Abstract
Cutaneous malignant melanoma is an aggressive disease of poor prognosis. Clinical and experimental studies have provided major insight into the pathogenesis of the disease, including the functional interaction between melanoma cells and surrounding keratinocytes, fibroblasts and immune cells. Nevertheless, patients with metastasized melanoma have a very poor prognosis and are largely refractory to clinical therapies. Hence, novel diagnostic tools to monitor melanoma development as well as therapeutic targets are urgently needed. We investigated the expression pattern of the kallikrein-related peptidase 6 (KLK6) in human melanoma tissue sections throughout tumor development. Although, KLK6 was not detectable in tumor cells, we found strong KLK6 protein expression in keratinocytes and stromal cells located adjacent to benign nevi, primary melanomas and cutaneous metastatic lesions, suggesting a paracrine function of extracellular KLK6 during neoplastic transformation and malignant progression. Accordingly, recombinant Klk6 protein significantly induced melanoma cell migration and invasion accompanied by an accelerated intracellular Ca2+-flux. We could further demonstrate that KLK6-induced intracellular Ca2+-flux and tumor cell invasion critically depends on the protease-activated receptor PAR1. Our data provide experimental evidence that specific inhibition of the KLK6-PAR1 axis may interfere with the deleterious effect of tumor-microenvironment interaction and represent a potential option for translational melanoma research.
Keywords: Ca2+-flux, keratinocytes, KLK, PAR, proteases
Introduction
The skin is the largest human organ, being composed of the epidermis and the dermis separated by the basement membrane. The epidermis forms a stratified epithelium with several layers of keratinocytes at different stages of differentiation. Beside keratinocytes the interfollicular epidermis contains melanocytes, which are located in the stratum basale. Their proliferation and differentiation are regulated by cells of their environment, especially the keratinocytes (Haass et al., 2005). Keratinocytes regulate melanocyte functions through a complex system of paracrine growth factors and cell-cell adhesion molecules. Alteration of this homeostatic balance can trigger a continuous proliferation of melanocytes supporting neoplastic transformation and may lead to the development of melanoma (Haass et al., 2004). Human melanomas are malignant and strongly pigmented tumors, whose development is divided into four steps: the dysplastic nevus, primary melanoma in the radial and vertical growth phase, and finally, metastatic melanoma (Gaggioli and Sahai, 2007; Hsu et al., 2002). A prerequisite for malignant progression of melanoma is the invasion of tumor cells in the surrounding tissue, characterized by reduced cell-cell adhesion, enhanced migration activity and degradation of the extracellular matrix (ECM) (Kuphal et al., 2005). It is well accepted that tumor-induced proteases are involved in all three processes (Hsu et al., 1996; Kapadia et al., 2004; Shinoda et al., 2007).
Recently, enhanced expression of the Kallikrein-related peptidase 6 (Klk6) was identified in advanced tumor stages of the chemically induced tumor model of mouse back skin and its human orthologue KLK6 was correlated with malignant progression of human skin cancer (Breitenbach et al., 2001; Klucky et al., 2007). KLK6 belongs to a large family of Kallikrein-related peptidases, representing secreted chymotrypsin- and trypsin-like serine proteases with diverse expression under physiological and pathological conditions, including common types of human cancer (Yousef and Diamandis, 2001). Ectopic Klk6 expression in a mouse keratinocyte cell line induced spindle-like cell morphology with impaired cell-cell adhesion and accelerated cell proliferation, migration, as well as invasion (Klucky et al., 2007). Alterations in keratinocyte cell physiology were partly due to KLK6-induced E-cadherin ectodomain shedding, which critically depends on the presence of matrix-metalloprotease (MMP) activity (Klucky et al., 2007). Here we focused on KLK6 protein expression in the development and progression of melanoma skin cancer and investigated the impact of recombinant rat Klk6 protein on human melanoma cell migration and invasion.
Results
KLK6 expression during human melanoma development
In order to identify the KLK6 expression pattern at distinct stages of human melanoma development, human tissue sections of dysplastic nevi (n = 10), primary melanoma (n = 11), and metastatic melanoma (n = 12) were subjected to immunohistochemistry (IHC) analysis. KLK6 protein expression was not detectable in tumor cells of primary and metastatic melanoma, and specific KLK6 staining was also absent from atypic melanocytes of dysplastic nevi (Figure S1 and Figure 2). However, melanin production in some of these cells hampered, at least in part, a final conclusion. Hence, we confirmed the absence of KLK6 protein expression in atypic melanocytes of dysplastic nevi, and tumor cells of primary and metastatic melanoma using the red AEC+ substrate chromogen for IHC staining (Figure 1). While KLK6 expression was absent in tumor cells, both staining procedures revealed positive staining for KLK6 protein expression in endothelial cells (Figure 1 and Figure S1). However, KLK6-positive endothelial cells were not restricted to blood vessels within the tumor or the adjacent tissue, but were also present in dermal tissue of normal skin. KLK6 protein expression in CD34-positive endothelial cells was further confirmed by immunofluorescence analysis, while no co-expression was detected for CD68-positive macrophages, CD45-positive lymphocytes, CD1a-positive dentritic cells, or mast cells (Figure S2).
Figure 2. KLK6 protein expression in human metastatic melanoma.
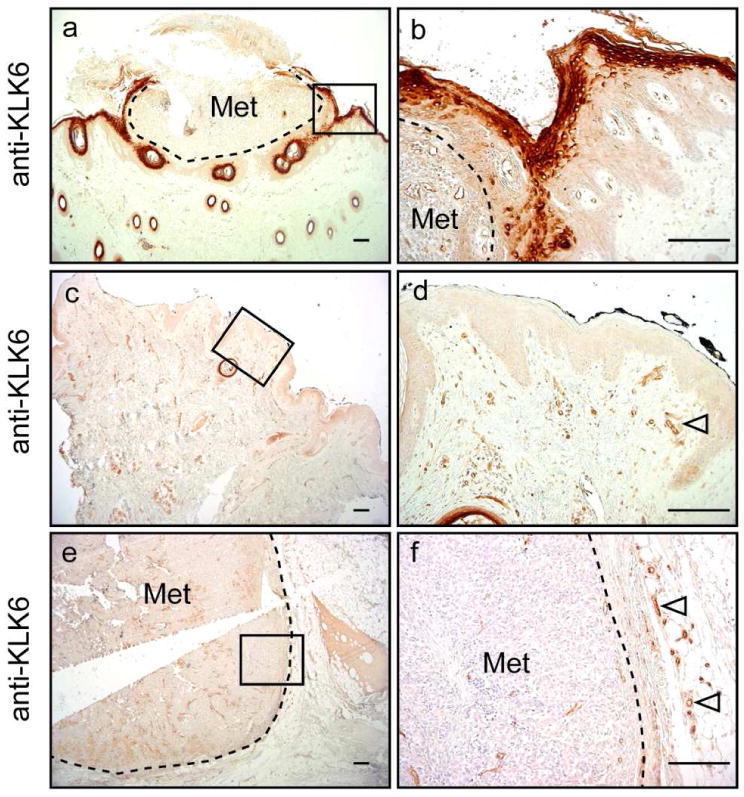
IHC analysis was performed as described in Figure 1 and revealed strong KLK6 expression in keratinocytes adjacent to metastatic epidermotropic melanoma (a-b), but not in samples of metastatic dermal melanoma (c-f). Dashed lines mark the tumor border, KLK6 expression in blood vessels is indicated by open arrowheads. Scale bars, 40 μm. Met, Metastasis. Right panels show a higher magnification of areas marked by black boxes.
