Abstract
Telomeres form the ends of eukaryotic chromosomes and are vital in maintaining genetic integrity. Telomere dysfunction is associated with cancer and several chronic diseases. Patterns of genetic variation across individuals can provide keys to further understanding the evolutionary history of genes. We investigated patterns of differentiation and population structure of 37 telomere maintenance genes among 53 worldwide populations. Data from 898 unrelated individuals were obtained from the genome-wide scan of the Human Genome Diversity Panel (HGDP) and from 270 unrelated individuals from the International HapMap Project at 716 single-nucleotide polymorphism (SNP) loci. We additionally compared this gene set to HGDP data at 1396 SNPs in 174 innate immunity genes. The majority of the telomere biology genes had low to moderate haplotype diversity (45–85%), high ancestral allele frequencies (>60%) and low differentiation (FST <0.10). Heterozygosity and differentiation were significantly lower in telomere biology genes compared with the innate immunity genes. There was evidence of evolutionary selection in ACD, TERF2IP, NOLA2, POT1 and TNKS in this data set, which was consistent in HapMap 3. TERT had higher than expected levels of haplotype diversity, likely attributable to a lack of linkage disequilibrium, and a potential cancer-associated SNP in this gene, rs2736100, varied substantially in genotype frequency across major continental regions. It is possible that the genes under selection could influence telomere biology diseases. As a group, there appears to be less diversity and differentiation in telomere biology genes than in genes with different functions, possibly due to their critical role in telomere maintenance and chromosomal stability.
Keywords: telomere, selection, single-nucleotide polymorphism, HGDP, HapMap
Introduction
Telomeres consist of (TTAGGG)n nucleotide repeats and an associated protein complex located at chromosome ends. They are essential for maintaining chromosomal integrity. Telomere-associated proteins include the telomerase reverse transcriptase (TERT) and its RNA component (TERC), plus an ordered protein complex, or shelterin, consisting of six proteins: TERF1, TERF2, TINF2, TERF2IP, ACD and POT1 (Collins and Mitchell, 2002; de Lange, 2005). This telomere complex, and many other associated proteins, are responsible for preserving chromosome ends, and thus genomic stability, by protecting chromosomes from end-to-end fusion, atypical recombination and degradation (Moon and Jarstfer, 2007). Many of the components of the telomeric complex are highly conserved across species in comparative sequence and functional investigations (Nakamura and Cech, 1998; Li et al., 2000; Kanoh and Ishikawa, 2003; de Lange, 2004; Savage et al., 2005). It was also shown that seven of these genes (TERT, POT1, TNKS, TERF1, TINF2, TERF2 and TERF2IP) had lower nucleotide diversity compared with other gene families; they were also highly conserved and the most common allele was ancestral (Savage et al., 2005).
Telomere nucleotide repeats progressively shorten with each cell division due to incomplete replication of the 3′ end by DNA polymerases. When they become critically short, cellular senescence or cellular crisis is induced in normal cells but in malignant cells this pathway is bypassed through the activation of telomerase or the alternative pathways (Gilley et al., 2005; Rodier et al., 2005). Short telomeres induce genetic instability and thereby promote the initiation and development of cancer (Blasco et al., 1997; Rudolph et al., 1999, 2001; Wu et al., 2003; Plentz et al., 2003, 2004). Telomere attrition has also been associated with aging, many diseases (including diabetes mellitus and cardiovascular disease), inflammation, oxidative stress, an unhealthy lifestyle and smoking (von Zglinicki, 2002; Wong and Collins, 2003; Morlá et al., 2006; Aubert and Lansdorp, 2008; Mirabello et al., 2009). Several disorders are associated with mutations in telomere biology genes (Crabbe et al., 2004; Blasco, 2007; Vulliamy et al., 2008; Armanios, 2009; Savage and Alter, 2009). Patients with dyskeratosis congenita, a heterogeneous inherited bone marrow failure and cancer predisposition syndrome, have extremely short telomeres and germline mutations in genes important in the maintenance of telomeres (DKC1, TERC, TERT, NOLA3 (alias NOP10), TINF2 or NOLA2 (alias NHP2)) (Crabbe et al., 2004; Vulliamy et al., 2008; Armanios, 2009; Savage and Alter, 2009). In addition, recent genome-wide association studies found that genetic variation at 5p15.33 (TERT-CLPTM1L locus) was associated with risk of glioma (Shete et al., 2009), basal cell carcinoma (Stacey et al., 2008, 2009), testicular cancer (Turnbull et al., 2010), pancreatic cancer (Petersen et al., 2010) and lung cancer (McKay et al., 2008; Jin et al., 2009; Landi et al., 2009); an association study of multiple tumor types suggest that this region may contain important markers of overall cancer risk (Rafnar et al., 2009).
The extent to which disease-associated alleles differ in frequency between populations and the evolutionary forces responsible for the observed degree of population differentiation may provide keys to further understanding disease pathogenesis. Allele frequencies for many genetic variants differ by geographical regions (Guthery et al., 2007; Lan et al., 2007; Myles et al., 2008), possibly the result of several factors including natural selection and neutral genetic drift. There may be functional consequences of a particular variant that leads to a more favorable response and thus certain variants may be under selective pressure. Searching for a signature of selection has the potential to identify functional and disease related variants (Bamshad and Wooding, 2003; Hurst, 2009).
We examined patterns of differentiation, allele frequencies and the haplotype structure of 37 genes important in telomere biology among 53 worldwide populations from Africa, the Middle East, Europe, Central/South (C/S) Asia, East Asia, Oceania and the Americas. Data from 1168 unrelated individuals were obtained from the genome-wide scan of the Human Genome Diversity Panel (HGDP-CEPH) (Cann et al., 2002; Li et al., 2008) and from the International HapMap Project (The International HapMap Consortium, 2003) at 716 single-nucleotide polymorphism (SNP) loci. We additionally compared our telomere gene set to HGDP-CEPH data that we had on 174 innate immunity genes at 1396 SNPs, this allowed us to determine if two sets of genes grouped by function have similar genetics. We hypothesized that genetic variation in telomere biology genes may be constrained because of both the high degree of sequence similarity previously observed across species and the critical roles their protein products have in chromosomal stability.
