Abstract
Two distinct nuclear factor κB (NFκB) signaling pathways have been described; the canonical pathway that mediates inflammatory responses, and the non-canonical pathway that is involved in immune cell differentiation and maturation and secondary lymphoid organogenesis. The former is dependent on the IκB kinase adaptor molecule NEMO, the latter is independent of it. Here, we review the molecular mechanisms of regulation in each signaling axis and attempt to relate the apparent regulatory logic to the physiological function. Further, we review the recent evidence for extensive cross-regulation between these two signaling axes and summarize them in a wiring diagram. These observations suggest that NEMO-dependent and -independent signaling should be viewed within the context of a single NFκB signaling system, which mediates signaling from both inflammatory and organogenic stimuli in an integrated manner. As in other regulatory biological systems, a systems approach including mathematical models that include quantitative and kinetic information will be necessary to characterize the network properties that mediate physiological function, and that may break down to cause or contribute to pathology.
Keywords: NFκB, inflammation, immune response, immune development, mathematical model, signaling crosstalk
Introduction
Nuclear factor κB (NFκB) is a critical transcription factor involved in a broad range of biological processes, including immune responses, cell survival, stress responses and maturation of various cell types. While NFκB activation is required to protect organisms from environmental effects, misregulated NFκB activity is often observed in various diseases including chronic inflammation and cancer. Thus, understanding the regulation of NFκB signaling is important for maintaining human health.
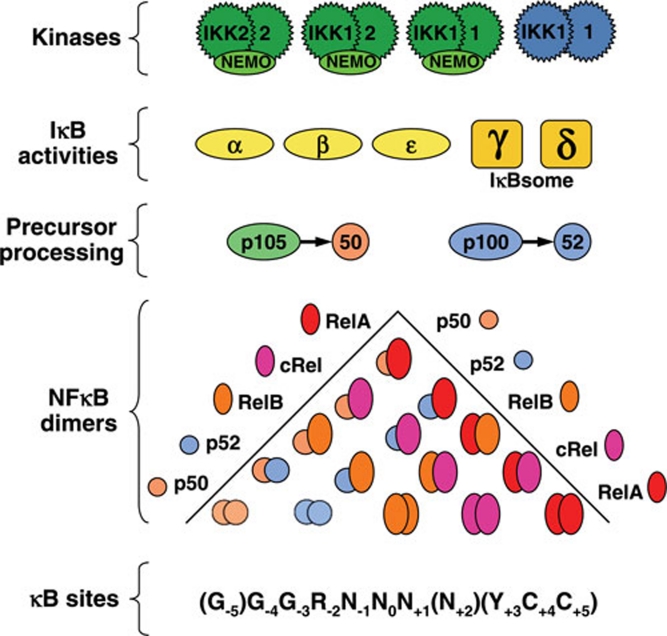
The NFκB signaling system consists of NFκB heterodimers or homodimers of Rel homology domain (RHD)-containing polypeptides and their stoichiometric inhibitor proteins, IκBs (Figure 1). The RHD within the five monomers – RelA, RelB, cRel, p50 and p52 – mediates dimerization, DNA binding, interaction with IκBs and nuclear translocation. The five monomers form 15 potential dimers. Generation of all monomers is transcriptionally regulated, but p50 and p52 are also regulated by processing of precursor proteins p105 and p100, respectively. Thus, different NFκB dimeric complexes are expressed cell type- and stimulus-specifically; some of the physiologically important dimers are RelA:p50, cRel:p50 and RelB:p52.
Figure 1.
Components of the IKK-IκB-NFκB signaling system. The IKK form canonical NEMO-containing (green) complexes and non-canonical (blue) complexes, which control the degradation of IκB proteins and precursor processing. IκBα, IκBβ, IκBɛ and the IκB activities within the IκBsome IκBγ and IκBδ are able to sequester NFκB dimers. The p50 and p52 NFκB proteins are generated from the processing of newly synthesized precursor proteins p105 and p100, respectively. The five NFκB family members (RelA/p65, cRel, RelB, p52 and p50) can potentially form 15 heterodimers and homodimers that can bind to a large number of κB sites in DNA, which are characterized by a remarkably broad sequence consensus.
The classical inhibitor proteins in the NFκB signaling system consist of the single polypeptide IκBs: IκBα, IκBβ and IκBɛ. In resting cells, IκB binds and sequesters NFκB dimer and prevents DNA binding and transcriptional activation. Stimulus-responsive activation of the IκB kinase (IKK) results in the degradation of the IκBs to release and activate NFκB (Figure 2). Synthesis of IκBs may be dependent on NFκB activity, and the inducible activation of IκBs results in negative feedback. Classical IκBs are characterized by their ankyrin repeats domain (ARD) that sequesters NFκB into a latent state. Recently, it was found that p100, when present in a multimeric complex, may also mediate NFκB inhibition in trans; this activity was termed IκBδ 1. Moreover, size exclusion chromatography analyses suggest that IκBδ activity is mediated by a ∼650 kDa high molecular weight complex, termed the IκBsome, which also contains IκBγ activity from p105 protein 2. The pathways that govern IκBsome assembly and degradation are critical for regulating NFκB activity 3.
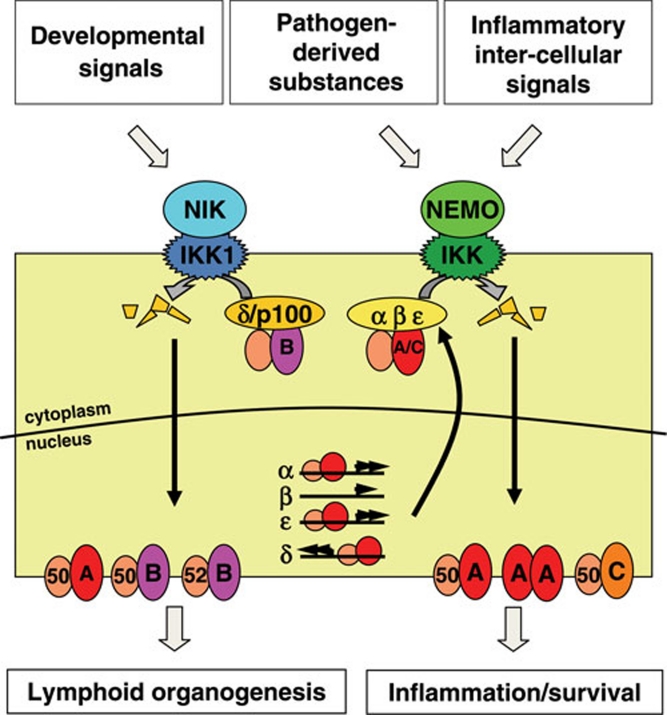
Figure 2.
The NFκB signaling module consisting of canonical and non-canonical pathways. The non-canonical pathway is activated through developmental signals activating NIK/IKK1. This activation results in the degradation of IκBδ and processing of p100 and allows the nuclear translocation of RelA:p50, RelB:p50 and RelB:p52 dimers, which activate genes responsible for organ development. The canonical pathway is activated through pathogen and inflammatory signals activating NEMO/IKK. This activation results in the degradation of IκBα/IκBβ/IκBɛ, allowing for the nuclear translocation of RelA:p50, RelA:RelA and cRel:p50 dimers, which then activate genes responsible for inflammation and survival.
Activation of NFκB results in the induction of a variety of inflammatory, developmental and survival genes. The rapid and reversible inflammatory and immune response typically occurs through the activation of the canonical pathway, while the slower and irreversible developmental response typically occurs through the non-canonical pathway. These two pathways are thought to be fundamentally distinct. While the canonical pathway is mediated through the activation of a NEMO (IKKγ)-dependent IKK, the non-canonical pathway is classically defined as being mediated through the activation of a NEMO-independent kinase complex involving IKK1 and the NFκB-inducing kinase (NIK) 4. In the canonical pathway, pre-existing, latent NFκB dimers are released from classical IκBs. In the non-canonical pathway, new synthesis of p100 and RelB allows for generation of RelB:p52 which is insensitive to IκB control and thus localized to the nucleus.
Though canonical and non-canonical pathways are generally thought to be distinct, recent studies have revealed numerous crosstalk mechanisms that connect them. This crosstalk involves expression control of NFκB monomers, interdependent proteolytic processing events of precursors and the newly identified IκBδ activity that is inducibly expressed by one pathway and inducibly degraded by another.
This review first describes the distinct characteristics of the canonical and non-canonical pathways and then discusses extensive cross-regulatory mechanisms that suggest that the regulation of NFκB dimers is in fact mediated by a single NFκB signaling system, and ought to be investigated through systems-level analysis.
The canonical pathway
The canonical NFκB pathway is defined as being mediated by a NEMO-dependent IKK; in contrast, the non-canonical pathway is defined as not requiring NEMO. Genetic deletion of NEMO resulted in embryonic lethality as a result of massive apoptosis in the fetal liver 5. The fact that the IKK2 knockout 6 and the RelA knockout 7 showed a similar phenotype led to the broadly accepted model of a signaling axis of NEMO-IKK2-RelA, termed the canonical pathway. However, recent work suggests that IKK1 plays a role in the canonical pathway as well, and may compensate for the loss of IKK2 within the NEMO-dependent kinase 8. Conversely, NEMO-independent IKK2 functions have also been suggested 9. We therefore suggest that only NEMO dependence strictly defines the canonical pathway. However, given the close functional association of IKK2 with NEMO in the literature, our current understanding of canonical activation is in large part limited to the activation of IKK2. Similarly, RelA was recently shown to be activated by the non-canonical activation mechanism 1 and thus should not be viewed as the exclusive effector of the canonical pathway; however, our current understanding of RelA function is largely limited to canonical signaling.
