Abstract
BACKGROUND
Gastroesophageal reflux symptoms (GERD), higher body mass index (BMI), smoking, and genetic variants in angiogenic pathway genes have been individually associated with increased risk of esophageal adenocarcinoma (EA). However, how angiogenic gene polymorphisms and environmental factors jointly affect EA development remains unclear.
METHODS
Using a case-only design (n = 335), we examined interaction between 141 functional/tagging angiogenic SNPs and environmental factors (GERD, BMI, smoking) in modulating EA risk. Gene-environment interactions were assessed by a two-step approach. First, we applied random forest (RF) to screen for important SNPs that had either main or interaction effects. Second, we used case-only logistic regression (LR) to assess the effects of gene-environment interactions on EA risk, adjusting for covariates and false-discovery rate (FDR).
RESULTS
RF analyses identified three sets of SNPs (17 SNPs-GERD, 26 SNPs-smoking, and 34 SNPs-BMI) that had the highest importance scores. In subsequent LR analyses, interactions between 3 SNPs (rs2295778 of HIF1AN, rs133376 of TSC2, and rs2519757 of TSC1) and GERD, 2 SNPs (rs2295778 of HIF1AN, rs2296188 (VEGFR1) and smoking, and 7 SNPs (rs2114039 of PDGRFA, rs2296188 of VEGFR1, rs11941492 of VEGFR1, rs3756309 of PDGFRB, rs7324547 of VEGFR1, rs17619601 of VEGFR1, and rs17625898 of VEGFR1) and BMI were significantly associated with EA development (all FDR ≤0.10). Moreover, these interactions tended to have a SNP dose-response effects for increased EA risk with increasing number of combined risk genotypes.
CONCLUSIONS
These findings suggest that genetic variations in angiogenic genes may modify EA susceptibility through interactions with environmental factors in a SNP dose-response manner.
Keywords: Esophageal adenocarcinoma, angiogenesis pathway genes, gene-environment interaction, case-only analysis
Esophageal adenocarcinoma (EA) has the fastest rising incidence of all solid tumors in the US and in Western Europe over the last four decades1, 2. The rapidly increasing incidence and sporadically development feature of EA suggest that environmental influences dominate its etiology3. Over recent years, it has been established that gastroesophageal reflux disease (GERD), smoking, higher body mass index (BMI) or obesity, and certain genetic variations are individually associated with EA risk4, 5. However, no single factor can fully explain the rising incidence of EA. It has been suggested that gene-environment interactions play more important role in cancer etiology than do individual factors alone6.
Angiogenesis is a process of new blood vessel formation from endothelial precursors. This process facilitates tumor growth by providing oxygenation and nutrition to tumor cells7–9. Conversely, inhibition of angiogenesis results in suppression of tumor growth10, 11. The importance of angiogenesis specifically in EA has been supported by a large body of research, including in vitro, in vivo, and clinical studies9. Overexpression of angiogenic mediators, such as vascular endothelial growth factor (VEGF), inducible nitric oxide synthase (iNOS), and cyclooxygenase (COX)-2 have been associated with EA development 8, 9, 12 and prognosis9, 13.
Previously, we and others have reported that functional genetic variations that modulate angiogenic mediator gene expression, protein production, and angiogenesis processes are associated with increased risk of developing malignancies, including EA5, 14–17. However, these associations only partly explain the etiology of EA, suggesting that studying one or a few SNPs or genes may not be sufficient to explain complex diseases such as EA because they are unlikely to result from the effect of only one or a few genes. In the present study, we examine further whether interactions between multiple SNPs in the angiogenesis pathway genes and environmental factors affect EA susceptibility. We investigated 143 functional and tagging SNPs in major angiogenesis genes in 335 EA cases. We used a case-only design and a two-step analytical approach to improve statistical power for interaction estimations18. Our findings highlight the importance of complex gene-environment interactions in the development of EA.
MATERIALS AND METHODS
Patients
A total of 335 histologically confirmed EA patients were recruited at Massachusetts General Hospital (MGH) from 1999 to 2005 and at Dana Farber Cancer Institute (DFCI) between 2004 and 2005. This study was approved by the Human Subjects Committees of MGH, DFCI, and the Harvard School of Public Health (HSPH). Written informed consent was obtained from all subjects prior to study participation. Since 96% of cases were Caucasians, we selected only Caucasians for analysis19.
Upon enrollment, a trained research assistant conducted a personal interview with each participant to collect baseline clinical data that served as important covariates in the analysis. Variables of interest included: demographic information (age, gender, adult height and weight), information regarding smoking exposure and lifetime GERD symptoms up to 1 year prior to diagnosis. Body mass index (BMI) was calculated using self-reported height and weight when the patients were in their twenties. Smoking history was grouped into “never smokers” and “ever smokers”. To assess the presence of GERD, participants were asked to indicate whether they experienced symptoms, such as heartburn, acid reflux, or regurgitation17.
SNP Selection and Genotyping
DNA was extracted from whole blood using the Puregene DNA isolation Kit (Gentra System). Candidate genes were selected based on their roles in angiogenesis and on published evidence of their relationship to carcinogenesis19. Angiogenesis pathway was defined based on information in KEGG and GeneGO databases. SNPs were either functional SNPs with recorded functional information in the literature, or tagging SNPs with pairwise r2≥0.8 and minor allele frequency (MAF) ≥5% in Caucasians. A total of 141 SNPs in 13 angiogenic genes (FGFR4, HIF1, HIF1AN, MMP1, MMP3, PDGFRA, PDGFRB, THBD, TSC1, TSC2, VEGF, VEGFR1, VEGFR2) were selected. Genotyping was determined using the Illumina Goldengate assay on BeadChip by laboratory personnels blinded to the subjects’ clinical information. Ten percent of duplicate samples were randomly included in the testing plates for quality controls.