Materials and methods
Data set
We obtained SNP data for each gene, including 20 kbp upstream and 10 kbp downstream, from the HGDP-CEPH (Cann et al., 2002) genome-wide scan of 650 000 common SNPs (Li et al., 2008) and the HapMap Phase 2 (The International HapMap Consortium, 2003) public database. Genotype data were retrieved for 37 gene regions: ACD, ATM, BLM, DDX1, DDX11, DKC1, MRE11A, NBN, NOLA1, NOLA2, NOLA3, PARP1, PARP2, PINX1, POT1, PRKDC, RAD50, RAD51AP1, RAD51C, RAD51L1, RAD51L3, RAD54L, RECQL, RECQL4, RECQL5, RTEL1, TEP1, TERC, TERF1, TERF2, TERF2IP, TERT, TINF2, TNKS, TNKS2, WRN and XRCC6, as well as the region between PARP2/TEP1. Genes were chosen based on their involvement in telomere biology or presumed interaction with telomeres as reported in the literature. Telomerase complex genes include TERC, TERT, DKC1, TEP1, NOLA1, NOLA2 and NOLA3; shelterin genes include TERF1, TERF2, TERF2IP, POT1, TINF2 and ACD; DNA repair genes include XRCC6, NBN, RAD50, ATM, RAD54L, RAD51L3, RAD51C, RAD51AP1, RAD51L1 and MRE11A; helicase genes include WRN, BLM, RECQL, RECQL4, RECQL5, DDX1 and DDX11; and, other telomere-associated genes include PRKDC, PARP1, PARP2, PINX1, TNKS, TNKS2 and RTEL1. All SNPs, regardless of minor allele frequency, were included in the analysis as for many of these genes there were only a few SNPs available. Data were retrieved for all individuals in the 52 populations (952 individuals) included in the HGDP-CEPH 952 panel and the four populations (270 individuals) of the HapMap project for the same 716 SNPs. Atypical and related individuals were removed (Rosenberg, 2006), which resulted in 898 individuals from the HGDP-CEPH panel and 270 from the HapMap project. The final data set included 1168 unrelated individuals from 53 unique populations. We did not limit the SNP data to SNPs only within exons, introns, promoters or 3′areas because the goal of this study was to understand the gene regions, including upstream and downstream regions.
We also obtained genotype data for 174 genes involved in innate immunity as a comparison set for our telomere biology genes. SNP data for each gene were acquired from the HGDP-CEPH (Cann et al., 2002) genome-wide scan for all individuals in the 52 populations, and cleaned as described above. The immune gene set was chosen as a comparison gene set as these genes are often highly variable. Additional comparisons were made with data reported in the literature. Supplementary Table 1 lists the 174 innate immune gene regions evaluated.
HapMap phase 3 (The International HapMap Consortium, 2003) SNP data for 11 populations (1115 individuals) were also retrieved for a subset of the telomere maintenance genes that were potential candidates for evolutionary selection (defined in results): ACD, NOLA2, RECQL4, POT1, TERF2IP and TNKS, as well as for TERT. Individuals in this phase do not overlap with HapMap phase 2 participants.
Data analysis
Haplotype and SNP frequencies were estimated using a Bayesian algorithm implemented in PHASE version 2.1 (Stephens et al., 2001; Stephens and Scheet, 2005). Haplotypes determined by PHASE were used as input for all other analyses. The package ARLEQUIN version 3.11 (Excoffier et al., 2005) was used to compute haplotype diversity, FST values, Mantel test, analysis of molecular variance (AMOVA) and heterozygosity. FST values based on allele frequencies were calculated as a measure of population differentiation and significance was estimated with 10 000 permutations. A Mantel test was used to test the significance of the regression of genetic distance on geographic distance between population pairs with 10 000 permutations. In order to apportion the fraction of the genetic variance due to differences between and within continental groups and infer the genetic structure of the populations, AMOVA was performed with 10 000 permutations. Mega version 4.0 (Tamura et al., 2007) was used to construct a neighbor-joining tree based on genetic differentiation. Population structure was inferred by a Bayesian clustering analysis performed with structure version 2.2 (Pritchard et al., 2000; Falush et al., 2007) using the following settings: admixture model, correlated markers, K=1–10, a length of 100 000 for the burn-in period, and 100 000 repetitions following the burn-in period. Haploview version 4.1 (Barrett et al., 2005) was used to determine the degree of linkage disequilibrium (LD) and minor allele frequencies (MAF). LD P values (with s.e.) were estimated by Monte Carlo approximation with 10 000 steps in the Markov Chain using ARLEQUIN. This LD calculation is an extension of Fisher exact probability test on contingency tables, and the results are given as a significance level of LD for each pair of loci with a small P value (<0.05) indicating high LD (Excoffier et al., 2005; Santos-Lopes et al., 2007). Differences between the telomere and immune gene set results were tested for significance with parametric (t-test) and non-parametric tests (Mann–Whitney U-test).
We retrieved ancestral (chimpanzee) alleles for 98.2% of the SNPs using the UCSC Genome Browser (March 2006 Assembly: http://genome.ucsc.edu/) and/or Ensembl (release 50, Jul 2008: http://www.ensembl.org/index.html). In cases where neither human allele corresponded to the chimpanzee allele or when the chimpanzee allele was unknown we excluded these SNPs from the analysis. Pairwise geographic distance between populations and distance from Addis Ababa, Ethiopia (the putative point of origin of modern humans (White et al., 2003)) was estimated in kilometers (km) following the likely colonization route (shortest path through landmasses) as in Prugnolle et al. (2005).