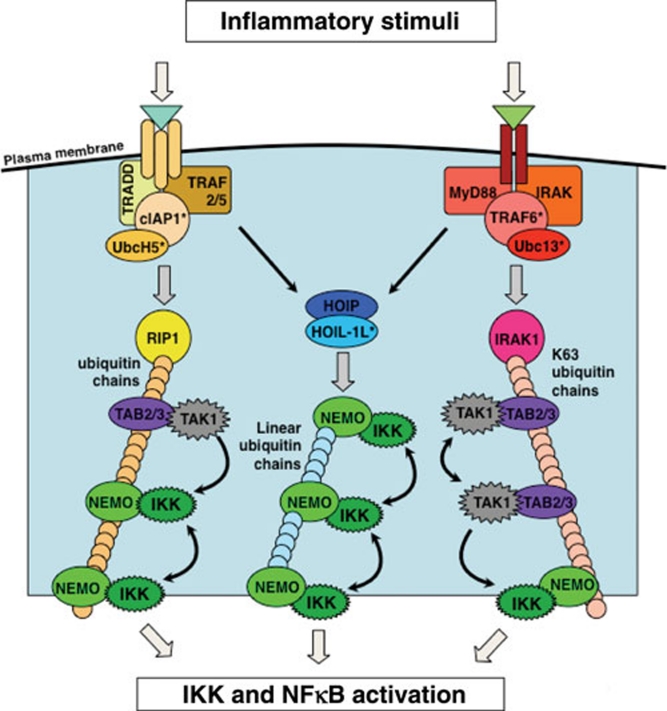
The canonical NFκB pathway is activated by pro-inflammatory signals such as cytokines, pathogen-associated molecular patterns (PAMPs), and some danger-associated molecular patterns (DAMPs). Upon cellular exposure to these agents, extracellular and intracellular receptors trigger the activation of NEMO-containing kinase complexes through the recruitment of various proteins involved in ubiquitin chain formation. IKK2 is activated by phosphorylation at serine (Ser) 177 and Ser181 10. Emerging evidence indicates that three possible pathways for IKK2 activation are utilized to varying degrees by different inflammatory receptors (Figure 3). In the case of the TNF receptor I, associated proteins recruit the E2/E3 ligase complex consisting of UbcH5 and cIAP1, which subsequently forms ubiquitin chains of various linkages to RIP1 11, 12. The TAB/TAK1 and IKK complexes are able to bind these ubiquitin chains, allowing the activated TAK1 to phosphorylate and activate IKK2 13. Additionally, oligomerization of the NEMO-IKK2 complex upon mixed ubiquitin chain binding can allow for TAK1-independent trans-autophosphorylation and activation of the IKK2 complex 12. Another pathway of IKK2 activation revolves around linear ubiquitin chain formation. In the context of TNF signaling, the receptor-associated proteins TRADD, tumor necrosis factor receptor-associated factor (TRAF) 2 and cIAP1/2 recruit LUBAC, an ubiquitin ligase complex consisting of HOIP and HOIL-1L 14. LUBAC conjugates linear-linked ubiquitin chains to NEMO, which results in IKK activation, possibly through trans-autophosphorylation 15. In the third pathway of IKK2 activation, ligand engagement of the IL-1R and TLRs leads to the recruitment of the E2/E3 ligase complex made up of Ubc13 and TRAF6 16. This complex conjugates K63-linked ubiquitin chains to IRAK1, allowing for TAK1 and IKK2 complex binding and subsequent TAK1 activation of IKK2 through phosphorylation 12. While a molecular description of these three pathways has advanced, the stimulus specificity of each pathway remains to be delineated.
Figure 3.
Mechanisms of canonical IKK activation. Several distinct pathways of canonical IKK activation have been described: LEFT, upon TNFR engagement, receptor-associated proteins such as TRADD and TRAF2/5 recruit the E2/E3 ligase complex consisting of cIAP1 and UbcH5. cIAP1/UbcH5 subsequently conjugate ubiquitin chains of various types of linkages to RIP1, which allows for TAK1 complex and IKK binding. The binding of Ub by the TAK1 complex activates TAK1 and allows it to activate IKK; in addition, Ub binding by the IKK complex induces IKK activation, allowing IKK to trans-autophosphorylate. Center, upon ligand binding of the TNFR, the ubiquitin ligase complex consisting of HOIP and HOIL-1L, known as LUBAC, is recruited by TRADD, TRAF2 and cIAP1. LUBAC conjugates linear-linked ubiquitin to NEMO in the IKK complex, resulting in IKK activation via trans-autophosphorylation. Right, when members of the TLR/IL-1R family are engaged, receptor-associated proteins Myd88 and IRAK recruit the E2/E3 ligase complex consisting of TRAF6 and Ubc13, which conjugates K63-linked ubiquitin chains to IRAK1. The TAK1 complex binds these K63 Ub chains, leading to TAK1 activation, presumably by trans-autophosphorylation. TAK1 subsequently activates nearby IKK. How these and possibly other mechanisms that result in canonical IKK activation combine to transduce signaling from different receptors remains an active area of investigation.
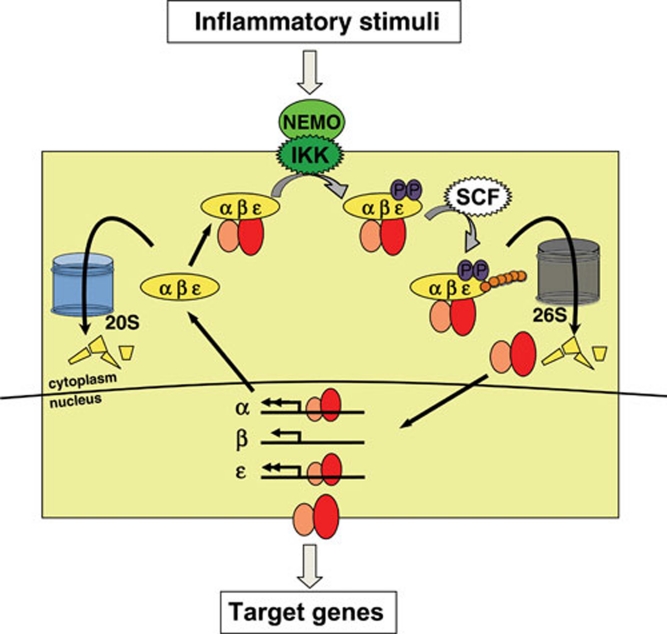
Once canonical IKK is activated, it phosphorylates and designates the classical IκBs (IκBα, IκBβ and IκBɛ) for degradation releasing NFκB into the nucleus (Figure 4). The phosphorylation of the canonical IκBα on the N-terminal serines (Ser32/36) allows for the recruitment of the E3 ubiquitin ligase SCF/βTRCP, thus marking it for degradation via the 26S proteasome 17. This ubiquitination and degradation of IκBα releases NFκB into the nucleus and allows transcriptional activation to occur. Similar phosphorylations occur on IκBβ and IκBɛ upon canonical IKK activation resulting in degradation of these IκBs and release of NFκB into the nucleus. Interestingly, the molecular determinants for the enzymatic specificity of the canonical IKK complex for classical IκB proteins are not yet understood.
Figure 4.
The canonical IKK-NFκB pathway. Upon canonical IKK activation, the classical IκBs, IκBα, -β, -ɛ, which sequester NFκB to the cytoplasm in a stoichiometric manner, are phosphorylated on specific N-terminal residues that function as docking sites for the E3 ubiquitin ligase complex SCF/βTRCP. Ubiquitinated IκBα is degraded by the 26S proteasome, allowing for translocation of NFκB into the nucleus. Nuclear NFκB activity induces the expression of IκBα and IκBɛ, providing for negative feedback. Free IκB proteins are rapidly degraded in a ubiquitin-independent manner by the 20S proteasome, presumably in conjunction with alternate proteasome targeting or activating proteins.
The primary NFκB effectors of the canonical pathway are transactivation domain-containing polypeptides RelA/p65 and cRel, which form either homodimers or heterodimers with the transactivation domain-lacking p50. Thus, four transcriptional activators (RelA:RelA, RelA:p50, cRel:cRel, cRel:p50) are potentially activated in response to stimuli of the canonical pathway. The phenotypes of genetic ablation studies of the constituent polypeptides are largely correlated with their tissue distribution. Thus, these studies showed an essential role for cRel in B- and T-lymphocyte functions, cell types in which it is highly expressed. crel−/− mice are not lethal but show several defects, including defects in the cell cycle progression and survival in B cells, defects in CD4 and CD8 T-cell responses, and impaired cytokine production 18, 19. Further, dendritic cells lacking cRel:p50 are defective in CD40L-induced cell survival 20 and displayed reduced maturation phenotype 21. However, the results from domain mutation studies are not fully explained: removal of the C-terminal activation domain of cRel results in enlarged lymph nodes and lymphoid hyperplasia 22.
In contrast, deficiency of the ubiquitously expressed RelA/p65 protein in rela−/− mice results in embryonic lethality due to massive apoptosis in the fetal liver 7, 23. Disrupting TNF signaling prevents this lethality, and generation of the rela−/−tnfr1−/− or rela−/−tnf−/− genotype allows for these mice to survive embryogenesis so that the absence of RelA can be studied 24, 25. The rela−/−tnfr1−/− genotype results in defects in the immune system, confirming the requirement of RelA for a functioning immune and inflammatory response in cells. By delaying embryonic death (compare rela−/−, crel−/−rela−/−), cRel partially compensates for the loss of RelA in rela−/− mice, suggesting that cRel and RelA have overlapping functions in mediating survival signals in fetal liver cells 26. As crel−/−rela−/− immune system shows more severe phenotypes than either of the single gene knockouts 27, it also appears that these two effectors have overlapping functions in the survival of lymphocyte precursors.
