Abstract
Summary: Antimicrobials are valuable therapeutics whose efficacy is seriously compromised by the emergence and spread of antimicrobial resistance. The provision of antibiotics to food animals encompasses a wide variety of nontherapeutic purposes that include growth promotion. The concern over resistance emergence and spread to people by nontherapeutic use of antimicrobials has led to conflicted practices and opinions. Considerable evidence supported the removal of nontherapeutic antimicrobials (NTAs) in Europe, based on the “precautionary principle.” Still, concrete scientific evidence of the favorable versus unfavorable consequences of NTAs is not clear to all stakeholders. Substantial data show elevated antibiotic resistance in bacteria associated with animals fed NTAs and their food products. This resistance spreads to other animals and humans—directly by contact and indirectly via the food chain, water, air, and manured and sludge-fertilized soils. Modern genetic techniques are making advances in deciphering the ecological impact of NTAs, but modeling efforts are thwarted by deficits in key knowledge of microbial and antibiotic loads at each stage of the transmission chain. Still, the substantial and expanding volume of evidence reporting animal-to-human spread of resistant bacteria, including that arising from use of NTAs, supports eliminating NTA use in order to reduce the growing environmental load of resistance genes.
INTRODUCTION
For many decades, antibiotic resistance has been recognized as a global health problem. It has now been escalated by major world health organizations to one of the top health challenges facing the 21st century (40, 65). Some of its causes are widely accepted, for example, the overuse and inappropriate use of antibiotics for nonbacterial infections such as colds and other viral infections and inadequate antibiotic stewardship in the clinical arena (109). But the relationship of drug-resistant bacteria in people to antibiotic use in food animals continues to be debated, particularly in the United States (11, 14, 38, 44, 48, 96, 124).
Many have delved into this question, producing volumes of direct and indirect evidence linking animal use to antibiotic resistance confronting people. Among these are a number of studies which unequivocally support the concern that use of antibiotics in food animals (particularly nontherapeutic use) impacts the health of people on farms and, more distantly, via the food chain (69, 88, 90, 111). While it was hoped by many that the years of experience following the bans on nontherapeutic use of antimicrobials in Europe would clearly signal an end to this practice, arguments continue, largely along the lines of a cost/benefit ratio and perceived deficits in solid scientific evidence. Action in the United States continues to lag far behind that of the European Union, which has chosen to operate proactively based on the “precautionary principle,” a guiding tenet of public health. This principle states that “when evidence points toward the potential of an activity to cause significant widespread or irreparable harm to public health or the environment, options for avoiding that harm should be examined and pursued even if the harm is not yet fully understood or proven” (103).
This communication summarizes a large number of studies on the links between antimicrobials used for growth promotion, in particular, as well as other nontherapeutic antimicrobial (NTA) use in animal husbandry and aquaculture, and the emergence of antibiotic-resistant bacteria in humans. The FAAIR Report (Facts about Antibiotics in Animals and the Impact on Resistance) of the Alliance for the Prudent Use of Antibiotics (APUA) cites areas where antibiotic use can be curtailed and proposes several viable recommendations that could be utilized to reduce the burden of resistance genes created by nontherapeutic antibiotic use in animals (22).
Lastly, we consider whether knowledge gaps exist that need addressing in order to answer persisting questions that fuel the controversy over NTA use in food animals.
ANTIMICROBIAL USE IN ANIMALS: EFFECTS ON ANTIBIOTIC RESISTANCE EMERGENCE
Antimicrobials are delivered to animals for a variety of reasons, including disease treatment, prevention, control, and growth promotion/feed efficiency. Antimicrobial growth promotants (AGPs) were first advocated in the mid-1950s, when it was discovered that small, subtherapeutic quantities of antibiotics such as procaine penicillin and tetracycline (1/10 to 1/100 the amount of a therapeutic dose), delivered to animals in feed, could enhance the feed-to-weight ratio for poultry, swine, and beef cattle (142). For many years, the positive effects of this practice were championed, while the negative consequences went undetected. But microbiologists and infectious disease experts facing antibiotic resistance questioned the possible harm from this use (74, 89, 109, 136). They found that farms using AGPs had more resistant bacteria in the intestinal floras of the farm workers and farm animals than in those for similar people and animals on farms not using AGPs. A prospective in vivo/in situ study in 1975 was performed to evaluate the effect of introducing low-dose in-feed oxytetracycline as an AGP on the intestinal floras of chickens and farm dwellers (111). The results showed not only colonization of the chickens with tetracycline-resistant and other drug-resistant Escherichia coli strains but also acquisition of resistance in E. coli in the intestinal flora of the farm family. Other studies over the ensuing 3 decades further elucidated the quantitative and qualitative relationships between the practice of in-feed antimicrobials for animals and the mounting problem of hard-to-treat, drug-resistant bacterial infections in humans (83, 162).
Nontherapeutic Agents and Practices
The chief agricultural NTAs, used extensively in the United States and also used in Europe until the 1970s, include drugs that have likewise been employed widely in human medicine. In the absence of complete, unbiased data, this NTA use in the United States is estimated to be equal to (159) or as much as eight times greater than (67, 117) the quantity administered for therapeutic use.
More recently, concerns have arisen over the extensive use of antimicrobials in the burgeoning aquaculture industry, which more than doubled between 1994 and 2004 (36, 84). Eighty to 90 percent of total production occurs in Asia, with 67% occurring in China alone (64). In many parts of the world, fish farming is integrated with sewage or industrial wastewater or with land agriculture, as manure and other agricultural residues are commonly employed in fish feed (123). The overcrowding, unhygienic measures, and other manipulations in this intensive, industrial-scale production act as stressors to the fish and promote an increased use of antibiotic prophylaxis, particularly in the shrimp and carnivorous fish (such as salmon) industries. Moreover, even though the aquaculture use of AGPs in Western Europe and North America has been discontinued, therapeutic treatment of fish generally occurs en masse via inclusion in fish food, which results in exposure of the entire body of water to the antibiotic. The broad application of antibiotics in fish food leads to leaching from unconsumed food and feces into the water and pond sediments, where it not only exerts selective pressures on the sediment and water microflora but also can be washed to more distant sites, exposing wild fish and shellfish to trace antimicrobials (36). In this environment, the role of transduction (infection by bacterial phages) is considered highly important in facilitating lateral gene transfer (71). Sorum suggested that, historically, the transfer and emergence of resistance have occurred faster from aquatic bacteria to humans than from terrestrial animal bacteria to humans (141).
In the United States, the total fish industry use of antibiotics was estimated to be 204,000 to 433,000 pounds in the mid-1990s (25) (about 2% of the nonmedical use in cattle, swine, and poultry [117]). In much of the world, however, antibiotics are unregulated and used indiscriminately, and use statistics are rarely collected (25, 157). Although the total quantities of antibiotics employed in aquaculture are estimated to be smaller than those used in land animal husbandry, there is much greater use of antibiotic families that are also used in human medicine (Table 1 ). In Chile, for example, ∼100 metric tons of quinolones are used annually (10-fold greater than the amount used in human medicine), mostly in aquaculture (35). At least 13 different antimicrobials are reportedly used by farmers along the Thai coast (75).
Table 1.