Selection was evaluated with the following analyses: (1) between population differentiation (FST), which can be inflated due to environmental pressures on populations causing local adaptation and allele frequency changes (positive directional selection), and negative or balancing selection can decrease the differentiation of selected loci (Akey et al., 2002; Nielsen, 2005; Sabeti et al., 2007); (2) genetic diversity, a significant decrease points to positive selection when a particular allele is favored, and increases could be balancing selection with more diversity being potentially adaptive; (3) LD across populations, selection can increase LD; and (4) MAF and derived allele frequency (DAF) tests (Walsh et al., 2006). The iHS for HapMap phase 2 data was retrieved with HAPLOTTER (The International HapMap Consortium, 2005; Voight et al., 2006).
Results
We analyzed SNP data from the HGDP-CEPH (Cann et al., 2002) genome-wide scan for 37 gene regions involved in telomere biology and from Phase 2 of the International HapMap project (The International HapMap Consortium, 2003). The telomere data set consisted of a total of 716 SNPs in 1168 individuals from 53 worldwide populations. Supplementary Tables 1 and 2 give summary statistics for the telomere biology and innate immune gene regions analyzed (for example, alleles, MAF, heterozygosity, variation components).
Population structure and differentiation
Bayesian cluster analysis and a distance-based neighbor-joining tree segregated individuals into five genetic clusters: Africa, Eurasia (Middle East, Europe, Utah, C/S Asia), East Asia, Oceania and America (Supplementary Figure 1). The Utah, USA population clustered within Eurasia, with high genetic similarity to the European populations. We tested for isolation by distance using a Mantel test on FST estimates and found a significant positive correlation between the degree of genetic divergence and the pairwise geographical distance (correlation coefficient (r)=0.64, P<0.001).
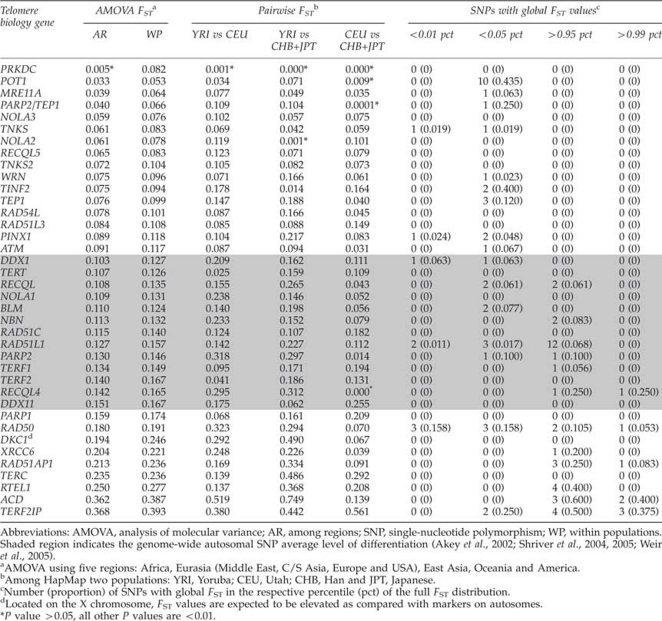
Table 1 shows the levels of differentiation (FST) by gene sorted in descending order by among regions differentiation. PRKDC and POT1 have the lowest levels of differentiation, and nearly half (0.44) of the POT1 SNPs have global FST estimates less than the 0.05 percentile of the overall FST distribution. ACD and TERF2IP had the highest levels of differentiation, and a large portion of their SNPs were above 0.95 (0.60 and 0.50, respectively) and 0.99 (0.40 and 0.38, respectively) percentiles. ACD had very high levels of differentiation observed between HapMap Yoruba and Utah populations and between Yoruba and Chinese/Japanese populations (0.52 and 0.75, respectively), and TERF2IP between Utah and Chinese/Japanese populations (0.56).
Table 1. Levels of differentiation (FST) by gene using HapMap 2 and HGDP data.
Overall, TERT had average levels of differentiation among regions and HapMap populations (Table 1 and Supplementary Table 3). Limiting the SNPs to only those within the TERT gene (introns, exons and UTRs, n=4 SNPs), the levels of differentiation were lower among regions (Fst=0.072) and within populations (Fst=0.089). We further evaluated the recently identified TERT SNP, rs2736100 (localized to intron 2: at chromosome 5p15.33, position 1 339 516) as it appears to be associated with risk of lung cancer, testicular cancer and glioma (McKay et al., 2008; Jin et al., 2009; Landi et al., 2009; Shete et al., 2009; Turnbull et al., 2010). rs2736100 had variable levels of differentiation among geographical regions. Its genotype frequencies varied among regions and levels of pairwise differentiation were particularly high among Oceania and all other regions, as well as among America and Eurasia, and low among Africa, East Asia and Eurasia (Supplementary Table 3 and Supplementary Figure 2).
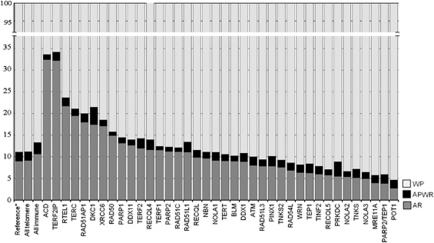
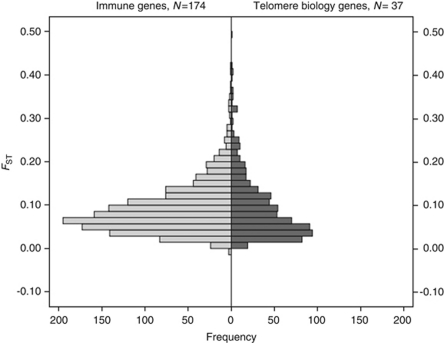
As a gene set, the AMOVA partitions variation among the seven geographical regions similar to that observed for the entire 650k autosomal SNP panel (Li et al., 2008) with within-population variation accounting for the majority of the genetic diversity (Figure 1). There was some disparity in how variation is partitioned among individual genes. There was substantially higher among-regions variation observed in ACD and TERF2IP and the least in POT1 (Figure 1). There was significantly lower differentiation observed among geographical regions for the telomere biology genes compared with the innate immune genes (P=0.0002) (Figure 1), and the distributions of FST values among all the HGDP populations showed a shift down towards lower FST for the telomere biology genes (Figure 2). AMOVA variation components and differentiation by locus are shown in Supplementary Tables 1 and 2 for the innate immune and telomere biology genes, respectively. Grouping the genes by function showed that telomerase complex genes had the lowest FST values (among regions=0.07), followed by helicase genes, and other telomere associated genes compared with the genome-wide average for autosomal SNPs (0.10–0.15 (Akey et al., 2002; Shriver et al., 2004, 2005; Weir et al., 2005)) and the innate immune gene set, as well as among HapMap 2 populations in comparison to other gene sets (Table 2).
