulated by tumor necrosis factor-{alpha} and lymphotoxin-{beta} receptor activation: critical roles for p100. J Biol Chem. 2003;278:23278–23284. doi: 10.1074/jbc.M300106200. [DOI] [PubMed] [Google Scholar]
- Lin L, DeMartino GN, Greene WC. Cotranslational biogenesis of NF-kappaB p50 by the 26S proteasome. Cell. 1998;92:819–828. doi: 10.1016/s0092-8674(00)81409-9. [DOI] [PubMed] [Google Scholar]
- Betts JC, Nabel GJ. Differential regulation of NF-kappaB2 (p100) processing and control by amino-terminal sequences. Mol Cell Biol. 1996;16:6363–6371. doi: 10.1128/mcb.16.11.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-kappaB2 in maturing B cells. Nat Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- Dejardin E, Droin NM, Delhase M, et al. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Yan M, Seshasayee D, et al. BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-kappaB2. Immunity. 2002;17:515–524. doi: 10.1016/s1074-7613(02)00425-9. [DOI] [PubMed] [Google Scholar]
- Novack DV, Yin L, Hagen-Stapleton A, et al. The IkappaB function of NF-kappaB2 p100 controls stimulated osteoclastogenesis. J Exp Med. 2003;198:771–781. doi: 10.1084/jem.20030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong A, Sun S-C. Genetic evidence for the essential role of beta-transducin repeat-containing protein in the inducible processing of NF-κB2/p100. J Biol Chem. 2002;277:22111–22114. doi: 10.1074/jbc.C200151200. [DOI] [PubMed] [Google Scholar]
- Liang C, Zhang M, Sun SC. Beta-TrCP binding and processing of NF-kappaB2/p100 involve its phosphorylation at serines 866 and 870. Cell Signal. 2006;18:1309–1317. doi: 10.1016/j.cellsig.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Mordmuller B, Krappmann D, Esen M, Wegener E, Scheidereit C. Lymphotoxin and lipopolysaccharide induce NF-kappaB-p52 generation by a co-translational mechanism. EMBO J. 2003;4:82–87. doi: 10.1038/sj.embor.embor710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- Amir RE, Haecker H, Karin M, Ciechanover A. Mechanism of processing of the NF-kappa B2 p100 precursor: identification of the specific polyubiquitin chain-anchoring lysine residue and analysis of the role of NEDD8-modification on the SCF(beta-TrCP) ubiquitin ligase. Oncogene. 2004;23:2540–2547. doi: 10.1038/sj.onc.1207366. [DOI] [PubMed] [Google Scholar]
- Scherer DC, Brockman JA, Chen A, Maniatis T, Ballard DW. Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong A, Zhang M, Neely J, Sun SC. S9: a 19S proteasome subunit interacting with ubiquitinated NF-kB2/p100. J Biol Chem. 2002;277:40697–40702. doi: 10.1074/jbc.M205330200. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatsyayan J, Qing G, Xiao G, Hu J. SUMO1 modification of NF-kappaB2/p100 is essential for stimuli-induced p100 phosphorylation and processing. EMBO Rep. 2008;9:885–890. doi: 10.1038/embor.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusch M, Lin L, Geleziunas R, Greene WC. The generation of nfkb2 p52: mechanism and efficiency. Oncogene. 1999;18:6201–6208. doi: 10.1038/sj.onc.1203022. [DOI] [PubMed] [Google Scholar]
- Liao G, Sun SC. Regulation of NF-kappaB2/p100 processing by its nuclear shuttling. Oncogene. 2003;22:4868–4874. doi: 10.1038/sj.onc.1206761. [DOI] [PubMed] [Google Scholar]
- Blondel M, Galan JM, Chi Y, et al. Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4. EMBO J. 2001;19:6085–6097. doi: 10.1093/emboj/19.22.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassot I, Segeral E, Berlioz-Torrent C, et al. ATF4 degradation relies on a phosphorylation-dependent interaction with the SCF(betaTrCP) ubiquitin ligase. Mol Cell Biol. 2001;21:2192–2202. doi: 10.1128/MCB.21.6.2192-2202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadot E, Simcha I, Iwai K, et al. Differential interaction of plakoglobin and beta-catenin with the ubiquitin-proteasome system. Oncogene. 2001;19:1992–2001. doi: 10.1038/sj.onc.1203519. [DOI] [PubMed] [Google Scholar]