Statistical Analysis
We restricted our data analysis to Caucasians since most EA cases (>90%) in our study cohort were Caucasians. We applied a case-only study design which is considered a more powerful method for analyzing gene-environment interaction than a case-control study of the same size18. We used a two-step approach to investigate gene-environment interactions. In the first step, we applied random forest (RF) to screen for SNPs most likely to be associated with environmental factors. RF consists of a collection of classification or regression trees grown from a random selection of variables on boot-strapped samples (bagging). The importance of a variable (average importance, Avlmp) can be estimated by the increase in misclassification for the left-out (out-of bag) samples when using a data vector containing the original variable values and a vector with randomly permuted values. It has been shown by simulation studies that RF importance measures are able to simultaneously detect single-locus heterogeneity models and multiplicative interaction models20, 21. RF can also capture joint variable effects by a recently introduced joint-importance measure22, which extends the concept of single importance by jointly permutating the values of variables of interest. The aim of the second-step was to obtain valid parameter estimates for the selected variables. Case-only logistic regression model23, 24 was used to evaluate the associations of gene-environment interaction markers with EA development, adjusting for covariates, such as, age, gender. False-discovery rate (FDR) was assessed to account for multiple comparisons. Polymorphisms were categorized into homozygous wild-type, and variant allele-containing genotypes. GERD, smoking, and BMI were dichotomized into 0 and 1.
RESULTS
Patients
A total of 335 patients were included in the analyses. Demographic and tumor characteristics are listed in Table 1. The majority of EA cases (88.0%) were male. The mean age at the time of diagnosis for the entire cohort was 62.9 years (±SD11.8). 52% of patients reported to have GERD symptoms and 80% of EA cases were ever-smokers. About 31% cases were overweight (BMI≥25) at the age of 18. Twenty-nine percent were stage I–IIA and seventy-one percent were stage IIB–IV. All cases were without a prior EA diagnosis at enrollment.
Table 1.
Clinical and Demographic Characteristics of EA Cases
| Characteristics | EA Cases (n = 335) |
|---|---|
| Age (years, mean ± SD) | 62.9 ± 11.8 |
| Gender | |
| Male (%) | 295 (88%) |
| Female (%) | 40 (12%) |
| GERD symptoms* | |