Whereas the importance of the RelA- and cRel-containing NFκB dimers in the immune and inflammatory response demonstrated by genetic ablation studies was anticipated based on prior biochemical studies in various cell types, the survival defect in rela−/− genotypes and the role of NFκB in regulating apoptosis and possibly other forms of cell death – though now well established – was initially surprising. Studies have shown that RelA is crucial in preventing TNF-mediated apoptosis in many different cell types, including macrophages, hepatocytes and T cells 23, 28, 29. An interesting question is how critical the stimulus-responsive activation of the canonical pathway is for this survival function of NFκB or whether it is mediated by some other regulatory mechanism that controls RelA activity. The fact that deficiency of TBK1, which does not participate in the canonical pathway but may mediate phosphorylation of RelA, also results in massive fetal liver apoptosis 30 may indicate that the latter is the case.
It remains surprisingly unclear whether specificity between NFκB dimers and DNA sequences plays an important role in NFκB-dependent gene activation. In macrophages, IL12 p40 was reported to specifically require cRel for its LPS-induced gene expression 31. A systematic genetic analysis of the RelA-containing NFκB dimers showed that some NFκB target genes had different activation requirements with regard to specific NFκB proteins 32. Further, swapping promoter-binding sites between NFκB-dependent genes altered NFκB dimer specificity, suggesting that single nucleotides within the κB site may be determinants for dimer specificity 33. RelA/cRel chimeras identified amino acids within the DNA-binding domain as mediating the specificity of cRel for IL12 transcriptional control 34. Interestingly, there are indications that dimer specificity may not be mediated by large affinity differences in dimer-κB site interactions, but may be mediated by alternate conformations of dimers triggered by alternate κB site sequences 33, 35. According to this model, DNA-triggered allostery of the NFκB dimer may be “read” by co-activators or transcriptional activators, such as Bcl3, CBP, or IRF3, interacting with the NFκB dimer.
Physiologically, dimer specificity is suggested by distinct phenotypes of knockouts; however, phenotypic differences may be due to distinct tissue-specific expression of the proteins (e.g., cRel is abundant in the lymphoid system) or pathway-specific activation of dimers (e.g., RelA/cRel vs RelB). Thus, the extent of NFκB dimer specificity in regulating NFκB target genes and the underlying mechanisms for such specificity remain to be addressed. Contemporary tools of genome-wide expression and location studies, and bioinformatic analysis, should prove useful in this endeavor.
Kinetic control of the canonical pathway
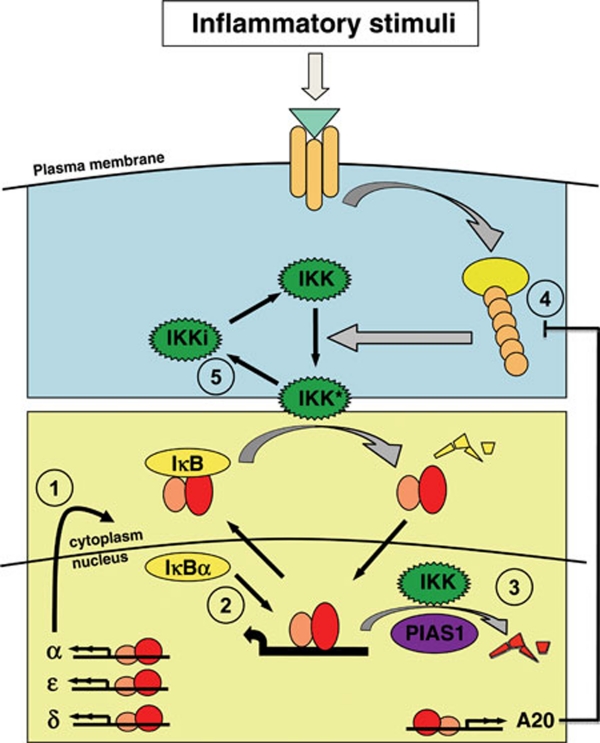
The activation of the canonical pathway results in expression of inflammatory and immune response genes. These signals must be transient and properly controlled, as prolonged NFκB activation can lead to aberrant gene expression, and the misregulation of NFκB activation has been implicated in pathologies including chronic inflammation and cancer. Hence, it is not surprising that numerous mechanisms have evolved to provide for post-induction attenuation or termination of signaling (Figure 5).
Figure 5.
Mechanisms of canonical pathway attenuation. The best characterized attenuation mechanism is negative feedback synthesis of IκB proteins (1). IκBα was recently shown to have the ability to enhance the dissociation rate of NFκB from DNA, which may facilitate its negative feedback control function (2). In addition, DNA-bound RelA NFκB, when phosphorylated on S536 by nuclear IKK complexes, was shown to be subjected to ubiquitination by the E3 ligase PIAS1, which targets NFκB to degradation (3). The ubiquitin protease A20, a highly inducible NFκB target gene, attenuates the IKK activation pathway by counteracting E3 ligases involved in the formation of ubiquitin chains that are critical signaling scaffolds (4). In addition, canonical IKK was proposed to undergo autophosphorylation that results in an inactivated kinase (5); a recycling step is necessary to return IKK back to the activatable state.
The biological importance of the IκBs as negative feedback regulators in NFκB signaling has been established. Both IκBα and IκBɛ are inducibly expressed and function as negative feedback regulators of NFκB. IκBα knockouts result in lethality 7-10 days after birth due to hyperinflammation 36. In addition, mice with mutated κB enhancers of the IκBα promoter display shortened life span (13-15 months), hypersensitivity to septic shock and abnormal T-cell development/activation 37. IκBɛ knockout mice are not lethal, but result in the increased expression of cytokines 38. Mice deficient in both IκBα and IκBɛ result in neonatal lethality and almost complete absence of both B and T cells and reduced NK cell numbers 39, indicating overlapping functions of these two IκBs that may therefore be considered fail-safe duplicates. Indeed, IκBɛ is upregulated in resting IκBα-deficient cells. This upregulation is reversible upon IκBα reconstitution in the deficient cells 40.
Interestingly, it was recently shown that spontaneous dissociation of NFκB RelA:p50 from DNA can be enhanced by IκBα, but not by IκBβ 41. Kinetic enhancement of NFκB-DNA complex dissociation by IκBα may facilitate negative feedback-mediated NFκB post-induction repression, particularly in the transcriptional deactivation of genes that have multiple κB sites. However, further studies are required to test this prediction.
An additional IκB negative feedback loop was recently characterized. Newly synthesized nfκb2 protein p100 may form multimers in the absence of sufficient IKK1 signaling that catalyzes its degradation or processing into p52. These multimers may exhibit IκB activity when a p100 ARD traps RelA:p50 dimers in trans. Because p100 expression is inducible by RelA:p50, this forms a negative feedback loop that has slow kinetics, yet is not degraded when canonical IKK activity persists. Computational simulations-directed experimental studies revealed that IκBδ forms a negative feedback loop to regulate RelA:p50 activity in a stimulus-specific manner, such that it is effective to LPS but not to TNF 3. Indeed, the work revealed that IκBα provides effective negative feedback only to cytokine stimuli that produce transient canonical IKK activity. Perinatal lethality observed in IκBα-deficient mice could be rescued when the inducing stimulus TNF was ablated.
IκBs function as stoichiometric inhibitors that quickly sequester NFκB in the cytoplasm. A20, on the other hand, inhibits signaling upstream of IKK via its enzymatic function as a protease of signaling-associated K63-linked ubiquitin chains 42. While both A20 and IκBα are inducible negative regulators, only IκBα functions as a dynamic negative feedback regulator 43. Instead, the evidence indicates that A20 may be thought of as a tunable rheostat that determines IKK and NFκB responsiveness to subsaturating stimuli.
Degradation of promoter-bound RelA dimers may also be a stimulus-induced post-induction attenuation mechanism. RelA degradation was proposed to be essential for the transient activity of NFκB RelA dimers 44. It has also been shown that genes activated by RelA dimers can be negatively regulated by the E3 ligase PIAS1, a member of the protein inhibitor of activated STAT (PIAS) family 45. Pias1−/− mice showed significantly elevated RelA binding to κB sites in genes that also showed hyper-expression 46. A second line of research provided evidence that promoter-bound RelA and cRel can be phosphorylated by IKK activity thereby catalyzing their degradation 47. This would constitute a negative feed-forward mechanism, a potential timing device that ensures that the released NFκB activity is transient. While IKK results in the phosphorylation of IκBs and the nuclear translocation and activation of NFκB, it may also limit the duration of NFκB activity through the direct phosphorylation of RelA and cRel that targets them to the ubiquitin proteasome pathway.
Not only does negative feedback limit the duration of stimulus-induced NFκB activity and the magnitude of the gene expression response, it may also mediate the transduction of stimulus-specific information via a “temporal signaling code” that determines which of the many possible target genes are activated in response to a specific stimulus. IκBα deficiency for example, which alters the TNF-induced dynamic controls, results in inappropriate gene expression 48. Further, IκBɛ appears to dampen IκBα-mediated NFκB oscillations, because its mRNA induction is delayed to occur in anti-phase with that of IκBα 49. In addition, newly synthesized IκBδ forms a negative feedback loop to regulate RelA:p50 activity in a stimulus-specific manner, as described above 3. Single-cell studies with fluorescent fusion proteins of RelA and IκBα have drawn attention to the potential for oscillatory control of NFκB, suggesting that frequency of successive NFκB peaks is a critical feature of the temporal code 50, 51. However, it remains unclear whether inappropriate expression of the fusion protein may impact the physiological relevance of these observations and whether frequency of NFκB peaks is in fact a function of the stimulus. An alternate view may be that cell-to-cell variability of periodic NFκB activities (potentially the result of transcriptional bursting of the short-lived IκBα inhibitor) may impart functional robustness at the organ or tissue level 52. The temporal code may thus be better described as amplitude modulated (AM) rather than frequency modulated (FM). However, the information carrier of AM signals may involve periodic or irregularly spaced activity peaks.
