One of the most important scientific discoveries in the recent past concerns RNA interference (RNAi), which is a post-transcriptional gene-silencing mechanism induced by small interfering RNA (siRNA) and micro-RNA (miRNA).[1] RNAi has opened up new avenues in the development of siRNA and miRNA as therapeutic agents for various diseases.[2] The reason for the large number of reports about chemically modified siRNA is their potential to enhance nuclease resistance, to prevent immune activation, to decrease off-target effects, and to improve pharmacokinetic and pharmacodynamic properties, all of which are important for the application of siRNA as therapeutic agents.[3] Another substantial challenge is siRNA delivery, because these reagents cannot easily traverse cell membranes because of their size and negative charge.[4] To date, the most promising therapeutic approach based on RNAi involves chemically modified siRNA that can resolve some of the issues mentioned above.
Chemical siRNA modifications belong to four classes—backbone, ribose, nucleobase, and terminal modifications—with ribose modifications being the most common.[2b] Structurally simple alterations, such as 2′-OCH3 and 2′-F, lead to significantly enhanced performance of siRNA with diverse target genes, provided that they are positioned in a site-specific manner.[3] In particular, 2′-F modifications possess the extraordinary property of being very well accepted onto the guide (antisense) strand, while most other modifications are much better tolerated by the passenger (sense) strand.[5] The guide strand is incorporated into the crucial functional particle, the RNA-induced silencing complex (RISC); thus RNA recognition and discrimination from non-native counterparts is very stringent.[6]
We postulated that siRNA with specific 2′-azido groups (2′-N3) should have the potential for enhanced performance, because this functional group is small, polar, and supports the C3′-endo ribose pucker[7] that is characteristic for an A-form RNA double helix. Interestingly, this modification has not yet been explored for siRNA technologies. It is even more surprising that, to the best of our knowledge, the solid-phase chemical synthesis of 2′-azido-modified RNA has not yet been described.[8] The prospect of potential siRNA applications, and also of promising applications in modern bioconjugation chemistry (such as Staudinger ligation and click chemistry)[9] prompted us to take up the challenge of synthesizing these RNA derivatives (Scheme 1).
Scheme 1.
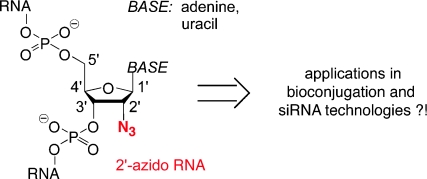
Solid-phase chemical synthesis of RNA with site-specifically 2′-azido-modified nucleosides, and the potential of these novel RNA derivates for bioconjugation reactions and siRNA technologies.
Nucleosides that carry an azido group—no matter at which position—cannot be isolated in the form of stable phosphoramidite building blocks, the most convenient form for use in advanced RNA solid-phase synthesis approaches.[10] This is due to the inherent reactivity between phosphor-III species and azides according to the Staudinger reaction. However, we had experimental indications that this reactivity becomes prevalent only at very high concentrations of azido-modified nucleoside phosphoramidites (e.g., upon evaporation of the solvents to isolate these compounds). This led us to consider the possibility that strand assembly by phosphoramidite chemistry might still be feasible (at reasonable phosphoramidite concentrations) once the azide moiety has been incorporated into the RNA. Additionally, two very recent independent studies, one on the chemical synthesis of 4′-azidomethyl-thymidine modified DNA and the other on the circularization of DNA by using a solid-support bearing an azido linker, encouraged us to target 2′-azido-RNA synthesis.[11a,b]
We decided to focus on the 2′-azido-2′-deoxyuridine and the 2′-azido-2′-deoxyadenosine building blocks as 2-chlorophenyl phosphodiester derivatives, 1 and 2, with the aim of employing them later in a single cycle of standard RNA phosphotriester coupling for the incorporation into RNA, while strand assembly should follow standard phosphoramidite chemistry.
For building block 1, we started the synthesis with 2,2′-anhydrouridine 3 (Scheme 2) which is commercially available or can be obtained from uridine by a single transformation by using diphenyl carbonate in DMF.[12] Nucleophilic ring-opening with in situ-generated lithium azide in DMF furnished the 2′-azido-2′-deoxyuridine derivative 4.[13] Subsequently, the 5′-hydroxyl group was protected as dimethoxytrityl (DMT) ether to form compound 5.[14] Conversion into the corresponding phosphodiester 1 was achieved in good yield by reaction with in situ-generated 2-chlorophenyl chlorophosphorotriazolide, analogously to a general procedure in the literature.[15]
Scheme 2.