Antimicrobials used in food animal productiona
| Antibiotic | Purpose | Antimicrobial class | Spectrum of activity | Use in human medicine | Structurally related antibiotic(s)/antibiotic(s) with shared cross-resistance | Commentsc |
|---|---|---|---|---|---|---|
| Ardacin | Bovine AGP | Glycopeptides | Gram-positive organisms | No | Vancomycin, teicoplanin | Withdrawn from EU in 1997; not licensed in U.S. |
| Amoxicillin,b ampicillinb | Aquaculture, oral treatment of swine colibacillosis, treatment of bovine bacterial enteritis and subclinical mastitis | Aminopenicillins | Moderate | Yes | All penicillins | |
| Avilamycin | AGP for broilers | Orthosomysins | Gram-positive organisms | No | Everninomycin | Withdrawn from EU; not licensed in U.S. |
| Avoparcinb | AGP | Glycopeptides | Gram-positive organisms | No | Vancomycin, teicoplanin | Withdrawn from EU in 1997; not licensed in U.S. |
| Bacitracin/zinc bacitracin | AGP for poultry, beef cattle, and swine; control of swine dysentery and bacterial enteritis; control of poultry enteritis | Polypeptides | Gram-positive organisms | Yes (zinc bacitracin) | Actinomycin, colistin, polymyxin B | Withdrawn from EU in 1999; available in U.S.; used in Japan |
| Bambermycin | AGP for poultry, swine, and cattle | Phosphoglycolipids | Gram-positive organisms | No | Vancomycin, teicoplanin | Withdrawn from EU in 2006; available in U.S. |
| Carbadox | Control of swine dysentery | Quinoxalines | Gram-positive and -negative organisms | No | Other quinoxolines | Withdrawn due to worker toxicity in EU and Canada; available in U.S. and Mexico |
| Carbomycinb | Respiratory disease prevention and treatment in poultry | Macrolides | Gram-positive organisms | No | Other macrolides | |
| Chloramphenicol | Aquaculture (oral/bath/injection) | Amphenicols | Broad | Yes | All amphenicols | Chloramphenicol approved in U.S. for dogs only |
| Colistin | Broiler, swine, and cattle feed | Cyclopolypeptides | Gram-negative organisms | Yes | All polymyxins | Used in Japan |
| Efrotomycin | AGP for swine | Elfamycins | Gram-positive organisms | No | Other elfamycins only | Not marketed |
| Enrofloxacinb | Therapy for bovine and swine respiratory disease, use in aquaculture (oral/bath) | Fluoroquinolones | Broad | No | All quinolones | Banned for use in poultry by FDA in 2005; not approved for aquaculture in U.S. |
| Erythromycinb | Aquaculture (oral/bath/injection); AGP for poultry, cattle, and swine; therapy for poultry respiratory disease and bovine mastitis | Macrolides | Gram-positive organisms | Yes | Oleandomycin and other macrolides and lincosamides | |
| Florfenicol | Respiratory disease treatment of cattle and swine | Amphenicols | Broad | No | All amphenicols | |
| Flumequinb | Aquaculture (oral) | Fluoroquinolones | Broad | No | Fluoroquinolones and quinolones | Not approved in U.S. |
| Furazolidone | Aquaculture (oral/bath) | Nitrofurans | Broad | Yes | Banned in U.S. food animals in 2005 | |
| Gentamicinb | Therapy for swine colibacillosis and dysentery, prevention of early poultry mortality, turkey egg dip | Aminoglycosides | Gram-positive and -negative organisms | Yes | Other aminoglycosides | |
| Lasalocid | AGP for cattle, poultry, sheep, and rabbits; coccidiosis prevention in poultry and sheep | Ionophores | Gram-positive organisms | No | Not demonstrated | Approved in EU and U.S. |
| Lincomycin | AGP for chickens and swine; therapy for swine dysentery, pneumonia, chicken necrotic enteritis, and respiratory disease | Lincosamides | Gram-positive organisms | Rare | Erythromycin and other macrolides and lincosamides, clindamycin | Approved in U.S. |
| Maduramycin | Poultry coccidiostat | Ionophores | Coccidia, Gram-positive organisms | No | Not demonstrated | Approved in EU and U.S. |
| Monensin | Bovine AGP; prevention/control of coccidiosis in bovines, poultry, and goats | Ionophores | Coccidia, Gram-positive organisms | No | Not demonstrated | Withdrawn from EU as bovine AGP but authorized as poultry coccidiostat |
| Neomycinb | AGP for swine and poultry; treatment/control of swine enteritis and pneumonia; control of mortality from E. coli in turkeys, bovines, swine, sheep, and goats; control of respiratory and other poultry diseases; aquaculture (bath) | Aminoglycosides | Gram-positive and -negative organisms | Yes | Gentamicin and other aminoglycosides | Approved in U.S. |
| Narasin | Poultry feed coccidiostat, prevention of necrotic enteritis in chickens, AGP for cattle | Ionophores | Coccidia, Gram-positive organisms | No | Not demonstrated | Approved in U.S. |
| Nourseothricin | Swine AGP | Streptothricins | Gram-negative organisms | No | None | Withdrawn from EU; never used in U.S. |
| Novobiocin | Treatment of staph infections, treatment and control of fowl cholera, treatment of bovine mastitis | Aminocoumarins | Gram-positive organisms | Yes | ||
| Olaquindox | Swine AGP, control of swine dysentery/enteritis | Quinoxalines | Gram-positive and -negative organisms | No | Other quinoxolines | Withdrawn due to worker toxicity in EU and Canada; available in U.S. |
| Oleandomycinb | Poultry and swine AGP | Macrolides | Gram-positive organisms | Yes | Erythromycin and other macrolides | |
| Ormetoprim | Poultry AGP, prevention of fowl cholera and other infections | Diaminopyrimidines | Broad | No | Trimethoprim | |
| Oxolinic acidb | Aquaculture (oral) | Quinolones | Broad | No | Other quinolones | Not approved in U.S. |
| Pristinamycin | AGP | Streptogramins | Gram-positive organisms | Yes | Other streptogramins (virginiamycin, quinupristin/ dalfopristin) | |
| Procaine penicillinb | AGP in poultry and swine | Beta-lactams | Gram-positive organisms | Yes | Other beta-lactams | Withdrawn in EU; available in U.S. |
| Roxarsone | AGP for poultry and swine, poultry coccidiostat, treatment of swine dysentery | Arsenicals | Coccidia | No | Other arsenicals | |
| Salinomycin | Swine AGP, prevention/control of swine dysentery and porcine intestinal adenomatosis, control of Clostridium perfringens in growers | Ionophores | Gram-positive organisms | No | Not demonstrated | Withdrawn from EU |
| Spiramycinb | Swine AGP, treatment of bovine mastitis | Macrolides | Gram-positive organisms | Yes | Erythromycin and other macrolides and lincosamides | AGP use withdrawn from EU in 1999; not approved in U.S. |
| Streptomycinb | Aquaculture (bath) | Aminoglycosides | Broad | Yes | All aminoglycosides | |
| Sulfonamides | Aquaculture (sulfamerazine [oral] and sulfadimethoxine [oral]), swine AGP (sulfamethazine), chicken AGP (sulfadimethoxine) | Sulfonamides | Broad | Yes | All sulfonamides | Sulfamerazine authorized for U.S. aquaculture, but not marketed |
| Tetracyclines (oxy- and chlor-)b | AGP for poultry, swine, and cattle; treatment and control of multiple livestock diseases; aquaculture (oral/bath/injection); control of fish and lobster disease | Tetracylines | Broad | Yes | All tetracyclines | Withdrawn from EU; authorized in U.S. |
| Tiamulin | Swine AGP; treatment of swine enteritis, dysentery, and pneumonia | Pleuromutilins | Gram-positive organisms, mycoplasmas, spirochetes | No | Tylosin, erythromycin, and other macrolides | Used for disease prevention and treatment in chickens outside the U.S. |
| Tylosinb | Swine AGP, therapeutic treatment of mastitis | Macrolides | Gram-positive organisms | No | Erythromycin and other macrolides and lincosamides | AGP use withdrawn from EU; available in U.S. |
| Virginiamycin | AGP for broilers | Streptogramins | Gram-positive organisms | Yes | Quinupristin/ dalfopristin and other streptogramins | Withdrawn from EU; available in U.S. |
Salmonella and the Swann Report
Alarmed by the rise in multidrug-resistant Salmonella in the 1960s, the United Kingdom's Swann Report of 1969 recognized the possibility that AGPs were contributing largely to the problem of drug-resistant infections (144). It concluded that growth promotion with antibiotics used for human therapy should be banned. The recommendation was implemented first in England and then in other European countries and Canada. The practice continued unchanged, however, in the United States and ultimately also continued in Europe, but with agents that were not used therapeutically in humans. Antibiotics such as bacitracin, avoparcin, bambermycins, virginiamycin, and tylosin gained in popularity as narrower-spectrum substitutes that had a smaller impact on the broad range of gut flora. Unforeseen, however, was the structural relationship between some of these agents and agents used clinically in humans (Table 1). This similarity meant that they shared a single bacterial target and that use of one agent could produce cross-resistance to the other.
