Abstract
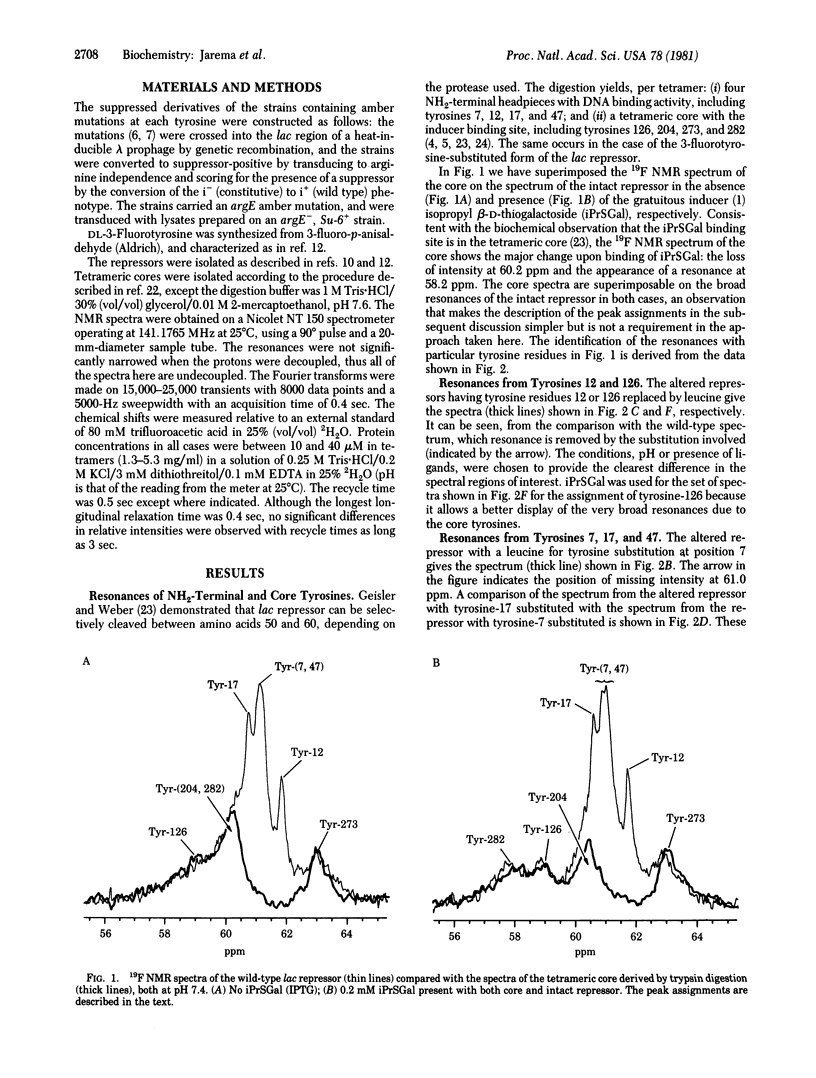
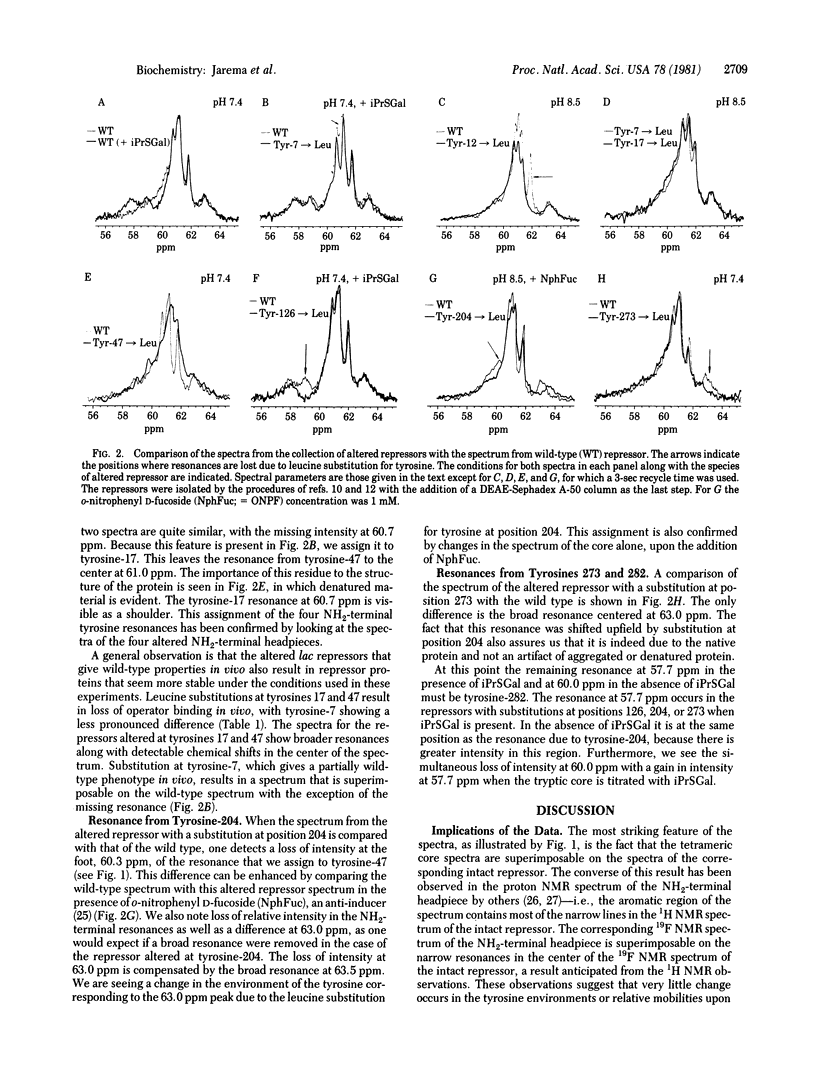
By using a systematic genetic approach, the resonances in the 19F NMR spectrum of 3-fluorotyrosine-substituted lac repressor protein have been assigned. The NMR data indicate that each monomer of the repressor consists of two distinct and independent domains. One domain, the NH2-terminal sixth of the primary sequence, which has been shown to be very important for DNA binding, is very mobile. The remaining COOH-terminal sequence is more rigid. Ligands of the repressor, which affect its DNA binding capability, lead to conformational changes in the COOH-terminal domain. The approach to the assignment of spectral features taken here can be extended to other systems.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexander M. E., Burgum A. A., Noall R. A., Shaw M. D., Matthews K. S. Modification of tyrosine residues of the lactose repressor protein. Biochim Biophys Acta. 1977 Aug 23;493(2):367–379. doi: 10.1016/0005-2795(77)90193-3. [DOI] [PubMed] [Google Scholar]
- Anderson R. A., Nakashima Y., Coleman J. E. Chemical modifications of functional residues of fd gene 5 DNA-binding protein. Biochemistry. 1975 Mar 11;14(5):907–917. doi: 10.1021/bi00676a006. [DOI] [PubMed] [Google Scholar]
- Bourgeois S., Pfahl M. Repressors. Adv Protein Chem. 1976;30:1–99. doi: 10.1016/s0065-3233(08)60478-7. [DOI] [PubMed] [Google Scholar]
- Buck F., Rüterjans H., Beyreuther K. 1H NMR study of the lactose repressor from Escherichia coli. FEBS Lett. 1978 Dec 15;96(2):335–338. doi: 10.1016/0014-5793(78)80430-x. [DOI] [PubMed] [Google Scholar]
- Cassels R., Dobson C. M., Poulsen F. M., Williams R. J. Study of the tryptophan residues of lysozyme using 1H nuclear magnetic resonance. Eur J Biochem. 1978 Dec 1;92(1):81–97. doi: 10.1111/j.1432-1033.1978.tb12725.x. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran R., Jardetzky T. S., Jardetzky O. The delta helix--a possible left-handed stable polypeptide structure in the N-terminal segment of the lac repressor. FEBS Lett. 1979 May 1;101(1):11–14. [PubMed] [Google Scholar]
- Cohen J. S., Hayes M. B. Nuclear magnetic resonance titration curves of histidine ring protons. V. Comparative study of cytochrome c from three species and the assignment of individual proton resonances. J Biol Chem. 1974 Sep 10;249(17):5472–5477. [PubMed] [Google Scholar]
- Coleman J. E., Armitage I. M. Tyrosyl-base-phenylalanyl intercalation in gene 5 protein-DNA complexes: proton nuclear magnetic resonance of selectively deuterated gene 5 protein. Biochemistry. 1978 Nov 14;17(23):5038–5045. doi: 10.1021/bi00616a028. [DOI] [PubMed] [Google Scholar]
- Dobson C. M., Ferguson S. J., Poulsen F. M., Williams R. J. Complete assignment of aromatic 1H nuclear magnetic resonances of the tyrosine residues of hen lysozyme. Eur J Biochem. 1978 Dec 1;92(1):99–103. doi: 10.1111/j.1432-1033.1978.tb12726.x. [DOI] [PubMed] [Google Scholar]