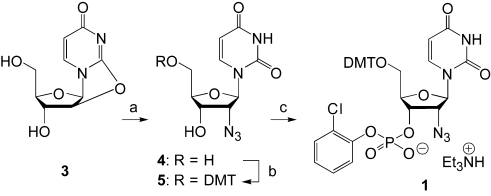
Synthesis of the 2′-azido-2′-deoxyuridine 3′-phosphodiester building block 1 for RNA solid-phase synthesis. Reagents and conditions: a) 1.8 equiv LiF, 1.8 equiv (CH3)3SiN3, in N,N,N′,N′-tetramethylethylenediamine (TMEDA)/DMF 1:1, 110 °C, 48 h, 66 %; b) 1.5 equiv 4,4′-dimethoxytriphenylmethyl chloride (DMT-Cl), in pyridine, RT, 24 h, 69 %; c) i: 4 equiv N-methylimidazole, in THF, RT, 5 min; ii: 2.5 equiv 2-chlorophenyl phosphorodichloridate, 5.5 equiv 1,2,4-triazole, 5 equiv Et3N, in THF, RT, 10 min, 86 %.
For building block 2, we started the synthesis with the triflated adenosine derivative 6, which was readily obtained from adenosine according to the previously described synthesis to 2′-methylseleno adenosine (Scheme 3).[16] Treatment of 6 with lithium azide in DMF furnished key compound 7, followed by cleavage of the silyl protecting group to give the diol derivative 8. The 5′-hydroxyl group was then protected as the DMT ether to form 9. Conversion into the corresponding phosphodiester 2 was achieved in good yield by treatment with 2-chlorophenyl chlorophosphorotriazolide.
Scheme 3.
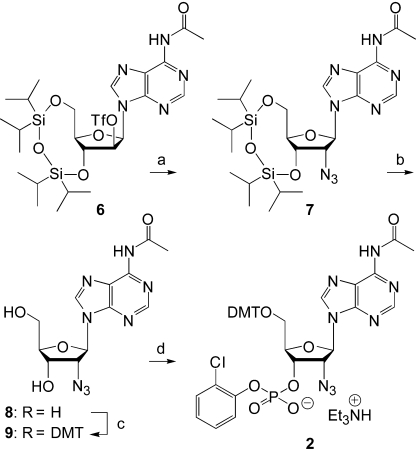
Synthesis of the 2′-azido-2′-deoxyadenosine 3′-phosphodiester building block 2 for RNA solid-phase synthesis. Reagents and conditions: a) 5 equiv LiN3, in DMF, RT, 20 h, 86 %; b) 1 m TBAF, 0.5 m CH3COOH, in THF, RT, 2 h, 71 %; c) 1.5 equiv DMT-Cl, in pyridine, RT, 16 h, 64 %; d) i: 4 equiv N-methylimidazole, in THF, RT, 5 min; ii: 2.5 equiv 2-chlorophenyl phosphorodichloridate, 5.5 equiv 1,2,4-triazole, 5 equiv Et3N, in THF, RT, 10 min, 79 %.
For the incorporation of the 2′-azido-modified nucleoside phosphodiester building blocks 1 and 2 into RNA, we conducted strand assembly by automated standard RNA solid-phase synthesis with 2′-O-[(Triisopropylsilyl)oxy]methyl (2′-O-TOM)-protected nucleoside phosphoramidites[17] up to the position of the intended azide modification. The synthesis was interrupted after the detritylation step that liberated the terminal 5′-hydroxyl group. Coupling of the phosphodiester building block, 1 or 2, was achieved by activation with 1-(mesitylene-2-sulfonyl)-3-nitro-1,2,4-triazole (MSNT) by using two syringes attached to the column for reagent delivery and mixing under argon.[18] By using this simple setup (see the Supporting Information), coupling yields higher than 95 % were achieved. After manual capping, strand elongation was continued by standard automated RNA phosphoramidite chemistry. With sequences of up to about 25 nucleotides, we had no indication (from inspection of oligonucleotide by-products by LC-ESI mass spectrometry; see also refs. [10b] and [11a, c]) that the ribose 2′-azido group had reacted with the phosphoramidite moiety under the coupling conditions. For deprotection, the oligoribonucleotides containing 2′-azido groups were first treated with syn-2-pyridine aldoxime/tetramethylguanidine in dioxane/water to cleave the 2-chlorophenyl phosphate protecting groups.[18] Then, standard deprotection conditions were applied by using CH3NH2 in ethanol/water followed by treatment with 1 m tetra-n-butylammonium fluoride (TBAF) in THF. After purification by anion-exchange HPLC, the expected molecular weights were confirmed by liquid chromatography ESI (LC-ESI) mass spectrometry (Table 1, Figure 1).