| Yes (%) | 173 (52%) |
| No (%) | 162(48%) |
| Smoking status | |
| Ever smoker (%) | 269 (80%) |
| Never smoker (%) | 66 (20%) |
| Pack-years, median (min ~ max) | 24.4 (0 ~ 212.0) |
| BMI | |
| BMI at age 18, (kg/m2, mean ± SD) | 23.6 ± 3.8 |
| BMI ≥ 25 at age 18 (%) | 104 (31%) |
| BMI <25 at age 18 (%) | 231 (69%) |
| Stage | |
| I–IIA | 98 (29%) |
| IIB–IV | 237 (71%) |
7 (2%) missing and imputed
Examination of the Independence between Environmental Exposures and Genetic Factors
In case-only analysis, the odds ratio (OR) can be interpreted as the multiplicative interaction between gene and environment under the assumption that the gene and environment are independent25. To determine the independence assumption, the relationship between the gene and environment in a sample of controls from the same base population as the cases should be examined26. A lack of association between the exposures and SNPs among the controls would indicate that the exposures and SNPs are independent27. To determine whether environmental factors and SNPs were independent in our study, we examined their correlations in a sample of 313 controls from the same base population as the cases15. We found no significant correlations between any environmental factors and SNPs studied, suggesting that the independence assumption in the present study was not violated.
RF Analysis
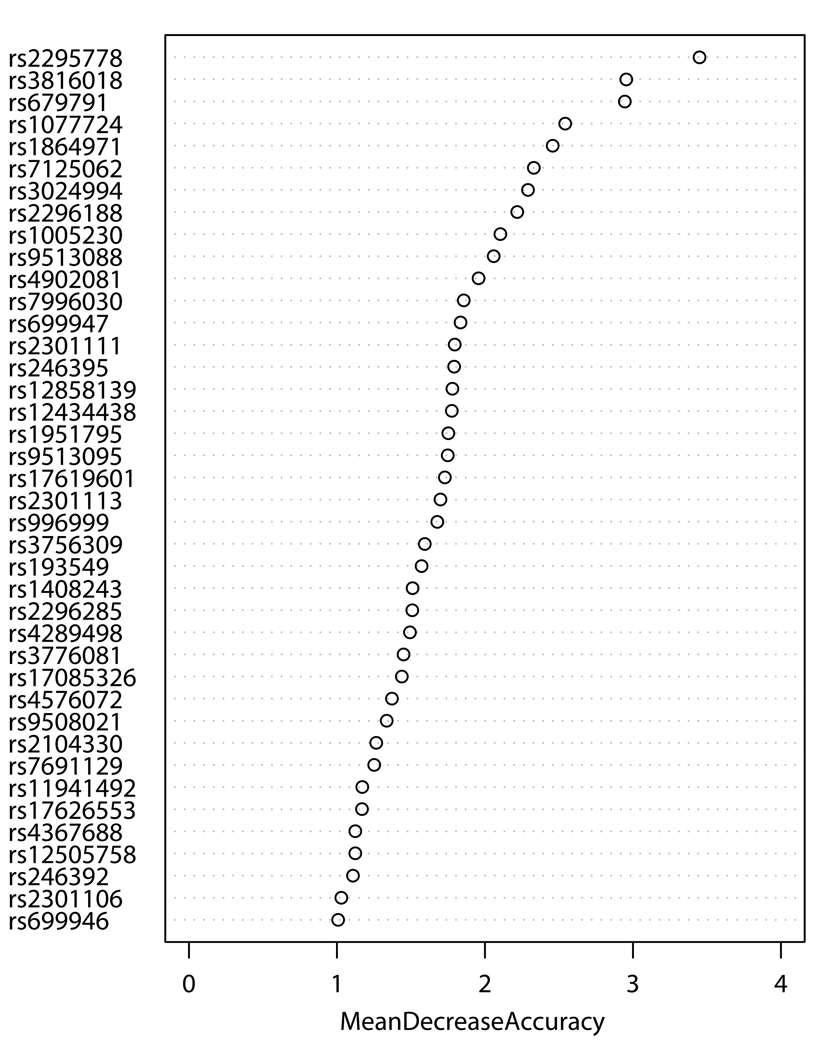
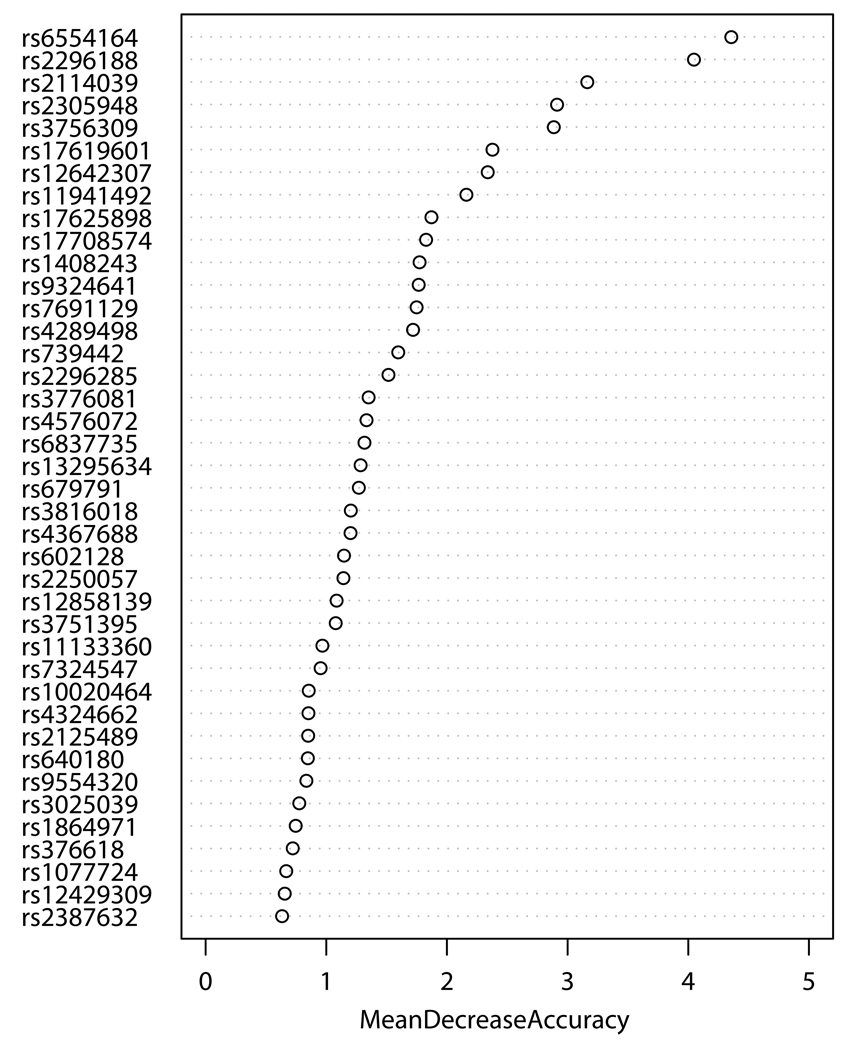
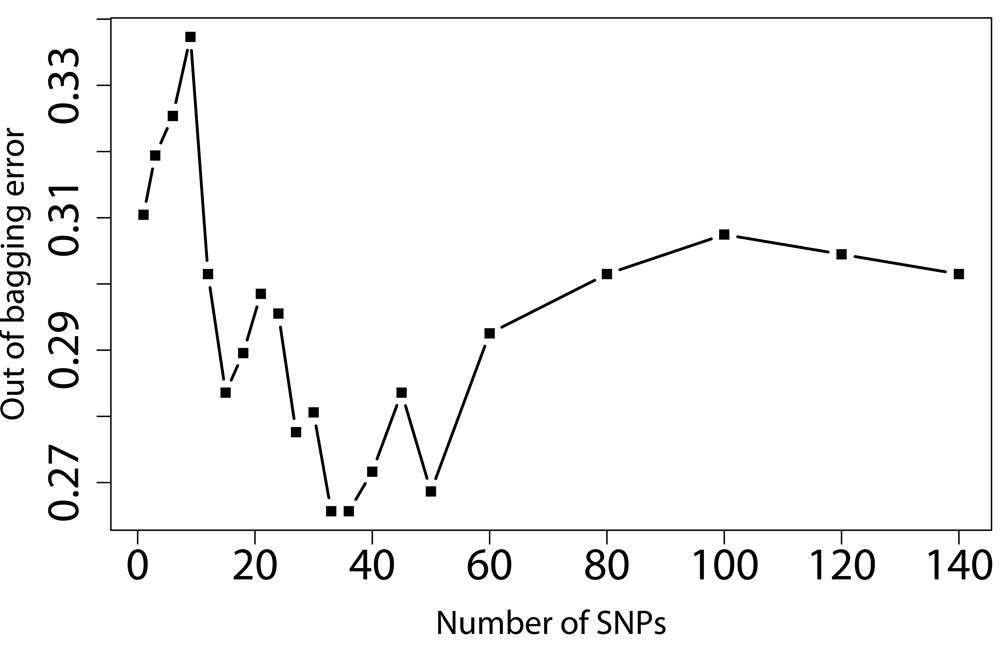
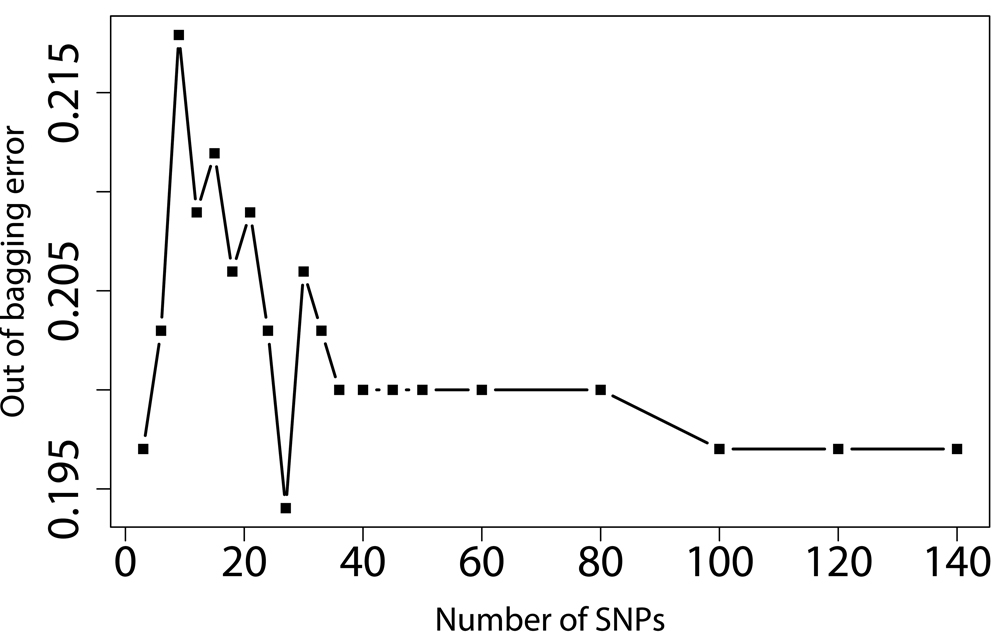
SNPs-GERD, SNPs-smoking, and SNPs-BMI interactions were screened by RF, respectively. The RF algorithm generates an importance score for each variable that quantifies the relative contribution of that variable to the prediction accuracy (Figure 1). After ranking the importance score in each of three runs of RF using 100,000 trees, the classification errors of each RF run were estimated by sliding window sequential forward feature selection (SWSFS) algorithm 28. The SWSFS analysis identified three sets of SNPs (17 SNPs with GERD, 26 SNPs with smoking, and 34 SNPs with BMI, respectively) that had the lowest classification error rates (Figure 2). These three sets of SNPs were chosen for subsequent SNP-GERD, SNP-smoking, and SNP-BMI interaction analyses, respectively, in case-only logistic regression models,
Figure 1.
Importance score plots of SNP-GERD (A), SNP-smoking (B), and SNP-BMI (C) interactions ranked by case-only random forest analysis. Mean decrease accuracy (MDA) was estimated from 100,000 tests (trees), respectively. Higher MDA means higher contribution of the interaction to EA risk.