Figure 1. KLK6 protein expression in human melanoma tissue sections.

IHC analysis with tissue sections from dysplastic nevi (a-b), primary melanoma (SSM, c-d), and metastatic epidermotropic melanoma (e-f) was performed with an anti-KLK6 antibody and revealed specific staining (red signal) in epidermal keratinocytes (black arrowheads) and blood vessels (open arrowheads). Right panels show a higher magnification of areas marked by black boxes. Sections were counterstained with hematoxylin. Dashed lines mark the tumor border. Scale bars, 40 μm. Mel, Melanoma and Met, Metastasis.
The most striking difference in KLK6 expression between tissue sections of unaffected skin (no or weak staining) and tumor samples was observed in keratinocytes adjacent to tumor cells (Figure 1, Figure 2 and Figure S1). Increased KLK6 staining in epidermal keratinocytes was found in six (medium staining) and a strong KLK6 staining in five out of eleven melanoma sections. Tumor cells were identified using an anti-S100 antibody, a well-established marker for melanoma cells. These data suggested that the presence of melanoma cells or melanoma cell-derived factors induce KLK6 expression in keratinocytes. In line with this assumption, we observed enhanced KLK6 protein levels in keratinocytes nearby metatstatic melanomas growing within the epidermis (epidermotropic metastasis; Figure 2a and b), but not on tissue sections from metastatic melanomas located in the dermis or subcutis (dermal metastasis; Figure 2c-f).
KLK5 and KLK7 are two abundant Kallikrein-related peptidases in human skin that are critically involved in skin barrier function and homeostasis, and part of a KLK proteolytic activation cascade (Eissa and Diamandis, 2008). In contrast to increased KLK6 expression in keratinocytes adjacent to melanoma cells, we found no major increase in KLK5 and KLK7 protein levels that showed strong expression in terminally differentiated keratinocytes of both unaffected skin and tumor tissue sections (Figure S3). However, we observed a broadened expression pattern of KLK5 and KLK7 in the epidermis of some tissue sections of epidermotropic metastasis (Figure S3).
In summary, the IHC analysis of tissue sections from human melanoma patients indicate the existence of melanoma cell-derived factors, which may trigger molecular pathways in early stages of melanoma pathogenesis resulting in increased levels of KLK6 expression in adjacent keratinocytes, but not of other abundant Kallikrein-related peptidases in human epidermis.
Recombinant Klk6 induces cell migration and invasion of human melanoma cells
Our findings that strong KLK6 expression in epidermal keratinocytes is a characteristic feature of human melanoma raised the hypothesis that enhanced levels of extracellular KLK6 may contribute to neoplastic transformation and/or malignant progression of melanoma cells. In order to test this assumption and to identify potential tumor-relevant processes affected by KLK6, we treated two established human melanoma cell lines, MeWo and SKmel23, with recombinant rat Klk6 protease. Measuring cell proliferation by quantification of BrdU incorporation using FACS analysis revealed no significant induction of BrdU-positive MeWo or SKmel23 melanoma cells upon Klk6 stimulation (Figure S4), demonstrating that the protease does not affect melanoma growth by induction of tumor cell proliferation.
Next, we determined tumor cell migration in the presence or absence of recombinant rat Klk6 protein using a scratch wounding assay. We observed a significant increase in MeWo and SKmel23 cell migration 12 hours post Klk6 addition compared to control cells that persisted until 48 hours or 96 hours, respectively (Figure 3a and b). The invasion capacity of control and Klk6-treated SKmel23 cells was additionally examined in a matrigel invasion assay. Quantification of fluorescent-labeled tumor cells revealed a significantly enhanced invasion capacity of Klk6-treated SKmel23 cells in comparison to untreated controls (Figure 3c).
Figure 3. Recombinant Klk6 induces melanoma cell migration and invasion.
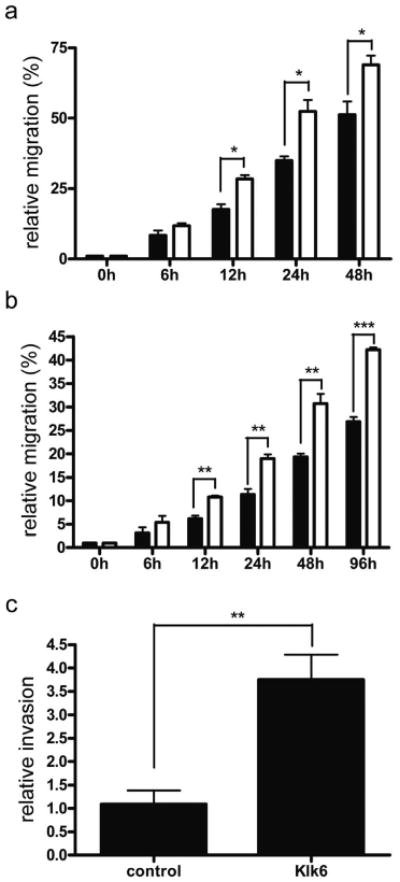
Relative migration of MeWo (a) and SKmel23 (b) cells was quantified in a scratch wounding assay at indicated time points. The distance between two migrating cell borders was measured and relative migration is indicated in percentage with closed wounds set to 100%. Black bars represent untreated cells and white bars cells treated with 40 nM recombinant rat Klk6. (c) Relative invasion of SKmel23 cells was measured using BD BioCoat™ Matrigel™ invasion chambers. Invading Klk6-treated (Klk6) and untreated cells (control) were stained with Calcein Fluorescent Dye and measured with the BMG FLUOSTAR Optima device. The number of invading cells in a control experiment was set to one and relative invasion capacity was determined. * P<0.05; ** P≤0.007; *** P<0.0005.