Stimulus-specific temporal control of NFκB is a result of the stimulus-specific temporal control of the canonical IKK activity 53. It is interesting to note that TNF-induced IKK activity rises rapidly and is rapidly attenuated. Though the negative regulator A20 plays some role in determining this temporal profile, it remains unknown what the mechanisms are, which ensure that IKK activity undergoes rapid post-induction repression. Early studies identified C-terminal residues on the IKK2 protein that dampen its kinase activity, suggesting a cyclical control cycle 10. However, this model has not been substantiated. In contrast, LPS-induced IKK activity shows a mode of slower activation but of longer duration. Interestingly, the longer duration is in part due to co-opting the TNFR pathway at late time points through the expression of TNF at early time points 53, 54.
Stimulus-specific temporal profiles of IKK and NFκB are correlated with the expression of stimulus-specific genes. Although inactivation of the TNF autocrine loop provided a means to manipulate the LPS-specific IKK temporal profiles 53, this intervention may also affect a parallel pathway. Thus, whether the stimulus-specific temporal profiles of NFκB activity encode information about the stimulus that is critical for stimulus-specific gene expression remains to be tested rigorously. Furthermore, it remains unknown how promoters of activated genes, or associated gene regulatory networks, distinguish between different temporal profiles. Understanding the mechanisms by which the temporal activity profile is decoded by target genes and elucidating what features of the temporal profile of NFκB activity carry stimulus-specific information remain the pressing questions in the field 55.
Finally, studies of the cellular steady state have shown that a short half-life of free classical IκB proteins (< 10 min) necessitates a surprisingly high constitutive synthesis rate 40. Subsequent work showed that the short half-life of free IκBα is mediated by the ubiquitin-independent proteasome pathway and is likely due to incompletely folded regions of C-terminal ankyrin repeats that have evolved away from the consensus 56, 57. Further studies showed that the high apparently futile flux of IκB metabolism ensures relative resistance to metabolic and ribotoxic stress agents, such as UV and UPR, ensuring that the NFκB signaling module is dedicated to sensors of the extracellular environments such as TLR and cytokine receptors that produce significant changes in canonical IKK activity 58. Thus, the distinct degradation pathways between the free and bound IκBs result in highly dynamic homeostatic control of the NFκB signaling module, which imparts functional robustness to this signaling system.
The non-canonical pathway
Activation of the non-canonical NFκB pathway (Figure 6) is mediated through a NEMO-independent IKK1 kinase complex 59. Recent reports indicated that the regulated assembly of the upstream signaling complex that involves cellular inhibitor of apoptosis (cIAP1 and cIAP2), TRAF2 and TRAF3, and NIK is crucial for the activation of this pathway. Upon receptor engagement, TRAF2-dependent recruitment of cIAP1 and cIAP2 results in activation of cIAPs and K63-linked ubiquitination by TRAF2. TRAF3 serves as an adaptor molecule that allows for the assembly of a complex involving NIK, TRAF2 and activated cIAPs, which causes TRAF3 itself to be modified by K48-linked ubiquitination by cIAPs 60, 61. As NIK protein expression is regulated by TRAF3, signal-induced proteosomal degradation of TRAF3 leads to accumulation of NIK protein and activation of the downstream pathway 62, 63. Increase in NIK protein results in phosphorylation of p100 at Ser866 and Ser870, and activation of IKK1 64, 65. Activated IKK1-containing kinase complex further phosphorylates p100 at Ser99, Ser108, Ser115, Ser123 and Ser872. Phosphorylation of these residues provides a signal for p100 to be recognized by the SCF/βTRCP ubiquitin ligase complex, and this results in partial degradation of the C-terminal ARD of p100 by the 26S proteosome 66. Two mechanisms of NFκB activation ensue: first IκBδ within the p100-containing IκBsome is inactivated through degradation of the C-terminal ARD of p100, leading to the release of associated RelB:p50 and RelA:p50 dimers. Second, co-translational processing of p100 that requires de novo protein synthesis leads to nuclear translocation of the major RelB-containing dimer, RelB:p52, and RelA:p52 as a minor species 1, 67, 68.
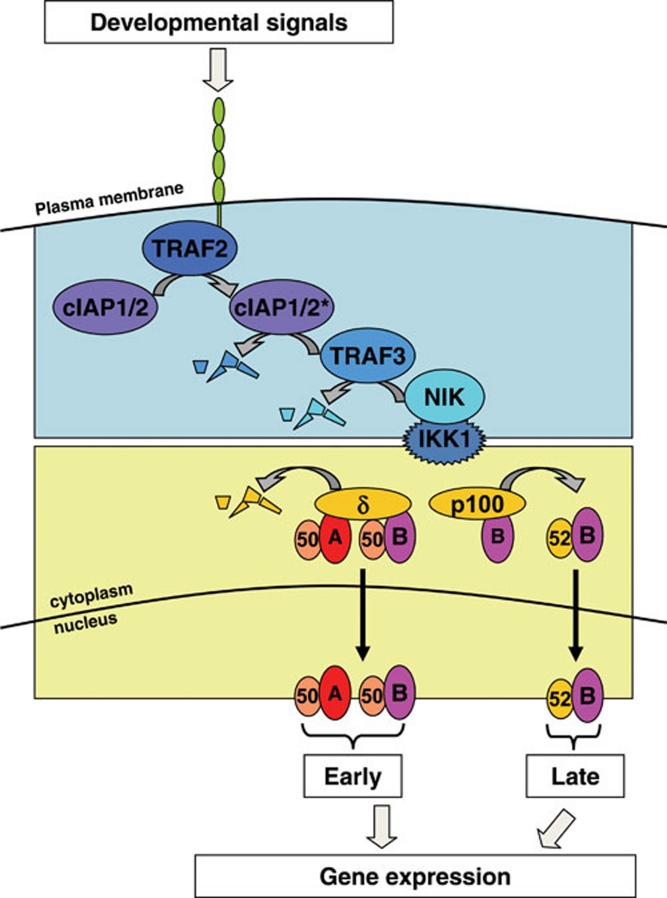
Figure 6.
The non-canonical NFκB pathway. Receptor engagement leads to recruitment and activation of cIAP1/2 mediated by TRAF2 resulting in the degradation of TRAF3. Decreased levels of TRAF3 stabilize NIK, which in turn activates IKK1 activity. Degradation of IκBδ following non-canonical IKK activation releases RelA:p50 or RelB:p50 dimers sequestered by IκBδ. Processing of newly synthesized p100 also occurs and generates RelB:p52 complex, whose levels slowly build up to provide sustained activity of the RelB:p52 dimer. Translocation of NFκB dimers to the nucleus activates gene expression program.
The sequence of molecular events that result in stimulus-induced RelB:p52 dimers remains surprisingly poorly understood. RelB associates with p100 through multiple protein-protein contacts. This multidomain interaction results in protein co-stabilization, as nfκb2−/− cells showed reduced level of RelB protein and relb−/− cells have decreased amounts of p100 protein 69, 70. Counterintuitively, this interaction also inhibits p100 processing and RelB:p52 formation, suggesting that RelB:p52 dimer formation may occur after p100 processing, though it may involve alternate, transient interactions with p100.
Unlike the canonical NFκB pathway, there are few reports for negative feedback control of the non-canonical pathway. NFκB-inducible expression of TRAF3, which plays a negative role in non-canonical signaling, may impede RelB activation by degrading NIK. Phosphorylation of NIK on Ser809, Ser812 and Ser815 by IKK1 destabilizes NIK and provides a way to fine-tune non-canonical signaling 71. MicroRNAs – miR-223, miR-15a, miR-16 – that have been implicated in targeting IKK1 during macrophage differentiation may modulate the strength of non-canonical signaling, if upregulated upon stimulation 72. In primary thymocytes, phosphorylation of RelB stimulated by TPA/ionomysin leading to degradation of RelB may serve as a mechanism to control signal-inducible protein turnover 73. In general, however, dynamic control of RelB activity has not been observed. It seems unlikely that there is a mechanism analogous to IκB-mediated removal of RelA:p50 from κB sites or the nucleus that is applicable to nuclear RelB-containing dimers.
The non-canonical pathway is activated by the engagement of a subset of tumor necrosis factor receptors that are involved in development. Given roles of the non-canonical pathway in developmental processes that require sustained signaling, its kinetics involve a slow build-up of activity and long-lasting activity, as opposed to the acute activation of RelA-containing dimers induced by inflammatory stimuli. Stimuli that have been reported to activate non-canonical signaling include lymphotoxin β (LTβ), B cells activating factor (BAFF), CD40 ligand and receptor activator of NFκB ligand (RANKL), etc. Studies using genetically deficient cells or mice have shown the essential role of non-canonical NFκB pathway in various biological processes. Mice deficient in components of non-canonical pathway – relb−/−, nfκb2−/−, nik−/− and NIKaly/aly mice that bear a point mutation on nik alleles – showed defective development of lymph nodes and Peyer's Patch 74, 75, 76, 77, 78. p100 and RelB are required for maintaining proper splenic architecture as nfκb2−/− and relb−/− mice lack germinal centers and follicular dendritic cells network 79, 80, 81. Reduced number of marginal zone B cells was observed in both nfκb2−/− and relb−/−, indicating the importance of non-canonical signaling in B-cell development 81. Involvement of the non-canonical pathway in mammary gland development has also been implicated. IKK1AA mice that do not have functional IKK1 kinase activity are defective in lactation during pregnancy 82. RelB:p52 activation was proposed to rescue the delay in the early mammary gland development observed in transgenic mice overexpressing the IκBα super-repressor 83. Defective osteoclastogenesis observed in nik−/− mice can be restored by overexpressing RelB, but not RelA, indicating a specific function of RelB in osteoclast differentiation 84. Further, specific expression of relb transcripts was found in antigen-presenting cells, and requirement of RelB for CD4+CD8α− dendritic cell development was reported 85, 86, 87. Even though the physiological function of RelB in DC and MZB cell development has been established, the mechanistic aspects of its regulation and the downstream target gene expression programs have not been revealed.