Figure 1.
Analysis of molecular variance by gene using HapMap 2 and HGDP data. Partitioning variation into three components: within population (WP), among region (AR) and among-population-within-region (APWR). Populations are assigned to the seven main geographic regions from the HGDP-CEPH panel; *HGDP panel at 650k autosomal SNPs (Li et al., 2008).
Figure 2.
Differentiation among the HGDP populations for the telomere biology and innate immune gene sets.
Table 2. Genetic differentiation (FST) among major continental groups and HapMap 2 populations in comparison to other data sets.
| Gene sets or SNP data sets |
Among HapMap 2 populations |
Among regions | ||
|---|---|---|---|---|
| YRI vs CEU | YRI vs CHB+JPT | CEU vs CHB+JPT | ||
| Telomere biology genes (37 genes, 716 SNPs, N=1168)b | 0.136 | 0.189 | 0.095 | 0.095a |
| Shelterin genes | 0.140 | 0.224 | 0.148 | 0.109 |
| Telomerase complex genes | 0.130 | 0.161 | 0.071 | 0.072 |
| DNA repair genes | 0.154 | 0.209 | 0.099 | 0.107 |
| Helicase genes | 0.139 | 0.196 | 0.071 | 0.085 |
| Other telomere-associated genes | 0.098 | 0.144 | 0.093 | 0.086 |
| CVD genes (364 genes, 15 559 SNPs, N=270)c | 0.139 | 0.158 | 0.095 | — |
| Blood circulation and gas exchange genes | 0.129 | 0.158 | 0.062 | — |
| Lipoprotein metabolism genes | 0.144 | 0.165 | 0.082 | — |
| Insulin/IGF-mitogen-activated protein kinase kinase/ MAP kinase cascade genes | 0.194 | 0.174 | 0.138 | — |
| Disease associated SNPs (25 SNPs, N=952)d | — | — | — | 0.10a |
| Innate immune genes (174 genes, 1396 SNPs, N=917) | — | — | — | 0.11 |
| Genome-wide autosomal SNP averagee | — | — | — | 0.10–0.15 |
Abbreviations: CEU, Utah; CHB, Han; CVD, cardiovascular disease; IGF, insulin-like growth factor; JPT, Japanese; MAP, mitogen-activated protein kinase; N=number of individuals; SNP, single-nucleotide polymorphism; YRI, Yoruba.
Among the seven geographic regions represented in the CEPH-HGDP panel.
See methods for a description of genes included in each gene set.
Haplotype diversity and LD
The number of haplotypes and diversity estimates by gene and region are shown in Table 3 and Supplementary Table 4. Overall, the haplotype diversity was highest in Africa (0.844) and lowest in Oceania (0.634; Table 3). The majority of genes had very low to moderate haplotype diversity (Supplementary Table 4). DKC1, TERC and XRCC6 had very low haplotype diversity of less than 50%, and TINF2, NOLA2 and RECQL4 had low diversity estimates between 60–70%. Surprisingly, TERT had high haplotype diversity (ranging from 81 to 96%). Using only the SNPs within the TERT gene, the haplotype diversity was lower in East Asia, Oceania and America (ranging from 37 to 74%), and higher in Eurasia and Africa (88% and 90%, respectively) (Supplementary Figure 2). The mean haplotype diversity for all of the 53 populations was negatively correlated with geographic distance from Ethiopia (r=−0.89, slope=−1.17e−5). Heterozygosity in the telomere gene set was significantly lower than in the immune gene set by geographical region (P=0.014; Table 3).
Table 3. Telomere biology genes average diversity of the major continental regions from the HapMap 2 project and HGDP-CEPH panel.
| Region | N | 2N | Average hd | Het telomere gene set | Het immune gene set | Pa |
|---|---|---|---|---|---|---|
| Africa | 185 | 370 | 84.37 | 0.299 | 0.286 | 0.059 |
| Middle East | 161 | 322 | 76.91 | 0.297 | 0.324 | 0.00005 |
| Europe | 156 | 312 | 75.04 | 0.309 | 0.330 | 0.0017 |
| C/S Asia | 198 | 396 | 73.04 | 0.300 | 0.324 | 0.0003 |
| USAb | 90 | 180 | 70.55 | — | — | — |
| East Asia | 316 | 632 | 72.19 | 0.295 | 0.303 | 0.284 |
| Oceania | 21 | 42 | 63.35 | 0.313 | 0.301 | 0.203 |
| Americac | 41 | 82 | 65.51 | 0.287 | 0.264 | 0.0057 |
Abbreviations: hd, haplotype diversity (%); het, heterozygosity in each region using HGDP-CEPH data; HGDP, Human Genome Diversity Panel; N, number of individuals; 2N, number of chromosomes.
for differences between the heterozygosity of the telomere and immune gene sets
Includes individuals from Utah.
Includes populations in Brazil, Colombia and Mexico.
LD P values were estimated for all marker pairs for each gene and a summary of the proportion of SNPs with significant LD (P<0.05) is presented in Table 4. The proportion of marker pairs with significant LD varied among geographic regions and genes. A low proportion of marker pairs with LD was often observed in Oceania. The lowest LD was observed in RAD51L1 (⩽0.4 of marker pairs in all populations) and the highest in POT1 (>0.9). However, this analysis is limited by the small number of SNPs in many of these genes and the limited population sizes in Oceania and America.