Recombinant Klk6 regulates PAR signaling and Ca2+-release in melanoma cell lines
In order to identify the mechanism of how KLK6 promotes melanoma cell migration and invasion we focused on the recently described function of KLK6 as an endogenous activator of protease-activated receptors (Angelo et al., 2006; Oikonomopoulou et al., 2006). Protease-activated receptors (PARs) are G-protein-coupled receptors that trigger numerous cellular responses, including intracellular Ca2+-release in response to extracellular protease activity (Steinhoff et al., 2005). We focused our analysis on PAR1, since high expression was shown on tumor cells of human melanoma lesions and PAR1 seems to contribute to the acquisition of the malignant phenotype by facilitating tumor cell invasion and metastasis (Melnikova et al., 2008). Indeed, PAR1 expression was detectable in various melanoma cell lines as measured by Western blot and FACS analysis (Figure 4a and b). For further analysis SKmel23 cells were used because of the described low invasive phenotype in vitro and in vivo (Klose et al., 2006). Stimulation of SKmel23 cells with a PAR1 specific agonist peptide (TFLLRamide) revealed a strong increase in intracellular Ca2+-flux, which was monitored by FACS analysis (Figure 4c). A comparable increase in intracellular Ca2+-flux was also measured after treatment of SKmel23 cells with recombinant rat Klk6 (Figure 4d), suggesting that accelerated tumor cell migration and invasion is, at least in part, mediated by KLK6-induced PAR1 signaling.
Figure 4. PAR-1 expression and Ca2+-release in melanoma cell lines.
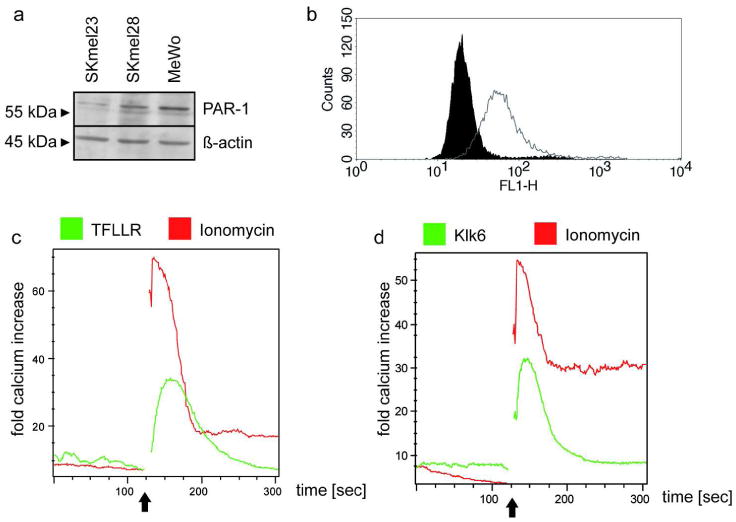
(a) Western blot analysis of whole cell lysates with an anti-PAR-1 antibody. β-actin protein levels served as control for cell extract quantity and quality. (b) FACS measurement of PAR-1 receptor expression at the SKmel23 cell surface (white) in comparison to control staining with the secondary antibody (black). (c, d) Intracellular calcium levels were assessed by FACS analysis using the calcium-indicative dyes fluo-4 and FuraRed. One representative measurement (n=3) of calcium release after stimulation with 1 μg/ml Ionomycin and 100 μM of PAR-1 (TFLLRamide) is shown in (c). Calcium release after stimulation with 1 μg/ml Ionomycin and 40 nM recombinant rat Klk6 is shown in (d). The arrows indicate treatment with the stimulus.
To test the impact of PAR1 signaling on KLK6 mediated tumor cell migration and invasion, we silenced PAR1 expression in SKmel23 cells using a lentiviral shRNA strategy. Efficient down-regulation of PAR1 expression was confirmed for SKmel23 cells infected with shRNA2152 (SKmel23#1 and SKmel23#2) by quantitative RT-PCR, Western blot and FACS analysis (Figure 5a-c). In contrast, SKmel23 cells infected with shRNA1349 (SKmel23#3) or a scrambled shRNA (SKmel23#4) showed comparable PAR1 transcript and protein levels (Figure 5 and data not shown), and were used as controls for further experiments. Finally, Ca2+-flux measurement after treatment with TFLLRamide confirmed impaired PAR1 activation in SKmel23#1 and SKmel23#2 compared to control cells (Figure 5d and data not shown).
Figure 5. PAR-1 silencing in SKmel23 cells.
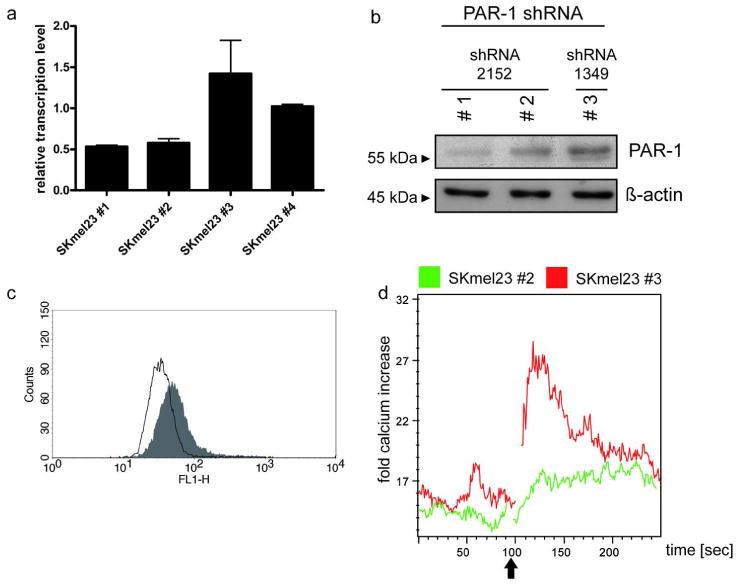
(a) Relative PAR-1 transcript levels in SKmel23 #1-4 cells were analyzed using real-time RT-PCR. All expression levels were normalized to Lamin B1 transcript levels. (b) Western blot analysis of whole cell lysate using anti-PAR-1 antibody. β-actin protein levels served as control for cell extract quantity and quality. (c) FACS measurement to quantify PAR-1 receptor expression at the cell surface of SKmel23 cells (#2 white and #3 grey). (d) Calcium measurement was performed as described in Figure 4. One representative (n = 3) measurement of calcium release is shown in PAR-1 knock down SKmel23 cells (#2) in comparison to control SKmel23 cells (#3) upon stimulation with 100 μM of PAR-1 agonistic peptide (TFLLRamide). The arrow indicates treatment with the stimulus.