Table 1.
Selection of synthesized 2′-azido-2′-deoxyuridine (U*) and 2′-azido-2′-deoxyadenosine (A*) containing oligoribonucleotides
| Sequence (5′→3′) | Amount [nmol] | MW calcd [amu] | MW obs [amu] |
|---|---|---|---|
| GGU*CGACC | 125 | 2549.6 | 2549.2 |
| GGUCGA*CC | 196 | 2549.6 | 2549.2 |
| CCAGGCCU*GG | 87 | 3200.1 | 3200.2 |
| CCA*GGCCUGG | 79 | 3200.1 | 3200.2 |
| GAAGGGCAACCU*UCG | 173 | 4839.0 | 4838.4 |
| GAA*GGGCAACCUUCG | 161 | 4839.0 | 4838.3 |
| G2UCUCU*GCCA2UA2GACATT | 298 | 6678.1 | 6677.8 |
| UGUCU2A*U2G2CAGAGAC2TdG | 120 | 6697.1 | 6696.8 |
| UGUCU*UAUU*G2CAGAGAC2TdG | 55 | 6722.1 | 6721.5 |
| UGUCU2AUU*G2CA*GAGAC2TdG | 68 | 6722.1 | 6721.2 |
Figure 1.
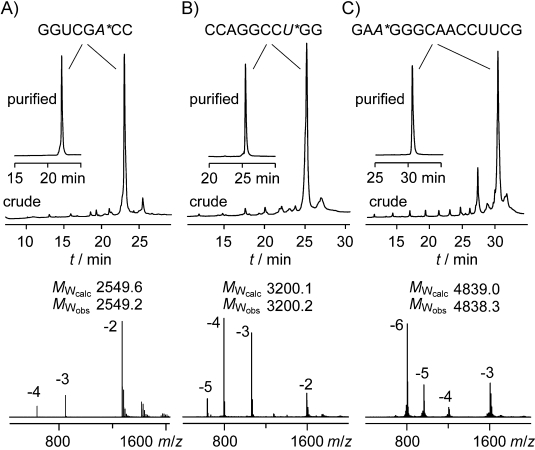
Characterization of 2′-azido-modified oligoribonucleotides by anion-exchange HPLC and LC-ESI mass spectrometry. A) 8 nucleotide (nt) RNA containing 2′-azido-2′-deoxyadenosine. B) 10 nt RNA containing 2′-azido-2′-deoxyuridine. C) 15 nt RNA containing 2′-azido-2′-deoxyadenosine. For conditions see the Supporting Information.
Next we investigated the influence of the 2′-azido group on the thermal stability of RNA double helices by using temperature-dependent UV spectroscopy (Figure 2A). At 150 mm NaCl and 10 mm Na2HPO4 (pH 7.0), the unmodified hairpin 5′-GAAGGGCAACCUUCG melted at 72.5±0.5 °C while the 2′-azido-modified counterparts, 5′-GAAGGGCAACCU*UCG and 5′-GAA*GGGCAACCUUCG, displayed Tm values of 71.7±0.5 °C and 71.6±0.5 °C, respectively. This demonstrates that the 2′-azido group is modestly destabilizing, which might arise from a slight steric interference of the moiety with the 3′-phosphate.[7a] Additionally, we examined this hairpin system with CD spectroscopy: the spectra clearly indicate that the overall conformation of a typical A-form double helical geometry is retained in the 2′-azido-modified RNA (Figure 2B).
Figure 2.
