Abstract
Deg/HtrA proteases are a large group of ATP-independent serine endoproteases found in almost every organism. Their usual domain arrangement comprises a trypsin-type protease domain and one or more PDZ domains. All Deg/HtrA proteases form homo-oligomers with trimers as the basic unit, where the active protease domain mediates the interaction between individual monomers. Among the members of the Deg/HtrA protease family, the plant protease DEG7 is unique since it contains two protease domains (one active and one degenerated) and four PDZ domains. In the present study, we investigated the oligomerization behaviour of this unusual protease using yeast two-hybrid analysis in vivo and with recombinant protein in vitro. We show that DEG7 forms trimeric complexes, but in contrast with other known Deg/HtrA proteases, it shows a new principle of oligomerization, where trimerization is based on the interactions between degenerated protease domains. We propose that, during evolution, a duplicated active protease domain degenerated and specialized in protein–protein interaction and complex formation.
Keywords: Arabidopsis thaliana, complex formation, degenerated protease domain, DEG7, serine protease, taxonomic distribution
Abbreviations: AD, activation domain; BD, DNA-binding domain; 2ME, 2-mercaptoethanol; IPTG, isopropyl β-D-thiogalactopyranoside; NTA, nitrilotriacetic acid; SC, synthetic complete; SEC, size-exclusion chromatography; Y2H, yeast two-hybrid
INTRODUCTION
Deg/HtrA (for degradation of periplasmic proteins/high temperature requirement A) proteases are ATP-independent serine proteases found in almost every organism [1,2]. They are generally defined by a trypsin-type protease domain (S1B, glutamylpeptidase I subfamily according to the nomenclature proposed by the MEROPS database [3], http://merops.sanger.ac.uk/) and one or more PDZ domains (originally identified in the postsynaptic density 95 protein, the Drosophila tumour-suppressor protein Discs large and the tight-junction protein zonula occludens 1) responsible for protein–protein interaction. Members of this family without a PDZ domain have also been described [4–6].
Deg/HtrA proteases form homo-oligomeric complexes, with trimers as the basic unit and oligomerization mediated by the protease domain [7–9]. Exceptions are plant DEG5 and DEG8 which form a heterohexameric complex in the thylakoid lumen of chloroplasts [6]. Biochemical and crystallographic analysis indicated that purified DegP from Escherichia coli [9,10] or HtrA from Thermotoga maritima [11] mainly exist as proteolytically inactive hexamers, where two homotrimers are stacked in a face-to-face manner. In solution, these hexamers assemble into large catalytically active spherical structures around their substrates forming 12- or 24-mers composed of four or eight homotrimers respectively [12,13].
Deg/HtrA proteases are involved in a variety of processes, including signalling [14,15], degradation of damaged proteins and housekeeping [16,17], apoptosis [18] and protein processing [4,5,19]. The Arabidopsis thaliana genome contains 16 genes encoding Deg/HtrA proteases [20]. Phylogenetic comparison of Deg/HtrA proteases from various organisms (including plants, animals, fungi and bacteria) revealed that this family is divided into four distinct groups [4]. DEG7 was the only protease from A. thaliana that clustered with Deg/HtrA proteases from fungi, forming a group of Deg/HtrA enzymes with an unusual domain arrangement. All members of this group are twice as long as other Deg/HtrA proteases, possess two protease domains (one degenerated) and up to four PDZ domains [2,4]. The best examined protease from this group is the Ynm3p protein (also called Nma111p, for nuclear mediator of apoptosis) from Saccharomyces cerevisiae [21–23]. It is a nuclear protein [21,22] interacting with the nuclear core complex [22] and long-chain acyl-CoA synthetases [23], as assayed by Y2H (yeast two-hybrid) screens. In contrast with the yeast protease, DEG7 from A. thaliana (At3g03380) was identified as a chloroplast protein involved in the degradation of photodamaged D1 protein, a core protein of the Photosystem II reaction centre [24].
In the present study, we investigated the taxonomic distribution of DEG7-like proteases and analysed how the unusual domain arrangement affects the oligomerization of DEG7 from A. thaliana. We demonstrated that DEG7 orthologues are restricted to fungi and plants, including various algae and land plants. Using Y2H assays and SEC (size-exclusion chromatography) with recombinant DEG7 purified from E. coli, we show that trimerization of DEG7 monomers is mediated by the second (degenerated) protease domain.
EXPERIMENTAL
Bioinformatics
We searched annotated protein databases for the presence of DEG7 orthologues using the BLAST algorithm [25] with default parameters as implemented on the web pages of the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/blast), The Gene Index Project (TGI, http://compbio.dfci.harvard.edu/tgi/), and the Joint Genome Institute (JGI, http://genomeportal.jgi-psf.org/). A list of DEG7 orthologues with their respective accession numbers is presented in Supplementary Table S1 (at http://www.BiochemJ.org/bj/435/bj4350167add.htm). Conserved protease and PDZ domains were identified using the InterProScan platform at the European Molecular Biology Laboratory/European Bioinformatics Institute (http://www.ebi.ac.uk/Tools/InterProScan) [26]. Secondary-structure prediction and comparison was performed using the HHpred platform (Max-Planck Institute for Developmental Biology, http://toolkit.tuebingen.mpg.de/hhpred/) [27]. Multiple sequence alignments of full-length and protease domain sequences were generated by the M-Coffee web server using standard settings [28]. Phylogenetic trees were constructed using the Phylogeny.fr platform (http://www.phylogeny.fr) [29]. Gaps in the alignment were removed with the built-in curation method, and phylogenetic trees were constructed using the Maximum-Likelihood, Parsimony or Neighbour-Joining method. All methods resulted in essentially the same tree. Human HtrA2, E. coli DegP and DEG1 from A. thaliana, representing the best examined Deg/HtrA proteases from animals, bacteria and plants respectively, were chosen as outgroups, since they do not belong to the DEG7 group [4]. Models of the first and second half of DEG7 were created using the MODELLER 9v6 program [30] with multiple template alignment, using the structure files 2pzd.pdb [31], 1lcy.pdb [7], 3cs0.pdb [12] and 1ky9.pdb [9] as templates. Manual assembly of the obtained structures into a DEG7 trimer was performed using the PyMOL program (http://www.pymol.org/).