Impacts of Nontherapeutic Use
Therapeutics applied properly for the treatment of individual animals tend to control the emergence and propagation of antimicrobial-resistant strains, in large part due to their relatively short-term application and relatively small numbers of animals treated. The resistant strains which may appear are generally diluted out by the return of normal, drug-susceptible commensal competitors (110). In contrast, any extended antibiotic applications, such as the use of AGPs, which are supplied for continuous, low-dose application, select for increasing resistance to the agent. Their use in large numbers of animals, as in concentrated animal feeding operations (CAFOs), augments the “selection density” of the antibiotic, namely, the number (density) of animals producing resistant bacteria. An ecological imbalance results—one that favors emergence and propagation of large numbers of resistance genes (113). The selection is not linked merely to the total amount of antibiotic used in a particular environment but to how many individuals are consuming the drug. Each animal feeding on an antibiotic becomes a “factory” for the production and subsequent dispersion of antibiotic-resistant bacteria. NTA uses are also clearly linked to the propagation of multidrug resistance (MDR), including resistance against drugs that were never used on the farm (10, 52, 59, 60, 92, 107, 111, 132, 141, 153, 154, 164). The chronic use of a single antibiotic selects for resistance to multiple structurally unrelated antibiotics via linkage of genes on plasmids and transposons (111, 143).
Studies on the impact of NTA use on resistance in land food animals have focused primarily on three bacterial genera—Enterococcus, Escherichia, and Campylobacter—and, to a lesser extent, on Salmonella and Clostridium. All of the above may be members of the normal gut flora (commensals) of food animals but possess the potential to become serious human pathogens. The prospective farm study by Levy in 1975 (111) and studies of others in the following decades clearly demonstrated the selective nature of low-dose, nontherapeutic AGPs on both the pathogenic and commensal flora of food animals such as poultry, swine, and cattle (8, 16, 18, 90, 98, 146, 149). Likewise, in the past decade, studies have demonstrated the selective nature of mass treatment with antimicrobials in aquaculture (36, 62, 84). In the latter, studies have focused on Aeromonas pathogens of both fish and humans and the subsequent high-frequency transfer of their resistance plasmids to E. coli and Salmonella (36).
Aarestrup and Carstensen found that resistance derived from use of one NTA (tylosin) was not confined to swine gut bacteria only but could cross species and appear in staphylococci isolated from the skin. While the conversion of gut enterococci to erythromycin (a related human therapeutic) resistance occurred rapidly (within 1 week) the skin-derived resistant organism Staphylococcus hyicus appeared more gradually, escalating to a 5-fold increase over 20 days (5).
The finding of bacterial cross-resistance between NTAs used in food animals and human drugs was aptly demonstrated with avoparcin (an AGP) and its close relative vancomycin (an important human therapeutic) when vancomycin-resistant enterococci (VRE) emerged as a serious human pathogen. A connecting link between resistance in animals and humans was revealed when Bates et al. found avoparcin- and vancomycin-coresistant enterococci in pigs and small animals from two separate farms. Ribotyping methods showed that some of the patterns from farms and sewage exactly matched those of Enterococcus spp. from the hospital (24). The structures of the two drugs are similar: they are both members of the glycopeptide family (24).
Since that time, numerous studies have examined the impacts of newer NTAs on the floras of animals. The use of tylosin and virginiamycin in Norwegian swine and poultry led to high prevalences of resistance to both these agents in Enterococcus faecium (75% to 82% for tylosin and 49 to 70% for virginiamycin) (1). Avilamycin resistance, while significantly associated with avilamycin use, has been observed on both exposed and unexposed farms and was significantly higher in isolates from poultry than in those from swine, despite its use in both these species (4). These findings suggest that other selective agents may be present in the environment or that substances related to avilamycin were not recognized. As described above, not only the drug choice and amount but also the number of animals treated can affect the consequence of its use.
Other findings suggest that complex ecologic and genetic factors may play a role in perpetuating resistance (63). Resistance (particularly to tetracycline, erythromycin, and ampicillin) has been found inherently in some antibiotic-free animals, (10, 45, 93, 130), suggesting that its emergence is related to other factors, such as diet, animal age, specific farm type, cohort variables, and environmental pressures (26). While Alexander et al. found MDR (tetracycline plus ampicillin resistance) in bacteria in control animals, the strains that emerged after AGP use were not related to these (10). In addition, resistance to tetracycline was higher for a grain-based diet than a silage-based one. Costa et al. found non-AGP-related resistances in enterococci, most likely derived from previous flocks, i.e., the farm environment and the feed source appeared to be responsible for the emergence of the unrelated resistances (45). Khachatryan et al. found an MDR phenotype (streptomycin, sulfonamide, and tetracycline [SSuT] resistance phenotype) propagated by oxytetracycline in a feed supplement, but upon removal of the drug, the phenotype appeared to be maintained by some unknown component of the unmedicated feed supplement, possibly one that selects for another gene that is linked to a plasmid bearing the SSuT resistance phenotype (100). The persistence may also relate to the stability of the plasmid in its host and the fact that expression of tetracycline resistance is normally silent until it is induced by tetracycline. Thus, the energy demands exerted on the host by tetracycline resistance are lower. One can conclude that removal of the antibiotic may not lead to rapid loss of the resistant strain or plasmid.
EFFECTS OF BANNING GROWTH PROMOTANTS IN ANIMAL FEEDS IN EUROPE
One of the first bans on AGP use was that imposed on tetracycline by the European Common Market in the mid-1970s (39). Prior to institution of the ban in the Netherlands (1961 to 1974), Van Leeuwen et al. had tracked a rise in tetracycline-resistant Salmonella spp. Following the ban, however, they observed a decline in tetracycline resistance in both swine and humans (150).