- Dobson C. M., Moore G. R., Williams R. J. Assignment of aromatic amino acid PMR resonances of horse ferricytochrome c. FEBS Lett. 1975 Mar 1;51(1):60–65. doi: 10.1016/0014-5793(75)80854-4. [DOI] [PubMed] [Google Scholar]
- Fanning T. G. Iodination of Escherichia coli lac repressor. Effect of tyrosine modification on repressor activity. Biochemistry. 1975 Jun 3;14(11):2512–2520. doi: 10.1021/bi00682a034. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J. Sequence of the lacI gene. Nature. 1978 Aug 24;274(5673):765–769. doi: 10.1038/274765a0. [DOI] [PubMed] [Google Scholar]
- Fung L. W., Ho C. A proton nuclear magnetic resonance study of the quaternary structure of human homoglobins in water. Biochemistry. 1975 Jun 3;14(11):2526–2535. doi: 10.1021/bi00682a036. [DOI] [PubMed] [Google Scholar]
- Geisler N., Weber K. Isolation of amino-terminal fragment of lactose repressor necessary for DNA binding. Biochemistry. 1977 Mar 8;16(5):938–943. doi: 10.1021/bi00624a020. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Müller-Hill B. Isolation of the lac repressor. Proc Natl Acad Sci U S A. 1966 Dec;56(6):1891–1898. doi: 10.1073/pnas.56.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Müller-Hill B. The lac operator is DNA. Proc Natl Acad Sci U S A. 1967 Dec;58(6):2415–2421. doi: 10.1073/pnas.58.6.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini L. Informational suppression. Annu Rev Genet. 1970;4:107–134. doi: 10.1146/annurev.ge.04.120170.000543. [DOI] [PubMed] [Google Scholar]
- Hagen D. S., Weiner J. H., Sykes B. D. Fluorotyrosine M13 coat protein: fluorine-19 nuclear magnetic resonance study of the motional properties of an integral membrane protein in phospholipid vesicles. Biochemistry. 1978 Sep 5;17(18):3860–3866. doi: 10.1021/bi00611a028. [DOI] [PubMed] [Google Scholar]
- Hagen D. S., Weiner J. H., Sykes B. D. Investigation of solvent accessibility of the fluorotyrosyl residues of M13 coat protein in deoxycholate micelles and phospholipid vesicles. Biochemistry. 1979 May 15;18(10):2007–2012. doi: 10.1021/bi00577a026. [DOI] [PubMed] [Google Scholar]
- JACOB F., MONOD J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961 Jun;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Jayaraman K., Müller-Hill B., Rickenberg H. V. Inhibition of the synthesis of beta-galactosidase in Escherichia coli by 2-nitrophenyl-beta-D-fucoside. J Mol Biol. 1966 Jul;18(2):339–343. doi: 10.1016/s0022-2836(66)80251-6. [DOI] [PubMed] [Google Scholar]
- Jovin T. M., Geisler N., Weber K. Amino-terminal fragments of Escherichia coli lac repressor bind to DNA. Nature. 1977 Oct 20;269(5630):668–672. doi: 10.1038/269668a0. [DOI] [PubMed] [Google Scholar]
- Keller R. M., Pettigrew G. W., Wüthrich K. Structural studies by proton NMR of cytochrome C-557 from Crithidia oncopelti. FEBS Lett. 1973 Oct 15;36(2):151–156. doi: 10.1016/0014-5793(73)80358-8. [DOI] [PubMed] [Google Scholar]