Supporting a role of the non-canonical pathway in secondary lymphoid organ development, splenocytes from IKK1AA knock-in mice injected with LTβR agonist showed defective expression of chemokine genes, including secondary lymphoid tissue chemokine (SLC), EBI1 ligand chemokine (ELC), B lymphoblastoid cell chemokine (BLC), and stromal cell-derived factor 1α (SDF-1α) 88. Dependence on RelB for expression of these genes was further established by comparing molecular events downstream of LTβR signaling in wt and relb−/− MEF 70. Selective recruitment of RelB to the promoter region of these genes and in vitro binding assays between NFκB dimeric complexes and DNA probes suggested that RelB:p52 recognizes specific κB-binding sites that are different from those bound by RelA:p50 89. However, genome-wide gene expression analysis derived from LTβR-stimulated MEF revealed that the majority of the induced genes require both RelA- and RelB-containing dimers 90. RelB:p52 and RelA:p50 complexes derived from eukaryotic cells showed similar ability to bind to consensus binding sites 91. Further, B cells with constitutive IKK2 activity (IKK2ca) show rescue of the developmental block observed in BAFF-R-deficient B cells 92. It suggests that nuclear accumulation of RelA, cRel and p50 in IKK2ca+ B cells can compensate for the loss of RelB:p52 activation in B cells deficient in BAFF-R signaling. Interestingly, analysis of crystal structure of RelB-containing dimers suggested that RelB may recognize a broader range of κB sequences than other dimers 93, 94. In sum, studies performed so far have not been able to reach a definite conclusion if different NFκB dimers activated by the non-canonical pathway have specific or overlapping gene activation functions.
Crosstalk between canonical and non-canonical pathways
Activation of the canonical NFκB pathway is generally associated with inflammatory exposure, whereas activation of the non-canonical pathway is mostly related to developmental cues. These two pathways were first thought to transduce signals independently and have separate physiological functions 95. Interestingly, several studies have provided evidence that inter-connections between these two pathways exist.
Control of RelA:p50 by non-canonical signals
In early overexpression studies, RelA was reported to be associated with the C-terminal portion of p100 96. Mice expressing a mutant form of p100 that does not allow for processing show defective activation of RelA-containing dimers, impaired development of secondary lymphoid organs and B cells, and impaired osteoclastogenesis 97. Cytosolic accumulation of p100 in nik−/− osteoclast precursors leads to enhanced association of RelA with p100 and defective RANKL-induced osteoclastogenesis 98. Sequestration of RelA:p50 by p100 mediates a immunosuppressive phenotype observed in nik−/− mice through regulating the activation of naïve T cells 99. Developmental LTβR signaling disrupts the IκBδ inhibitory complex, which results in the release of canonical effector RelA:p50 into the nucleus. Further, increased amount of RelA associated with IκBδ in TNF-primed cells leads to enhanced LTβR-induced inflammatory gene expression 1. These studies indicated that IκBδ, capable of inhibiting RelA and responsive to non-canonical stimuli, serves as a mediator of crosstalk between canonical and non-canonical NFκB signaling. Moreover, regulation of IκBδ may thus modulate NFκB/RelA activity by balancing canonical and non-canonical signaling, when cells are physiologically exposed to multiple stimuli.
Given this emerging understanding of NFκB control (Figure 7A), chronic inflammatory mediators should not give rise to chronically elevated NFκB activation given the irreversible feedback control mediated by IκBδ. However, in a microenvironment where inflammatory exposure is accompanied by developmental factors that promote IκBδ degradation, prolonged RelA:p50 activity may be the outcome. Tonic developmental signals, which are not considered detrimental, may exacerbate chronic inflammation when IκBδ is constitutively degraded. Indeed, several studies have found that B- and T-cell leukemias/lymphomas have mutations in nfκb2 gene that result in the removal of its inhibitory domain 100, 101. Elevated expression of NIK and p100 processing, a hallmark of non-canonical pathway activation, were found in multiple myeloma cell lines and primary MM patients' samples 102, 103. However, it should be noted that RelA-containing dimer can be activated through a NIK-IKK2 axis when NIK protein accumulates in cells 60, 104. Clarifying the contribution of non-canonical pathway activation in the misregulation of canonical NFκB effectors will affect treatment strategies for inflammatory diseases and cancers.
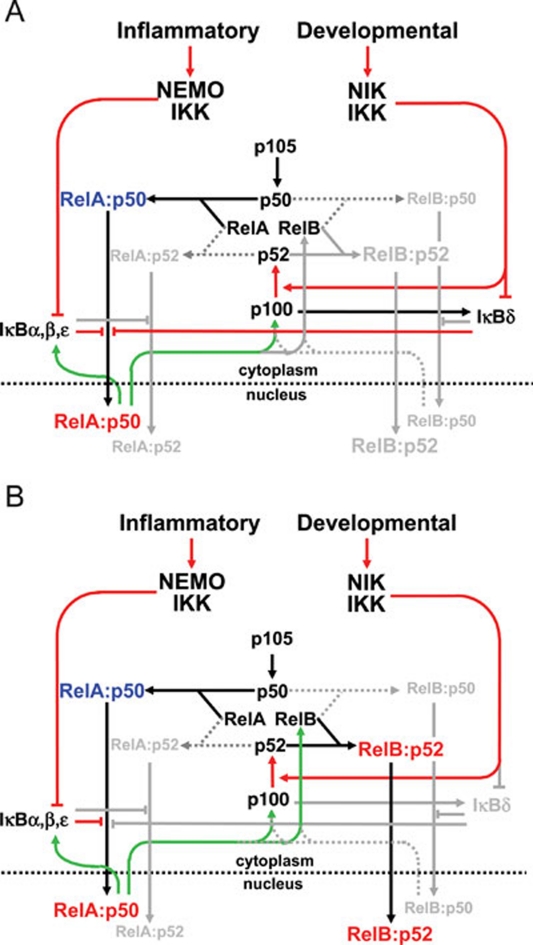
Figure 7.
Wiring diagrams of the NFκB signaling system to chart crosstalk between canonical and non-canonical signaling pathways. (A) Non-canonical control of RelA:p50. RelA:p50 is inhibited by not only IκBα, -β, -ɛ, but also IκBδ. Whereas inflammatory canonical signals lead to degradation of IκBα, -β, -ɛ, developmental signals engage the non-canonical pathway to disrupt IκBδ activity; however, both result in the nuclear translocation of RelA:p50. Interestingly, prior canonical signaling history results in an enhancement of the non-canonical-RelA:p50 axis, due to inducible expression of p100. (B) Canonical control of RelB:p52 activation. Expression of p100 and RelB are dependent on RelA:p50 activity and therefore canonical signals. The amount of basal RelB expression, controlled by constitutive canonical pathway activity, rather than the inducible p100 expression, is the main determinant of the strength of non-canonical signaling.
RelA:p50 control of non-canonical signaling
Another layer of interdependency between the canonical and non-canonical NFκB pathways is based on transcriptional control (Figure 7B). Early studies showed that the promoter regions of relb and nfκb2 genes contain κB-binding sites and transcriptional regulation of these two genes is dependent on RelA 105, 106. LTβR signaling was shown to activate RelA:p50 followed by RelB:p52 via two distinct pathways 107. Activation of RelB-containing dimers by LTβR signaling is deficient in rela−/− MEF, suggesting a requirement of RelA for transcription of relb and nfκb2. Reconstitution of relb or constitutive form of IKK2 into rela−/− MEF restored LTβR-induced RelB activation. However, overexpression of p100 in rela−/− MEF failed to restore LTβR-induced RelB activity 70. These genetic/biochemical analyses indicated that homeostatic relb transcription driven by basal RelA activity, rather than p100 synthesis induced by RelA activation, is the main determinant of non-canonical pathway responses. Interestingly, mice lacking RelA (TNFR1-deficiency background) mirrored the phenotypes observed in mice deficient in components of the non-canonical pathway. RelA-deficient mice showed an absence of lymph nodes, Peyer's Patch and disorganized splenic microarchitecture 108, supporting the cross-regulatory mechanism between canonical and non-canonical pathways provided by ex vivo studies. The requirement of canonical NFκB activity for providing non-canonical NFκB activation in lymph nodes formation represents a physiologically relevant interdependency between these two pathways.
Functional overlap between NFKB1 and NFKB2
NFκB gene deletion studies have shown that there is an unexpected interdependence involving functional overlap between nfκb1, which encodes p105/p50, and nfκb2, which encodes p100/p52. The interplay between these two genes and their importance in both the canonical and non-canonical pathways illustrate the interdependence of the two pathways.