More than 10 years have passed since the final 1999 European Union ban, during which a plethora of studies from multiple European countries, Canada, and Taiwan have examined antibiotic use and resistance trends subsequent to the removal of key AGP drugs, especially avoparcin, and the consequences on vancomycin resistance in Enterococcus (7, 15, 17, 21, 29, 30, 76, 85, 97, 102, 107, 121, 148, 150, 156a). Its structural relationship to and cross-resistance with avoparcin render vancomycin a drug of prime interest for determining the impact of avoparcin in triggering and promoting resistance in human infection.
Avoparcin
In many European countries, the use of avoparcin as a feed additive led to frequent isolation of VRE from farm animals and healthy ambulatory people (3, 18, 102). Since the emergence of the enterococcus as a major MDR pathogen, vancomycin has evolved as a key therapy, often as the drug of last resort. Following the 1995 ban on avoparcin, several investigators reported a decline in animal VRE. In Denmark, frequencies peaked at 73 to 80% and fell to 5 to 6% (7, 18) in poultry. In Italy, VRE prevalence in poultry carcasses and cuts decreased from 14.6% to 8% within 18 months of the 1997 ban (121), and in Hungary, a 4-year study showed not only a decline in prevalence of VRE among slaughtered cattle, swine, and poultry after removal of avoparcin but also a decrease in vancomycin MICs (97). In surveillance studies both before and after the German ban in 1996, Klare et al. showed a high frequency of VRE in 1994, followed by a very low frequency of just 25% of poultry food products in 1999 (102). Similar declines were reported in broiler farms following a ban on avoparcin in Taiwan in 2000 (107).
A dramatic reduction in human carriage of VRE also followed the ban on avoparcin. Parallel surveillances of the gut floras of healthy ambulatory people showed that VRE colonization in Germany declined from 13% in 1994 to 4% in 1998 (102), and in Belgium, it declined from 5.7% in 1996 to ∼0.7% in 2001 (68).
Virginiamycin and Other Antibiotics
Increased virginiamycin use in Danish broilers during the mid-1990s correlated with a rise in resistant E. faecium prevalence, from 27% to ∼70% (7). Following the ban, resistance declined to 34% in 2000. Likewise, in Denmark, the 1998 ban on the use of tylosin in swine resulted in a decline in erythromycin (a structurally related macrolide) resistance, from 66% to 30% (49). Avilamycin use in 1995 and 1996 increased resistance in broiler E. faecium strains, from 64% to 77%, while declining applications after 1996 lowered the prevalence to 5% in 2000 (7).
Some of these studies revealed a genetic linkage between bacterial macrolide and glycopeptide resistances in swine, such that neither resistance declined in prevalence until both avoparcin (a glycopeptide) and tylosin (a macrolide) use was limited. With a reduction in tylosin use, the prevalence of glycopeptide-resistant enterococci fell to 6% and macrolide resistance fell from nearly 90% to 47% in E. faecium and to 28% in Enterococcus faecalis (7). Notably, the first report of transfer of vancomycin resistance from Enterococcus to Staphylococcus aureus was demonstrated in laboratory mice because of its linkage to macrolide resistance on the same plasmid (119).
One concern voiced following the banning of NTAs was that the incidence of disease in animals would rise and result in a parallel increase in therapeutic use. This has become the subject of some debate. Some countries encountered rises in necrotic enteritis in chickens and colitis in swine soon after the institution of AGP bans (33, 159). In Norway, an abrupt increase in necrotizing enteritis (NE) in poultry broilers was reported following the removal of avoparcin, with a coincident rise in antibiotic therapy. When the ionophore feed additive narasin was approved, NE declined once again (77). It was concluded that the ban on avoparcin consumption produced a negligible effect on the need for antibiotic therapy (76). Likewise, in Switzerland, Arnold et al. reported a postban increase in overall antibiotic quantities used in swine husbandry but observed a stable therapy intensity (prescribed daily dose) (15). By 2003, total animal use of antibiotics in Denmark, Norway, and Sweden had declined by 36%, 45%, and 69%, respectively (76). The most thorough postban analysis of this phenomenon comes from Denmark. In a careful review of swine disease emergence, animal production, and antibiotic use patterns over the years 1992 to 2008, Aarestrup et al. reported no overall deleterious effects from the ban on finishers and weaners in the years 1998 and 2000, respectively. Despite an increase in total therapeutic antibiotic consumption immediately following the ban, no lasting negative effects were detected on mortality rate, average daily weight gain, or animal production (6). Moreover, even if therapeutic use increased, the numbers of animals treated would be reduced compared to those with growth promotion use, so selection density would be decreased (113).
In summary, the in-depth, retrospective analyses in Denmark shed a different perspective on postban concerns over increased therapeutic use. Over time, it appears that the negative after-effects of the ban have waned. As farmers modified their animal husbandry practices to accommodate the loss of banned NTAs, these disease outbreaks became less prominent. Improved immunity and reduced infection rates led to fewer demands for therapeutic antibiotics.
Interestingly, recent studies have shown that the original beneficial aspects observed with AGP use (i.e., weight gain and feed efficiency) appear to have diminished, although the results are mixed and depend upon the kind of animals and type of antibiotic involved. Diarra et al. found no effect on body weight or feed intake in poultry from five different AGPs, and feed efficiency was improved with penicillin only (52). In contrast, Dumonceaux et al. reported a significantly increased body weight (10%) and a 7% increase in feed efficiency with the AGP virginiamycin, but only for the first 15 days (55a). In short, improved farming practices and breeding programs, which may include reduced animal density, better hygiene, targeted therapy, and the use of enzymes, prebiotics, probiotics, and vaccines, appear to have at least partially replaced the beneficial aspects of antibiotic growth promoters (27, 158, 160).
EVIDENCE FOR ANIMAL-TO-HUMAN SPREAD OF ANTIBIOTIC RESISTANCE
Any use of antibiotics will select for drug-resistant bacteria. Among the various uses for antibiotics, low-dose, prolonged courses of antibiotics among food animals create ideal selective pressures for the propagation of resistant strains. Spread of resistance may occur by direct contact or indirectly, through food, water, and animal waste application to farm fields. It can be augmented greatly by the horizontal transfer of genetic elements such as plasmids via bacterial mating (conjugation). We summarize here the evidence for animal-to-human transfer of resistant bacteria on farms using antibiotics for treatment and/or nontherapeutic use.
Resistance Acquisition through Direct Contact with Animals
Farm and slaughterhouse workers, veterinarians, and those in close contact with farm workers are directly at risk of being colonized or infected with resistant bacteria through close contact with colonized or infected animals (Table 2). Although this limited transmission does not initially appear to pose a population-level health threat, occupational workers and their families provide a conduit for the entry of resistance genes into the community and hospital environments, where further spread into pathogens is possible (118, 155).
Table 2.