- Davis M, Hatzubai A, Andersen JS, et al. Pseudosubstrate regulation of the SCF(β-TrCP) ubiquitin ligase by hnRNP-U. Genes Dev. 2002;16:439–451. doi: 10.1101/gad.218702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Chiocca S. Nuclear ubiquitin ligases, NF-kappaB degradation, and the control of inflammation. Sci Signal. 2008;1:pe1. doi: 10.1126/stke.11pe1. [DOI] [PubMed] [Google Scholar]
- Qing G, Qu Z, Xiao G. Endoproteolytic processing of C-terminally truncated NF-kappaB2 precursors at kappaB-containing promoters. Proc Natl Acad Sci USA. 2007;104:5324–5329. doi: 10.1073/pnas.0609914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Marazzi I, Beg AA, Natoli G. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor κB response. J Exp Med. 2004;200:107–113. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinin NL, Boldin MP, Kovalenko AV, Wallach D. MAP3K-related kinase involved in NF-κB induction by TNF, CD95 and IL-1. Nature. 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- Shinkura R, Kitada K, Matsuda F, et al. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-kappa b-inducing kinase. Nat Genet. 1999;22:74–77. doi: 10.1038/8780. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu L, Wesche H, et al. Defective lymphotoxin-beta receptor-induced NF-kappaB transcriptional activity in NIK-deficient mice. Science. 2001;291:2162–2165. doi: 10.1126/science.1058453. [DOI] [PubMed] [Google Scholar]
- Senftleben U, Cao Y, Xiao G, et al. Activation of IKKα of a second, evolutionary conserved, NF-κB signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- Matsushima A, Kaisho T, Rennert PD, et al. Essential role of nuclear factor (NF)-kappaB-inducing kinase and inhibitor of kappaB (IkappaB) kinase alpha in NF-kappaB activation through lymphotoxin beta receptor, but not through tumor necrosis factor receptor I. J Exp Med. 2001;193:631–636. doi: 10.1084/jem.193.5.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Fong A, Sun SC. Induction of p100 processing by NF-kappaB-inducing kinase involves docking IkappaB kinase alpha (IKKalpha) to p100 and IKKalpha-mediated phosphorylation. J Biol Chem. 2004;279:30099–30105. doi: 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- Sun SC, Harhaj EW.Receptors and adaptors for NF-kB signalingIn: Liou HC, ed. NF-kB/Rel Transcription Factor FamilyNew York: Springer US, 200626–24.
- Munroe ME, Bishop GA. Role of tumor necrosis factor (TNF) receptor-associated factor 2 (TRAF2) in distinct and overlapping CD40 and TNF receptor 2/CD120b-mediated B lymphocyte activation. J Biol Chem. 2004;279:53222–53231. doi: 10.1074/jbc.M410539200. [DOI] [PubMed] [Google Scholar]
- Rauert H, Wicovsky A, Müller N, et al. Membrane tumor necrosis factor (TNF) induces p100 processing via TNF receptor-2 (TNFR2) J Biol Chem. 2010;285:7394–7404. doi: 10.1074/jbc.M109.037341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Nakayama M, Nakano H, et al. TWEAK induces NF-kappaB2 p100 processing and long lasting NF-kappaB activation. J Biol Chem. 2003;278:36005–36012. doi: 10.1074/jbc.M304266200. [DOI] [PubMed] [Google Scholar]
- Wicovsky A, Salzmann S, Roos C, et al. TNF-like weak inducer of apoptosis inhibits proinflammatory TNF receptor-1 signaling. Cell Death Differ. 2009;16:1445–1459. doi: 10.1038/cdd.2009.80. [DOI] [PubMed] [Google Scholar]
- Bishop GA, Xie P. Multiple roles of TRAF3 signaling in lymphocyte function. Immunol Res. 2007;39:22–32. doi: 10.1007/s12026-007-0068-1. [DOI] [PubMed] [Google Scholar]
- Sun SC. Controlling the fate of NIK: a central stage in noncanonical NF-kappaB signaling. Sci Signal. 2010;3:pe18. doi: 10.1126/scisignal.3123pe18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris PS, Ware CF. The LT beta R signaling pathway. Adv Exp Med Biol. 2007;597:160–172. doi: 10.1007/978-0-387-70630-6_13. [DOI] [PubMed] [Google Scholar]
- Crowley JE, Treml LS, Stadanlick JE, Carpenter E, Cancro MP. Homeostatic niche specification among naive and activated B cells: a growing role for the BLyS family of receptors and ligands. Semin Immunol. 2005;17:193–199. doi: 10.1016/j.smim.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Morrison MD, Reiley W, Zhang M, Sun SC. An atypical tumor necrosis factor (TNF) receptor-associated factor-binding motif of B cell-activating factor belonging to the TNF family (BAFF) receptor mediates induction of the noncanonical NF-kappaB signaling pathway. J Biol Chem. 2005;280:10018–10024. doi: 10.1074/jbc.M413634200. [DOI] [PubMed] [Google Scholar]