Treatment of PAR-1 down-regulated cells (SKmel23#1 and SKmel23#2) with recombinant rat Klk6 protein revealed an almost complete loss of intracellular Ca2+-flux and impaired tumor cell invasion compared to control cells (Figure 6). Finally, we performed a co-IHC staining on human primary melanoma and metastatic epidermotrophic melanoma sections and could confirm PAR1 protein expression in melanoma cells and KLK6 protein expression in adjacent epidermal keratinocytes (Figure 6d).
Figure 6. Impaired calcium release and invasion after Klk6 stimulation in PAR-1 silenced SKmel23 cells.

(a) Calcium measurement was performed as described in Figure 4. One representative measurement of calcium release is shown in PAR-1 knock down SKmel23 cells (#2) in comparison to control SKmel23 cells (#3) upon stimulation with 40 nM recombinant rat Klk6. The arrow indicates treatment with the stimulus. (b, c) Invasion assay was performed as described in Figure 3. Relative invasion capacity of Klk6-treated (Klk6) and control PAR-1 knock down SKmel23 cells (#2; b) and SKmel23 control cells (#3; c) is shown. * P<0,05. (d) Co-IHC analysis with tissue sections from unaffected skin, primary melanoma (SSM), and metastatic epidermotropic melanoma specimens was performed with an anti-KLK6 antibody (green signal) and an anti-PAR1 antibody (red signal). Sections were counterstained with hematoxylin. Scale bar = 200 μm. Mel, Melanoma and Met, Metastasis.
In summary, these data strongly suggest that the KLK6-PAR1 axis contributes to the active interaction between melanoma cells and their microenvironment and supports malignant progression by inducing melanoma cell invasion.
Discussion
Kallikreins are implicated in a vast range of normal and pathological conditions, where they either act independently or as part of proteolytic cascades (Borgono and Diamandis, 2004). Accumulating evidence support the notion that human tissue Kallikreins whose concentrations are often abnormal in common human malignancies, have not only prognostic and diagnostic value in various types of cancer but also contribute to tumor pathogenesis (Borgono and Diamandis, 2004; Obiezu and Diamandis, 2005). However, little attention has been paid to their expression and involvement in the development and malignant progression of human melanoma, the leading cause of skin cancer mortality worldwide. In the past, Winnepenninckx and colleagues identified a 254-gene signature by gene expression profiling including KLK4, KLK7, and KLK11 that is associated with metastatic dissemination and overall survival of patients with primary melanoma (Winnepenninckx et al., 2006). More recently, KLK6 and KLK7 protein expression was analyzed in human nevi and primary melanoma tissue sections, and both protein levels were higher in melanomas than in common nevi (Rezze et al., 2011). In line with this data, we found enhanced KLK6 transcript levels in malignant melanoma compared to unaffected skin samples (Klucky et al., 2007). However, the detailed immunohistochemical analyses on human melanoma tissue sections shown in this study demonstrate that enhanced KLK6 levels are not due to expression in melanoma cells, but rather due to its expression in adjacent keratinocytes and endothelial cells within the tumor microenvironment. These data suggest the presence of one or several tumor cell-derived factors that induce aberrant KLK6 expression in adjacent keratinocytes. However, we cannot exclude the possibility that induced KLK6 expression, which is most prominent in differentiated keratinocytes, is at least in part also due to aberrant epidermal homeostasis.
It is well established that melanoma cells actively interact with the tumor microenvironment by the secretion of a range of growth factors, cytokines and chemokines, and thereby modulate the malignant phenotype (Lee and Herlyn, 2007). These soluble factors could affect intracellular signaling pathways in adjacent keratinocytes that are known to activate KLK6 transcription in epithelial cells, such as ERK, PI3K, and AKT dependent cascades (Bourcier et al., 2006; Henkhaus et al., 2008a; Henkhaus et al., 2008b). Analysis of KLK6 expression in co-culture experiments with human keratinocyte and melanoma cells should unravel the existence of a melanoma cell-derived soluble factor.
The establishment of an inflammatory microenvironment that is often observed at the site of tumors, including melanoma, could also contribute to enhanced KLK6 expression in adjacent keratinocytes. In line with this assumption, we demonstrated induced expression of the murine orthologue of human KLK6 in epidermal keratinocytes after stimulation with the phorbol ester TPA, a potent inducer of skin inflammation (Breitenbach et al., 2001). Moreover, significantly higher levels of KLK6 were described in keratinocytes of psoriasis and other inflammatory skin disorders (Komatsu et al., 2007; Komatsu et al., 2005). It is worth noting that KLK6 is often co-expressed with other Kallikrein family members some of which can activate the KLK6 proenzyme by proteolytic cleavage (Blaber et al., 2007; Yoon et al., 2007). Although KLK5 and KLK7 are expressed by epidermal keratinocytes and play a pivotal role in the process of skin desquamation (Ovaere et al., 2009), their function in human melanoma development and progression has not been addressed so far.
Although the physiological function of KLK6 has been only partly elucidated, clinical studies and experimental model systems provide compelling evidence for its critical role in neoplastic transformation as well as malignant progression (Borgono and Diamandis, 2004). Recently, we showed that KLK6 induces E-cadherin ectodomain shedding thereby interfering with cell-cell adhesion and supporting migration and invasion of keratinocytes (Klucky et al., 2007). E-cadherin is a key player that mediates direct cell-cell adhesion between epidermal keratinocytes and melanocytes, which is pivotal for normal growth and behavior of melanocytes in the state of homeostasis. Melanoma cells escape from the control by keratinocytes via several mechanisms, including down-regulation of E-cadherin expression and function (Haass et al., 2005). Hence, it is tentative to speculate that increased KLK6 expression by keratinocytes may promote malignant progression due to functional inactivation of cell-cell adhesion by E-cadherin during early stages of melanoma development.