Figure 2.
Prediction error rate of angiogenic SNPs using GERD (A), BMI (B), and smoking (C) as outcomes. X-axis denotes the number of SNPs in the model, where the SNPs are included one by one based on their MDA ranks ranging from the most (left) to the least (right) importance. Y-axis is the error rate given by random forest (out of bag error estimation from 10, 000 trees). The SNP sets with the lowest error rates were selected for further case-only logistic regression analysis.
Case-only Logistic Regression Analysis
Table 2 shows the odds ratios (ORs) of SNPs-GERD interaction for ARDS risk, as determined by the case-only design. Significant associations were found between 2 SNPs (rs2295778 of HIF1N, rs13337626 of TSC2, respectively) and GERD combinations and EA risk. Interactions between 2 SNPs (rs2295778 of HIF1N, rs2296188 of VEGFR1, respectively) and smoking were also significantly involved in EA development. When we assessed ORs effects for higher BMI and angiogenic SNPs, significant associations were detected between 7 SNPs (rs2114039 of PDGRFA, rs2296188 of VEGFR1, rs17619601 of VEGFR1, rs7324547 of VEGFR1, rs11941492 of VEGFR1, rs1770857 of PDGFRB, rs1762598 of VEGFR1) and higher BMI, respectively.
Table 2.
Interaction Between Angiogenic SNPs and Environmental Factors (GERD, Smoking, and BMI) on EA Risk: Case-Only Analysis
| Interaction Markers | ORi (95% CI) | P | FDR |
|---|---|---|---|
| SNP-GERD# | |||
| rs2295778 (HIF1N)-GERD | 2.23 (1.41–3.50) | 0.0005 | 0.0080 |
| rs13337626 (TSC2)-GERD | 2.40 (1.27–4.51) | 0.0067 | 0.0512 |
| SNP-Smoking* | |||
| rs2295778 (HIF1AN)-Smoking | 0.44 (0.26–0.78) | 0.0043 | 0.0630 |
| rs2296188 (VEGFR1)-Smoking | 0.50 (0.28–0.87) | 0.0138 | 0.1035 |
| SNP-BMI& | |||
| rs2114039 (PDGRFA)-BMI | 2.06 (1.29–3.30) | 0.0026 | 0.0286 |
| rs2296188 (VEGFR1)-BMI | 0.41 (0.24–0.73) | 0.0023 | 0.0286 |
| rs17619601 (VEGFR1)-BMI | 0.26 (0.09–0.75) | 0.0130 | 0.0572 |
| rs7324547 (VEGFR1)-BMI | 1.84 (1.14–2.98) | 0.0129 | 0.0572 |
| rs11941492 (VEGFR1)-BMI | 0.52 (0.32–0.84) | 0.0083 | 0.0572 |
| rs1770857 (PDGFRB)-BMI | 1.89 (1.12–3.18) | 0.0164 | 0.0601 |
| rs17625898 (VEGFR1)-BMI | 0.38 (0.16–0.88) | 0.0232 | 0.0742 |
ORi: interaction odds ratio; BMI: BMI at age 18 (< 25 vs. ≥ 25); FDR: False discovery rate; FDR <0.10 was considered noteworthy.
adjusted for age, sex, BMI at age 18 (< 25 vs. ≥ 25) and smoking (+/−).
adjusted for age, sex, BMI at age 18 (< 25 vs. ≥ 25) and GERD (+/−).
adjusted for age, sex, GERD (+/−), and smoking (+/−).