Key evidence for transfer of antibiotic resistance from animals to humans
| Transfer type | Species tracked | Animal host(s) | Recipient host(s) | Resistance transferred | Evidence | Reference |
|---|---|---|---|---|---|---|
| Human colonization via direct or indirect animal contact | E. coli | U.S. chickens | Animal caretakers, farm family | Tetracycline | Following introduction of tetracycline on a farm, resistant E. coli strains with transferable plasmids were found in caretakers' gut floras, with subsequent spread to the farm family | 111 |
| S. aureus, Streptococcus spp., E. coli and other enterobacteria | French swine | Swine farmers | Erythromycin, penicillins, nalidixic acid, chloramphenicol, tetracycline, streptomycin, cotrimoxazole | Phenotypic antibiotic resistance was significantly higher in the commensal floras (nasal, pharyngeal, and fecal) of swine farmers than in those of nonfarmers | 16 | |
| E. coli | U.S. chickens | Poultry workers | Gentamicin | Increase in phenotypic gentamicin resistance in workers through direct contact with chickens receiving gentamicin prophylactically | 126 | |
| E. coli | Chinese swine and chickens | Farm workers | Apramycin (not used in human medicine) | Detection of aac(3)-IV apramycin resistance gene in humans, with 99.3% homology to that in animal strains | 164 | |
| MRSA ST398 | Dutch veal calves | Veal farmers | MDR | Human nasal carriage of the mecA gene was strongly associated with (i) greater intensity of animal contact and (ii) the number of MRSA-positive animals; animal carriage was related to animal antibiotic treatment | 78 | |
| Human infection via direct or indirect animal contact | Salmonella Newport | Beef cattle (ground beef) receiving chlortetracycline AGP | Salmonella-infected patients with diarrhea | Ampicillin, carbenicillin, tetracycline | Direct genetic tracking of resistance plasmid from hamburger meat to infected patients | 87 |
| E. coli | German swine (ill) | Swine farmers, family members, community members, UTI patients | Streptothricin | Identification of transferable resistance plasmids found only in human gut and UTI bacteria when nourseothricin was used as swine AGP | 90 | |
| E. coli, Salmonella enterica (serovar Typhimurium) | Belgian cattle (ill) | Hospital inpatients | Apramycin, gentamicin | Plasmid-based transfer of aac(3)-IV gene bearing resistance to a drug used only in animals (apramycin) | 42 | |
| Enterococcus faecium | Danish swine and chickens | Hospital patients with diarrhea | Vancomycin | Clonal spread of E. faecium and horizontal transmission of the vanA gene cluster (Tn1546) found between animals and humans | 80 | |
| E. coli | Spanish chickens (slaughtered) | Bacteremic hospital patients | Ciprofloxacin | Multiple molecular and epidemiological typing modalities demonstrated avian source of resistant E. coli | 95 |
The majority of studies examining the transmission of antibiotic-resistant bacteria from animals to farm workers document the prevalence of resistance among farmers and their contacts or among farmers before and after the introduction of antibiotics at their workplace. Direct spread of bacteria from animals to people was first reported by Levy et al., who found the same tetracycline-resistant E. coli strains in the gut flora of chicken caretakers as in the chickens receiving tetracycline-laced feed (112). The observation extended to the farm family as well and showed an increased frequency of tetracycline-resistant and multidrug-resistant E. coli after several months of use of AGP-laden feed. Studies such as this (which examined a variety of antibiotic classes and assorted pathogens) have consistently shown a higher prevalence of resistant gut bacteria among farm workers than in the general public or among workers on farms not using antibiotics (16, 90, 98, 149).
While gentamicin is not approved for growth promotion in the United States, it remains the most commonly used antibiotic in broiler production, being employed for prevention of early poultry mortality (115). A revelatory 2007 study found that the risk for carrying gentamicin-resistant E. coli was 32 times higher in poultry workers than in other members of the community: half of all poultry workers were colonized with gentamicin-resistant E. coli, while just 3% of nonpoultry workers were colonized. Moreover, the occupationally exposed population was at significantly greater risk for carriage of multidrug-resistant bacteria (126).
New gene-based methods of analysis provide even stronger evidence for the animal origin of bacteria that colonize or infect humans. Homologous relationships between bacterial resistance genes in humans and farm animals have been identified most commonly for food-borne pathogens such as Escherichia coli and Salmonella (see below) but have also been recorded for various species of Enterococcus and for methicillin-resistant Staphylococcus aureus (MRSA). Zhang and colleagues found E. coli strains resistant to apramycin (an antibiotic used in agriculture but not in human medicine) in a study of Chinese farm workers. All farms in the study that used apramycin as an AGP had workers that carried apramycin resistance genes. The same resistance gene, aac(3)-IV, was present in each swine, poultry, and human isolate, with some resistance profiles also matching across species (164). A group of French scientists found the same resistance gene [aac(3)-IV] in cow, pig, and human E. coli strains that bore resistance to apramycin and gentamicin (42). In another study, similar resistance patterns and genes were detected in E. faecalis and E. faecium strains from humans, broilers, and swine in Denmark (2). Lee sampled MRSA isolates from cattle, pigs, chickens, and people in Korea and found that 6 of the 15 animal isolates containing mecA (the gene responsible for methicillin resistance in S. aureus) were identical to human isolates (108).
Antibiotic Resistance Transmission through the Food Chain
Consumers may be exposed to resistant bacteria via contact with or consumption of animal products—a far-reaching and more complex route of transmission. There is undeniable evidence that foods from many different animal sources and in all stages of processing contain abundant quantities of resistant bacteria and their resistance genes. The rise of antibiotic-resistant bacteria among farm animals and consumer meat and fish products has been well documented (36, 108, 122, 162). Demonstrating whether such reservoirs of resistance pose a risk to humans has been more challenging as a consequence of the complex transmission routes between farms and consumers and the frequent transfer of resistance genes among host bacteria. Such correlations are becoming more compelling with the advent of molecular techniques which can demonstrate the same gene (or plasmid) in animal or human strains, even if the isolates are of different species.
For example, Alexander et al. showed that drug-resistant Escherichia coli was present on beef carcasses after evisceration and after 24 h in the chiller and in ground beef stored for 1 to 8 days (9). Others isolated ciprofloxacin-resistant Campylobacter spp. from 10% to 14% of consumer chicken products (79, 137). MRSA has been reported to be present in 12% of beef, veal, lamb, mutton, pork, turkey, fowl, and game samples purchased in the consumer market in the Netherlands (50), as well as in cattle dairy products in Italy (120). Likewise, extensive antibiotic resistance has been reported for bacteria, including human pathogens, from farmed fish and market shrimp (56, 84, 140).
Some of the antibiotic resistance genes identified in food bacteria have also been identified in humans, providing indirect evidence for transfer by food handling and/or consumption. In 2001, Sorensen et al. confirmed the risk of consuming meat products colonized with resistant bacteria, showing that glycopeptide-resistant Enterococcus faecium of animal origin ingested via chicken or pork lasted in human stool for up to 14 days after ingestion (139). Donabedian et al. found overlap in the pulsed-field gel electrophoresis (PFGE) patterns of gentamicin-resistant isolates from humans and pork meat as well as in those of isolates from humans and grocery chicken (55). They identified that when a gene conferring antibiotic resistance was present in food animals, the same gene was present in retail food products from the same species. Most resistant enterococci possessed the same resistance gene, aac(6′)-Ie-aph(2″)-Ia (55).
Emergence of Resistance in Human Infections
There is likewise powerful evidence that human consumption of food carrying antibiotic-resistant bacteria has resulted, either directly or indirectly, in acquisition of antibiotic-resistant infections (Table 2). In 1985, scientists in Arizona traced an outbreak of multidrug-resistant Salmonella enterica serovar Typhimurium, which included the death of a 72-year-old woman, to consumption of raw milk. Isolates from most patients were identical to the milk isolates, and plasmid analysis showed that all harbored the same resistance plasmid (145). A 1998 S. Typhimurium outbreak in Denmark was caused by strains with nalidixic acid resistance and reduced fluoroquinolone susceptibility. PFGE revealed that a unique resistance pattern was common to Salmonella strains from all patients, two sampled pork isolates, the swine herds of origin, and the slaughterhouse (118).