Plasmid construction
General molecular biological procedures were conducted using the method described in [32]. Sequences of primers used in this study are given in Supplementary Table S2 (at http://www.BiochemJ.org/bj/435/bj4350167add.htm). A DNA fragment encoding A. thaliana DEG7 (At3g03380) was generated by PCR with SALK cDNA clone U21730 [33] as a template, using primers 0734 and 0740. The resulting cDNA fragment was cloned into the pET151-D/TOPO plasmid (Invitrogen), resulting in plasmid pHS36. This plasmid was mutagenized with primers 0724 and 0725 using the QuikChange® II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions, resulting in plasmid pHS52 encoding DEG7Ser206Ala. This mutation was introduced to prevent downstream applications (purification, expression in yeast) from negative effects of potential uncontrolled proteolysis. A change of the active-site serine residue to alanine does not influence the oligomerization behaviour of the protease domain, as was shown for human HtrA2 and E. coli DegP [7,9,12]. Amplification of the DEG7 cDNA with primers 0740 and 0799, using pHS52 as a template, and ligation of the respective DNA into pET151-D/TOPO, created a vector for overexpression of full-length His6-tagged DEG7 in E. coli (pHS166). Plasmids for the overexpression of DEG7 cDNA fragments were constructed by amplifying DEG7Ser206Ala with the following primers: first half, primers 0740 and 0771; second half, primers 0747 and 0777; active protease domain, primers 0765 and 0774; degenerated protease domain, primers 0793 and 0797) and cloning the fragment into pET151-D/TOPO, resulting in pHS183 (active protease domain), pHS184 (first half), pHS185 (second half) and pHS186 (degenerated protease domain).
For the Y2H assay, vectors were created using gateway technology (Invitrogen). Entry vectors were created by a TOPO reaction using cDNA fragments coding for DEG7Ser206Ala full-length (DEG7-fl, pHS73, primers 0740 and 0747), DEG7Met1-Gln563 (first half, pHS81, primers 0740 and 0771), DEG7Glu527-Gln1097 (second half, pHS80, primers 0777 and 0747), DEG7Ser35-Lys256 (active protease domain, pHS77, primers 0765 and 0774), DEG7Gly581-Gly840 (degenerated protease domain, pHS 172, primers 0793 and 0797), DEG7Asp255-Ser373 (PDZ1, pHS78, primers 0775 and 0767) and DEG7Ser868-Gln1097 (PDZ3+4, pHS88, primers 0776 and 0747), which were amplified by PCR using pHS52 as template. Final vectors for the assay were created by performing a gateway reaction with the entry vectors and modified pAD-GAL4–2.1 (Stratagene), introducing the GAL4 AD (activation domain) and modified pBD-Gal4 Cam (Stratagene), introducing the GAL4 BD (DNA-binding domain) respectively, resulting in pHS82 (AD–DEG7-fl), pHS83 (BD–DEG7-fl), pHS96 (AD–first half), pHS91 (BD–first half), pHS95 (AD–second half), pH-S84 (BD–second half), pHS93 (AD–active protease domain), pHS86 (BD–active protease domain), pHS 176 (AD–degenerated protease domain), pHS 179 (BD–degenerated protease domain), pHS90 (AD–PDZ1), pHS85 (BD–PDZ1), pHS97 (AD–PDZ3+4) and pHS89 (BD–PDZ3+4). All primers were obtained from Operon Biotechnologies. The inserts of all plasmids were sequenced (GATC Biotech), confirming the correct nucleotide sequence and insert orientation.
Protein overexpression, purification and SEC
Recombinant DEG7 containing an N-terminal His6-tag was produced in E. coli strain BL21 Star™ (DE3) cells (Invitrogen) carrying plasmid pHS166. Cells were grown at 30 °C in 2 litres of 2×YT medium [1.6% (w/v) tryptone/1% (w/v) yeast extract/0.5% NaCl] [32] containing 2% (w/v) glucose and 100 μg·l−1 ampicillin to a D600 of 0.8 and cooled down to 20 °C. DEG7 overexpression was induced by adding 0.01 mM IPTG (isopropyl β-D-thiogalactopyranoside) (final concentration). Cells were harvested after 3 h by centrifugation at 6000 g for 10 min, resuspended in 2×PBS [32] supplemented with 20 mM 2ME (2-mercaptoethanol) and Complete™ EDTA-free protease inhibitor cocktail (Roche) according to the manufacturer's instructions, and lysed by ultrasonification for 20 s, followed by incubation on ice for 60 s, repeated ten times. After clarification of the lysate by centrifugation at 23000 rev./min for 1 h using a Beckman Coulter JS13.1 rotor, the supernatant was applied to a Ni2+-NTA (nitrilotriacetic acid) column using an ÄKTA FPLC system (GE Healthcare) and washed first with 2×PBS containing 20 mM 2ME, and then with 2×PBS containing 20 mM 2ME and 25% (v/v) glycerol. His6–DEG7 was eluted with 50 mM imidazole and concentrated using Vivaspin concentrator tubes (Vivascience), and subjected to SEC using a Superdex 200 16/300 GL column (GE Healthcare) and 2×PBS with 20 mM 2ME to remove any remaining impurities. For a more accurate identification of the elution volume, the main fraction containing DEG7 (as identified by immunoblotting) was concentrated as described above and subjected to a second run on a Superdex 200 16/300 GL column. To correlate elution volume and molecular mass, the column was calibrated with Blue Dextran, ferritin (440 kDa), aldolase (158 kDa) and ovalbumin (43 kDa) according to the manufacturer's instructions (GE Healthcare).