To investigate the cumulative effects of multiple risk SNPs, we analyzed the interactions between combined risk genotypes and environmental risk factors (GERD, smoking, and BMI), respectively, We found that increased evidence for gene-environment interaction was associated with increasing number of risk genotypes, suggesting the existence of a SNP dose-response in the gene-environment interactions in EA development (Table 3).
Table 3.
Cumulative Effects of Gene (Risk Genotypes*)-Environment (GERD, Smoking, and BMI) Interactions on EA Risk: Case-Only Analysis
| Risk Genotypes of rs2295778 and rs13337626 |
GERD | Cumulative ORi (95% CI) |
P | |
|---|---|---|---|---|
| No (n) | Yes (n) | |||
| No risk genotype | 95 | 63 | 1 (reference) | - |
| 1 risk genotype | 62 | 92 | 2.24 (1.42–3.52) | 4.91E-4 |
| 2 risk genotype | 6 | 17 | 4.27 (1.60–11.43) | 3.81E-3 |
| Trend | 2.16 (1.49–3.13) | 4.81E-5 | ||
|
Risk Genotypes of rs2295778 and rs2296188 |
Smoking | Cumulative ORi (95% CI) | P | |
| No (n) | Yes (n) | |||
| No risk genotype | 17 | 28 | 1 (reference) | - |
| 1 risk genotype | 34 | 122 | 2.18 (1.07–4.44) | 3.22E-2 |
| 2 risk genotypes | 15 | 119 | 4.82 (2.15–10.80) | 1.35E-4 |
| Trend | 2.20 (1.47–3.28) | 1.25E-4 | ||
|
Risk Genotypes of rs2114039, rs2296188, rs17619601, rs7324547, rs11941492, rs1770857, and rs17625898 |
BMI≥25 | Cumulative ORi (95% CI) | P | |
| No (n) | Yes (n) | |||
| Risk genotypes ≤3 | 88 | 12 | 1 (reference) | - |
| 4 risk genotypes | 80 | 32 | 2.93 (1.41–6.08) | 3.83E-3 |
| 5 risk genotypes- | 46 | 32 | 5.10 (2.40–10.83) | 2.24E-5 |
| >5 risk genotypes | 17 | 28 | 12.07 (5.15–28.32) | 1.02E-8 |
| Trend | 2.19 (1.70–2.82) | 1.23E-9 | ||
SNPs with FDR <0.10 were considered noteworthy.
DISCUSSION
Understanding the interplay between the genetic and environmental factors involved in the development of EA will help guide early detection and personalized treatment strategies for this devastating disease. One of the limitations that slows progress in this regard is the sample size requirement for statistical power in case-control studies29. This is particular difficult, if not impossible, for uncommon diseases such as EA. Recently, case-only design has been proved to be an effective and powerful approach in the assessment of interactions in the etiology of disease27, 30–32. In the present study, we used case-only analysis to examine gene-environment interactions in EA carcinogenesis. Our results indicated that specific genetic variations in angiogenic pathway genes modify EA risk through interactions with GERD, BMI, and smoking. In addition, we found that gene-environment interaction may contribute to EA risk in a dose-response fashion.
In many candidate gene approach of polygenic diseases, the effect of each individual SNP is generally limited, highlighting the need for a more compressive approach for association studies33. In this study, we grouped functional related SNPs and genes into pathway for analysis. The pathway-based multigenic approach combining multiple SNPs that interact in the same pathway may amplify the effect of individual SNP and enhance the predictive power. Analyzing SNP data at a pathway level may have several advantages over analysis of single SNPs or multiple SNPs within a single gene. Grouping the SNPs by biological pathway allows for a biologically meaningful interpretation of the results. In addition, it is often difficult to replicate the findings of individual SNPs or haplotypes due to different linkage disequilibrium patterns or different allele frequencies among different study populations. Thus, it may be more feasible and meaningful to perform replication at the level of biological pathways, though there have few studies using this type of pathway analyses.
It is biologically plausible that genetic variation in angiogenic pathway genes may modify the effect of GERD, BMI, and smoking on EA risk. GERD has been well documented to be able to cause esophageal inflammatory injuries34. Such injuries may lead to cancer through the inflammation-metaplasia-dysplasia-adenocarcinoma sequence in the esophagus by triggering a cascade of activation of oncogenes and/or inactivation of tumor suppressor genes, and DNA mutations34, 35. Inflammatory mediators can modulate angiogenesis directly, or indirectly through up-regulation of VEGF activities36. Angiogenic mediators, such as VEGF, can also enhance inflammatory processes, resulting in more severe inflammation37. Inflammatory inhibitors, such as corticosteroids have been shown to limit both inflammation and angiogenesis processes38. Cigarette smoke can induce 5-LOX expression which plays an important role in activation of MMP-2 and VEGF to induce angiogenic process and promotion of inflammation-associated adenoma formation in mice39. Additionally, it has been shown that smoke-induced VEGF expression and release were mediated by inflammatory responses40. Increased levels of angiogenic mediators (VEGF-C, VEGF-D, sVEGF-R2, Ang-2, HGF) as well as the angiogenesis inhibitor endostatin are present in overweight and obese subjects41, 42. Conversely, serum VEGF levels significantly decreased after weight loss following Roux-en-Y gastric bypass or low-fat diet treatment43.