Samples from gentamicin-resistant urinary tract infections (UTIs) and fecal E. coli isolates from humans and food animal sources in China showed that 84.1% of human samples and 75.5% of animal samples contained the aaaC2 gene for gentamicin resistance (86). Johnson et al. used PFGE and random amplified polymorphic DNA (RAPD) profiles of fluoroquinolone-resistant E. coli strains in human blood and fecal samples and in slaughtered chickens to determine that the two were virtually identical to resistant isolates from geographically linked chickens. Drug-susceptible human E. coli strains, however, were genetically distinct from poultry bacteria, suggesting that the ciprofloxacin-resistant E. coli strains in humans were imported from poultry rather than originating from susceptible human E. coli (94, 95).
Other reports demonstrate a broader linkage of resistance genes through the farm-to-fork food chain. A resistance-specifying blaCMY gene was found in all resistant isolates of Salmonella enterica serotype Newport originating from humans, swine, cattle, and poultry. The host plasmid, which conferred resistance to nine or more antimicrobials, was capable of transmission via conjugation to E. coli as well (165). An observed homology between CMY-2 genes in cephalosporin-resistant E. coli and Salmonella suggested that plasmids conferring resistance had moved between the two bacterial species. The authors found higher rates of CMY-2 in strains from animals than in those from humans, supporting an animal origin for the human pathogen (161). A 2000 study found matching PFGE profiles among vancomycin-resistant Enterococcus faecium isolates from hospitalized humans, chickens, and pigs in Denmark. Molecular epidemiology studies have also linked tetracycline resistance genes from Aeromonas pathogens in a hospital effluent to Aeromonas strains from a fish farm (127). These results support the clonal spread of resistant isolates among different populations (80).
Chronologic studies of the emergence of resistance across the food chain also strongly imply that reservoirs of resistance among animals may lead to increased resistance in consumers of animal food products. Bertrand et al. chronicled the appearance of the extended-spectrum beta-lactamase (ESBL) gene CTX-M-2 in Salmonella enterica in Belgium. This resistance element was identified first in poultry flocks and then in poultry meat and, finally, human isolates (28). A recent Canadian study also noted a strong correlation between ceftiofur-resistant bacteria (the pathogen Salmonella enterica serovar Heidelberg and the commensal E. coli) from retail chicken and human infections across Canada. The temporary withdrawal of ceftiofur injection from eggs and chicks dramatically reduced resistance in the chicken strains and the human Salmonella isolates, but the trend reversed when the antibiotic use was subsequently resumed (57).
In three countries (United States, Spain, and the Netherlands), a close temporal relationship has been documented between the introduction of fluoroquinolone (sarafloxacin and enrofloxacin) therapy in poultry and the emergence of fluoroquinolone-resistant Campylobacter in human infections. An 8- to 16-fold increase in resistance frequency was observed—from 0 to 3% prior to introduction to ∼10% in the United States and the Netherlands and to ∼50% in Spain—within 1 to 3 years of the licensure (61, 128, 137). In the Netherlands, this frequency closely paralleled an increase in resistant isolates from retail poultry products (61), while the U.S. study used molecular subtyping to demonstrate an association between the clinical human isolates and those from retail chicken products (137).
It is now theorized, from molecular and epidemiological tracking, that the resistance determinants found in salmonella outbreaks (strain DT104) in humans and animals in Europe and the United States likely originated in aquaculture farms of the Far East. The transmissible genetic element contains the florfenicol gene (floR) and the tetracycline class G gene, both of which were traced to Vibrio fish pathogens (Vibrio damsel and Vibrio anguillarum, respectively). Both drugs are used extensively in aquaculture (36).
In the above examples, the link to nontherapeutic antibiotic use in the farm animals is still circumstantial and largely implied, often because the authors do not report any statistics on farm use of antibiotics. Interpreting these studies is also difficult because of the widespread resistance to some drugs in bacteria of both animals and humans and the ubiquitous nature of resistance genes. Moreover, the same farmer may use antibiotics for both therapeutic and nontherapeutic purposes.
The complexities of the modern food chain make it challenging to perform controlled studies that provide unequivocal evidence for a direct link between antibiotic use in animals and the emergence of antibiotic resistance in food-borne bacteria associated with human disease. While this concrete evidence is limited, a small number of studies have been able to link antibiotic-resistant infection in people with bacteria from antibiotic-treated animals. While not necessarily involving NTAs, these studies substantiate the considerable ease with which bacteria in animals move to people. For example, a multidrug-resistant Salmonella enterica strain in a 12-year-old Nebraska boy was traced to his father's calves, which had recently been treated for diarrhea. Isolates from the child and one of the cows were determined to be the same strain of CMY-2-mediated ceftriaxone-resistant S. enterica (69). It is now believed that the 1992 multiresistant Vibrio cholerae epidemic in Latin America was linked to the acquisition of antibiotic-resistant bacteria arising from heavy antibiotic use in the shrimp industry of Ecuador (13, 156).
By comparing the plasmid profiles of MDR Salmonella Newport isolates from human and animal sources, Holmberg et al. provided powerful evidence that salmonella infections in 18 persons from 4 Midwestern states were linked directly to the consumption of hamburger meat from cattle fed subtherapeutic chlortetracycline. A plasmid which bore tetracycline and ampicillin resistance genes was present in the organisms causing serious illness in those persons who ate the hamburger meat and who were also consuming penicillin derivatives for other reasons (87).
One of the most compelling studies to date is still Hummel's tracking of the spread of nourseothricin resistance, reported in 1986. In Germany, nourseothricin (a streptogramin antibiotic) was used solely for growth promotion in swine. Resistance to it was rarely found and was never plasmid mediated. Following 2 years of its use as a growth promotant, however, resistance specified by plasmids appeared in E. coli, not only from the treated pigs (33%) but also in manure, river water, food, and the gut floras of farm employees (18%), their family members (17%), and healthy outpatients (16%) and, importantly, in 1% of urinary tract infections (90). Ultimately, the resistance determinant was detected in Salmonella and Shigella strains isolated from human diarrhea cases (146).
The movement of antibiotic resistance genes and bacteria from food animals and fish to people—both directly and indirectly—is increasingly reported. While nontherapeutic use of antibiotics is not directly implicated in some of these studies, there is concern that pervasive use of antimicrobials in farming and widespread antimicrobial contamination of the environment in general may be indirectly responsible. For instance, within the past 5 years, MRSA and MDR Staphylococcus aureus have been reported in 25 to 50% of swine and veal calves in Europe, Canada, and the United States (51, 78, 101, 114). Graveland et al. noted that this frequency was higher in veal calves fed antibiotics (78). Studies also show that colonization among farmers correlates significantly with MRSA colonization among their livestock (78, 101, 114, 138). In the Netherlands, colonization of swine farmers was found to be more than 760 times greater than that of patients admitted to Dutch hospitals (155). In a study of nasal swabs from veal and veal calf growers, family members, and employees at 102 veal calf farms in the Netherlands, Graveland et al. found that human MRSA sequence type ST398 carriage among the farmers was strongly associated with the degree of animal contact and the frequency of MRSA-colonized animals on the farm. When <20% of calves were carriers, the estimated prevalence in humans was ∼1%—similar to that in the general public. With >20% carriage in calves, the prevalence in humans was >10% (78).