Truncated versions of DEG7 with an N-terminal His6 tag were produced at 30 °C in E. coli strain BL21 Star™ (DE3) cells carrying plasmid pHS183, pHS184, pHS185 or pHS186. Cells were grown in 2 litres of 2×YT medium [32] containing 2% (w/v) glucose and 100 μg·l−1 ampicillin to a D600 of 0.8, and DEG7 overexpression was induced by adding IPTG to a final concentration of 0.3 mM. After 3 h, cells were harvested by centrifugation at 6000 g for 10 min and resuspended in 50 mM Hepes/KOH (pH 8.0), 300 mM NaCl and 50 mM imidazole, supplemented with 10 mM 2ME and Complete™ EDTA-free protease inhibitor cocktail. Cells were lysed and centrifuged as described above. Proteins were purified with Ni2+-NTA columns on an ÄKTA FPLC system, using an imidazole gradient from 50 mM to 500 mM. The fraction containing the maximum amount of His6-tagged proteins was subjected to SEC as described above, using a buffer containing 50 mM Hepes/KOH (pH 8.0), 300 mM NaCl and 10 mM 2ME.
Electrophoresis and immunoblotting
SDS/PAGE and immunoblotting were conducted using the method described in [32]. A DEG7-specific antibody was raised in rabbit against the peptide (Ac-)KGGSSGSPVIDWQGR(-COOH) (AgriSera AB) and used at 1:500 dilution.
Yeast two-hybrid assay
A Y2H assay was performed using pBD-Gal4 Cam as bait and pAD-Gal42.1 as the prey vector from the GAL4 phagemid vector kit (Stratagene). See the Plasmid construction section for details of cloning of the respective plasmids. pBD-WT and pAD-WT (Stratagene; both coding for fragment C of lambda cl repressor) were used as positive interaction control plasmids, pAD-WT and pLamin C (Stratagene; coding for human lamin C) were used as negative interaction control plasmids. S. cerevisiae strain YRG-2 was used for all Y2H assays. All experiments were performed at least in triplicate, starting each time from the transformation of the plasmids into the yeast cells. For composition of media and yeast transformation by the lithium acetate method, see http://home.cc.umanitoba.ca/~gietz/ and [34]. For HIS3 reporter gene assays, candidate clones were cultured overnight in liquid SC (synthetic complete) dropout medium lacking leucine, uracil and tryptophan, pelleted, and resuspended to give a D600 of 1.0. A 50 μl volume of the suspension, as well as 10×, 100×and 1000×dilutions were dropped on solid SC medium lacking either leucine, uracil and tryptophan (growth control) or histidine, leucine, uracil and tryptophan (reporter gene assay). Plates were grown for 2–3 days at 30 °C.
RESULTS AND DISCUSSION
Taxonomic distribution of DEG7 orthologues
We searched freely available genome databases for genes encoding DEG7-like proteases and retrieved hits for fungi and plant (including green, heterokont and haptophyte algae) genomes. All DEG7 orthologues share the unusual domain arrangement with two protease domains and up to four PDZ domains [2,4] (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/435/bj4350167add.htm for sequence alignment data of the full-length proteins). The second protease domain is degenerated and lacks the active-site residues, such as histidine, aspartic acid and serine (results not shown), but is still recognized when scanned against the InterPro database [35] and the HHpred program [27] (results not shown). In contrast, four PDZ domains are not detected in every DEG7 orthologue, although the overall length of the proteins is comparable. This indicates that, provided the gene models retrieved from the databases are correct, some PDZ domains are not conserved enough to be detected by the domain prediction programs used in the present study. The domain architecture suggests that DEG7-like proteases evolved from a whole-gene duplication/fusion event of a DegP-like protease (containing a protease domain and two PDZ domains), followed by a subsequent degeneration of the second protease domain (Figure 1, and Supplementary Figure S2 at http://www.BiochemJ.org/bj/435/bj4350167add.htm). No DEG7 orthologues were found in animals, since all hits have much shorter sequences and no hit contained a second protease domain (query coverage <30%; results not shown). Notably, genes coding for DEG7 orthologues were also absent from the genomes analysed of a primitive red alga (Cyanidioschyzon merolae), Cryptophyta (Guillardia theta and Hemiselmis andersenii) and diatoms (Phaeodactylum tricornutum and Thalassiosira pseudonana).
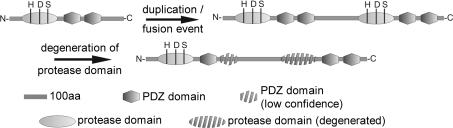
Figure 1. DEG7 in plant and fungi are the result of an internal whole-gene duplication/fusion event of an ancient Deg/HtrA gene.
An ancient Deg/HtrA gene encoding one protease domain and two PDZ domains was duplicated and the gene copy generated was fused within the same open reading frame, resulting in a gene encoding a protein with two protease and four PDZ domains. Subsequently, the second protease domain, containing a catalytic triad of histidine (H), aspartate (D) and serine (S), residues, degenerated. 100aa, 100-amino-acid stretch.