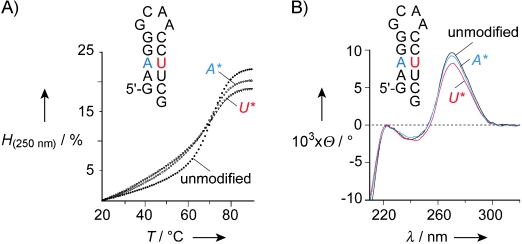
Spectroscopic characterization of 2′-azido-modified oligoribonucleotides. A) Overlay of UV melting profiles of GAAGGGCAACCUUCG, GAAGGGCAACCU*UCG, GAA*GGGCAACCUUCG, cRNA=5 μm; B) Overlay of CD spectra of GAAGGGCAACCUUCG, GAAGGGCAACCU*UCG, GAA*GGGCAACCUUCG, cRNA=2 μm. Conditions: 10 mm Na2HPO4, 150 mm NaCl, pH 7.0. U*=2′-azido-2′-deoxyuridine, A*=2′-azido-2′-deoxyadenosine, H=hyperchromicity.
To assess the potential of site-specifically 2′-azido-modified RNA for bioconjugation reactions[9] we attached a fluorescence quencher to the azido group to construct a molecular beacon (MB).[19] MBs are widely used probes for the specific detection of DNA and RNA targets, and their function is based on the interaction between a fluorescent (F) and a quenching (Q) moiety (Figure 3A). In the absence of the target, the MB adopts a hairpin structure that brings F and Q into close proximity resulting in quenching of fluorescence. In contrast, hybridization with the target of the MB leads to separation of F and Q and thus fluorescence emission. Here we wished to demonstrate that the attachment of a dye to the 2′-azido position can be performed post-synthetically by following one of the most widespread bioconjugation strategies, the Staudinger ligation.[9a, 20] We synthesized the Disperse Red 1 (DR1) phosphine derivative 10 (see Supporting Information) and then applied typical Staudinger conditions[20] in H2O/DMF for ligation to the 5′-fluorescein-labeled RNA 11 (Figure 3B). With a 100-fold excess of 10, we observed complete conversion of 11 into the amide- (12) and the O-methyl imidate (12 a) linked DR1-RNA conjugates (85 %), and a minor amount (15 %) of the reduced (nonconjugated) 2′-NH2 RNA derivative of 11 (see the Supporting Information).
Figure 3.
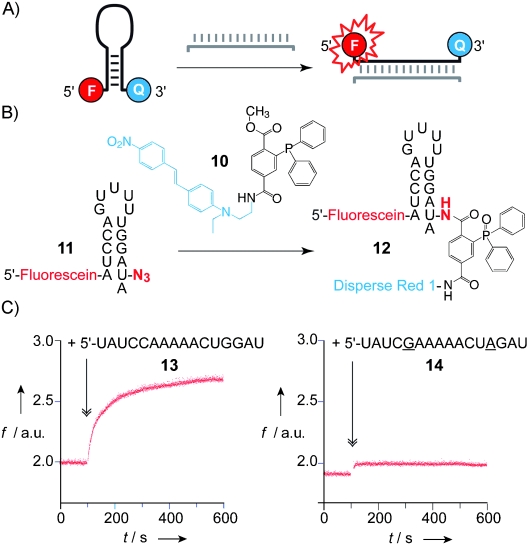
Attachment of a fluorescence quencher to RNA with an internal site-specific ribose 2′-azido group. A) Concept of molecular beacon (MB); B) Staudinger ligation of the phosphine derivative 10 to RNA 11; C) Timedependent fluorescence response of the MB 12 to addition of the target sequence 13 (left) and of the near-cognate sequence 14 (right) with double mismatch discrimination (mismatched nucleobases underlined). Conditions: cMB=0.5 μm, ctarget=2.5 μm, 100 mm MES⋅NaOH, 1.0 mm MgCl2, pH 6.5. λex=490 nm, λem=514 nm. f=fluorescence (emission), t=time.
This clearly shows that, despite the relatively high steric hindrance of the ribose 2′-azido group in RNA, its reactivity is still sufficient for efficient chemical ligation. Moreover, the fluorescence responses of 12 depicted in Figure 3C demonstrate the expected functionality for target recognition and for discrimination from near-cognate sequences.