Non-canonical signaling results in prominent RelB:p52 dimer activation that is thought to be important for lymph node organogenesis. Yet, nfκb2−/− mice do not phenocopy relb−/− mice in terms of lymph node formation and germinal center organization 109. Biochemical analysis did confirm that nfκb2-deficient cells show reduced responsiveness to LTβR signaling due to lack of p100, which forms the non-canonical regulator IκBδ and is a necessary precursor for p52-containing complexes formed upon LTβR engagement 70. However, mutant cells also revealed that the deficiency of the RelB-binding partner p52 results in formation of the RelB:p50 dimer. Thus, constitutive RelB:p50 NFκB activity, which is not controlled by classical IκB proteins, IκBα, -β and -ɛ, may compensate for the loss of non-canonical pathway-inducible RelB:p52 activity (Figure 8A). These observations provide explanations for the intermediate lymph node phenotypes in nfκb2−/− mice 109.
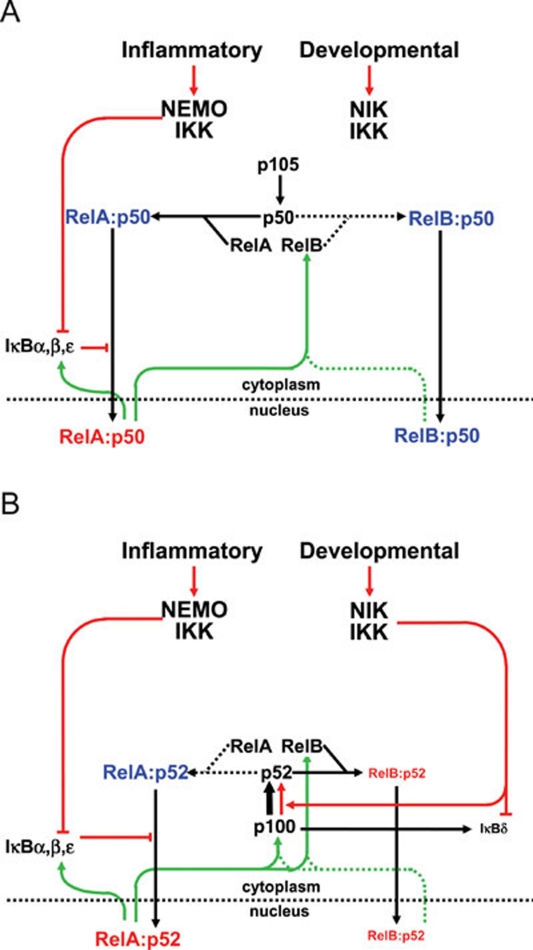
Figure 8.
Wiring diagrams of the NFκB signaling system to chart the interdependence and functional overlap of nfκb1 and nfκb2 gene products. (A) The NFκB signaling system in nfκb2−/− cells. Without its normal binding partner p52, RelB will bind p50 to form RelB:p50 dimers, which accumulate in the nucleus. As a result, constitutive RelB:p50 activity compensates for the loss of inducible RelB:p52 activity. (B) The NFκB signaling system in nfκb1−/− cells. Without its normal binding partner p50, RelA will bind p52 causing an increase in constitutive p100 processing and thus a depletion of the IκBδ and p100 pool, which is required for RelB:p52 activation. Though the canonical pathway is largely preserved in nfκb1−/− cells, compensation by p52 weakens the non-canonical pathway.
Canonical activation results in the ubiquitous RelA:p50 dimer. Yet nfκb1−/− mice show only mild phenotypes in inflammatory regulation, not nearly as severe as rela−/− mice 110. Biochemical analysis of mutant cells showed that loss of p50 results in the formation of RelA:p52, near normal RelA activation in response to inflammatory stimuli, and activation of several RelA target genes 32.
Further studies were prompted by the unexpected finding that nfκb1−/− mice also showed lymph node organogenesis defects 109. Biochemical analysis revealed that the compensatory generation of p52 via enhanced constitutive p100 processing to provide the binding partner for RelA resulted in reduced p100 levels in nfκb1−/− cells, thereby weakening the responsiveness of the non-canonical signaling pathway 70. Thus, upon stimulation by a non-canonical stimulus such as BAFF or LTβ, less p100 is available for processing to p52 to form a RelB:p52 dimer complex. Much of the p100 will have already been processed for the formation of the RelA:p52 dimer which is dedicated to canonical signaling (Figure 8B). This weakening of the non-canonical pathway provides a mechanistic explanation for the lymph node organogenesis phenotypes of nfκb1−/− mice 70.
In sum, p50 and p52 appear to have largely overlapping functions in dimer formation (despite preferences for RelA and RelB, respectively) and target gene activation. However, compensation comes at a price, as the precursors p105 and especially p100 have signaling functions. When their pool is diminished due to compensation in dimer formation, deficiencies in signaling can occur. Hence, functional overlap between p50 and p52 revealed interdependencies in precursor processing and nfκb1 and nfκb2 gene function.
Conclusions
The majority of previous NFκB review articles have focused on the existence of two separate signaling pathways mediated through two biological classes of receptors (inflammatory vs immune cell maturation) that result in activation of two temporally distinct NFκB dimers, namely the RelA/cRel- and RelB-containing dimers. Recent work has indicated that numerous interconnections between canonical and non-canonical NFκB pathways exist, and these may be crucial in various biological processes. Gene knockout studies also revealed functional overlap and interdependencies between canonical and non-canonical pathways. In sum, we propose the view that NFκB signaling mediated by NEMO-dependent or NEMO-independent IKK complexes should be considered within the context of a single regulatory or signaling system. Indeed, biological outcomes are the result of numerous stimuli and intracellular and intercellular signals. A systems view of NFκB signaling allows one to make sense of otherwise counterintuitive phenomena in physiology and pathology.
Within the NFκB signaling system, stimulus-responsive dynamic regulation of NFκB activity is achieved by a network of myriad biochemical reactions that govern the synthesis and degradation of NFκB dimers and IκB proteins, their interactions with each other, and subcellular localization of the components and complexes. All reactions are potentially regulated by signaling events that are triggered by extracellular stimuli. As reviewed here, in the canonical signaling axis, the focus has been on stimulus-induced degradation and feedback resynthesis of IκB proteins, though homeostatic control of free IκB synthesis and degradation has also been shown to play a critical role. In the non-canonical signaling axis, the focus has been on stimulus-induced generation of the RelB:p52 dimer, though inactivation of the IκBδ activity was also shown to be important. However, signaling crosstalk mechanisms demonstrate that biochemical reactions that are not usually associated with a particular signaling axis should also be considered.
A systems understanding of NFκB control may seem dauntingly complex: several different IKK kinase complexes, at least 5 IκBs and 15 potential NFκB dimeric complexes must be taken into account, as well as their interdependent regulation and temporal control through feedback. Indeed, to understand NFκB signaling in such a manner requires the development of mathematical models that represent the biochemical reactions. First of all, such models are useful as repositories of what is known about a regulatory system. Second, when the models are parameterized with rate constants from biophysical or cell biological experiments, they also allow for in silico computational simulations that can be helpful in interpreting experimental data and often directing experimental design by making interesting predictions. Just like model diagrams on paper, such models are a work in progress; however, they are better suited for quantitative and kinetic arguments and thus they can be a useful tool in ongoing research to characterize the functioning of a pathway. Third, when a mathematical model has been extensively validated, it may then in fact be considered a model system, not unlike a model organism or a HeLa or MEF cell line, with which results obtained have value in their own rights in the context of multipronged approaches to discover the molecular basis of human health and disease. Fourth, mathematical model reduction strategies allow one to characterize the regulatory design principles underlying specific regulatory phenomena. As described in this review, an experimentally validated mathematical model of the NFκB signaling system that accounts for the intricate molecular mechanisms pertaining to both canonical and non-canonical signaling will have broad applicability in characterizing the mechanisms underlying cellular regulation in numerous physiological and pathological states.