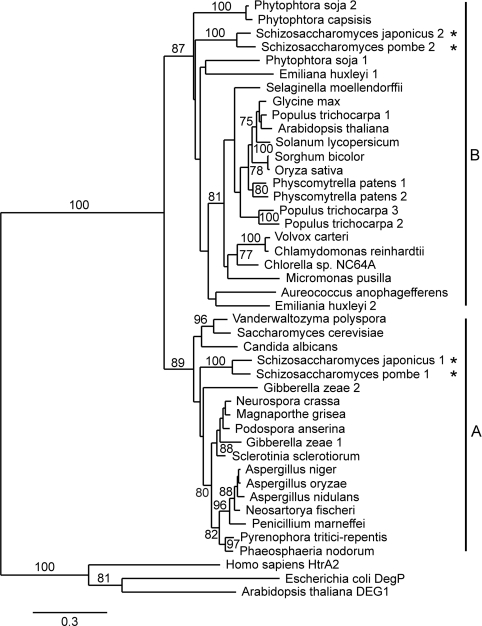
Phylogenetic comparison (see Supplementary Figure S3 at http://www.BiochemJ.org/bj/435/bj4350167add.htm for sequence alignment data) confirmed the close evolutionary relationship of DEG7 orthologues from various organisms (Figure 2). Two main clades of DEG7 proteases can be distinguished: clade A contains DEG7 proteases from fungi, whereas DEG7 proteins from green, heterokont and haptophyte algae, mosses and higher plants form clade B. Whereas most species investigated possess only one DEG7-like gene, the genomes of the yeasts Schizosaccharomyces pombe and Schizosaccharomyces japonicus, the ascomycete Giberella zeae, the oomycete Phytophtora soja, the haptophyte alga Emiliania huxleyi and the moss Physcomitrella patens contain two, whereas the higher plant Populus trichocarpa possesses three DEG7 paralogues (Figure 2). Expressed sequence tag data from S. pombe, P. patens and P. trichocarpa indicate that all paralogues are expressed in these organisms (TGI; results not shown), but whether they have overlapping/complementary physiological roles is still unknown.
Figure 2. Evolutionary relationship of DEG7 proteases and selected members from the Deg/HtrA family.
The Neighbour-Joining phylogenetic tree is based on the amino acid sequences of Deg/HtrA protease domains. The result of a bootstrap test (1000 replicates) is shown, and branches with confident values >75% are indicated. Maximum-Likelihood and Parsimony methods resulted in essentially the same trees (not shown). Human HtrA2, E. coli DegP and DEG1 from A. thaliana were chosen as outgroups, since they do not belong to the DEG7 group [4]. Group A contains DEG7 orthologues from fungi, Group B contains mainly DEG7 proteases from algae, mosses and higher plants. Asterisks mark the distribution of two DEG7 paralogues from S. japonicus und S. pombe within groups A and B.
The distribution of DEG7 proteases to two clades may reflect the different intracellular localizations for DEG7-like proteins from plants and fungi: DEG7 from A. thaliana was reported to be a plastidial [24] protein, whereas Ynm3p from yeast was found in the nucleus [21,22]. However, one of the two DEG7 paralogues from S. pombe and S. japonicus (Figure 2, marked with asterisks) is also present in clade B, together with their plant orthologues. Although the intracellular localization of these proteases has not been examined so far, a lack of plastids in yeast points to other than a plastidial localization.
DEG7 forms homotrimers
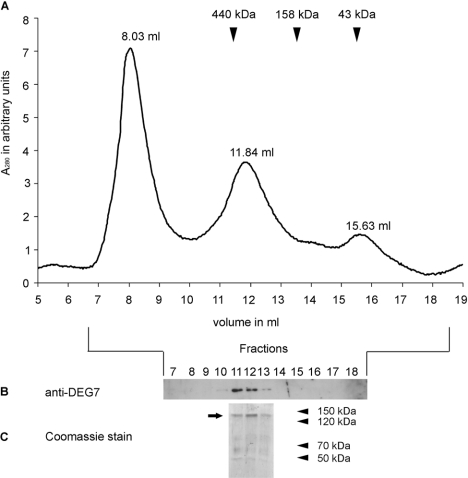
Almost all Deg/HtrA proteases examined so far are present as homo-oligomers, ranging from trimers, e.g. DegS from E. coli [8,36] and mammalian HtrA2 [7], to 24-mers, e.g. DegP from E. coli [12,13]. To investigate the oligomerization state of DEG7, we performed SEC with recombinant tagged DEG7 purified from E. coli (Figure 3). Three absorption peaks at 280 nm were observed during the elution (Figure 3A). DEG7 was present in one peak, as analysed by immunoblotting with an anti-DEG7 antibody and SDS/PAGE with subsequent Coomassie Blue staining (Figures 3B and 3C). The elution volume of 11.84 ml corresponds to an apparent molecular mass of 352 kDa, as identified by calibrating the size-exclusion column with proteins of known molecular mass. Since the recombinant tagged DEG7 has a calculated molecular mass of 120 kDa per monomer, this result indicates the formation of a trimeric complex.
Figure 3. Recombinant DEG7 forms trimers.
(A) Elution profile of purified recombinant DEG7. Three peaks were eluted from the SEC column with the indicated elution volumes. Approximate elution volumes of standard proteins are indicated by arrows. (B) Immunoblot analysis of proteins from marked fractions, using a DEG7 specific antibody, indicates the presence of DEG7 in the 11.84 ml elution peak. (C) Coomassie Blue staining of an SDS gel loaded with proteins from indicated elution fractions. DEG7 (arrow) is the main protein in the 11.84 ml elution peak. Molecular masses are indicated in kDa.