In order to evaluate the potential of 2′-azido-modified oligoribonucleotides in siRNA applications, we employed a model system for the knockdown of the brain acid soluble protein 1 (BASP1) gene by transient siRNA nucleofection in the chicken DF-1 cell line by using standard commercial siRNA duplexes as previously described.[21a] Expression of the BASP1 gene is specifically suppressed by the Myc oncoprotein, while the BASP1 protein is an efficient inhibitor of Myc-induced cell transformation.[21a] We synthesized ten siRNA duplexes for the Myc target BASP1 with a single 2′-azido-2′-deoxyuridine or 2′-azido-2′-deoxyadenosine modification and the sequence organisation depicted in Figure 4A (see also Supporting Information). All the modified siRNAs caused gene silencing to an extent that was comparable to that of the unmodified reference duplex (Figure 4B). Remarkably, the 2′-azido group is very well tolerated in the guide strand, even when the site of modification is located in the seed region or very close to the actual cleavage site. Moreover, double azido-labeled siRNAs, one embracing the site of cleavage (SIR Az-U9A13) and another with the modifications at the seed region (SIR Az-U5, U9), also showed efficient silencing activity (Figure 4B). These experiments demonstrate that the 2′-azido group adds to the limited selection of modifications (2′-F and 2′-OCH3) that are excellently tolerated in the guide strand.[3d, 5, 6a, 22]
Figure 4.
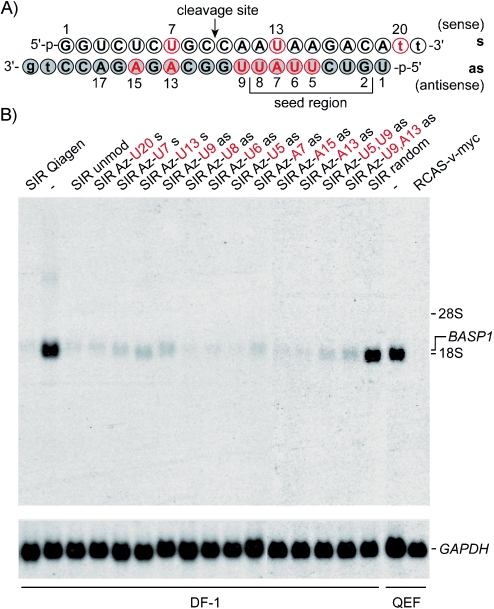
Gene silencing by 2′-azido-2′-deoxyuridine- and 2′-azido-2′-deoxyadenosine-modified siRNA. A) Sequence of the BASP1 siRNA used in this study;[21a] nucleosides in red indicate positions of 2′-azido modification. B) Biological activities of 2′-azido siRNAs directed against BASP1 mRNA. Chicken DF-1 cells grown on 60 mm dishes were transiently nucleofected with 0.12 nmol (∼1.5 μg) aliquots of unmodified commercial (Qiagen) or own (unmod) synthetic siRNAs (SIR) directed against the chicken BASP1 mRNA, or of siRNAs containing azido (Az) groups at the indicated nucleotides on sense (s) or antisense (as) strands. An equal aliquot of siRNA with a shuffled (random) nucleotide sequence was used as a control. Total RNAs were isolated two days after siRNA delivery and 5 μg-aliquots were analyzed by Northern hybridization by using DNA probes specific for the chicken BASP1 gene, or the quail housekeeping GAPDH gene. RNAs (5 μg) from normal quail embryo fibroblasts (QEF), and from QEF transfected with the retroviral construct RCAS-v-myc[21b] encoding the BASP1-suppressing v-Myc oncoprotein were used as controls. The electrophoretic positions of ribosomal RNA are indicated in the margin. All siRNAs depicted contain overhangs of 2′-deoxynucleosides (lower case letters). SIR random: 5′-UCUGGGUCUAAGCCAAACAUT/5′-UGUUUGGCUUAGACCCAGAUdG.