In this study, stronger interactions were observed between rs2295778 (HIF1)-GERD and rs2295778 (HIF1)-smoking. HIF is a transcription factor that, in hypoxia, induces angiogenesis activating angiogenic factors, such as VEGF and VEGF receptor44, 45. Increased tumor expression of HIF-1 and VEGF was correlated with more aggressive lesions on histological studies of human cancers46. Both GERD and smoking are known to be associated with inflammation34. Inflammatory cytokines increased HIF-1 expression through NF-kappaB pathway47. HIF-1 can also induce inflammatory responses48 by cell autonomous NF-kappaB activation49. One important mechanism underlying the cross-talk between NF-kappaB and HIF-1 is that NF-kappaB binds at a distinct element in the proximal promoter of the HIF-1 gene50. Overexpression of HIF1 has been seen in the Barrett's metaplasia-dysplasia-adenocarcinoma sequence51 and associated with inflammation in Barrett’s metaplasia52. Cigarette smoke exposure impairs angiogenesis by inhibiting VEGF through decreased expression of HIF-1alpha in hypoxic conditions53. HIF-1 alpha protein expression is associated with VEGF gene and protein expression in acetic acid-induced esophageal ulcers54.
Our study had several limitations. First, we only considered functional SNPs and tagging SNPs, rather than a comprehensive SNP approach that would capture most of the genetic variation in each gene. Therefore, based on our findings, we can not exclude potential interaction roles of those SNPs for which we did not included in the present study in EA risk. Additionally, there is no gold standard or pathway definition and different databases have different guidelines for their pathway construction. Consequently, the gene content of pathways representing the same biological process may vary between different databases, and this may have a major impact on the sensitivity and specificity of this approach. We tried to minimize this impact by selecting pathways from three commonly used and manually curetted resources. Third, we restricted our analyses to Caucasians, as most of the subjects in our cohort were white (96%). The results of this study may not be generalized to other ethnic populations.
In summary, our findings supported the hypothesis that genetic variations in the angiogenesis pathway genes can contribute to EA risk through interactions with GERD, smoking and BMI. Our results also provide further support for using pathway-based approach to identify the complex relationship between genetic polymorphisms and cancer susceptibility involving multiple factors.
Acknowledgements
We thank Andrea Shafer, Maureen Convery, and Salvatore Mucci for their research assistance.
Funding Sources: Supported by National Institute of Health (NIH) grants CA92824, CA74386, CA90578, and CA119650 (to D.C); Flight Attendant Medical Research Institute (FAMRI) grant 062459_YCSA (to RZ); the Kevin Jackson Memorial Fund and Alan Brown Chair of Molecular Genomics (to GL).
Footnotes
Conflicts of Interest Disclosures:
None.
REFRENCES
- 1.Brown LM, Devesa SS, Chow WH. Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J Natl Cancer Inst. 2008;100:1184–1187. doi: 10.1093/jnci/djn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pera M, Manterola C, Vidal O, Grande L. Epidemiology of esophageal adenocarcinoma. J Surg Oncol. 2005;92:151–159. doi: 10.1002/jso.20357. [DOI] [PubMed] [Google Scholar]
- 3.Fitzgerald RC. Molecular basis of Barrett's oesophagus and oesophageal adenocarcinoma. Gut. 2006;55:1810–1820. doi: 10.1136/gut.2005.089144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmes RS, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncol. 2007;17:2–9. doi: 10.1016/j.semradonc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Cheung WY, Liu G. Genetic variations in esophageal cancer risk and prognosis. Gastroenterol Clin North Am. 2009;38:75–91. doi: 10.1016/j.gtc.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Giarelli E, Jacobs LA. Modifying cancer risk factors: the gene-environment interaction. Semin Oncol Nurs. 2005;21:271–277. doi: 10.1016/j.soncn.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Islam A, Banerjee S, Kambhampati S, Baranda J, Banerjee S, Weston AP, Saxena NK, Banerjee SK. Angiogenic switch in Barrett's adenocarcinoma: the role of vascular endothelial growth factor. Front Biosci. 2006;11:2336–2348. doi: 10.2741/1973. [DOI] [PubMed] [Google Scholar]
- 8.Wilson KT. Angiogenic markers, neovascularization and malignant deformation of Barrett's esophagus. Dis Esophagus. 2002;15:16–21. doi: 10.1046/j.1442-2050.2002.00212.x. [DOI] [PubMed] [Google Scholar]
- 9.Kleespies A, Guba M, Jauch KW, Bruns CJ. Vascular endothelial growth factor in esophageal cancer. J Surg Oncol. 2004;87:95–104. doi: 10.1002/jso.20070. [DOI] [PubMed] [Google Scholar]
- 10.Kim KJ, Li B, Winer J, Armanini M, Gillett N, Phillips HS, Ferrara N. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362:841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 11.Asano M, Yukita A, Matsumoto T, Kondo S, Suzuki H. Inhibition of tumor growth and metastasis by an immunoneutralizing monoclonal antibody to human vascular endothelial growth factor/vascular permeability factor121. Cancer Res. 1995;55:5296–5301. [PubMed] [Google Scholar]