Table 4. Proportion of marker pairs with significant linkage disequilibriuma using HapMap 2 and HGDP data.
| Telomere biology geneb | n |
Populationc |
||||
|---|---|---|---|---|---|---|
| YRI (N=110) | CEU (N=90) | CHB (N=89) | Oceania (N=21) | America (N=41) | ||
| ATM | 15 | 0.818 | 0.836 | 0.806 | 1.000 | 0.600 |
| BLM | 26 | 0.659 | 0.717 | 0.709 | 0.695 | 0.529 |
| DDX1 | 16 | 0.733 | 0.697 | 0.581 | 0.308 | 0.552 |
| MRE11A | 16 | 0.783 | 0.825 | 1.000 | 0.982 | 0.582 |
| NBN | 24 | 0.593 | 0.575 | 0.721 | 0.577 | 0.559 |
| NOLA3 | 13 | 0.745 | 0.615 | 0.641 | 0.462 | 0.731 |
| PARP1 | 16 | 0.724 | 0.978 | 0.857 | 0.800 | 0.736 |
| PARP2 | 10 | 0.714 | 0.600 | 0.750 | 0.619 | 0.667 |
| PINX1 | 42 | 0.756 | 0.840 | 0.683 | 0.950 | 0.359 |
| POT1 | 23 | 0.929 | 1.000 | 1.000 | 1.000 | 0.925 |
| PRKDC | 18 | 0.508 | 0.581 | 0.562 | 0.622 | 0.533 |
| RAD50 | 19 | 0.634 | 0.756 | 0.864 | 0.652 | 0.527 |
| RAD51AP1 | 12 | 0.848 | 0.911 | 0.889 | 0.933 | 0.622 |
| RAD51L1 | 177 | 0.277 | 0.404 | 0.331 | 0.275 | 0.308 |
| RAD51L3 | 11 | 0.444 | 0.929 | 0.489 | 0.286 | 0.571 |
| RAD54L | 10 | 0.714 | 0.917 | 0.750 | NA | 0.750 |
| RECQL | 33 | 0.679 | 0.750 | 0.939 | 0.570 | 0.772 |
| RECQL5 | 7 | 0.714 | 1.000 | 1.000 | 0.533 | 0.762 |
| RTEL1 | 10 | 0.528 | 0.778 | 0.821 | 0.528 | 0.578 |
| TEP1 | 25 | 0.446 | 0.438 | 0.352 | 0.267 | 0.447 |
| TERF1 | 18 | 0.713 | 0.895 | 0.758 | 0.706 | 0.721 |
| TERF2 | 6 | 0.667 | 0.933 | 0.733 | NA | NA |
| TERF2IP | 8 | 0.667 | 0.429 | 0.571 | 0.467 | NA |
| TERT | 8 | 0.571 | 0.536 | 0.524 | 0.095 | 0.381 |
| TNKS | 52 | 0.813 | 0.797 | 0.879 | 0.703 | 0.606 |
| TNKS2 | 11 | 0.855 | 0.867 | 0.822 | 0.639 | 0.556 |
| WRN | 43 | 0.680 | 0.769 | 0.710 | 0.492 | 0.690 |
Abbreviations: CEU, Utah; CHB, Han; HGDP, Human Genome Diversity Panel; n, number of SNPs; N, number of individuals; NA, ⩽5 polymorphic pairs of loci; YRI, Yoruba.
See methods for a description of this analysis; significant LD refers to a significant P value for rejecting the null hypothesis of free recombination.
Only genes with >5 polymorphic pairs of loci are shown due to the uncertainty of the estimation with so few loci.
One population was chosen to represent Africa (YRI), Eurasia (CEU) and East Asia (CHB), due to limited sample sizes all of the populations in Oceania and America were combined.
Ancestral alleles
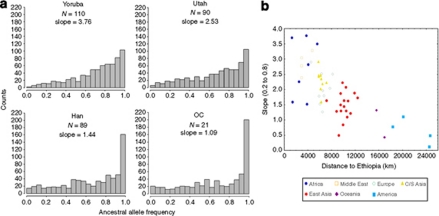
Comparing the ancestral allele frequency (AAF) spectrums among the HapMap 2 and HGDP populations, we found that populations in Africa had more SNPs with high AAFs and populations in America had the lowest (Figure 3). A steeper slope of SNP counts in the midrange of the distribution reflects more SNPs with high AAFs (Li et al., 2008). The slopes of SNP counts in the range of 0.2–0.8 AAF for all of the populations progressively declined moving away from Ethiopia (3.8–0.1; Figure 3b). This AAF pattern did not change after limiting the SNPs to only those in exons, introns and UTR regions. The average AAF was highest in African populations (0.735) and lowest in American populations (0.655) (data not shown). Average AAFs for the majority of our genes was high (>60%). For TERT, the average AAF was 64%, and for the TERT SNP, rs2736100, it was 49%. The AAF for rs2736100 (ancestral allele: T) was variable by geographic region, the highest AAF was observed in America (0.88) and the lowest in Oceania (0.045).
Figure 3.
Ancestral allele frequency spectrum using HapMap 2 and HGDP data. (a) Histograms of AAFs for four populations: Yoruba, USA, Han and OC (the two populations in Oceania were combined due to small sample sizes). N is the number of individuals and the slope is for the SNP counts in the range of 0.2–0.8 AAF. (b) Slopes of AAFs between 0.2 and 0.8 for all of the 53 populations versus geographic distance from Ethiopia.