Identification of oligomerization site by Y2H screen and SEC
In all Deg/HtrA proteases examined so far, the formation of the trimer as the basic unit is mediated by residues of the individual active protease domains [1]. Since DEG7 seems to be the result of a Deg/HtrA gene-duplication and fusion event (Figure 1), the DEG7 trimer may be compared with the hexameric state described for DegP from E. coli [9] rather than the trimers observed for DegS from E. coli [8,36] or human HtrA2 [7]. However, the unique domain arrangement of DEG7 raises the question of which domains mediate the observed trimerization (Figure 3). To address this question, a Y2H array was set up with protease and PDZ domains and combinations of these as baits and as preys: full-length inactive DEG7 with the catalytic serine residue mutated to alanine (DEG7-fl) was fused to the AD and BD domain of the yeast GAL4 protein respectively. Additionally, the first half (with the catalytic serine residue mutated to alanine) and the second half of this protein were also fused to the AD domain, as well as the BD domain. To analyse individual domains, the predicted active protease domain (with the catalytic serine residue mutated to alanine), the second (degenerated) protease domain, PDZ1 and PDZ3+4 domains were fused to the AD and the BD domain. Since the second PDZ domain in the first half of the protein was less defined (Figure 1), this construct was omitted. The resulting fusion constructs were co-expressed in S. cerevisiae in every possible AD/BD combination and tested for their ability to activate expression of the HIS3 reporter gene that indicates physical interaction of the constructs (summarized in Table 1 and Figure 4). No interaction was detected when BD- and AD-fusion constructs were co-transformed with the empty AD or the BD vector respectively (Table 1), thus excluding autoactivation of the HIS3 reporter gene. Similarly, no interaction was observed for constructs containing only the PDZ1 domain or PDZ3+4 domains, indicating that the PDZ domains are not essential for the assembly of the oligomeric complex. In all combinations that activated reporter gene expression, at least one of the interaction partners contained the second (degenerated) protease domain (Figure 4 and Table 1). No interaction was observed when this part was lacking in both fusion constructs (Figure 4 and Table 1). An additional interaction between the active and the degenerated protease domain cannot be ruled out, since some combinations (DEG7-fl/first half, DEG7-fl/active protease domain first half/second half, first half/degenerated protease domain, second half/active protease domain, active protease domain/degenerated protease domain, given as AD/BD combinations) also activated reporter gene expression (Table 1, see also Supplementary Figure S4 at http://www.BiochemJ.org/bj/435/bj4350167add.htm). This also indicates that the lack of reporter gene expression with the combinations first half/first half and active protease domain/active protease domain is not due to a lack of protein expression in the yeast, since some combinations including the AD–first half, the BD–first half, the AD–active protease domain, or the BD–active protease domain construct, promoted growth of the yeast strain on histidine-free medium (see Supplementary Figure S4). In summary, the Y2H data suggest that the second (degenerated) protease domain is the key mediator for the formation of the trimeric DEG7 complex in plants. The active protease domain does not seem to be sufficient for trimer formation, in contrast with all Deg/HtrA proteases investigated so far.
Table 1. Interaction of DEG7 domains assayed by Y2H.
Abbreviations: +/+, HIS3 reporter gene activation with both possible AD- and BD-fusion combinations; +/−, HIS3 reporter gene activation with one combination only, −/− no HIS3 reporter gene activation with either possible combination. Interacting domains are marked in bold.
| Constructs | Interaction |
|---|---|
| DEG7-fl/DEG7-fl | +/+ |
| DEG7-fl/first half | +/− |
| DEG7-fl/second half | +/+ |
| DEG7-fl/active protease domain | +/− |
| DEG7-fl/degenerated protease domain | +/− |
| DEG7-fl/PDZ1 | −/− |
| DEG7-fl/PDZ3+4 | −/− |
| DEG7-fl/empty | −/− |
| First half/first half | −/− |
| First half/second half | +/+ |
| First half/active protease domain | −/− |
| First half/degenerated protease domain | +/− |
| First half/PDZ1 | −/− |
| First half/PDZ3+4 | −/− |
| First half/empty | −/− |
| Second half/second half | +/+ |
| Second half/active protease domain | +/− |
| Second half/degenerated protease domain | +/+ |
| Second half/PDZ1 | −/− |
| Second half/PDZ3+4 | −/− |
| Second half/empty | −/− |
| Active protease domain/active protease domain | −/− |
| Active protease domain/degenerated protease domain | +/− |
| Active protease domain/PDZ1 | −/− |
| Active protease domain/PDZ3+4 | −/− |
| Active protease domain/empty | −/− |
| Degenerated protease domain/degenerated protease domain | +/+ |
| Degenerated protease domain/PDZ1 | −/− |
| Degenerated protease domain/PDZ3+4 | −/− |
| Degenerated protease domain/empty | −/− |
| PDZ1/PDZ1 | −/− |
| PDZ1/PDZ3+4 | −/− |
| PDZ1/empty | −/− |
| PDZ3+4/PDZ3+4 | −/− |
| PDZ3+4/empty | −/− |
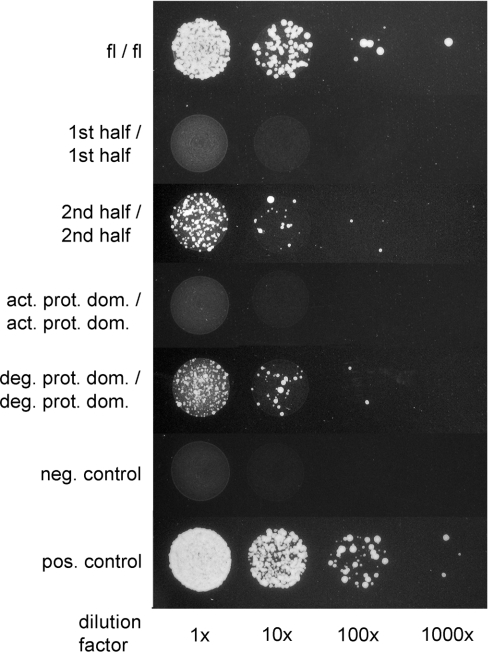
Figure 4. Y2H assay shows that oligomerization of DEG7 is mediated by the second (degenerated) protease domain.
Growth of S. cerevisiae YRG2 cells on histidine-free medium (indicative for interaction) is only visible when the combination of interaction partners contains the second (degenerated) protease domain (deg. prot. dom.), which is part of the second half (2nd half) and the full-length protein (fl). No interaction occurs with constructs containing only the first half (1st half) or first (active) protease domain (act. prot. dom.). The columns show different dilutions of the yeast cultures. A dilution factor of 1×corresponds to a D600 of 1.0. Controls: neg., negative; pos., positive.