Stimulation of human melanoma cells with recombinant Klk6 protein revealed a significant increase in tumor cell migration and invasion, accompanied by accelerated intracellular Ca2+-flux. Recent studies highlighted that KLK6 evokes intracellular Ca2+-flux by the activation of protease-activated receptors (PARs) (Angelo et al., 2006; Oikonomopoulou et al., 2006; Vandell et al., 2008). Several clinical and experimental observations suggested that PAR1 contributes to the acquisition of the malignant phenotype by facilitating tumor cell invasion and metastasis (Arora et al., 2007; Melnikova et al., 2008): (i) PAR1 overexpression was found predominantly in malignant melanoma and in metastatic lesions (Depasquale and Thompson, 2008; Massi et al., 2005; Tellez et al., 2007), (ii) PAR1 expression correlated with the metastatic potential of melanoma cell lines (Blackburn et al., 2009; Melnikova et al., 2009; Villares et al., 2009), and (iii) inhibition of PAR1 expression in a human melanoma xenograft model inhibited tumor growth and metastasis (Tellez and Bar-Eli, 2003; Tellez et al., 2003; Villares et al., 2008). In line with the assumption that KLK6 may promote malignant progression of melanoma by activation of PAR1, its silencing in melanoma cells impaired both, KLK6 induced intracellular Ca2+-flux as well as invasion. However, the role of intracellular Ca2+ signaling in melanoma development and malignant progression is not well established, and several other signaling cascades have been described downstream of PAR1 signaling that are critically involved in tumorigenesis, such as activation of mitogen-activated protein kinases (Steinhoff et al., 2005; Villares et al., 2011). Indeed, we found an increase in the activation of ERK1/2 and p38 MAPKs upon treatment of SKMel23 cells with recombinant rat Klk6, which was determined by Western blot analysis with phospho-specific antibodies, while JNK1/2 signaling seems to be unaffected (data not shown). Although, ERK1/2 and p38 MAPK signaling pathways exert several key functions in melanoma cell development and progression, including tumor cell migration and invasion, more detailed analysis will be required in the near future to unravel the role of both signaling cascades in the KLK6-mediated cross-talk between melanoma cells and its microenvironment.
In summary, our data provide experimental evidence for a potent role of extracellular KLK6 in neoplastic transformation and malignant progression of human melanoma. Further investigations focusing on the regulation of KLK6 expression and its impact on molecular pathways, such as PAR signaling will certainly contribute to a better understanding of the molecular nature of melanoma development. Finally, it will be a major challenge for the future to test the concept, whether specific inhibition of KLK6 expression and/or function could be used as a novel strategy for translational cancer research.
Materials and Methods
Human skin biopsies
Skin biopsies from patients with melanoma were obtained from surgical excisions of the affected areas at the Department of Dermatology, University of Cologne. The patient signed the informed consent from the Department of Dermatology, University of Cologne, approved by the Institutional Commission of Ethics (Az. 9645/96). The study was conducted according to Declaration of Helsinki Principles. Staging of primary melanomas that were used in this study is given as Supplemental Table S1.
Immunohistochemistry analysis and scoring
For IHC analysis, paraffin sections were stained according to the manufacturer's instructions (Vectastain Elite ABC Kit and ImmPRESS Kit, Vector Laboratories, Lörrach, Germany). Primary and secondary antibodies are listed in Table S2. As substrates we used liquid DAB+ Substrate or AEC+ Substrate Chromogen (Dako, Hamburg, Germany), or VectorRed and Histogreen (Vector Laboratories). All tissue sections were counterstained with hematoxylin (AppliChem, Darmstadt, Germany).
Scoring of tumor samples was based on the signal intensity and abundance in keratinocytes adjacent to the tumor tissue (score 1 = no or weak staining; score 2 = medium staining; score 3 = strong staining), and according to the staining in keratinocytes of the epidermis in control and unaffected skin (score 1 = no or weak staining).
Cell lines, culture conditions, and PAR1 silencing
Human melanoma MeWo cells were cultured in DMEM medium (PAA, Pasching, Austria), human SKmel23 and SKmel28 melanoma cells were cultured in RPMI medium (PAA) and HEK293T cells were cultured in IMEM medium (Invitrogen, Germany). Culture medium was supplemented with 10% FCS, 2 mM Glutamin (PAA), 100 U/ml Penicillin and 0.1 mg/ml Streptomycin (PAA), and the cells were cultured at 37°C in a humidified atmosphere of 8% CO2.
Lentiviral particles were produced by transient co-transfection of specific shRNA vectors (listed in Table S3) with pMDLg-pRRE and pRSV-REV packaging vectors, and the pMD2-VSVG envelope vector in HEK293T cells. pMDLg-pRRE and pRSV-REV packaging vectors, as well as the pMD2-VSVG envelope vector were kindly provided by Dr. Luigi Naldini (San Raffaele Telethon Institute for Gene Therapy, University Medical School, Italy). For the transfection of one dish 12.5 μg of pMDLg-pRRE plasmid, 6.25 μg of pRSV-REV, 9 μg of pMD2-VSVG and 32 μg of shRNA vector was mixed in 1 ml OPTIMEM medium containing 179.25 μg/ml Polyethylenimine (PEI). After 30 min incubation at room temperature the transfection mixture was added to the cells. 24 hours after transfection the virus was purified by ultra-centrifugation for 2 hours at 20°C.
For lentiviral transduction 1×104 SKmel23 cells were seeded in a 48-well plate and transduced with lentiviral particles. After 3 days, transduced cells were selected with Puromycin (0.5 μg/ml) for one week.
Scratch wounding assay
MeWo and SKmel23 cells were seeded in a 12-well plate and cultured for 24 hours to reach confluency. Cells were treated with 10 μg/ml mitomycin C (Sigma, Taufkirchen, München) for 1 hour and washed intensively with PBS. Subsequently the cell monolayer was scratched with a micropipette tip, and cells were cultivated in the absence or presence of 40 nM recombinant rat Klk6 (Blaber et al., 2002). Images were taken at indicated time points using light microscopy and a digital camera (Olympus, Hamburg, Germany). The distance between the migration fronts was measured using the UTHSCSA Image tool software (http://ddsdx.uthscsa.edu/dig/itdesc.html).
Cell invasion assay
The cell invasion assay was performed according to the manufacturer's instructions of the BD BioCoat™ Matrigel™Invasion chamber for 24-wells (BD Biosciences, Heidelberg, Germany). SKmel23 cells (150,000 cells) were seeded in the insert and incubated for 2 hours. Subsequently, 40 nM recombinant rat Klk6 (Blaber et al., 2002) was added and cell invasion was measured after 48 hours. Cells were stained with 4 μg/ml Calcein BD AM Fluorescent Dye (BD Biosciences) in HPBS for 1 hour at 37°C. The measurement was done using the BMG FLUOSTAR Optima device (BMG Labtech, Offenburg, Germany) with the bottom reading optic.
RNA isolation and qRT-PCR
RNA was isolated using Pure Link™ RNA Mini Kit (Invitrogen, Karlsruhe, Germany) and reverse transcribed as described previously (Gebhardt et al., 2008). Quantitative RT-PCR reactions were performed with equal amounts of cDNA using the AbiPrism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) in a volume of 30 μl using the Platinum SYBR Green qPCR SuperMix from Invitrogen. Primers are listed in Table S4.