- 12.Stein HJ, Mobius C. Angiogenesis and neoplastic transformation of Barrett's epithelium. Br J Surg. 2004;91:941–942. doi: 10.1002/bjs.4633. [DOI] [PubMed] [Google Scholar]
- 13.Saad RS, Lindner JL, Liu Y, Silverman JF. Lymphatic vessel density as prognostic marker in esophageal adenocarcinoma. Am J Clin Pathol. 2009;131:92–98. doi: 10.1309/AJCPKWUQSIPVG90H. [DOI] [PubMed] [Google Scholar]
- 14.Lurje G, Husain H, Power DG, et al. Genetic variations in angiogenesis pathway genes associated with clinical outcome in localized gastric adenocarcinoma. Ann Oncol. 2010;21:78–86. doi: 10.1093/annonc/mdp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhai R, Liu G, Asomaning K, et al. Genetic polymorphisms of VEGF, interactions with cigarette smoking exposure and esophageal adenocarcinoma risk. Carcinogenesis. 2008;29:2330–2334. doi: 10.1093/carcin/bgn210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradbury PA, Zhai R, Hopkins J, et al. Matrix metalloproteinase 1, 3 and 12 polymorphisms and esophageal adenocarcinoma risk and prognosis. Carcinogenesis. 2009;30:793–798. doi: 10.1093/carcin/bgp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung WY, Zhai R, Kulke MH, et al. Epidermal growth factor A61G gene polymorphism, gastroesophageal reflux disease and esophageal adenocarcinoma risk. Carcinogenesis. 2009;30:1363–1367. doi: 10.1093/carcin/bgp126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke GM, Morris AP. A comparison of sample size and power in case-only association studies of gene-environment interaction. Am J Epidemiol. 2010;171:498–505. doi: 10.1093/aje/kwp398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu CY, Wu MC, Chen F, et al. A Large Scale Genetic Association Study of Esophageal Adenocarcinoma Risk. Carcinogenesis. 2010;31:1259–1263. doi: 10.1093/carcin/bgq092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lunetta KL, Hayward LB, Segal J, Van Eerdewegh P. Screening large-scale association study data: exploiting interactions using random forests. BMC Genet. 2004;5:32. doi: 10.1186/1471-2156-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maenner MJ, Denlinger LC, Langton A, Meyers KJ, Engelman CD, Skinner HG. Detecting gene-by-smoking interactions in a genome-wide association study of early-onset coronary heart disease using random forests. BMC Proc. 2009;3 Suppl 7:S88. doi: 10.1186/1753-6561-3-s7-s88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bureau A, Dupuis J, Falls K, Lunetta KL, Hayward B, Keith TP, Van Eerdewegh P. Identifying SNPs predictive of phenotype using random forests. Genet Epidemiol. 2005;28:171–182. doi: 10.1002/gepi.20041. [DOI] [PubMed] [Google Scholar]
- 23.Prentice RL, Huang Y, Hinds DA, et al. Variation in the FGFR2 gene and the effect of a low-fat dietary pattern on invasive breast cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:74–79. doi: 10.1158/1055-9965.EPI-09-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lash TL, Bradbury BD, Wilk JB, Aschengrau A. A case-only analysis of the interaction between N-acetyltransferase 2 haplotypes and tobacco smoke in breast cancer etiology. Breast Cancer Res. 2005;7:R385–R393. doi: 10.1186/bcr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khoury MJ, Flanders WD. Nontraditional epidemiologic approaches in the analysis of gene-environment interaction: case-control studies with no controls! Am J Epidemiol. 1996;144:207–213. doi: 10.1093/oxfordjournals.aje.a008915. [DOI] [PubMed] [Google Scholar]
- 26.Piegorsch WW, Weinberg CR, Taylor JA. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat Med. 1994;13:153–162. doi: 10.1002/sim.4780130206. [DOI] [PubMed] [Google Scholar]
- 27.Pande M, Amos CI, Eng C, Frazier ML. Interactions between cigarette smoking and selected polymorphisms in xenobiotic metabolizing enzymes in risk for colorectal cancer: A case-only analysis. Mol Carcinog. 2010;49 doi: 10.1002/mc.20682. 974-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang R, Tang W, Wu X, Fu W. A random forest approach to the detection of epistatic interactions in case-control studies. BMC Bioinformatics. 2009;10 Suppl 1:S65. doi: 10.1186/1471-2105-10-S1-S65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155:478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 30.Pavanello S, Mastrangelo G, Placidi D, Campagna M, Pulliero A, Arici C, Porru S. CYP1A2 polymorphisms, occupational and environmental exposures and risk of bladder cancer. Eur J Epidemiol. 2010;25:491–500. doi: 10.1007/s10654-010-9479-8. [DOI] [PubMed] [Google Scholar]
- 31.Moorman PG, Iversen ES, Marcom PK, et al. Evaluation of established breast cancer risk factors as modifiers of BRCA1 or BRCA2: a multi-center case-only analysis. Breast Cancer Res Treat. 2010;124:441–451. doi: 10.1007/s10549-010-0842-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brevik A, Joshi AD, Corral R, et al. Polymorphisms in base excision repair genes as colorectal cancer risk factors and modifiers of the effect of diets high in red meat. Cancer Epidemiol Biomarkers Prev. 2010;19 doi: 10.1158/1055-9965.EPI-10-0606. 3167-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore JH, Asselbergs FW, Williams SM. Bioinformatics challenges for genome-wide association studies. Bioinformatics. 2010;26:445–455. doi: 10.1093/bioinformatics/btp713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdel-Latif MM, Duggan S, Reynolds JV, Kelleher D. Inflammation and esophageal carcinogenesis. Curr Opin Pharmacol. 2009;9:396–404. doi: 10.1016/j.coph.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 35.O'Riordan JM, Abdel-latif MM, Ravi N, et al. Proinflammatory cytokine and nuclear factor kappa-B expression along the inflammation-metaplasia-dysplasia-adenocarcinoma sequence in the esophagus. Am J Gastroenterol. 2005;100:1257–1264. doi: 10.1111/j.1572-0241.2005.41338.x. [DOI] [PubMed] [Google Scholar]