- Kimber B. J., Griffiths D. V., Birdsall B., King R. W., Scudder P., Feeney J., Roberts G. C., Burgen A. S. 19 Fnuclear magnetic resonance studies of ligand binding to 3-fluorotyrosine-and 6-fluorotryptophan-containing dihydrofolate reductase from Lactobacillus casei. Biochemistry. 1977 Jul 26;16(15):3492–3500. doi: 10.1021/bi00634a032. [DOI] [PubMed] [Google Scholar]
- Laiken S. L., Gross C. A., Von Hippel P. H. Equilibrium and kinetic studies of Escherichia coli lac repressor-inducer interactions. J Mol Biol. 1972 Apr 28;66(1):143–155. doi: 10.1016/s0022-2836(72)80012-3. [DOI] [PubMed] [Google Scholar]
- Leijenaar-van den Berg G., Migchelsen C., Beintema J. J. Proton nuclear magnetic resonance of histidine residues in reindeer pancreatic ribonuclease. FEBS Lett. 1974 Nov 15;48(2):218–221. doi: 10.1016/0014-5793(74)80471-0. [DOI] [PubMed] [Google Scholar]
- Lu P., Jarema M., Mosser K., Daniel W. E. lac repressor: 3-fluorotyrosine substitution for nuclear magnetic resonance studies. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3471–3475. doi: 10.1073/pnas.73.10.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markley J. L. Correlation proton magnetic resonance studies at 250 MHz of bovine pancreatic ribonuclease. I. Reinvestigation of the histidine peak assignments. Biochemistry. 1975 Aug 12;14(16):3546–3554. doi: 10.1021/bi00687a006. [DOI] [PubMed] [Google Scholar]
- Markley J. L., Finkenstadt W. R. Correlation proton magnetic resonance studies at 250 MHz of bovine pancreatic ribonuclease. III. Mutual electrostatic interaction between histidine residues 12 and 119. Biochemistry. 1975 Aug 12;14(16):3562–3566. doi: 10.1021/bi00687a008. [DOI] [PubMed] [Google Scholar]
- Matthews K. S., Matthews H. R., Thielmann H. W., Jardetzky G. Ultraviolet difference spectra of the lactose repressor protein. Biochim Biophys Acta. 1973 Jan 25;295(1):159–165. doi: 10.1016/0005-2795(73)90083-4. [DOI] [PubMed] [Google Scholar]
- Matthews K. S. Ultraviolet difference spectra of the lactose repressor protein. II. Trypsin core protein. Biochim Biophys Acta. 1974 Aug 8;359(2):334–340. doi: 10.1016/0005-2795(74)90232-3. [DOI] [PubMed] [Google Scholar]
- Maurizot J. C., Charlier M. Circular dichroism studies of the binding of o-nitrophenyl-beta-D-fucoside and o-nitrophenyl-beta-D-galactoside to lac repressor. Eur J Biochem. 1977 Oct 3;79(2):395–399. doi: 10.1111/j.1432-1033.1977.tb11821.x. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Coulondre C., Farabaugh P. J. Correlation of nonsense sites in the lacI gene with specific codons in the nucleotide sequence. Nature. 1978 Aug 24;274(5673):770–775. doi: 10.1038/274770a0. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Coulondre C., Hofer M., Schmeissner U., Sommer H., Schmitz A., Lu P. Genetic studies of the lac repressor. IX. Generation of altered proteins by the suppression of nonsence mutations. J Mol Biol. 1979 Jun 25;131(2):191–222. doi: 10.1016/0022-2836(79)90073-1. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Ganem D., Lu P., Schmitz A. Genetic studies of the lac repressor. I. Correlation of mutational sites with specific amino acid residues: construction of a colinear gene-protein map. J Mol Biol. 1977 Jan 15;109(2):275–298. doi: 10.1016/s0022-2836(77)80034-x. [DOI] [PubMed] [Google Scholar]
- Moore G. R., Williams R. J. Comparison of the structures of various eukaryotic ferricytochromes c and ferrocytochromes and their antigenic differences. Eur J Biochem. 1980 Feb;103(3):543–550. doi: 10.1111/j.1432-1033.1980.tb05978.x. [DOI] [PubMed] [Google Scholar]
- Moore G. R., Williams R. J. The solution structures of tuna and horse cytochromes c. Eur J Biochem. 1980 Feb;103(3):533–541. doi: 10.1111/j.1432-1033.1980.tb05977.x. [DOI] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. An amino-terminal fragment of lac repressor binds specifically to lac operator. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5851–5854. doi: 10.1073/pnas.75.12.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. DNA-binding site of lac repressor probed by dimethylsulfate methylation of lac operator. J Mol Biol. 1979 Aug 25;132(4):709–728. doi: 10.1016/0022-2836(79)90384-x. [DOI] [PubMed] [Google Scholar]
- Oldfield E., Norton R. S., Allerhand A. Studies of individual carbon sites of proteins in solution by natural abundance carbon 13 nuclear magnetic resonance spectroscopy. Strategies for assignments. J Biol Chem. 1975 Aug 25;250(16):6381–6402. [PubMed] [Google Scholar]
- Oshima Y., Matsuura M., Horiuchi T. Conformational change of the lac repressor induced with the inducer. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1444–1450. doi: 10.1016/0006-291x(72)90234-3. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. L., Bovey F. A. Reassignment of the active site histidines in ribonuclease A by selective deuteration studies. Biopolymers. 1975 May;14(5):987–997. doi: 10.1002/bip.1975.360140508. [DOI] [PubMed] [Google Scholar]
- Poulsen F. M., Hoch J. C., Dobson C. M. A structural study of the hydrophobic box region of lysozyme in solution using nuclear Overhauser effects. Biochemistry. 1980 Jun 10;19(12):2597–2607. doi: 10.1021/bi00553a011. [DOI] [PubMed] [Google Scholar]
- Richarz R., Sehr P., Wagner G., Wüthrich K. Kinetics of the exchange of individual amide protons in the basic pancreatic trypsin inhibitor. J Mol Biol. 1979 May 5;130(1):19–30. doi: 10.1016/0022-2836(79)90549-7. [DOI] [PubMed] [Google Scholar]
- Roberts G. C., Jardetzky O. Nuclear magnetic resonance spectroscopy of amino acids, peptides, and proteins. Adv Protein Chem. 1970;24:447–545. doi: 10.1016/s0065-3233(08)60246-6. [DOI] [PubMed] [Google Scholar]