Recently, MRSA ST398 has appeared in the community. A Dutch woman without any known risk factors was admitted to a hospital with endocarditis caused by MRSA ST398, suggesting a community reservoir which passed on to people (58). Voss et al. demonstrated animal-to-human and human-to-human transmission of MRSA between a pig and pig farmer, among the farmer's family members, and between a nurse and a patient in the hospital. All isolates had identical random amplified polymorphic DNA profiles (155). Examples of similar MRSA strains among animals and people are mounting (82, 108, 147, 151, 152, 163).
ADDRESSING KNOWLEDGE GAPS: RESERVOIRS OF ANTIBIOTIC RESISTANCE
Historically, considerable attention has been focused on a very small minority of bacterial species that actually cause disease. However, a vast “sea” of seemingly innocuous commensal and environmental bacteria continuously and promiscuously exchange genes, totally unnoticed (116). A staggeringly diverse group of species maintain a large capacity for carrying and mobilizing resistance genes. These bacteria constitute a largely ignored “reservoir” of resistance genes and provide multiple complex pathways by which resistance genes propagated in animals can directly, or more likely indirectly, make their way over time into human pathogens via food, water, and sludge and manure applied as fertilizer. Horizontal (or lateral) gene transfer studies have identified conjugal mating as the most common means of genetic exchange, and there appear to be few barriers that prevent this gene sharing across a multitude of dissimilar genera (104).
While colonic bacteria have received much focused study, water environments such as aquaculture, sludge, freshwater, and wastewaters are prime sites for gene exchange but have been examined minimally for their roles as “mixing pots” and transporters of genes from bacteria of antibiotic-fed animals to humans (116). Aside from the already described impacts of NTA use on bacterial resistance, food animal use of NTAs has broad and far-reaching impacts on these environmental bacteria. It is estimated that 75% to 90% of antibiotics used in food animals are excreted, largely unmetabolized, into the environment (43, 105). Antibiotics or resistant bacteria have been detected in farm dust (81), the air currents inside and emanating from swine feeding operations (41, 72, 129), the groundwater associated with feeding operations (31, 37), and the food crops of soils treated with antibiotic-containing manure (54). This leaching into the environment effectively exposes countless environmental organisms to minute quantities of antibiotic—enough to select bacteria with resistance mutations to promote the emergence and transfer of antibiotic resistance genes among diverse bacterial types (104). The potentially huge impact of all these residual antibiotics on the environmental bacteria that are directly or indirectly in contact with humans has scarcely been examined.
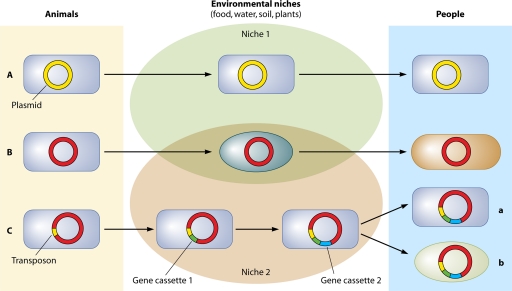
The multiple pathways and intricacies of gene exchange have so far thwarted attempts to qualitatively or quantitatively track the movement of these genes in vivo, and thus we are left with minimal direct evidence for linking resistance in animals to that in humans. With extensive gene movement between disparate hosts, it is less likely that the same bacterial hosts will be found in animals and humans and more probable that only the resistance genes themselves will be identifiable in the final pathogens that infect humans. Even these may be altered in their journey through multiple intermediate hosts (161) (Fig. 1). Mounting evidence exists in reports of complex gene “cassettes” which accumulate resistance genes and express multidrug resistance (106, 125).
Fig. 1.
Several scenarios may present themselves in the genetic transport that occurs as bacteria migrate from animal to human environments. (A) The same host and its indigenous genes in animals are transported unchanged to humans, with a resulting 100% match of the bacterial strain. (B) The genetic structure passes through one or more different hosts, ending in a new host (humans), with a resulting 100% match of DNA. (C) The host and its plasmid-borne genes pass through the environment, picking up gene cassettes en route, with a resulting 100% match for the host only (a) or a low-% match for DNA only (b). In both examples, the plasmid core remains the same.
A few investigators have undertaken the challenging task of developing mathematical models in order to predict the impacts of NTAs on human disease (12, 19, 20, 46, 91, 99, 134, 135). Models can be very useful in attempting to define the types of diverse data sets that are seen in this field. Some explore the entire “farm-to-fork” transmission process, while others tackle only portions of this extremely complex chain or adopt a novel backwards approach which looks first at human infections and then calculates the fraction that are potentially caused by NTA use in animals. Most models are deliberately simplified and admittedly omit many aspects of transmission and persistence. Moreover, current models are frequently based on multiple assumptions and have been challenged on the basis of certain shortcomings, such as limitation to single pathogens only, the determination of lethality while ignoring morbidity, and dependence on estimates of probabilities (19). Chief among these, however, is the lack of a complete understanding of the contribution made by commensals, which may play an important role in augmenting the link between animals and humans. Some models are driven by findings of dissimilar strains in animals and humans and therefore arrive at very low probabilities for a causal link between the two (47). A finding of dissimilar strains, however, overlooks two possibilities. First, it does not exclude the existence of small subpopulations of homologous strains that have gone undetected within the gut floras of animals. These may have been amplified temporarily by antibiotic selection and transferred their mobile genetic elements in multiple complex pathways. Subsequently, they may have declined to nondetectable levels or merely been outcompeted by other variants. Second, it overlooks dissimilarities that evolve as genes and their hosts migrate in very complex ways through the environment. Figure 1 illustrates the difficulties in tracking a resistance gene, since these genes are frequently captured in bacteria of different species or strains which no longer resemble the original host. Over time, even the genes themselves may undergo mutations or become entrapped in gene cassettes that alter their genetic landscape. State-of-the-art technology and thoughtful investigation are often necessary to identify and track the actual strains that link animals and humans. These are facets of modeling that have yet to be explored, and obtaining direct evidence for the origins of specific genes can be highly challenging.
In general, the weaknesses of present models lie in their simplicity and the lack of crucial knowledge of microbial loads at each stage of the “farm-to-fork” transmission chain. Many of the available studies that examine links between animals and humans suffer from a failure to examine the antibiotic use practices for the farm animals they investigate. More powerful evidence could have accumulated that would aid in modeling efforts if data on the quantities and uses of farm antibiotics had been reported. These oversights are often due to the lack of registries that record and report the utilization of antibiotics on food animal farms. It is widely advocated that surveillance studies of resistance frequencies at all levels of the transmission chain would aid greatly in reducing our knowledge deficits and would help to inform risk management deliberations (23, 34). A number of localized and international surveillance systems exist for the tracking of human pathogens. In the United States, the National Antimicrobial Resistance Monitoring System (NARMS) has become instrumental in the monitoring of resistance trends in pathogens found in food animals, retail meats, and humans (73). However, at the level of commensals, resistance monitoring is still in its infancy. The Reservoirs of Antibiotic Resistance (ROAR) database (www.roarproject.org) is a fledgling endeavor to promote the accumulation of data that specifically focus on commensal and environmental strains as reservoirs of antibiotic resistance genes. With advances in detection at the genetic level, the potential for tracking the emergence and spread of horizontally transmissible genes is improving rapidly. By capturing geographic, phenotypic, and genotypic data from global isolates from animal, water, plant, and soil sources, the ROAR project documents the abundance, diversity, and distribution of resistance genes and utilizes commensals as “barometers” for the emergence of resistance in human pathogens.