- Bishop GA, Hostager BS. The CD40-CD154 interaction in B cell-T cell liaisons. Cytokine Growth Factor Rev. 2003;14:297–309. doi: 10.1016/s1359-6101(03)00024-8. [DOI] [PubMed] [Google Scholar]
- Harnett MM. CD40: a growing cytoplasmic tale. Sci STKE. 2004;2004:pe25. doi: 10.1126/stke.2372004pe25. [DOI] [PubMed] [Google Scholar]
- Bishop GA, Moore CR, Xie P, Stunz LL, Kraus ZJ. TRAF proteins in CD40 signaling. Adv Exp Med Biol. 2007;597:131–151. doi: 10.1007/978-0-387-70630-6_11. [DOI] [PubMed] [Google Scholar]
- Theill LE, Boyle WJ, Penninger JM. RANK-L and RANK: T cells, bone loss, and mammalian evolution. Annu Rev Immunol. 2002;20:795–823. doi: 10.1146/annurev.immunol.20.100301.064753. [DOI] [PubMed] [Google Scholar]
- Vaira S, Johnson T, Hirbe AC, et al. RelB is the NF-kappaB subunit downstream of NIK responsible for osteoclast differentiation. Proc Natl Acad Sci USA. 2008;105:3897–3902. doi: 10.1073/pnas.0708576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley SR, Cassiano L, Lofton T, et al. A novel TNF receptor family member binds TWEAK and is implicated in angiogenesis. Immunity. 2001;15:837–846. doi: 10.1016/s1074-7613(01)00232-1. [DOI] [PubMed] [Google Scholar]
- Brown SA, Richards CM, Hanscom HN, Feng SL, Winkles JA. The Fn14 cytoplasmic tail binds tumour-necrosis-factor-receptor-associated factors 1, 2, 3 and 5 and mediates nuclear factor-kappaB activation. Biochem J. 2003;371:395–403. doi: 10.1042/BJ20021730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos C, Wicovsky A, Müller N, et al. Soluble and transmembrane TNF-like weak inducer of apoptosis differentially activate the classical and noncanonical NF-kappa B pathway. J Immunol. 2010;185:1593–1605. doi: 10.4049/jimmunol.0903555. [DOI] [PubMed] [Google Scholar]
- Varfolomeev E, Blankenship JW, Wayson SM, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- Nishikori M, Ohno H, Haga H, Uchiyama T. Stimulation of CD30 in anaplastic large cell lymphoma leads to production of nuclear factor-kappaB p52, which is associated with hyperphosphorylated Bcl-3. Cancer Sci. 2005;96:487–497. doi: 10.1111/j.1349-7006.2005.00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka M, Horie R, Itoh K, et al. Aberrant NF-kappaB2/p52 expression in Hodgkin/Reed-Sternberg cells and CD30-transformed rat fibroblasts. Oncogene. 2005;24:3976–3986. doi: 10.1038/sj.onc.1208564. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan P, Wang W, Wallach D. Receptor-specific signaling for both the alternative and the canonical NF-kappaB activation pathways by NF-kappaB-inducing kinase. Immunity. 2004;21:477–489. doi: 10.1016/j.immuni.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Borthakur A, Dudeja PK, Tobacman JK. Lipopolysaccharide-induced activation of NF-kappaB non-canonical pathway requires BCL10 serine 138 and NIK phosphorylations. Exp Cell Res. 2010;316:3317–27. doi: 10.1016/j.yexcr.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Zhang M, Harhaj EW, Sun SC. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- He JQ, Zarnegar B, Oganesyan G, et al. Rescue of TRAF3-null mice by p100 NF-kappa B deficiency. J Exp Med. 2006;203:2413–2418. doi: 10.1084/jem.20061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Calado DP, Derudder E, et al. NIK overexpression amplifies, whereas ablation of its TRAF3-binding domain replaces BAFF:BAFF-R-mediated survival signals in B cells. Proc Natl Acad Sci USA. 2008;105:10883–10888. doi: 10.1073/pnas.0805186105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S, Matsuzawa A, Zhang W, et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol. 2008;9:1364–1370. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vince JE, Wong WW, Khan N, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- Zarnegar BJ, Wang Y, Mahoney DJ, et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 2008;9:1371–1378. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P, Stunz LL, Larison KD, Yang B, Bishop GA. Tumor necrosis factor receptor-associated factor 3 is a critical regulator of B cell homeostasis in secondary lymphoid organs. Immunity. 2007;27:253–267. doi: 10.1016/j.immuni.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardam S, Sierro F, Basten A, Mackay F, Brink R. TRAF2 and TRAF3 signal adapters act cooperatively to control the maturation and survival signals delivered to B cells by the BAFF receptor. Immunity. 2008;28:391–401. doi: 10.1016/j.immuni.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Annunziata CM, Davis RE, Demchenko Y, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keats JJ, Fonseca R, Chesi M, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostager BS, Haxhinasto SA, Rowland SL, Bishop GA. Tumor necrosis factor receptor-associated factor 2 (TRAF2)-deficient B lymphocytes reveal novel roles for TRAF2 in CD40 signaling. J Biol Chem. 2003;278:45382–45390. doi: 10.1074/jbc.M306708200. [DOI] [PubMed] [Google Scholar]
- Lin X, Mu Y, Cunningham ETJ, et al. Molecular determinants of NF-kappaB-inducing kinase action. Mol Cell Biol. 1998;18:5899–5907. doi: 10.1128/mcb.18.10.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sedwick CE, Hu J, Altman A. Role for protein kinase Ctheta (PKCtheta) in TCR/CD28-mediated signaling through the canonical but not the non-canonical pathway for NF-kappaB activation. J Biol Chem. 2005;280:1217–1223. doi: 10.1074/jbc.M409492200. [DOI] [PubMed] [Google Scholar]
- Xiao G, Cvijic ME, Fong A, et al. Retroviral oncoprotein Tax induces processing of NF-kappaB2/p100 in T cells: evidence for the involvement of IKKalpha. EMBO J. 2001;20:6805–6815. doi: 10.1093/emboj/20.23.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Zhou XF, Yu J, Cheng X, Sun SC. Regulation of Th17 cell differentiation and EAE induction by the MAP3K NIK. Blood. 2009;113:6603–6610. doi: 10.1182/blood-2008-12-192914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B, Zarnegar B, Ytterberg AJ, et al. Negative feedback in non-canonical NF-κB signaling modulates NIK stability through IKKα-mediated phosphorylation. Sci Signal. 2010;3:ra41. doi: 10.1126/scisignal.2000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WH, Mo XM, Walters WM, Brambilla R, Bethea JR. TNAP, a novel repressor of NF-kappaB-inducing kinase, suppresses NF-kappaB activation. J Biol Chem. 2004;279:35975–35983. doi: 10.1074/jbc.M405699200. [DOI] [PubMed] [Google Scholar]
- Lich JD, Ting JP. Monarch-1/PYPAF7 and other CATERPILLER (CLR, NOD, NLR) proteins with negative regulatory functions. Microbes Infect. 2007;9:672–676. doi: 10.1016/j.micinf.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lich JD, Williams KL, Moore CB, et al. Monarch-1 suppresses non-canonical NF-kappaB activation and p52-dependent chemokine expression in monocytes. J Immunol. 2007;178:1256–1260. doi: 10.4049/jimmunol.178.3.1256. [DOI] [PubMed] [Google Scholar]
- Jin X, Jin HR, Jung HS, et al. An atypical E3 ligase Zinc Finger protein 91 stabilizes and activates NF-{kappa}B-inducing kinase via K63-linked ubiquitination. J Biol Chem. 2010;285:30539–30547. doi: 10.1074/jbc.M110.129551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruefli-Brasse AA, French DM, Dixit VM. Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science. 2003;302:1581–1584. doi: 10.1126/science.1090769. [DOI] [PubMed] [Google Scholar]
- Ruland J, Duncan GS, Wakeham A, Mak TW. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity. 2003;19:749–758. doi: 10.1016/s1074-7613(03)00293-0. [DOI] [PubMed] [Google Scholar]