- Robertson D. E., Kroon P. A., Ho C. Nuclear magnetic resonance and fluorescence studies of substrate-induced conformational changes of histidine-binding protein J of Salmonella typhimurium. Biochemistry. 1977 Apr 5;16(7):1443–1451. doi: 10.1021/bi00626a032. [DOI] [PubMed] [Google Scholar]
- Schmitz A., Schmeissner U., Miller J. H. Mutations affecting the quaternary structure of the lac repressor. J Biol Chem. 1976 Jun 10;251(11):3359–3366. [PubMed] [Google Scholar]
- Shindo H., Hayes M. B., Cohen J. S. Nuclear magnetic resonance titration curves of histidine ring protons. A direct assignment of the resonances of the active site histidine residues of ribonuclease. J Biol Chem. 1976 May 10;251(9):2644–2647. [PubMed] [Google Scholar]
- Sommer H., Lu P. Lac Repressor. Fluorescence of the two tryptophans. J Biol Chem. 1976 Jun 25;251(12):3774–3779. [PubMed] [Google Scholar]
- Sykes B. D., Hull W. E. Fluorine nuclear magnetic resonance studies of proteins. Methods Enzymol. 1978;49:270–295. doi: 10.1016/s0076-6879(78)49015-9. [DOI] [PubMed] [Google Scholar]
- Sykes B. D., Weingarten H. I., Schlesinger M. J. Fluorotyrosine alkaline phosphatase from Escherichia coli: preparation, properties, and fluorine-19 nuclear magnetic resonance spectrum. Proc Natl Acad Sci U S A. 1974 Feb;71(2):469–473. doi: 10.1073/pnas.71.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viggiano G., Wiechelman K. J., Chervenick P. A., Ho C. Proton nuclear magnetic resonance studies of hemoglobins Osler (beta145HC2 Tyr replaced by Asp) and McKee Rocks (beta145HC2 Tyr replaced by term): an assignment for an important tertiary structural probe in hemoglobin. Biochemistry. 1978 Mar 7;17(5):795–799. doi: 10.1021/bi00598a007. [DOI] [PubMed] [Google Scholar]
- Wade-Jardetzky N., Bray R. P., Conover W. W., Jardetzky O., Geisler N., Weber K. Differential mobility of the N-terminal headpiece in the lac-repressor protein. J Mol Biol. 1979 Feb 25;128(2):259–264. doi: 10.1016/0022-2836(79)90129-3. [DOI] [PubMed] [Google Scholar]
- Wagner G., Wüthrich K. Correlation between the amide proton exchange rates and the denaturation temperatures in globular proteins related to the basic pancreatic trypsin inhibitor. J Mol Biol. 1979 May 5;130(1):31–37. doi: 10.1016/0022-2836(79)90550-3. [DOI] [PubMed] [Google Scholar]
- Wiechelman K. J., Fairbanks V. F., Ho C. Proton nuclear magnetic resonance studies of hemoglobin Malmö: implications of mutations at homologous positions of the alpha and beta chains. Biochemistry. 1976 Apr 6;15(7):1414–1420. doi: 10.1021/bi00652a010. [DOI] [PubMed] [Google Scholar]
- Wu F. Y., Bandyopadhyay P., Wu C. W. Conformational transitions of the lac repressor from Escherichia coli. J Mol Biol. 1976 Feb 5;100(4):459–472. doi: 10.1016/s0022-2836(76)80040-x. [DOI] [PubMed] [Google Scholar]
- Wüthrich K., Wagner G. Nuclear magnetic resonance of labile protons in the basic pancreatic trypsin inhibitor. J Mol Biol. 1979 May 5;130(1):1–18. doi: 10.1016/0022-2836(79)90548-5. [DOI] [PubMed] [Google Scholar]