Protein isolation and Western blot analysis
Protein isolation and Western blot analysis were done as described previously (Klucky et al., 2007). Antibodies are listed in Table S2.
FACS analysis
For detection of membrane proteins by FACS analysis, melanoma cells were detached from the cell culture dish using accutase (PAA, Austria). Subsequently, 5×105 cells were washed once with PBS and stained with the primary antibody diluted in 200 μl PBS/ 1% BSA for 30 min at room temperature. After three washing steps, the cells were incubated with the secondary, fluorescence-labeled antibody for 30 min on ice and light protected. Afterwards the cells were washed three times and measured with the FACSCalibur (BD Biosciences). Antibodies are listed in Table S2.
Measurements of intracellular calcium release
For measurements of intracellular calcium release SKmel23 cells were incubated for 1 hour at 37°C with the calcium-sensitive dyes Fluo-4 and FuraRed (Invitrogen, Karlsruhe, Germany). Cells were resuspended in RPMI medium without FCS. During flow cytometric-based calcium measurement (FACSCalibur; BD Biosciences), SKmel23 cells were stimulated by addition of 1 μg/ml Ionomycin (Invitrogen), 100 nM of PAR-1 agonistic peptide TFLLR (Bachem, Bubendorf, Switzerland), 100 nM of PAR-2 agonistic peptide SLIGKV (Bachem) or 40 nM recombinant rat Klk6 (Blaber et al., 2002). The fold calcium increase was determined by using FCSpress and FlowJo (Treestar, Ashland) software.
Cell proliferation Assay
The cell proliferation assay was performed using the APC BrdU Flow Kit (BD Biosciences) according to the manufacturer's instruction.
Statistical procedures
Result represents three independent experiments, which were performed in triplicates. Statistical-data analysis was made with a t-test using GraphPad PRISM version 4 software (GraphPad, San Diego, USA). Bars represent mean values ± SEM.
Supplementary Material
Supplemental Figure S1. KLK6 protein expression in human melanoma. IHC analysis with tissue sections from unaffected skin (a-b), dysplastic nevi (c-f) and primary melanoma (g-j) was performed with an anti-KLK6 antibody and revealed specific staining (brown signal) in epidermal keratinocytes (black arrowheads) and blood vessels (open arrowheads). Right panels show a higher magnification of areas marked by black boxes. Staining with an anti-S100 antibody was used as a control to detect transformed melanoma cells (c-d, g-h). Sections were counterstained with hematoxylin. Scale bars, 40 μm. Mel, Melanoma.
Supplemental Figure S2. KLK6 protein expression in endothelial cells of human melanoma. Co-immunofluorescense analysis with tissue section from primary melanoma were performed using anti-KLK6 antibody (b, e, h, k, n, red staining), anti-CD68 antibody (c, green staining), anti-CD45 (f, green staining), anti-CDla (i, green staining), anti-CD34 (1, green staining), and anti-Mast cell tryptase (o, green staining). Left panels show the merge of the co-staining. Scale bars, 40 μm.
Supplemental Figure S3. KLK5 and KLK7 protein expression in epidermal keratinocytes of human melanoma tissue sections. IHC analysis with tissue sections of unaffected skin, dysplastic nevi, primary melanoma, and epidermotropic metastasis was performed with antibodies raised against KLK5 and KLK7 proteins and revealed specific staining (red signal) mainly in terminally differentiated epidermal keratinocytes (black arrowheads). Sections were counterstained with hematoxylin. Scale bars, 40 μm. Mel, Melanoma and Met, Metastasis.
Supplemental Figure S4. Measurement of BrdU positive cells. Cells were seeded in a 12-well plate and stimulated for 4 (a, c) and 8 hours (b, d) with 40 nM recombinant rat Klk6 and BrdU incorporation was quantified by FACS analysis. BrdU incorporation of MeWo cells (a, b) and Skmel23 cells (c, d) were quantified in the absence (control) and presence (Klk6) of 40 nM recombinant rat Klk6.
Supplemental Table S1: Summary of the patient characteristics
SSM: Superficial spreading melanoma, ALM: Acral lentigous melanoma, ulc: ulceration AJCC: American Joint Committee on Cancer Staging System for Cutaneous Melanoma m: male, f: female
Supplemental Table S2: Summary of the antibodies used in the study
Supplemental Table S3: shRNA against human PAR-1 cloned into the lentivirus vector pLKO.1-puro were purchased from Sigma Aldrich
Supplemental Table S4: Summary of primers used for qRT-PCR
Acknowledgments
We thank Dr. Luigi Naldini (San Raffaele Telethon Institute for Gene Therapy, University Medical School, Italy) for providing the lentivirus producing plasmids, and Gerald Bendner, Alexander Strecker, Melanie Sator-Schmitt, Nataly Henfling and Andre Nollert for technical assistance. Special thanks to Friederike Herbst for the great support during the production of lentiviral particles. We gratefully acknowledge Tobias Nübel, Marina Schorpp-Kistner, Maike Hildenbrand and Moritz Durchdewald for critical discussion and reading of the manuscript.
Financial support: This work was supported by the German Ministry for Education and Research National Genome Research Network NGFN-Plus (01GS0883 to P.A. and J.H.), the Dietmar-Hopp Foundation (to J.H.), Center of Molecular Medicine, University of Cologne (BMFT/IDZ 10, grant 01 GB 950/4 to C.M.) and U.S.P.H.S./N.I.H. grant (1R15NS057771-01 to M.B.).