- Tusche MW, Ward LA, Vu F, et al. Differential requirement of MALT1 for BAFF-induced outcomes in B cell subsets. J Exp Med. 2009;206:2671–2683. doi: 10.1084/jem.20091802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang H, Xue L, et al. Emu-BCL10 mice exhibit constitutive activation of both canonical and noncanonical NF-kappaB pathways generating marginal zone (MZ) B-cell expansion as a precursor to splenic MZ lymphoma. Blood. 2009;114:4158–4168. doi: 10.1182/blood-2008-12-192583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadiminty N, Lou W, Lee SO, et al. Stat3 activation of NF-{kappa}B p100 processing involves CBP/p300-mediated acetylation. Proc Natl Acad Sci USA. 2006;103:7264–7269. doi: 10.1073/pnas.0509808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Morgan MJ, Choksi S, et al. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol. 2010;11:799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonizzi G, Bebien M, Otero DC, et al. Activation of IKKalpha target genes depends on recognition of specific kappaB binding sites by RelB:p52 dimers. EMBO J. 2004;23:4202–4210. doi: 10.1038/sj.emboj.7600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova LV, Makeev VJ, Kuprash DV. In vitro selection of optimal RelB/p52 DNA-binding motifs. Biochem Biophys Res Commun. 2008;365:583–588. doi: 10.1016/j.bbrc.2007.10.200. [DOI] [PubMed] [Google Scholar]
- Tando T, Ishizaka A, Watanabe H, et al. Requiem protein links RelB/p52 and the Brm-type SWI/SNF complex in a noncanonical NF-kappaB pathway. J Biol Chem. 2010;285:21951–21960. doi: 10.1074/jbc.M109.087783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Carrasco D, Claudio E, Ryseck RP, Bravo R. Gastric hyperplasia and increased proliferative responses of lymphocytes in mice lacking the COOH-terminal ankyrin domain of NF-kappaB2. J Exp Med. 1997;186:999–1014. doi: 10.1084/jem.186.7.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko YN, Glebov OK, Zingone A, et al. Classical and/or alternative NF-kappaB pathway activation in multiple myeloma. Blood. 2010;115:3541–3552. doi: 10.1182/blood-2009-09-243535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar B, Yamazaki S, He JQ, Cheng G. Control of canonical NF-kappaB activation through the NIK-IKK complex pathway. Proc Natl Acad Sci USA. 2008;105:3503–3508. doi: 10.1073/pnas.0707959105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh Y, Yamamoto N, Dewan MZ, et al. Overexpressed NF-kappaB-inducing kinase contributes to the tumorigenesis of adult T-cell leukemia and Hodgkin Reed-Sternberg cells. Blood. 2008;111:5118–5129. doi: 10.1182/blood-2007-09-110635. [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7:270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- Sun SC, Yamaoka S. Activation of NF-κB by HTLV-I and implications for cell transformation. Oncogene. 2005;24:5952–5964. doi: 10.1038/sj.onc.1208969. [DOI] [PubMed] [Google Scholar]
- Sun SC, Cesarman E.NF-kB as a target for oncogenic viruses Curr Top Microbiol Immunol 2010. Sep 16. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- Bagnéris C, Ageichik AV, Cronin N, et al. Crystal structure of a vFlip-IKKgamma complex: insights into viral activation of the IKK signalosome. Mol Cell. 2008;30:620–631. doi: 10.1016/j.molcel.2008.04.029. [DOI] [PubMed] [Google Scholar]
- Soni V, Cahir-McFarland E, Kieff E. LMP1 TRAFficking activates growth and survival pathways. Adv Exp Med Biol. 2007;597:173–187. doi: 10.1007/978-0-387-70630-6_14. [DOI] [PubMed] [Google Scholar]
- de Jong SJ, Albrecht JC, Schmidt M, Müller-Fleckenstein I, Biesinger B. Activation of noncanonical NF-kappaB signaling by the oncoprotein Tio. J Biol Chem. 2010;285:16495–16503. doi: 10.1074/jbc.M110.102848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmae T, Hirata Y, Maeda S, et al. Helicobacter pylori activates NF-kappaB via the alternative pathway in B lymphocytes. J Immunol. 2005;175:7162–7169. doi: 10.4049/jimmunol.175.11.7162. [DOI] [PubMed] [Google Scholar]
- Peek RMJ, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- Lamb A, Yang XD, Tsang YH, et al. Helicobacter pylori CagA activates NF-kappaB by targeting TAK1 for TRAF6-mediated Lys 63 ubiquitination. EMBO Rep. 2009;10:1242–1249. doi: 10.1038/embor.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Xu H, Li T, et al. A Legionella type IV effector activates the NF-kappaB pathway by phosphorylating the IkappaB family of inhibitors. Proc Natl Acad Sci USA. 2009;106:13725–13730. doi: 10.1073/pnas.0907200106. [DOI] [PMC free article] [PubMed] [Google Scholar]