In conclusion, the solid-phase synthesis approach presented here efficiently yields RNA with site-specific 2′-azido groups, and provides a reliable foundation for a wide range of applications in modern bioconjugation strategies and siRNA technologies. Clearly, to evaluate the full potential of this type of modification, systematic studies of 2′-azido-modified siRNA on nuclease resistance, immune activation, off-target effects, toxicity, membrane permeability, and delivery will be required. We are confident that, in particular, the small size and the high polarity of the azido group will provide improvements for at least some of these effects. Moreover, the 2′-azido group in siRNA is available for the attachment of hydrophobic tags (such as cholesterols, lipids, cofactors, cell-penetrating peptides) to enhance delivery properties. Additionally, the attachment of fluorescent dyes to the azido anchor (as exemplified here in the context of MBs) facilitates the localization and tracking of siRNA in the cell. Another feature that has to be explored in future studies of 2′-azido-RNA (and in particular of 2′-azido-siRNA) is photoreactivity, and whether it can be exploited for crosslinking to peptide and protein targets. All these aspects represent the focus of ongoing studies in our laboratory together with the expansion of the concept towards 2′-azido-2′-deoxycytidine- and 2′-azido-2′-deoxyguanosine-containing RNA. At the current state of research, the 2′-azido modification might prove superior to the more than 35 other types of chemical modifications that have been tested for their effects on siRNA performance.[2] It might also prove useful in the manipulation of other types of noncoding RNA in the cell.[23]
Acknowledgments
M.A. thanks the Austrian Academy of Science and UNESCO for a l′Oréal fellowship. Funding by the Austrian Science Foundation FWF (P21641, I317 to R.M.; P18148 to M.H.; P17041 to K.B.) and the Ministry of Science and Research (GenAU III, “non-coding RNAs” P0726-012-012 to R.M.) is acknowledged. M.A. and R.M. thank Claudia Höbartner (MPI Göttingen) for stimulating discussions and critical comments on the manuscript.
Supplementary material
References
- 1a.Lee RC, Feinbaum RL, Ambros V. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 1b.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 1c.Kurreck J. Angew. Chem. 2009;121:1404–1426. Angew. Chem. Int. Ed.2009, 48, 1378–1398. [Google Scholar]
- 2a.Croce CM. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2b.Shukla S, Sumaria CS, Pradeepkumar PI. ChemMedChem. 2010;5:328–349. doi: 10.1002/cmdc.200900444. [DOI] [PubMed] [Google Scholar]
- 3a.Chernolovskaya EL, Zenkova MA. Curr. Opin. Mol. Ther. 2010;12:158–167. [PubMed] [Google Scholar]
- 3b.Behlke MA. Oligonucleotides. 2008;18:305–320. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 3c.Watts JK, Deleavey GF, Damha MJ. Drug Discovery Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 3d.Li F, Pallan PS, Maier MA, Rajeev KG, Mathieu SL, Kreutz C, Fan Y, Sanghvi J, Micura R, Rozners E, Manoharan M, Egli M. Nucleic Acids Res. 2007;35:6424–6438. doi: 10.1093/nar/gkm664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh SK, Hajeri PB. Drug Discovery Today. 2009;14:859–865. doi: 10.1016/j.drudis.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 5a.Prakash TP, Allerson CR, Dande P, Vickers TA, Sioufi N, Jarres R, Baker BF, Swayze EE, Griffey RH, Bhat B. J. Med. Chem. 2005;48:4247–4253. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 5b.Muhonen P, Tennilä T, Azhayeva E, Parthasarathy RN, Janckila AJ, Väänänen HK, Azhayev A, Laitala-Leinonen T. Chem. Biodiversity. 2007;4:858–873. doi: 10.1002/cbdv.200790073. [DOI] [PubMed] [Google Scholar]