Test for selection
For evaluating FST, we concentrated on regions that show high or low values among multiple markers, as individual SNPs show considerable variation. According to the cut-points estimated by Akey et al. (Akey et al., 2002), ACD and TERF2IP had the highest proportion of SNPs with high FST (⩾0.45) (0.6 and 0.5, respectively), and TNKS, RAD51L1 and RECQL all had very low FST (two SNPs with an FST=0 and one SNP with an FST ⩽0.005). We also plotted the average FST versus the average heterozygosity by region. There were three outliers with high FST and low heterozygosity, ACD, TERF2IP and TERF2, and two outliers with low FST and high heterozygosity, POT1 and NOLA2 (data not shown). There were three outliers in the DAF test with a large amount of derived alleles (>80%): ACD (in CEU and Han populations), NOLA2 (in CEU and America populations) and RECQL4 (in Oceania) (data not shown). All of the loci in TERC (n=3) and XRCC6 (n=5) had DAFs of <20% in all populations and in CEU and Han populations, respectively. The MAF test suggests RECQL4 (Han), POT1 (Han and CEU) and RAD54L (America) with an excess of SNPs with high MAF (>40%) and ACD (Han), TERF2IP (CEU), and RAD51L3 (America) with an excess of SNPs with low MAF (<10%). The strong LD observed for POT1 supports the existence of balancing selection. Overall, TERT did not show evidence of selection.
HapMap 3 data for select genes
HapMap 3 (The International HapMap Consortium, 2003) SNP data for 11 populations were retrieved for genes identified as potential evolutionary selection candidates in this study (based on at least two tests: ACD, NOLA2, RECQL4, POT1 TERF2IP), and previous studies (Savage et al., 2005 and the HapMap (The International HapMap Consortium, 2007): TNKS), to confirm our findings in an additional data set with a more dense SNP coverage. We also retrieved SNP data for TERT. Assigning the populations to the main geographic regions (identified by a distance-based neighbor-joining tree, Supplementary Figure 3), the AMOVA partitions the majority of the genetic diversity to within-population variation (91%); there is less variance attributable to among regions in NOLA2, POT1 and TNKS (<5%), more variance among regions in TERF2IP and ACD (>20%) and average in RECQL4 (14%), as observed in the HapMap 2 and HGDP data set.
TERT had high haplotype diversity (96–99%) and heterozygosity (0.26–0.35) in these 11 populations. There was average differentiation among geographical regions and within populations based on allele frequencies (FST=0.118 and 0.138, respectively), similar to the HapMap 2 and HGDP data set. However, population comparisons based on haplotype frequencies were all low, with FST <0.025.
There was evidence of positive selection (high FST, low heterozygosity, high or low DAFs and low MAFs) in ACD and TERF2IP, and evidence of balancing selection (low FST, high heterozygosity and high MAFs) in POT1, NOLA2 and regions of TNKS (Supplementary Figure 4). There were mixed signals in RECQL4, with a proportion of SNPs with extreme high and low FST, heterozygosity and MAFs. The patterns of variance attributable to among regions are also consistent, with extremely high values in ACD and TERF2IP and low in POT1, NOLA2 and TNKS.
Discussion
In this study, we examined allele frequency distributions, diversity, differentiation, LD and population structure among 53 worldwide populations by combining HapMap 2 and HGDP-CEPH genome-wide scan data of 37 genes vital for telomere stability. This extensive data set allowed us to create a comprehensive catalog of worldwide genetic variation for these genes. Overall, most telomere biology genes had low to moderate diversity and less than average differentiation. There was significantly lower differentiation among HGDP populations and heterozygosity in the telomere biology genes compared with innate immunity genes. Differentiation among geographical regions in the telomere biology genes grouped by function showed the lowest values in the telomerase complex genes compared with other gene sets and the genome average. These genes are required for telomere elongation and maintaining chromosomal stability.
As a gene set, there is a specific population structure; cluster analyses segregated individuals into five genetic clusters, concordant with larger analyses with the HGDP-CEPH panel (Rosenberg et al., 2002; Jakobsson et al., 2008; Li et al., 2008). The significant positive correlation between the degree of genetic divergence and the pairwise geographical distance suggests that the observed genetic differentiation can be partially explained by isolation by geographic distance, which agrees with previous data (Ramachandran et al., 2005; Jakobsson et al., 2008). As expected, the mean haplotype diversity and AAFs were highest in Sub-Saharan Africa. For all populations, diversity and AAF slopes were negatively correlated with geographic distance from Addis Ababa, Ethiopia, consistent with a serial founder model during a spatial expansion from Africa (Ramachandran et al., 2005). The AAFs for the majority of telomere maintenance genes were high, with most having an average AAF>60%, and the AAF slopes were much higher (range of 0.1–3.8) than observed by Li et al. (2008) (range of 0.001–0.004).
The high AAF, low diversity and differentiation in many of these genes and gene sets suggest that they may be constrained, possibly because of their essential roles in chromosomal stability. Several telomere maintenance genes have been previously shown to be highly conserved across species (Nakamura and Cech, 1998; Li et al., 2000; Kanoh and Ishikawa, 2003; de Lange, 2004; Savage et al., 2005). This conservation can be explained by a low mutation rate and/or negative selection, however, distinguishing the two is a difficult task as both result in little sequence change (Hurst, 2009). Savage et al., 2005 also found that seven of these genes had more synonymous compared with non-synonymous mutations per site. A plausible explanation for the lower levels of diversity and differentiation observed in many telomere maintenance genes is that negative selection acts to maintain the status quo of these essential genes. Perhaps these genes were highly conserved during evolution because of their important function and the accumulation of new mutations was not tolerable.