References
- Basak S, Kim H, Kearns JD, et al. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savinova OV, Hoffmann A, Ghosh G. The Nfkb1 and Nfkb2 proteins p105 and p100 function as the core of high-molecular-weight heterogeneous complexes. Mol Cell. 2009;34:591–602. doi: 10.1016/j.molcel.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih VF, Kearns JD, Basak S, et al. Kinetic control of negative feedback regulators of NF-kappaB/RelA determines their pathogen- and cytokine-receptor signaling specificity. Proc Natl Acad Sci USA. 2009;106:9619–9624. doi: 10.1073/pnas.0812367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidereit C. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene. 2006;25:6685–6705. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- Rudolph D, Yeh WC, Wakeham A, et al. Severe liver degeneration and lack of NF-kappaB activation in NEMO/IKKgamma-deficient mice. Genes Dev. 2000;14:854–862. [PMC free article] [PubMed] [Google Scholar]
- Li Q, Van Antwerp D, Mercurio F, Lee KF, Verma IM. Severe liver degeneration in mice lacking the IkappaB kinase 2 gene. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- Luedde T, Heinrichsdorff J, de Lorenzi R, et al. IKK1 and IKK2 cooperate to maintain bile duct integrity in the liver. Proc Natl Acad Sci USA. 2008;105:9733–9738. doi: 10.1073/pnas.0800198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinke S, Robinson MJ, Hugunin M, Ley SC. Lipopolysaccharide activation of the TPL-2/MEK/extracellular signal-regulated kinase mitogen-activated protein kinase cascade is regulated by IkappaB kinase-induced proteolysis of NF-kappaB1 p105. Mol Cell Biol. 2004;24:9658–9667. doi: 10.1128/MCB.24.21.9658-9667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhase M, Hayakawa M, Chen Y, Karin M. Positive and negative regulation of IkappaB kinase activity through IKKbeta subunit phosphorylation. Science. 1999;284:309–313. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009;36:302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyet J-L, Srinivasula SM, Lin J-h, et al. Activation of the IκB kinases by RIP via IKKγ/NEMO-mediated oligomerization. J Biol Chem. 2000;275:37966–37977. doi: 10.1074/jbc.M006643200. [DOI] [PubMed] [Google Scholar]
- Haas TL, Emmerich CH, Gerlach B, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- Lamhamedi-Cherradi SE, Zheng S, Hilliard BA, et al. Transcriptional regulation of type I diabetes by NF-kappa B. J Immunol. 2003;171:4886–4892. doi: 10.4049/jimmunol.171.9.4886. [DOI] [PubMed] [Google Scholar]
- Mason NJ, Liou HC, Hunter CA. T cell-intrinsic expression of c-Rel regulates Th1 cell responses essential for resistance to Toxoplasma gondii. J Immunol. 2004;172:3704–3711. doi: 10.4049/jimmunol.172.6.3704. [DOI] [PubMed] [Google Scholar]
- Ouaaz F, Arron J, Zheng Y, Choi Y, Beg AA. Dendritic cell development and survival require distinct NF-kappaB subunits. Immunity. 2002;16:257–270. doi: 10.1016/s1074-7613(02)00272-8. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang X, Hussain S, et al. Distinct roles of different NF-kappa B subunits in regulating inflammatory and T cell stimulatory gene expression in dendritic cells. J Immunol. 2007;178:6777–6788. doi: 10.4049/jimmunol.178.11.6777. [DOI] [PubMed] [Google Scholar]
- Carrasco D, Cheng J, Lewin A, et al. Multiple hemopoietic defects and lymphoid hyperplasia in mice lacking the transcriptional activation domain of the c-Rel protein. J Exp Med. 1998;187:973–984. doi: 10.1084/jem.187.7.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- Doi TS, Marino MW, Takahashi T, et al. Absence of tumor necrosis factor rescues RelA-deficient mice from embryonic lethality. Proc Natl Acad Sci USA. 1999;96:2994–2999. doi: 10.1073/pnas.96.6.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcamo E, Mizgerd JP, Horwitz BH, et al. Targeted mutation of TNF receptor I rescues the RelA-deficient mouse and reveals a critical role for NF-kappa B in leukocyte recruitment. J Immunol. 2001;167:1592–1600. doi: 10.4049/jimmunol.167.3.1592. [DOI] [PubMed] [Google Scholar]
- Grossmann M, Metcalf D, Merryfull J, et al. The combined absence of the transcription factors Rel and RelA leads to multiple hemopoietic cell defects. Proc Natl Acad Sci USA. 1999;96:11848–11853. doi: 10.1073/pnas.96.21.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugasyan R, Voss A, Varigos G, et al. The transcription factors c-rel and RelA control epidermal development and homeostasis in embryonic and adult skin via distinct mechanisms. Mol Cell Biol. 2004;24:5733–5745. doi: 10.1128/MCB.24.13.5733-5745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senftleben U, Li ZW, Baud V, Karin M. IKKbeta is essential for protecting T cells from TNFalpha-induced apoptosis. Immunity. 2001;14:217–230. doi: 10.1016/s1074-7613(01)00104-2. [DOI] [PubMed] [Google Scholar]
- Prendes M, Zheng Y, Beg AA. Regulation of developing B cell survival by RelA-containing NF-kappa B complexes. J Immunol. 2003;171:3963–3969. doi: 10.4049/jimmunol.171.8.3963. [DOI] [PubMed] [Google Scholar]
- Bonnard M, Mirtsos C, Suzuki S, et al. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kappaB-dependent gene transcription. EMBO J. 2000;19:4976–4985. doi: 10.1093/emboj/19.18.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjabi S, Hoffmann A, Liou HC, Baltimore D, Smale ST. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc Natl Acad Sci USA. 2000;97:12705–12710. doi: 10.1073/pnas.230436397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Leung TH, Baltimore D. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. EMBO J. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung TH, Hoffmann A, Baltimore D. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Sanjabi S, Williams KJ, Saccani S, et al. A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes Dev. 2005;19:2138–2151. doi: 10.1101/gad.1329805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Park FE, Huang DB, Noro B, Thanos D, Ghosh G. The kappa B DNA sequence from the HIV long terminal repeat functions as an allosteric regulator of HIV transcription. J Biol Chem. 2002;277:24701–24708. doi: 10.1074/jbc.M200007200. [DOI] [PubMed] [Google Scholar]
- Beg AA, Sha WC, Bronson RT, Baltimore D. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- Peng B, Ling J, Lee AJ, et al. Defective feedback regulation of NF-kappaB underlies Sjogren's syndrome in mice with mutated kappaB enhancers of the IkappaBalpha promoter. Proc Natl Acad Sci USA. 2010;107:15193–15198. doi: 10.1073/pnas.1005533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memet S, Laouini D, Epinat JC, et al. IkappaBepsilon-deficient mice: reduction of one T cell precursor subspecies and enhanced Ig isotype switching and cytokine synthesis. J Immunol. 1999;163:5994–6005. [PubMed] [Google Scholar]
- Samson SI, Memet S, Vosshenrich CA, et al. Combined deficiency in IkappaBalpha and IkappaBepsilon reveals a critical window of NF-kappaB activity in natural killer cell differentiation. Blood. 2004;103:4573–4580. doi: 10.1182/blood-2003-08-2975. [DOI] [PubMed] [Google Scholar]
- O'Dea EL, Barken D, Peralta RQ, et al. A homeostatic model of IkappaB metabolism to control constitutive NF-kappaB activity. Mol Syst Biol. 2007;3:111. doi: 10.1038/msb4100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergqvist S, Alverdi V, Mengel B, et al. Kinetic enhancement of NF-κB·DNA dissociation by IκBα. Proc Natl Acad Sci USA. 2009;106:19328–19333. doi: 10.1073/pnas.0908797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Werner SL, Kearns JD, Zadorozhnaya V, et al. Encoding NF-kappaB temporal control in response to TNF: distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 2008;22:2093–2101. doi: 10.1101/gad.1680708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Marazzi I, Beg AA, Natoli G. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor kappaB response. J Exp Med. 2004;200:107–113. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Yang R, Wong KA, et al. Negative regulation of NF-kappaB signaling by PIAS1. Mol Cell Biol. 2005;25:1113–1123. doi: 10.1128/MCB.25.3.1113-1123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahk S, Liu B, Chernishof V, et al. Control of specificity and magnitude of NF-kappa B and STAT1-mediated gene activation through PIASy and PIAS1 cooperation. Proc Natl Acad Sci USA. 2007;104:11643–11648. doi: 10.1073/pnas.0701877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature. 2005;434:1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- Kearns JD, Basak S, Werner SL, Huang CS, Hoffmann A. IkappaBepsilon provides negative feedback to control NF-kappaB oscillations, signaling dynamics, and inflammatory gene expression. J Cell Biol. 2006;173:659–664. doi: 10.1083/jcb.200510155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Ihekwaba AE, Elliott M, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- Ashall L, Horton CA, Nelson DE, et al. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science. 2009;324:242–246. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek P, Jackson DA, White MR.Oscillatory control of signalling molecules Curr Opin Genet Dev 2010. doi: 10.1016/j.gde.2010.08.004 [DOI] [PubMed]
- Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- Covert MW, Leung TH, Gaston JE, Baltimore D. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science. 2005;309:1854–1857. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- Behar M, Hoffmann A.Understanding the temporal codes of intra-cellular signals Curr Opin Genet Dev 2010. Oct 16; doi: 10.1016/j.gde.2010.09.007 [DOI] [PMC free article] [PubMed]