- 5c.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 6a.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6b.Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6c.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Ikehara M, Takatsuka Y, Uesugi S. Chem. Pharm. Bull. 1979;27:1830–1835. [Google Scholar]
- 7b.Vasileva SV, Abramova TV, Ivanova TM, Shishkin GV, Silnikov VN. Russ. J. Bioorg. Chem. 2004;30:234–242. [Google Scholar]
- 8. In the early 1970s, the enzymatic production of 2′-azido RNA derivatives was described. This used polynucleotide phosphorylase leading to homopolymers that had been recognized to induce interferon activity:
- 8a.Hobbs J, Sternbach H, Sprinzl M, Eckstein F. Biochemistry. 1973;12:5138–5145. doi: 10.1021/bi00749a018. [DOI] [PubMed] [Google Scholar]
- 8b.Torrence PF, Bobst AM, Waters JA, Witkop B. Biochemistry. 1973;12:3962–3972. doi: 10.1021/bi00744a028. [DOI] [PubMed] [Google Scholar]
- 8c.de Clercq E, Torrence PF, Stollar BD, Hobbs J, Fukui T, Kakiuchi N, Ikehara M. Eur. J. Biochem. 1978;88:341–349. doi: 10.1111/j.1432-1033.1978.tb12455.x. More recently, Sousa and co-workers demonstrated the incorporation of 2′-azido-modified UTP and CTP into RNA by engineered polymerases. [DOI] [PubMed] [Google Scholar]
- 8d.Padilla R, Sousa R. Nucleic Acids Res. 2002;30:e138. doi: 10.1093/nar/gnf138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Sletten EM, Bertozzi CR. Angew. Chem. 2009;121:7108–7133. doi: 10.1002/anie.200900942. Angew. Chem. Int. Ed.2009, 48, 6974–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b.Gramlich PME, Wirges CT, Manetto A, Carell T. Angew. Chem. 2008;120:8478–8487. doi: 10.1002/anie.200802077. Angew. Chem. Int. Ed.2008, 47, 8350–8358. [DOI] [PubMed] [Google Scholar]
- 10a.Sylvers LA, Wower J. Bioconjugate Chem. 1993;4:411–418. doi: 10.1021/bc00024a001. [DOI] [PubMed] [Google Scholar]
- 10b.Wada T, Mochizuki A, Higashiya S, Tsuruoka H, Kawahara S-i, Ishikawa M, Sekine M. Tetrahedron Lett. 2001;42:9215–9219. [Google Scholar]
- 10c.Efimov VA, Aralov AV, Klykov VN, Chakhmakhcheva OG. Nucleosides Nucleotides Nucleic Acids. 2009;28:846–865. doi: 10.1080/15257770903170286. [DOI] [PubMed] [Google Scholar]
- 10d.Polushin NN, Smirnov IP, Verentchikov AN, Coull JM. Tetrahedron Lett. 1996;37:3227–3230. [Google Scholar]
- 10e.El-Sagheer AH, Brown T. Chem. Soc. Rev. 2010;39:1388–1405. doi: 10.1039/b901971p. [DOI] [PubMed] [Google Scholar]
- 10f.El-Sagheer AH, Brown T. Proc. Natl. Acad. Sci. USA. 2010;107:15329–15334. doi: 10.1073/pnas.1006447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Kiviniemi A, Virta P, Lönnberg H. Bioconjugate Chem. 2008;19:1726–1734. doi: 10.1021/bc800221p. [DOI] [PubMed] [Google Scholar]
- 11b.Pourceau G, Meyer A, Vasseur J-J, Morvan F. J. Org. Chem. 2009;74:6837–6842. doi: 10.1021/jo9014563. [DOI] [PubMed] [Google Scholar]
- 11c.Steger J, Graber D, Moroder H, Geiermann A-S, Aigner M, Micura R. Angew. Chem. 2010;122:7632–7634. doi: 10.1002/anie.201003424. Angew. Chem. Int. Ed.2010, 49, 7470–7472. [DOI] [PubMed] [Google Scholar]
- 12a.Höbartner C, Micura R. J. Am. Chem. Soc. 2004;126:1141–1149. doi: 10.1021/ja038481k. [DOI] [PubMed] [Google Scholar]
- 12b.McGee DPC, Vaughn-Settle A, Vargeese C, Zhai Y. J. Org. Chem. 1996;61:781–785. doi: 10.1021/jo9510548. [DOI] [PubMed] [Google Scholar]
- 13a.Wnuk SF, Chowdhury SM, Garcia PI, Jr., Robins MJ. J. Org. Chem. 2002;67:1816–1819. doi: 10.1021/jo010899i. [DOI] [PubMed] [Google Scholar]