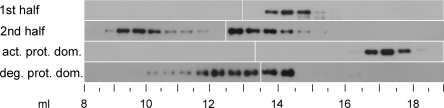
To confirm these findings by a method independent of the yeast system, the oligomerization status of the first and the second half of DEG7, as well as the first (active) and the second (degenerated) protease domain alone was analysed by SEC with recombinant proteins expressed in E. coli. Such truncated DEG7 constructs were affinity-purified as His6-tagged recombinant soluble proteins, and subsequently analysed by SEC and immunoblotting (Figure 5). Although the constructs representing the first and second halves of DEG7 have similar calculated molecular masses (65.1 and 66.2 kDa respectively), they exhibit totally different elution patterns. The first half of DEG7 elutes as one peak in fractions between 14.0 and 14.5 ml (corresponding to an apparent molecular mass of approx. 75–100 kDa), whereas the second half of DEG7 is present in two distinct peaks (fractions between 9.5 and 10.0 ml, and 12.5 and 13.0 ml, corresponding to apparent molecular masses of 980–1300 and 170–240 kDa respectively). Similar differences in the elution patterns are also observed for the active protease domain as compared with the degenerated protease domain (Figure 5). Whereas the active protease domain (calculated molecular mass of 27.9 kDa) is present in the fractions between 17.0 and 17.5 ml (corresponding to an apparent molecular mass of 13–18 kDa) the degenerated protease domain (calculated molecular mass of 31.5 kDa) elutes as two poorly resolved peaks between 12.0 and 12.5 ml, and 14.0 and 14.5 ml, corresponding to apparent molecular masses of 230–315 and 75–100 kDa respectively). Although the estimation of apparent molecular masses of oligomers is too inaccurate to determine the exact stoichiometry of the complexes, these data suggest that the first half of DEG7 and the active protease domain are eluted as monomers, whereas the second half of DEG7 and the degenerated protease domains form at least two types of oligomer, possibly a trimer and a higher oligomeric complex. The inaccuracy in the estimation of molecular masses of monomers and oligomers could be due to sterical factors and the fact that DEG7 deletion constructs are probably far from a globular shape, which might be different for the intact protein. For the deletion construct containing the active protease domain, this might also explain the difference between the molecular mass according to the SEC and that estimated by SDS/PAGE, which matched the calculated values (results not shown). In summary, we can conclude from these data that only DEG7 deletion constructs with the degenerated protease domain are able to form oligomeric complexes, thus confirming the data obtained from the Y2H screen (Figure 4 and Table 1, and see Supplementary Figure S4).
Figure 5. Only recombinant DEG7 containing the second (degenerated) protease domain forms oligomers.
Immunoblot (anti-His5-tag antibody) showing the distribution of truncated DEG7 after SEC. Fractions are indicated as vertical lines at the bottom, and numbers (in ml) indicate the respective elution volume. Truncated DEG7 proteins containing the second (degenerated) protease domain (2nd half, deg. prot. dom.) are eluted in two peaks at lower buffer volumes as compared with truncated proteins with the first (active) protease domain (1st half, act. prot. dom.), which are eluted in a single peak at the higher buffer volume.
Modelling of DEG7 structure
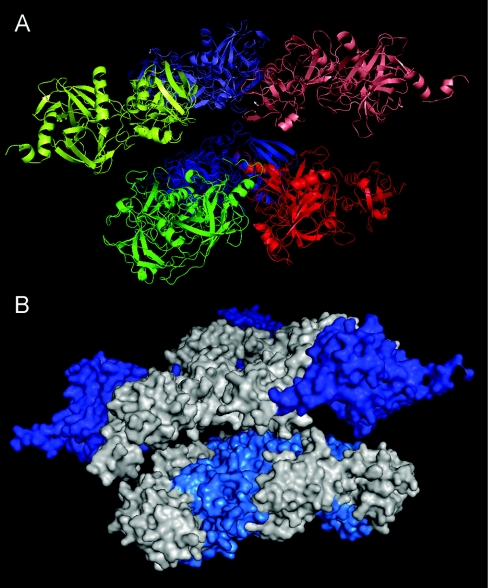
To illustrate how DEG7 might form oligomers, we built a model based on our findings that DEG7 forms trimers mediated by interactions of second (degenerated) protease domains, and the available crystal structures of E. coli DegP [9,12] and human HtrA2 [7] (Figure 6). For clarity, the approx. 100-amino-acid stretch between the second PDZ domain and the degenerated protease domain (Figure 1) was omitted. The oligomerization via the second (degenerated) protease domain might enable the protein to keep the catalytic subunits in close proximity, without the actual formation of a proteolytic active complex. This might ensure a very rapid initiation of proteolysis after binding of a substrate. However, details in this model, especially the relative orientations of the active protease domains, need to be resolved further.
Figure 6. Homology modelling of a DEG7 trimer.
The putative structure of a DEG7 trimer was modelled on the hexameric structure of E. coli DegP [9]. The approx. 100-amino-acid stretch between the second PDZ and the degenerated protease domain (Figure 1) was omitted for clarity. Relative orientations of the first half of the protein are not supported. (A) Cartoon model, side view. Monomeric subunits are coloured blue, green and red respectively. The second halves of the protein are at the bottom, coloured darker than the first halves. (B) Surface model, side view. The active protease domains are col-oured blue, the second (degenerated) protease domains involved in oligomerization are coloured light blue. This model might represent a protease oligomer in an inactive conformation.
Conclusions
We have shown that the DEG7 protease from A. thaliana has orthologues in green, heterokont and haptophyte algae, in fungi, mosses and higher plants, but no orthologues can be found in animals. The unusual domain arrangement of DEG7 with two protease domains (one degenerated) and four PDZ domains requires a different mode of oligomerization than the one which was described for all other Deg/HtrA proteases. Although trimerization through an active protease domain is a common feature of Deg/HtrA proteases in bacteria and mammals, the active protease domain of plant DEG7 is not essential for this process. Instead, the second (degenerated) protease domain is responsible for the trimerization of DEG7 in plants.