- 36.Jackson JR, Seed MP, Kircher CH, Willoughby DA, Winkler JD. The codependence of angiogenesis and chronic inflammation. Faseb J. 1997;11:457–465. [PubMed] [Google Scholar]
- 37.Angelo LS, Kurzrock R. Vascular endothelial growth factor and its relationship to inflammatory mediators. Clin Cancer Res. 2007;13:2825–2830. doi: 10.1158/1078-0432.CCR-06-2416. [DOI] [PubMed] [Google Scholar]
- 38.Feltis BN, Wignarajah D, Reid DW, Ward C, Harding R, Walters EH. Effects of inhaled fluticasone on angiogenesis and vascular endothelial growth factor in asthma. Thorax. 2007;62:314–319. doi: 10.1136/thx.2006.069229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye YN, Liu ES, Shin VY, Wu WK, Cho CH. Contributory role of 5-lipoxygenase and its association with angiogenesis in the promotion of inflammation-associated colonic tumorigenesis by cigarette smoking. Toxicology. 2004;203:179–188. doi: 10.1016/j.tox.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Edirisinghe I, Yang SR, Yao H, et al. VEGFR-2 inhibition augments cigarette smoke-induced oxidative stress and inflammatory responses leading to endothelial dysfunction. Faseb J. 2008;22:2297–2310. doi: 10.1096/fj.07-099481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silha JV, Krsek M, Sucharda P, Murphy LJ. Angiogenic factors are elevated in overweight and obese individuals. Int J Obes (Lond) 2005;29:1308–1314. doi: 10.1038/sj.ijo.0802987. [DOI] [PubMed] [Google Scholar]
- 42.Rehman J, Considine RV, Bovenkerk JE, Li J, Slavens CA, Jones RM, March KL. Obesity is associated with increased levels of circulating hepatocyte growth factor. J Am Coll Cardiol. 2003;41:1408–1413. doi: 10.1016/s0735-1097(03)00231-6. [DOI] [PubMed] [Google Scholar]
- 43.Gomez-Ambrosi J, Catalan V, Rodriguez A, et al. Involvement of serum vascular endothelial growth factor family members in the development of obesity in mice and humans. J Nutr Biochem. 2010;21:774–780. doi: 10.1016/j.jnutbio.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Brahimi-Horn C, Pouyssegur J. The role of the hypoxia-inducible factor in tumor metabolism growth and invasion. Bull Cancer. 2006;93:E73–E80. [PubMed] [Google Scholar]
- 45.Yamakawa M, Liu LX, Date T, et al. Hypoxia-inducible factor-1 mediates activation of cultured vascular endothelial cells by inducing multiple angiogenic factors. Circ Res. 2003;93:664–673. doi: 10.1161/01.RES.0000093984.48643.D7. [DOI] [PubMed] [Google Scholar]
- 46.Otrock ZK, Hatoum HA, Awada AH, Ishak RS, Shamseddine AI. Hypoxia-inducible factor in cancer angiogenesis: structure, regulation and clinical perspectives. Crit Rev Oncol Hematol. 2009;70:93–102. doi: 10.1016/j.critrevonc.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Kim J, Shao Y, Kim SY, et al. Hypoxia-induced IL-18 increases hypoxia-inducible factor-1alpha expression through a Rac1-dependent NF-kappaB pathway. Mol Biol Cell. 2008;19:433–444. doi: 10.1091/mbc.E07-02-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholas SA, Sumbayev VV. The involvement of hypoxia-inducible factor 1 alpha in Toll-like receptor 7/8-mediated inflammatory response. Cell Res. 2009;19:973–983. doi: 10.1038/cr.2009.44. [DOI] [PubMed] [Google Scholar]
- 49.Scortegagna M, Cataisson C, Martin RJ, Hicklin DJ, Schreiber RD, Yuspa SH, Arbeit JM. HIF-1alpha regulates epithelial inflammation by cell autonomous NFkappaB activation and paracrine stromal remodeling. Blood. 2008;111:3343–3354. doi: 10.1182/blood-2007-10-115758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorlach A, Bonello S. The cross-talk between NF-kappaB and HIF-1: further evidence for a significant liaison. Biochem J. 2008;412:e17–e19. doi: 10.1042/BJ20080920. [DOI] [PubMed] [Google Scholar]
- 51.Griffiths EA, Pritchard SA, McGrath SM, Valentine HR, Price PM, Welch IM, West CM. Increasing expression of hypoxia-inducible proteins in the Barrett's metaplasia-dysplasia-adenocarcinoma sequence. Br J Cancer. 2007;96:1377–1383. doi: 10.1038/sj.bjc.6603744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ling FC, Khochfar J, Baldus SE, et al. HIF-1alpha protein expression is associated with the environmental inflammatory reaction in Barrett's metaplasia. Dis Esophagus. 2009;22:694–699. doi: 10.1111/j.1442-2050.2009.00957.x. [DOI] [PubMed] [Google Scholar]
- 53.Michaud SE, Menard C, Guy LG, Gennaro G, Rivard A. Inhibition of hypoxia-induced angiogenesis by cigarette smoke exposure: impairment of the HIF-1alpha/VEGF pathway. Faseb J. 2003;17:1150–1152. doi: 10.1096/fj.02-0172fje. [DOI] [PubMed] [Google Scholar]
- 54.Baatar D, Jones MK, Tsugawa K, et al. Esophageal ulceration triggers expression of hypoxia-inducible factor-1 alpha and activates vascular endothelial growth factor gene: implications for angiogenesis and ulcer healing. Am J Pathol. 2002;161:1449–1457. doi: 10.1016/s0002-9440(10)64420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]