- Mathes E, O'Dea EL, Hoffmann A, Ghosh G. NF-kappaB dictates the degradation pathway of IkappaBalpha. EMBO J. 2008;27:1357–1367. doi: 10.1038/emboj.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathes E, Wang L, Komives E, Ghosh G. Flexible regions within I{kappa}B{alpha} create the ubiquitin-independent degradation signal. J Biol Chem. 2010;285:32927–36. doi: 10.1074/jbc.M110.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dea EL, Kearns JD, Hoffmann A. UV as an amplifier rather than inducer of NF-kappaB activity. Mol Cell. 2008;30:632–641. doi: 10.1016/j.molcel.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-kappa B2 in maturing B cells. Nat Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- Zarnegar BJ, Wang Y, Mahoney DJ, et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 2008;9:1371–1378. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S, Matsuzawa A, Zhang W, et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol. 2008;9:1364–1370. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JQ, Zarnegar B, Oganesyan G, et al. Rescue of TRAF3-null mice by p100 NF-kappa B deficiency. J Exp Med. 2006;203:2413–2418. doi: 10.1084/jem.20061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Zhang M, Harhaj EW, Sun SC. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- Senftleben U, Cao Y, Xiao G, et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- Xiao G, Harhaj EW, Sun SC. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- Xiao G, Fong A, Sun SC. Induction of p100 processing by NF-kappaB-inducing kinase involves docking IkappaB kinase alpha (IKKalpha) to p100 and IKKalpha-mediated phosphorylation. J Biol Chem. 2004;279:30099–30105. doi: 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- Mordmuller B, Krappmann D, Esen M, Wegener E, Scheidereit C. Lymphotoxin and lipopolysaccharide induce NF-kappaB-p52 generation by a co-translational mechanism. EMBO Rep. 2003;4:82–87. doi: 10.1038/sj.embor.embor710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derudder E, Dejardin E, Pritchard LL, et al. RelB/p50 dimers are differentially regulated by tumor necrosis factor-alpha and lymphotoxin-beta receptor activation: critical roles for p100. J Biol Chem. 2003;278:23278–23284. doi: 10.1074/jbc.M300106200. [DOI] [PubMed] [Google Scholar]
- Fusco AJ, Savinova OV, Talwar R, et al. Stabilization of RelB requires multidomain interactions with p100/p52. J Biol Chem. 2008;283:12324–12332. doi: 10.1074/jbc.M707898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak S, Shih VF, Hoffmann A. Generation and activation of multiple dimeric transcription factors within the NF-kappaB signaling system. Mol Cell Biol. 2008;28:3139–3150. doi: 10.1128/MCB.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B, Zarnegar B, Ytterberg AJ, et al. Negative feedback in noncanonical NF-kappaB signaling modulates NIK stability through IKKalpha-mediated phosphorylation. Sci Signal. 2010;3:ra41. doi: 10.1126/scisignal.2000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Morgan MJ, Choksi S, et al. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol. 11:799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marienfeld R, Berberich-Siebelt F, Berberich I, et al. Signal-specific and phosphorylation-dependent RelB degradation: a potential mechanism of NF-kappaB control. Oncogene. 2001;20:8142–8147. doi: 10.1038/sj.onc.1204884. [DOI] [PubMed] [Google Scholar]
- Weih F, Caamano J. Regulation of secondary lymphoid organ development by the nuclear factor-kappaB signal transduction pathway. Immunol Rev. 2003;195:91–105. doi: 10.1034/j.1600-065x.2003.00064.x. [DOI] [PubMed] [Google Scholar]
- Yilmaz ZB, Weih DS, Sivakumar V, Weih F. RelB is required for Peyer's patch development: differential regulation of p52-RelB by lymphotoxin and TNF. EMBO J. 2003;22:121–130. doi: 10.1093/emboj/cdg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wu L, Wesche H, et al. Defective lymphotoxin-beta receptor-induced NF-kappaB transcriptional activity in NIK-deficient mice. Science. 2001;291:2162–2165. doi: 10.1126/science.1058453. [DOI] [PubMed] [Google Scholar]
- Shinkura R, Kitada K, Matsuda F, et al. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-kappa b-inducing kinase. Nat Genet. 1999;22:74–77. doi: 10.1038/8780. [DOI] [PubMed] [Google Scholar]
- Fagarasan S, Shinkura R, Kamata T, et al. Alymphoplasia (aly)-type nuclear factor kappaB-inducing kinase (NIK) causes defects in secondary lymphoid tissue chemokine receptor signaling and homing of peritoneal cells to the gut-associated lymphatic tissue system. J Exp Med. 2000;191:1477–1486. doi: 10.1084/jem.191.9.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caamano JH, Rizzo CA, Durham SK, et al. Nuclear factor (NF)-kappa B2 (p100/p52) is required for normal splenic microarchitecture and B cell-mediated immune responses. J Exp Med. 1998;187:185–196. doi: 10.1084/jem.187.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzoso G, Carlson L, Poljak L, et al. Mice deficient in nuclear factor (NF)-kappa B/p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J Exp Med. 1998;187:147–159. doi: 10.1084/jem.187.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weih DS, Yilmaz ZB, Weih F. Essential role of RelB in germinal center and marginal zone formation and proper expression of homing chemokines. J Immunol. 2001;167:1909–1919. doi: 10.4049/jimmunol.167.4.1909. [DOI] [PubMed] [Google Scholar]
- Cao Y, Bonizzi G, Seagroves TN, et al. IKKalpha provides an essential link between RANK signaling and cyclin D1 expression during mammary gland development. Cell. 2001;107:763–775. doi: 10.1016/s0092-8674(01)00599-2. [DOI] [PubMed] [Google Scholar]
- Demicco EG, Kavanagh KT, Romieu-Mourez R, et al. RelB/p52 NF-kappaB complexes rescue an early delay in mammary gland development in transgenic mice with targeted suppressor IkappaB-alpha expression and promote carcinogenesis of the mammary gland. Mol Cell Biol. 2005;25:10136–10147. doi: 10.1128/MCB.25.22.10136-10147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaira S, Johnson T, Hirbe AC, et al. RelB is the NF-kappaB subunit downstream of NIK responsible for osteoclast differentiation. Proc Natl Acad Sci USA. 2008;105:3897–3902. doi: 10.1073/pnas.0708576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, D'Amico A, Winkel KD, et al. RelB is essential for the development of myeloid-related CD8alpha− dendritic cells but not of lymphoid-related CD8alpha+ dendritic cells. Immunity. 1998;9:839–847. doi: 10.1016/s1074-7613(00)80649-4. [DOI] [PubMed] [Google Scholar]
- Carrasco D, Ryseck RP, Bravo R. Expression of relB transcripts during lymphoid organ development: specific expression in dendritic antigen-presenting cells. Development. 1993;118:1221–1231. doi: 10.1242/dev.118.4.1221. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Walsh PT, Walsh MC, et al. TRAF6 is a critical factor for dendritic cell maturation and development. Immunity. 2003;19:353–363. doi: 10.1016/s1074-7613(03)00230-9. [DOI] [PubMed] [Google Scholar]
- Dejardin E, Droin NM, Delhase M, et al. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- Bonizzi G, Bebien M, Otero DC, et al. Activation of IKKalpha target genes depends on recognition of specific kappaB binding sites by RelB:p52 dimers. EMBO J. 2004;23:4202–4210. doi: 10.1038/sj.emboj.7600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovas A, Radke D, Albrecht D, et al. Differential RelA- and RelB-dependent gene transcription in LTbetaR-stimulated mouse embryonic fibroblasts. BMC Genomics. 2008;9:606. doi: 10.1186/1471-2164-9-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova LV, Makeev VJ, Kuprash DV. In vitro selection of optimal RelB/p52 DNA-binding motifs. Biochem Biophys Res Commun. 2008;365:583–588. doi: 10.1016/j.bbrc.2007.10.200. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Derudder E, Hobeika E, et al. Canonical NF-kappaB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity. 2006;24:729–739. doi: 10.1016/j.immuni.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Moorthy AK, Huang DB, Wang VY, Vu D, Ghosh G. X-ray structure of a NF-kappaB p50/RelB/DNA complex reveals assembly of multiple dimers on tandem kappaB sites. J Mol Biol. 2007;373:723–734. doi: 10.1016/j.jmb.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco AJ, Huang DB, Miller D, et al. NF-kappaB p52:RelB heterodimer recognizes two classes of kappaB sites with two distinct modes. EMBO Rep. 2009;10:152–159. doi: 10.1038/embor.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz JL, Baltimore D. Two pathways to NF-kappaB. Mol Cell. 2002;10:693–695. doi: 10.1016/s1097-2765(02)00697-4. [DOI] [PubMed] [Google Scholar]
- Mercurio F, DiDonato JA, Rosette C, Karin M. p105 and p98 precursor proteins play an active role in NF-kappa B-mediated signal transduction. Genes Dev. 1993;7:705–718. doi: 10.1101/gad.7.4.705. [DOI] [PubMed] [Google Scholar]
- Tucker E, O'Donnell K, Fuchsberger M, et al. A novel mutation in the Nfkb2 gene generates an NF-kappa B2 “super repressor”. J Immunol. 2007;179:7514–7522. doi: 10.4049/jimmunol.179.11.7514. [DOI] [PubMed] [Google Scholar]
- Novack DV, Yin L, Hagen-Stapleton A, et al. The IkappaB function of NF-kappaB2 p100 controls stimulated osteoclastogenesis. J Exp Med. 2003;198:771–781. doi: 10.1084/jem.20030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru N, Kishimoto H, Hayashi Y, Sprent J. Regulation of naive T cell function by the NF-kappaB2 pathway. Nat Immunol. 2006;7:763–772. doi: 10.1038/ni1351. [DOI] [PubMed] [Google Scholar]
- Courtois G, Gilmore TD. Mutations in the NF-kappaB signaling pathway: implications for human disease. Oncogene. 2006;25:6831–6843. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- Neri A, Fracchiolla NS, Migliazza A, Trecca D, Lombardi L. The involvement of the candidate proto-oncogene NFKB2/lyt-10 in lymphoid malignancies. Leuk Lymphoma. 1996;23:43–48. doi: 10.3109/10428199609054800. [DOI] [PubMed] [Google Scholar]
- Annunziata CM, Davis RE, Demchenko Y, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keats JJ, Fonseca R, Chesi M, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinin NL, Boldin MP, Kovalenko AV, Wallach D. MAP3K-related kinase involved in NF-kappaB induction by TNF, CD95 and IL-1. Nature. 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- Bren GD, Solan NJ, Miyoshi H, et al. Transcription of the RelB gene is regulated by NF-kappaB. Oncogene. 2001;20:7722–7733. doi: 10.1038/sj.onc.1204868. [DOI] [PubMed] [Google Scholar]
- Liptay S, Schmid RM, Nabel EG, Nabel GJ. Transcriptional regulation of NF-kappa B2: evidence for kappa B-mediated positive and negative autoregulation. Mol Cell Biol. 1994;14:7695–7703. doi: 10.1128/mcb.14.12.7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller JR, Siebenlist U. Lymphotoxin beta receptor induces sequential activation of distinct NF-kappa B factors via separate signaling pathways. J Biol Chem. 2003;278:12006–12012. doi: 10.1074/jbc.M210768200. [DOI] [PubMed] [Google Scholar]
- Alcamo E, Hacohen N, Schulte LC, et al. Requirement for the NF-kappaB family member RelA in the development of secondary lymphoid organs. J Exp Med. 2002;195:233–244. doi: 10.1084/jem.20011885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo JC, Basak S, James ES, et al. Coordination between NF-kappaB family members p50 and p52 is essential for mediating LTbetaR signals in the development and organization of secondary lymphoid tissues. Blood. 2006;107:1048–1055. doi: 10.1182/blood-2005-06-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]