Inactive protease domains were reported from the proteolytic active oligomers of other proteases. Clp proteases in bacteria, for example, are oligomeric serine proteases with a proteolytic core constituted by two heptameric rings, with each heptamer composed of identical proteolytically active ClpP subunits [37]. In chloroplasts of plants and in cyanobacteria, however, the Clp protease core is a heteroheptamer, containing three proteolytically active ClpP subunits and four inactive ClpR variants, which show sequence similarity to ClpP on the amino acid level, but lack the catalytic triad of serine type proteases [38,39]. A high sequence similarity between the inactive ClpR and the active ClpP variants indicates a common ancestor [40]. This is supported by the fact that loss of the catalytic triad appears to be ongoing process since some cyanobacterial ClpR variants still retain one of the active-site residues [39].
A similar situation was reported for the 20S proteolytically active core unit of the 26S proteasome. This core unit is composed of four stacked heptameric rings. In Archaea, the two inner rings are homo-oligomers of proteolytically active β-subunits, whereas, in eukaryotes, the β-subunit has diverged into seven different β-proteins (reviewed in [41,42]). Of these seven β-subunits, only three contain the active-site residues and are proteolytically active. Despite the lack of enzymatic activity, the inactive β-subunits are essential for the correct assembly of the eukaryotic proteasome.
In the case of ClpP from photosynthetic organisms and the proteasome from eukaryotes, formerly active protease domains (or, in these cases, one-domain subunits) evolved into domains with tasks other than proteolysis, e.g. mediating subunit contacts in the oligomeric protease complex. The second (degenerated) protease domain of DEG7, which is described in the present paper, might be yet another example, where a formerly active enzymatic entity is now involved in the oligomerization of the protease complex. For Deg/HtrA proteases, inactive protease domains involved in oligomer formation have not been described so far. In contrast with ClpP and the proteasome, however, this inactive domain is not encoded by a separate gene, but is a part of the same polypeptide chain as the active protease domain.
Online data
AUTHOR CONTRIBUTION
Holger Schuhmann and Ulrike Mogg performed research, analysed data and wrote the draft of the paper. Iwona Adamska conceived the research and edited the paper before submission.
ACKNOWLEDGEMENTS
We are grateful to Silvia Kuhn for technical assistance in the Y2H assays, and Dr Dietmar Funck for helpful advice. We thank the Nottingham Arabidopsis Stock Centre (NASC) for providing the cDNA clones.
FUNDING
This work was supported by grants from the Deutsche Forschungsgemeinschaft [grant numbers AD92/8-2 and AD92/8-3] and Konstanz University (to I.A.).
References
- 1.Kim D. Y., Kim K. K. Structure and function of HtrA family proteins, the key players in protein quality control. J. Biochem. Mol. Biol. 2005;38:266–274. doi: 10.5483/bmbrep.2005.38.3.266. [DOI] [PubMed] [Google Scholar]
- 2.Clausen T., Southan C., Ehrmann M. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell. 2002;10:443–455. doi: 10.1016/s1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- 3.Rawlings N. D., Morton F. R., Kok C. Y., Kong J., Barrett A. J. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36:D320–D325. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helm M., Luck C., Prestele J., Hierl G., Huesgen P. F., Fröhlich T., Arnold G. J., Adamska I., Gorg A., Lottspeich F., Gietl C. Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11501–1156. doi: 10.1073/pnas.0704733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuhmann H., Huesgen P. F., Gietl C., Adamska I. The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis thaliana. Plant Physiol. 2008;148:1847–1856. doi: 10.1104/pp.108.125377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun X., Peng L., Guo J., Chi W., Ma J., Lu C., Zhang L. Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell. 2007;19:1347–1361. doi: 10.1105/tpc.106.049510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W., Srinivasula S. M., Chai J., Li P., Wu J. W., Zhang Z., Alnemri E. S., Shi Y. Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat. Struct. Biol. 2002;9:436–441. doi: 10.1038/nsb795. [DOI] [PubMed] [Google Scholar]
- 8.Wilken C., Kitzing K., Kurzbauer R., Ehrmann M., Clausen T. Crystal structure of the DegS stress sensor: how a PDZ domain recognizes misfolded protein and activates a protease. Cell. 2004;117:483–494. doi: 10.1016/s0092-8674(04)00454-4. [DOI] [PubMed] [Google Scholar]
- 9.Krojer T., Garrido-Franco M., Huber R., Ehrmann M., Clausen T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature. 2002;416:455–459. doi: 10.1038/416455a. [DOI] [PubMed] [Google Scholar]
- 10.Jomaa A., Damjanovic D., Leong V., Ghirlando R., Iwanczyk J., Ortega J. The inner cavity of Escherichia coli DegP protein is not essential for molecular chaperone and proteolytic activity. J. Bacteriol. 2007;189:706–716. doi: 10.1128/JB.01334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim D. Y., Kim D. R., Ha S. C., Lokanath N. K., Lee C. J., Hwang H. Y., Kim K. K. Crystal structure of the protease domain of a heat-shock protein HtrA from Thermotoga maritima. J. Biol. Chem. 2003;278:6543–6551. doi: 10.1074/jbc.M208148200. [DOI] [PubMed] [Google Scholar]
- 12.Krojer T., Sawa J., Schäfer E., Saibil H. R., Ehrmann M., Clausen T. Structural basis for the regulated protease and chaperone function of DegP. Nature. 2008;453:885–890. doi: 10.1038/nature07004. [DOI] [PubMed] [Google Scholar]