Abbreviations
- IF
immunofluorescense
- IHC
immunohistochemistry
- KLK
Kallikrein-related peptidase
- PAR
Protease-activated receptor
- TPA
12-O-tetradecanoylphorbol-13-acetate
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- Angelo PF, Lima AR, Alves FM, et al. Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects. J Biol Chem. 2006;281:3116–26. doi: 10.1074/jbc.M510096200. [DOI] [PubMed] [Google Scholar]
- Arora P, Ricks TK, Trejo J. Protease-activated receptor signalling, endocytic sorting and dysregulation in cancer. J Cell Sci. 2007;120:921–8. doi: 10.1242/jcs.03409. [DOI] [PubMed] [Google Scholar]
- Blaber SI, Scarisbrick IA, Bernett MJ, et al. Enzymatic properties of rat myelencephalon-specific protease. Biochemistry. 2002;41:1165–73. doi: 10.1021/bi015781a. [DOI] [PubMed] [Google Scholar]
- Blaber SI, Yoon H, Scarisbrick IA, et al. The autolytic regulation of human kallikrein-related peptidase 6. Biochemistry. 2007;46:5209–17. doi: 10.1021/bi6025006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn JS, Liu I, Coon CI, et al. A matrix metalloproteinase-1/protease activated receptor-1 signaling axis promotes melanoma invasion and metastasis. Oncogene. 2009;28:4237–48. doi: 10.1038/onc.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgono CA, Diamandis EP. The emerging roles of human tissue kallikreins in cancer. Nat Rev Cancer. 2004;4:876–90. doi: 10.1038/nrc1474. [DOI] [PubMed] [Google Scholar]
- Bourcier C, Jacquel A, Hess J, et al. p44 mitogen-activated protein kinase (extracellular signal-regulated kinase 1)-dependent signaling contributes to epithelial skin carcinogenesis. Cancer Res. 2006;66:2700–7. doi: 10.1158/0008-5472.CAN-05-3129. [DOI] [PubMed] [Google Scholar]
- Breitenbach U, Tuckermann JP, Gebhardt C, et al. Keratinocyte-specific onset of serine protease BSSP expression in experimental carcinogenesis. J Invest Dermatol. 2001;117:634–40. doi: 10.1046/j.0022-202x.2001.01437.x. [DOI] [PubMed] [Google Scholar]
- Depasquale I, Thompson WD. Prognosis in human melanoma: PAR-1 expression is superior to other coagulation components and VEGF. Histopathology. 2008;52:500–9. doi: 10.1111/j.1365-2559.2008.02978.x. [DOI] [PubMed] [Google Scholar]
- Eissa A, Diamandis EP. Human tissue kallikreins as promiscuous modulators of homeostatic skin barrier functions. Biol Chem. 2008;389:669–80. doi: 10.1515/BC.2008.079. [DOI] [PubMed] [Google Scholar]
- Gaggioli C, Sahai E. Melanoma invasion - current knowledge and future directions. Pigment Cell Res. 2007;20:161–72. doi: 10.1111/j.1600-0749.2007.00378.x. [DOI] [PubMed] [Google Scholar]
- Gebhardt C, Riehl A, Durchdewald M, et al. RAGE signaling sustains inflammation and promotes tumor development. J Exp Med. 2008;205:275–85. doi: 10.1084/jem.20070679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass NK, Smalley KS, Herlyn M. The role of altered cell-cell communication in melanoma progression. J Mol Histol. 2004;35:309–18. doi: 10.1023/b:hijo.0000032362.35354.bb. [DOI] [PubMed] [Google Scholar]
- Haass NK, Smalley KS, Li L, et al. Adhesion, migration and communication in melanocytes and melanoma. Pigment Cell Res. 2005;18:150–9. doi: 10.1111/j.1600-0749.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- Henkhaus RS, Gerner EW, Ignatenko NA. Kallikrein 6 is a mediator of K-RAS-dependent migration of colon carcinoma cells. Biol Chem. 2008a;389:757–64. doi: 10.1515/BC.2008.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkhaus RS, Roy UK, Cavallo-Medved D, et al. Caveolin-1-mediated expression and secretion of kallikrein 6 in colon cancer cells. Neoplasia. 2008b;10:140–8. doi: 10.1593/neo.07817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MY, Meier F, Herlyn M. Melanoma development and progression: a conspiracy between tumor and host. Differentiation. 2002;70:522–36. doi: 10.1046/j.1432-0436.2002.700906.x. [DOI] [PubMed] [Google Scholar]
- Hsu MY, Wheelock MJ, Johnson KR, et al. Shifts in cadherin profiles between human normal melanocytes and melanomas. J Investig Dermatol Symp Proc. 1996;1:188–94. [PubMed] [Google Scholar]
- Kapadia C, Ghosh MC, Grass L, et al. Human kallikrein 13 involvement in extracellular matrix degradation. Biochem Biophys Res Commun. 2004;323:1084–90. doi: 10.1016/j.bbrc.2004.08.206. [DOI] [PubMed] [Google Scholar]
- Klose A, Wilbrand-Hennes A, Zigrino P, et al. Contact of high-invasive, but not low-invasive, melanoma cells to native collagen I induces the release of mature cathepsin B. Int J Cancer. 2006;118:2735–43. doi: 10.1002/ijc.21700. [DOI] [PubMed] [Google Scholar]
- Klucky B, Mueller R, Vogt I, et al. Kallikrein 6 induces e-cadherin shedding and promotes cell proliferation, migration, and invasion. Cancer Res. 2007;67:8198–206. doi: 10.1158/0008-5472.CAN-07-0607. [DOI] [PubMed] [Google Scholar]
- Komatsu N, Saijoh K, Kuk C, et al. Aberrant human tissue kallikrein levels in the stratum corneum and serum of patients with psoriasis: dependence on phenotype, severity and therapy. Br J Dermatol. 2007;156:875–83. doi: 10.1111/j.1365-2133.2006.07743.x. [DOI] [PubMed] [Google Scholar]
- Komatsu N, Saijoh K, Toyama T, et al. Multiple tissue kallikrein mRNA and protein expression in normal skin and skin diseases. Br J Dermatol. 2005;153:274–81. doi: 10.1111/j.1365-2133.2005.06754.x. [DOI] [PubMed] [Google Scholar]
- Kuphal S, Bauer R, Bosserhoff AK. Integrin signaling in malignant melanoma. Cancer Metastasis Rev. 2005;24:195–222. doi: 10.1007/s10555-005-1572-1. [DOI] [PubMed] [Google Scholar]
- Lee JT, Herlyn M. Microenvironmental influences in melanoma progression. J Cell Biochem. 2007;101:862–72. doi: 10.1002/jcb.21204. [DOI] [PubMed] [Google Scholar]
- Massi D, Naldini A, Ardinghi C, et al. Expression of protease-activated receptors 1 and 2 in melanocytic nevi and malignant melanoma. Hum Pathol. 2005;36:676–85. doi: 10.1016/j.humpath.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Melnikova VO, Balasubramanian K, Villares GJ, et al. Crosstalk between protease-activated receptor 1 and platelet-activating factor receptor regulates melanoma cell adhesion molecule (MCAM/MUC18) expression and melanoma metastasis. J Biol Chem. 2009;284:28845–55. doi: 10.1074/jbc.M109.042150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova VO, Villares GJ, Bar-Eli M. Emerging Roles of PAR-1 and PAFR in Melanoma Metastasis. Cancer Microenviron. 2008;1:103–11. doi: 10.1007/s12307-008-0002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obiezu CV, Diamandis EP. Human tissue kallikrein gene family: applications in cancer. Cancer Lett. 2005;224:1–22. doi: 10.1016/j.canlet.2004.09.024. [DOI] [PubMed] [Google Scholar]