Negative, positive and balancing selection can each leave a specific signature on allele frequency patterns and LD (Walsh et al., 2006; Hurst, 2009). Using these patterns, we found evidence suggestive of positive selection in two separate data sets for ACD and TERF2IP, and evidence of balancing selection in POT1, NOLA2 and TNKS. Regions of low recombination, and thus long-range LD, as observed in POT1 and regions of TNKS, could be the result of balancing selection; alleles under balancing selection can drag linked alleles with them and cause increased LD (Hurst, 2009). Two additional studies also found evidence of selection in POT1, TNKS and TERF2IP. POT1 and TNKS were found to have significantly positive Tajima's D (Tajima, 1989) using sequence data (Savage et al., 2005), POT1 in non-Hispanic Caucasians and TNKS in individuals of Pacific Rim ancestry, suggestive of balancing selection. POT1 (in Europeans), TERF2IP and TNKS (both in East Asian and African populations) were also identified as candidate regions for recent selection with the powerful long-range haplotype and iHS tests in the HapMap genome-wide study based on over 3.1 million SNPs (The International HapMap Consortium, 2007).
Allele-frequency-based tests are not considered the most powerful methods to detect a recent selective sweep (Hanchard et al., 2006) and there is no statistical significance associated with these results. However, they highlight regions that might justify further investigation. There is also the possibility of SNP ascertainment bias that may result in false positive signals. Some of the genome-wide platform-selected SNPs are chosen based on their location in and around specific genes as well as based on haplotype-tagging SNPs in the region. However, we did not limit our analyses to only SNPs within gene exons, introns and UTRs because the goal of the study was to understand the gene and its surrounding region. The HGDP data were generated based on common SNPs and the HapMap data are also skewed toward common alleles making it more difficult to detect an excess of rare or derived alleles near fixation. However, the identification of these genes as candidates for selection in other studies suggests that selection may indeed be present. It has been (Barreiro et al., 2008) suggested that positive selection has ensured the regional adaptation of human populations by increasing population differentiation in gene regions, and that these loci likely contribute to disease-related phenotypic diversity among these different human populations.
We further explored genetic variation in TERT because several studies have identified both SNPs and mutations in TERT as important in cancer and telomere biology disorders (Armanios, 2009; Rafnar et al., 2009; Savage and Alter, 2009). Others have observed a high degree of TERT sequence similarity across species, hence we hypothesized that there would be limited genetic variation in these populations. However, we observed high haplotype diversity and heterozygosity in TERT. The level of TERT differentiation among populations was average or lower than average (genome-wide average for autosomal SNPs), which may reflect a lack of LD and likely a high recombination rate in this region. The cancer-associated SNP, rs2736100, varied substantially in genotype frequency across major continental regions, which could correlate to varying disease risk.
In conclusion, this study suggests that, as a group, telomere biology genes have less diversity and differentiation than genes with different functions. Data suggest that TERT may be an exception to this hypothesis. The identification of telomere biology genes under selection (for example, ACD, TERF2IP, POT1 and TNKS) might provide clues to their roles in telomere and chromosomal stability. It is possible that higher levels of genetic variation may not be tolerated in these genes, possibly due to their critical role in telomere maintenance.
Acknowledgments
This project has been funded by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, and with federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government. We thank the staff of the NCI core genotyping facility, Dr Stephen Chanock, NCI and Elliott Richards, NCI, for valuable assistance and helpful discussions.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Akey J, Zhang G, Zhang K, Jin L, Shriver M. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M. Syndromes of telomere shortening. Annu Rev Genomics Hum Genet. 2009;10:45–61. doi: 10.1146/annurev-genom-082908-150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert G, Lansdorp P. Telomeres and aging. Physiol Rev. 2008;88:557–579. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- Bamshad M, Wooding S. Signatures of natural selection in the human genome. Nat Rev Genet. 2003;4:99–111. doi: 10.1038/nrg999. [DOI] [PubMed] [Google Scholar]
- Barreiro L, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40:340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- Barrett J, Fry B, Maller J, Daly M. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Blasco M. Telomere length, stem cells and aging. Nat Chem Biol. 2007;3:640–649. doi: 10.1038/nchembio.2007.38. [DOI] [PubMed] [Google Scholar]
- Blasco M, Lee H, Hande M, Samper E, Lansdorp PM, DePinho RA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Cann H, de Toma C, Cazes L, Legrand M, Morel V, Piouffre L, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Collins K, Mitchell J. Telomerase in the human organism. Oncogene. 2002;21:564–579. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- Crabbe L, Verdun R, Haggblom C, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard J. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D, Tanaka H, Herbert B. Telomere dysfunction in aging and cancer. Int J Biochem Cell Biol. 2005;37:1000–1013. doi: 10.1016/j.biocel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Guthery S, Salisbury B, Pungliya M, Stephens J, Bamshad M. The structure of common genetic variation in United States populations. Am J Hum Genet. 2007;81:1221–1231. doi: 10.1086/522239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanchard N, Rockett K, Spencer C, Coop G, Pinder M, Jallow M, et al. Screening for recently selected alleles by analysis of human haplotype similarity. Am J Hum Genet. 2006;78:153–159. doi: 10.1086/499252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst L. Genetics and the understanding of selection. Nat Rev Genet. 2009;10:83–93. doi: 10.1038/nrg2506. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Scholz S, Scheet P, Gibbs J, VanLiere J, Fung H, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Jin G, Xu L, Shu Y, Tian T, Liang J, Xu Y, et al. Common genetic variants on 5p15.33 contribute to risk of lung adenocarcinoma in a Chinese population. Carcinogenesis. 2009;30:987–990. doi: 10.1093/carcin/bgp090. [DOI] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F. Composition and conservation of the telomeric complex. Cell Mol Life Sci. 2003;60:2295–2302. doi: 10.1007/s00018-003-3245-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullo I, Ding K. Patterns of population differentiation of candidate genes for cardiovascular disease. BMC Genetics. 2007;8:48. doi: 10.1186/1471-2156-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q, Shen M, Garcia-Rossi D, Chanock S, Zheng T, Berndt S, et al. Genotype frequency and FST analysis of polymorphisms in immunoregulatory genes in Chinese and Caucasian populations. Immunogenetics. 2007;59:839–852. doi: 10.1007/s00251-007-0253-3. [DOI] [PubMed] [Google Scholar]