- 13b.Kirschenheuter GP, Zhai Y, Pieken WA. Tetrahedron Lett. 1994;35:8517–8520. [Google Scholar]
- 14.Catry MA, Madder A. Molecules. 2007;12:114–129. doi: 10.3390/12010114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Belsito E, Liguori A, Napoli A, Siciliano C, Sindona G. Nucleosides Nucleotides Nucleic Acids. 1999;18:2565–2580. doi: 10.1081/NCN-120022624. [DOI] [PubMed] [Google Scholar]
- 15b.El-Sagheer AH, Brown T. J. Am. Chem. Soc. 2009;131:3958–3964. doi: 10.1021/ja8065896. [DOI] [PubMed] [Google Scholar]
- 16a.Puffer B, Moroder H, Aigner M, Micura R. Nucleic Acids Res. 2008;36:970–983. doi: 10.1093/nar/gkm880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16b.Höbartner C, Rieder R, Kreutz C, Puffer B, Lang K, Polonskaia A, Serganov A, Micura R. J. Am. Chem. Soc. 2005;127:12035–12045. doi: 10.1021/ja051694k. [DOI] [PubMed] [Google Scholar]
- 17a.Micura R. Angew. Chem. 2002;114:2369–2373. Angew. Chem. Int. Ed.2002, 41, 2265–2269. [Google Scholar]
- 17b.Pitsch S, Weiss PA, Jenny L, Stutz A, Wu X. Helv. Chim. Acta. 2001;84:3773–3795. [Google Scholar]
- 17c.Wachowius F, Höbartner C. ChemBioChem. 2010;11:469–480. doi: 10.1002/cbic.200900697. [DOI] [PubMed] [Google Scholar]
- 17d.Beaucage SL. Curr. Opin. Drug Discovery Dev. 2008;11:203–216. [PubMed] [Google Scholar]
- 18.Micura R. Chem. Eur. J. 1999;5:2077–2082. [Google Scholar]
- 19a.Tyagi S, Kramer FR. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 19b.Grossmann TN, Röglin L, Seitz O. Angew. Chem. 2007;119:5315–5318. doi: 10.1002/anie.200700289. Angew. Chem. Int. Ed.2007, 46, 5223–5225. [DOI] [PubMed] [Google Scholar]
- 19c.Häner R, Biner SM, Langenegger SM, Meng T, Malinovskii VL. Angew. Chem. 2010;122:1249–1252. doi: 10.1002/anie.200905829. Angew. Chem. Int. Ed.2010, 49, 1227–1230. [DOI] [PubMed] [Google Scholar]
- 19d.Bratu DP, Cha B.-J, Mhlanga MM, Kramer F. Russell, Tyagi S. Proc. Natl. Acad. Sci. USA. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19e.Silverman AP, Kool ET. Trends Biotechnol. 2005;23:225–230. doi: 10.1016/j.tibtech.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 20a.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Proc. Natl. Acad. Sci. USA. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20b.Baccaro A, Weisbrod SH, Marx A. Synthesis. 2007;13:1949–1954. [Google Scholar]
- 20c.Wang C. C.-Y, Seo TS, Li Z, Ruparel H, Ju J. Bioconjugate Chem. 2003;14:697–701. doi: 10.1021/bc0256392. [DOI] [PubMed] [Google Scholar]
- 20d.Restituyo JA, Comstock LR, Petersen SG, Stringfellow T, Rajski SR. Org. Lett. 2003;5:4357–4360. doi: 10.1021/ol035635s. [DOI] [PubMed] [Google Scholar]
- 20e.Kosiova I, Janicova A, Kois P. Beilstein J. Org. Chem. 2006;2:23. doi: 10.1186/1860-5397-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Hartl M, Nist A, Khan MI, Valovka T, Bister K. Proc. Natl. Acad. Sci. USA. 2009;106:5604–5609. doi: 10.1073/pnas.0812101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21b.Hartl M, Mitterstiller A-M, Valovka T, Breuker K, Hobmayer B, Bister K. Proc. Natl. Acad. Sci. USA. 2010;107:4051–4056. doi: 10.1073/pnas.0911060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Bramsen JB, et al. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22b.Bramsen JB, Pakula MM, Hansen TB, Bus C, Langkjær N, Odadzic D, Smicius R, Wengel SL, Chattopadhyaya J, Engels JW, Herdewijn P, Wengel J, Kjems J. Nucleic Acids Res. 2010;38:5761–5773. doi: 10.1093/nar/gkq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, Hartmann G, Patel D. Nat. Struct. Mol. Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