- Oikonomopoulou K, Hansen KK, Saifeddine M, et al. Proteinase-activated Receptors, Targets for Kallikrein Signaling. J Biol Chem. 2006;281:32095–112. doi: 10.1074/jbc.M513138200. [DOI] [PubMed] [Google Scholar]
- Ovaere P, Lippens S, Vandenabeele P, et al. The emerging roles of serine protease cascades in the epidermis. Trends Biochem Sci. 2009;34:453–63. doi: 10.1016/j.tibs.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Rezze GG, Fregnani JH, Duprat J, et al. Cell adhesion and communication proteins are differentially expressed in melanoma progression model. Hum Pathol. 2011;42:409–18. doi: 10.1016/j.humpath.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Shinoda Y, Kozaki K, Imoto I, et al. Association of KLK5 overexpression with invasiveness of urinary bladder carcinoma cells. Cancer Sci. 2007;98:1078–86. doi: 10.1111/j.1349-7006.2007.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhoff M, Buddenkotte J, Shpacovitch V, et al. Proteinase-activated receptors: transducers of proteinase-mediated signaling in inflammation and immune response. Endocr Rev. 2005;26:1–43. doi: 10.1210/er.2003-0025. [DOI] [PubMed] [Google Scholar]
- Tellez C, Bar-Eli M. Role and regulation of the thrombin receptor (PAR-1) in human melanoma. Oncogene. 2003;22:3130–7. doi: 10.1038/sj.onc.1206453. [DOI] [PubMed] [Google Scholar]
- Tellez C, McCarty M, Ruiz M, et al. Loss of activator protein-2alpha results in overexpression of protease-activated receptor-1 and correlates with the malignant phenotype of human melanoma. J Biol Chem. 2003;278:46632–42. doi: 10.1074/jbc.M309159200. [DOI] [PubMed] [Google Scholar]
- Tellez CS, Davis DW, Prieto VG, et al. Quantitative analysis of melanocytic tissue array reveals inverse correlation between activator protein-2alpha and protease-activated receptor-1 expression during melanoma progression. J Invest Dermatol. 2007;127:387–93. doi: 10.1038/sj.jid.5700539. [DOI] [PubMed] [Google Scholar]
- Vandell AG, Larson N, Laxmikanthan G, et al. Protease-activated receptor dependent and independent signaling by kallikreins 1 and 6 in CNS neuron and astroglial cell lines. J Neurochem. 2008;107:855–70. doi: 10.1111/j.1471-4159.2008.05658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villares GJ, Dobroff AS, Wang H, et al. Overexpression of protease-activated receptor-1 contributes to melanoma metastasis via regulation of connexin 43. Cancer Res. 2009;69:6730–7. doi: 10.1158/0008-5472.CAN-09-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villares GJ, Zigler M, Dobroff AS, et al. Protease activated receptor-1 inhibits the Maspin tumor-suppressor gene to determine the melanoma metastatic phenotype. Proc Natl Acad Sci U S A. 2011;108:626–31. doi: 10.1073/pnas.1006886108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villares GJ, Zigler M, Wang H, et al. Targeting melanoma growth and metastasis with systemic delivery of liposome-incorporated protease-activated receptor-1 small interfering RNA. Cancer Res. 2008;68:9078–86. doi: 10.1158/0008-5472.CAN-08-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnepenninckx V, Lazar V, Michiels S, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–82. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- Yoon H, Laxmikanthan G, Lee J, et al. Activation profiles and regulatory cascades of the human kallikrein-related peptidases. J Biol Chem. 2007;282:31852–64. doi: 10.1074/jbc.M705190200. [DOI] [PubMed] [Google Scholar]
- Yousef GM, Diamandis EP. The new human tissue kallikrein gene family: structure, function, and association to disease. Endocr Rev. 2001;22:184–204. doi: 10.1210/edrv.22.2.0424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. KLK6 protein expression in human melanoma. IHC analysis with tissue sections from unaffected skin (a-b), dysplastic nevi (c-f) and primary melanoma (g-j) was performed with an anti-KLK6 antibody and revealed specific staining (brown signal) in epidermal keratinocytes (black arrowheads) and blood vessels (open arrowheads). Right panels show a higher magnification of areas marked by black boxes. Staining with an anti-S100 antibody was used as a control to detect transformed melanoma cells (c-d, g-h). Sections were counterstained with hematoxylin. Scale bars, 40 μm. Mel, Melanoma.
Supplemental Figure S2. KLK6 protein expression in endothelial cells of human melanoma. Co-immunofluorescense analysis with tissue section from primary melanoma were performed using anti-KLK6 antibody (b, e, h, k, n, red staining), anti-CD68 antibody (c, green staining), anti-CD45 (f, green staining), anti-CDla (i, green staining), anti-CD34 (1, green staining), and anti-Mast cell tryptase (o, green staining). Left panels show the merge of the co-staining. Scale bars, 40 μm.
Supplemental Figure S3. KLK5 and KLK7 protein expression in epidermal keratinocytes of human melanoma tissue sections. IHC analysis with tissue sections of unaffected skin, dysplastic nevi, primary melanoma, and epidermotropic metastasis was performed with antibodies raised against KLK5 and KLK7 proteins and revealed specific staining (red signal) mainly in terminally differentiated epidermal keratinocytes (black arrowheads). Sections were counterstained with hematoxylin. Scale bars, 40 μm. Mel, Melanoma and Met, Metastasis.
Supplemental Figure S4. Measurement of BrdU positive cells. Cells were seeded in a 12-well plate and stimulated for 4 (a, c) and 8 hours (b, d) with 40 nM recombinant rat Klk6 and BrdU incorporation was quantified by FACS analysis. BrdU incorporation of MeWo cells (a, b) and Skmel23 cells (c, d) were quantified in the absence (control) and presence (Klk6) of 40 nM recombinant rat Klk6.
Supplemental Table S1: Summary of the patient characteristics
SSM: Superficial spreading melanoma, ALM: Acral lentigous melanoma, ulc: ulceration AJCC: American Joint Committee on Cancer Staging System for Cutaneous Melanoma m: male, f: female
Supplemental Table S2: Summary of the antibodies used in the study
Supplemental Table S3: shRNA against human PAR-1 cloned into the lentivirus vector pLKO.1-puro were purchased from Sigma Aldrich
Supplemental Table S4: Summary of primers used for qRT-PCR