- 13.Jiang J., Zhang X., Chen Y., Wu Y., Zhou Z. H., Chang Z., Sui S. F. Activation of DegP chaperone-protease via formation of large cage-like oligomers upon binding to substrate proteins. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11939–11944. doi: 10.1073/pnas.0805464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ades S. E., Connolly L. E., Alba B. M., Gross C. A. The Escherichia coli σE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-σ factor. Genes Dev. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alba B. M., Leeds J. A., Onufryk C., Lu C. Z., Gross C. A. DegS and YaeL participate sequentially in the cleavage of RseA to activate the σE-dependent extracytoplasmic stress response. Genes Dev. 2002;16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipinska B., Fayet O., Baird L., Georgopoulos C. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J. Bacteriol. 1989;171:1574–1584. doi: 10.1128/jb.171.3.1574-1584.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strauch K. L., Johnson K., Beckwith J. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 1989;171:2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verhagen A. M., Silke J., Ekert P. G., Pakusch M., Kaufmann H., Connolly L. M., Day C. L., Tikoo A., Burke R., Wrobel C., et al. HtrA2 promotes cell death through its serine protease activity and its ability to antagonize inhibitor of apoptosis proteins. J. Biol. Chem. 2002;277:445–454. doi: 10.1074/jbc.M109891200. [DOI] [PubMed] [Google Scholar]
- 19.Kurochkin I. V., Mizuno Y., Konagaya A., Sakaki Y., Schonbach C., Okazaki Y. Novel peroxisomal protease Tysnd1 processes PTS1- and PTS2-containing enzymes involved in β-oxidation of fatty acids. EMBO J. 2007;26:835–845. doi: 10.1038/sj.emboj.7601525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huesgen P. F., Schuhmann H., Adamska I. The family of Deg proteases in cyanobacteria and chloroplasts of higher plants. Physiol. Plant. 2005;123:413–420. [Google Scholar]
- 21.Padmanabhan N., Fichtner L., Dickmanns A., Ficner R., Schulz J. B., Braus G. H. The yeast HtrA orthologue Ynm3 is a protease with chaperone activity that aids survival under heat stress. Mol. Biol. Cell. 2009;20:68–77. doi: 10.1091/mbc.E08-02-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fahrenkrog B., Sauder U., Aebi U. The S. cerevisiae HtrA-like protein Nma111p is a nuclear serine protease that mediates yeast apoptosis. J. Cell Sci. 2004;117:115–126. doi: 10.1242/jcs.00848. [DOI] [PubMed] [Google Scholar]
- 23.Tong F., Black P. N., Bivins L., Quackenbush S., Ctrnacta V., DiRusso C. C. Direct interaction of Saccharomyces cerevisiae Faa1p with the Omi/HtrA protease orthologue Ynm3p alters lipid homeostasis. Mol. Genet. Genomics. 2006;275:330–343. doi: 10.1007/s00438-005-0089-1. [DOI] [PubMed] [Google Scholar]
- 24.Sun X., Fu T., Chen N., Guo J., Ma J., Zou M., Lu C., Zhang L. The stromal chloroplast Deg7 protease participates in the repair of photosystem II after photoinhibition in Arabidopsis. Plant Physiol. 2010;152:1263–1273. doi: 10.1104/pp.109.150722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zdobnov E. M., Apweiler R. InterProScan: an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 27.Soding J., Biegert A., Lupas A. N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moretti S., Armougom F., Wallace I. M., Higgins D. G., Jongeneel C. V., Notredame C. The M-Coffee web server: a meta-method for computing multiple sequence alignments by combining alternative alignment methods. Nucleic Acids Res. 2007;35:W645–W648. doi: 10.1093/nar/gkm333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M., et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 2007;Chapter 2 doi: 10.1002/0471140864.ps0209s50. Unit 29. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y., Appleton B. A., Wu P., Wiesmann C., Sidhu S. S. Structural and functional analysis of the ligand specificity of the HtrA2/Omi PDZ domain. Protein Sci. 2007;16:1738–1750. doi: 10.1110/ps.072833207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: a Lab-oratory Manual. 2nd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Yamada K., Lim J., Dale J. M., Chen H., Shinn P., Palm C. J., Southwick A. M., Wu H. C., Kim C., Nguyen M., et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 34.Gietz R. D., Woods R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 35.Hunter S., Apweiler R., Attwood T. K., Bairoch A., Bateman A., Binns D., Bork P., Das U., Daugherty L., Duquenne L., et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeth K. Structural analysis of DegS, a stress sensor of the bacterial periplasm. FEBS Lett. 2004;569:351–358. doi: 10.1016/j.febslet.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 37.Wang J., Hartling J. A., Flanagan J. M. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 38.Peltier J. B., Ripoll D. R., Friso G., Rudella A., Cai Y., Ytterberg J., Giacomelli L., Pillardy J., van Wijk K. J. Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J. Biol. Chem. 2004;279:4768–4781. doi: 10.1074/jbc.M309212200. [DOI] [PubMed] [Google Scholar]
- 39.Andersson F. I., Tryggvesson A., Sharon M., Diemand A. V., Classen M., Best C., Schmidt R., Schelin J., Stanne T. M., Bukau B., et al. Structure and function of a novel type of ATP-dependent Clp protease. J. Biol. Chem. 2009;284:13519–13532. doi: 10.1074/jbc.M809588200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-Lorenzo M., Sjodin A., Jansson S., Funk C. Protease gene families in Populus and Arabidopsis. BMC Plant Biol. 2006;6:30. doi: 10.1186/1471-2229-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallastegui N., Groll M. The 26S proteasome: assembly and function of a destructive machine. Trends Biochem. Sci. 2010;35:634–642. doi: 10.1016/j.tibs.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Groll M., Bochtler M., Brandstetter H., Clausen T., Huber R. Molecular machines for protein degradation. ChemBioChem. 2005;6:222–256. doi: 10.1002/cbic.200400313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.