- Landi M, Chatterjee N, Yu K, Goldin L, Goldstein A, Rotunno M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- Li J, Absher D, Tang H, Southwick A, Casto A, Ramachandran S, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L, Huang W, Wong J, Chatterjee N, Reding D, Crawford E, et al. The association between leukocyte telomere length and cigarette smoking, dietary and physical variables, and risk of prostate cancer. Aging Cell. 2009;8:405–413. doi: 10.1111/j.1474-9726.2009.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon I, Jarstfer M. The human telomere and its relationship to human disease, therapy, and tissue engineering. Front Biosci. 2007;12:4595–4620. doi: 10.2741/2412. [DOI] [PubMed] [Google Scholar]
- Morlá M, Busquets X, Pons J, Sauleda J, MacNee W, Agustí A. Telomere shortening in smokers with and without COPD. Eur Respir J. 2006;27:525–528. doi: 10.1183/09031936.06.00087005. [DOI] [PubMed] [Google Scholar]
- Myles S, Davison D, Barrett J, Stoneking M, Timpson N. Worldwide population differentiation at disease-associated SNPs. BMC Med Genomics. 2008;1:22. doi: 10.1186/1755-8794-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Cech T. Reversing time: Origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Petersen G, Amundadottir L, Fuchs C, Kraft P, Stolzenberg-Solomon R, Jacobs K, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plentz R, Caselitz M, Bleck J, Gebel M, Flemming P, Kubicka S, et al. Hepatocellular telomere shortening correlates with chromosomal instability and the development of human hepatoma. Hepatology. 2004;40:80–86. doi: 10.1002/hep.20271. [DOI] [PubMed] [Google Scholar]
- Plentz R, Wiemann S, Flemming P, Meier P, Kubicka S, Kreipe H, et al. Telomere shortening of epithelial cells characterises the adenoma-carcinoma transition of human colorectal cancer. Gut. 2003;52:1304–1307. doi: 10.1136/gut.52.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F, Manica A, Balloux F. Geography predicts neutral genetic diversity of human populations. Curr Biol. 2005;15:R159–R160. doi: 10.1016/j.cub.2005.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, Sigurdsson A, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Deshpande O, Roseman C, Rosenberg N, Feldman M, Cavalli-Sforza L. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F, Kim S, Nijjar T, Yaswen P, Campisi J. Cancer and aging: the importance of telomeres in genome maintenance. Int J Biochem Cell Biol. 2005;37:977–990. doi: 10.1016/j.biocel.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Rosenberg N. Standardized subsets of the HGDP-CEPH human genome diversity cell line panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg N, Pritchard J, Weber J, Cann H, Kidd K, Zhivotovsky L, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Rudolph K, Chang S, Lee H, Blasco M, Gottlieb GJ, Greider C, et al. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- Rudolph K, Millard M, Bosenberg M, DePinho R. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat Genet. 2001;28:155–159. doi: 10.1038/88871. [DOI] [PubMed] [Google Scholar]
- Sabeti P, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Lopes SS, Pereira RW, Wilson IJ, Pena SDJ. A worldwide phylogeography for the human X chromosome. PLoS ONE. 2007;2:e557. doi: 10.1371/journal.pone.0000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage S, Alter B. Dyskeratosis congenita. Hematol Oncol Clin North Am. 2009;23:215–231. doi: 10.1016/j.hoc.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage S, Stewart B, Eckert A, Kiley M, Liao J, Chanock S. Genetic variation, nucleotide diversity, and linkage disequilibrium in seven telomere stability genes suggest that these genes may be under constraint. Hum Mutat. 2005;26:343–350. doi: 10.1002/humu.20226. [DOI] [PubMed] [Google Scholar]
- Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver M, Kennedy G, Parra E, Lawson H, Sonpar V, Huang J, et al. The genomic distribution of population substructure in four populations using 8525 autosomal SNPs. Hum Genomics. 2004;1:274–286. doi: 10.1186/1479-7364-1-4-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver M, Mei R, Parra E, Sonpar V, Halder I, Tishkoff S, et al. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum Genomics. 2005;2:81–89. doi: 10.1186/1479-7364-2-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey S, Gudbjartsson D, Sulem P, Bergthorsson J, Kumar R, Thorleifsson G, et al. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat Genet. 2008;40:1313–1318. doi: 10.1038/ng.234. [DOI] [PubMed] [Google Scholar]
- Stacey S, Sulem P, Masson G, Gudjonsson S, Thorleifsson G, Jakobsdottir M, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41:909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–862. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C, Rapley E, Seal S, Pernet D, Renwick A, Hughes D, et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42:604–607. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B, Kudaravalli S, Wen X, Pritchard J. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem Sci. 2002;27:339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- Vulliamy T, Beswick R, Kirwan M, Marrone A, Digweed M, Walne A, et al. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc Natl Acad Sci USA. 2008;105:8073–8078. doi: 10.1073/pnas.0800042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E, Sabeti P, Hutcheson H, Fry B, Schaffner S, de Bakker P, et al. Searching for signals of evolutionary selection in 168 genes related to immune function. Hum Genet. 2006;119:92–102. doi: 10.1007/s00439-005-0090-0. [DOI] [PubMed] [Google Scholar]
- Weir B, Cardon L, Anderson A, Nielsen D, Hill W. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 2005;15:1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, Asfaw B, DeGusta D, Gilbert H, Richards G, Suwa G, et al. Pleistocene omo sapiens from Middle Awash, thiopia. Nature. 2003;423:742–747. doi: 10.1038/nature01669. [DOI] [PubMed] [Google Scholar]
- Wong J, Collins K. Telomere maintenance and disease. Lancet. 2003;362:983–988. doi: 10.1016/S0140-6736(03)14369-3. [DOI] [PubMed] [Google Scholar]
- Wu X, Amos CI, Zhu Y, Zhao H, Grossman BH, Shay JW, et al. Telomere dysfunction: a potential cancer predisposition factor. J Natl Cancer Inst. 2003;95:1211–1218. doi: 10.1093/jnci/djg011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.