CONCLUSIONS
Data gaps continue to fuel the debate over the use of NTAs in food animals, particularly regarding the contribution and quantitation of commensal reservoirs of resistance to resistance in human disease. Nonetheless, it has been argued reasonably that such deficits in surveillance or indisputable demonstrations of animal-human linkage should not hinder the implementation of a ban on the use of nontherapeutic antibiotics (23). Food animals produce an immense reservoir of resistance genes that can be regulated effectively and thus help to limit the negative impacts propagated by this one source. In the mathematical model of Smith et al., which specifically evaluates opportunistic infections by members of the commensal flora, such as enterococci, it was concluded that restricting antibiotic use in animals is most effective when antibiotic-resistant bacteria remain rare. They suggest that the timing of regulation is critical and that the optimum time for regulating animal antibiotic use is before the resistance problem arises in human medicine (134).
A ban on nontherapeutic antibiotic use not only would help to limit additional damage but also would open up an opportunity for better preservation of future antimicrobials in an era when their efficacy is gravely compromised and few new ones are in the pipeline. Although the topic has been debated for several decades without definitive action, the FDA has recently made some strides in this direction. Officially, the organization now supports the conclusion that the use of medically important antimicrobials for nontherapeutic use in food animal production does not protect and promote public health (131). Although not binding, a guidance document was released in 2010 that recommended phasing in measures that would limit use of these drugs in animals and ultimately help to reduce the selection pressures that generate antimicrobial resistance (66).
The Danish experience demonstrated that any negative disease effects resulting from the ban of NTAs were short-lived and that altering animal husbandry practices could counter expected increases in disease frequency (6). For aquaculture, also, it has been demonstrated that alternative processes in industry management can be instituted that will reduce antibiotic use without detrimental financial effects (141). Still, it has been argued by some in animal husbandry that the different situation in the United States will result in increased morbidity and mortality, projected to cost $1 billion or more over 10 years. Again, however, the Danish postban evaluation found that costs of production increased by just 1% for swine and were largely negligible for poultry production due to the money saved on antibiotics themselves. Models also showed that Danish swine production decreased by just 1.4% (1.7% for exports), and poultry production actually increased, by 0.4% (0.5% for exports) (158). Such calculations still fail to consider the negative externalities that are added by the burden of antibiotic resistance and the antibiotic residue pollution generated by concentrated animal feeding operations.
Opponents of restriction of NTA use argue that a comprehensive risk assessment is lacking, but such an analysis is impossible without the kind of data that would come out of surveillance systems. Although surveillance systems have been advocated repeatedly (23, 70), such systems are sparse and extremely limited in their scope.
In 2002, working with the accumulated evidence and an assessment of knowledge deficits in the area of animal antibiotic use, the APUA developed a set of guidelines that are still viable today and can be used to guide both policy and research agendas. In summary, APUA recommended that antimicrobials should be used only in the presence of disease, and only when prescribed by a veterinarian; that quantitative data on antimicrobial use in agriculture should be made available; that the ecology of antimicrobial resistance in agriculture should be a research priority and should be considered by regulatory agencies in assessing associated human health risks; and that efforts should be invested in improving and expanding surveillance programs for antimicrobial resistance. Suitable alternatives to NTAs can be implemented, such as vaccination, alterations in herd management, and other changes, such as targeted use of antimicrobials with a more limited dosage and duration so as not to select for resistance to critical human therapeutics (23).
There is no doubt that human misuse and overuse of antibiotics are large contributors to resistance, particularly in relation to bacteria associated with human infection. Interventions in medical settings and the community are clearly needed to preserve the efficacy of antibiotics. Efforts in this area are being pursued by the Centers for Disease Control and Prevention, the Alliance for the Prudent Use of Antibiotics, the American Medical Association, the American Academy of Pediatrics, the Infectious Diseases Society of America, and other professional groups. Still, given the large quantity of antibiotics used in food animals for nontherapeutic reasons, some measure of control over a large segment of antibiotic use and misuse can be gained by establishing guidelines for animals that permit therapeutic use only and by then tracking use and health outcomes.
The current science provides overwhelming evidence that antibiotic use is a powerful selector of resistance that can appear not only at the point of origin but also nearly everywhere else (104). The latter phenomenon occurs because of the enormous ramifications of horizontal gene transfer. A mounting body of evidence shows that antimicrobial use in animals, including the nontherapeutic use of antimicrobials, leads to the propagation and shedding of substantial amounts of antimicrobial-resistant bacteria—both as pathogens, which can directly and indirectly infect humans, and as commensals, which may carry transferable resistance determinants across species borders and reach humans through multiple routes of transfer. These pathways include not only food but also water and sludge and manure applications to food crop soils. Continued nontherapeutic use of antimicrobials in food animals will increase the pool of resistance genes, as well as their density, as bacteria migrate into the environment at large. The lack of species barriers for gene transmission argues that the focus of research efforts should be directed toward the genetic infrastructure and that it is now imperative to take an ecological approach toward addressing the impacts of NTA use on human disease. The study of animal-to-human transmission of antibiotic resistance therefore requires a greater understanding of the genetic interaction and spread that occur in the larger arena of commensal and environmental bacteria.
ACKNOWLEDGMENTS
B. M. Marshall was supported in part by The Pew Charitable Trusts. S. B. Levy is a consultant.
We thank Amadea Britton for research help.
Biographies
 Bonnie Marshall is a Senior Research Associate in the Center for Adaptation Genetics and Drug Resistance in the Department of Microbiology and Molecular Biology at Tufts University School of Medicine in Boston, MA. After obtaining a B.A. in Microbiology at the University of New Hampshire, she did work on herpesviruses at Harvard's New England Regional Primate Research Center and then returned to school to complete a degree in medical technology. In 1977, she joined the laboratory of Dr. Stuart Levy, from which she has published over 23 peer-reviewed publications on the ecology and epidemiology of resistance genes in human and animal clinical and commensal bacteria and environmental strains of water, soils, and plants. Ms. Marshall has also been engaged actively for 30 years with the Alliance for the Prudent Use of Antibiotics, where she is Staff Scientist and serves on the Board of Directors.
Bonnie Marshall is a Senior Research Associate in the Center for Adaptation Genetics and Drug Resistance in the Department of Microbiology and Molecular Biology at Tufts University School of Medicine in Boston, MA. After obtaining a B.A. in Microbiology at the University of New Hampshire, she did work on herpesviruses at Harvard's New England Regional Primate Research Center and then returned to school to complete a degree in medical technology. In 1977, she joined the laboratory of Dr. Stuart Levy, from which she has published over 23 peer-reviewed publications on the ecology and epidemiology of resistance genes in human and animal clinical and commensal bacteria and environmental strains of water, soils, and plants. Ms. Marshall has also been engaged actively for 30 years with the Alliance for the Prudent Use of Antibiotics, where she is Staff Scientist and serves on the Board of Directors.
 Stuart B. Levy is a Board-Certified Internist at Tufts Medical Center, a Professor of Molecular Biology and Microbiology and of Medicine at Tufts University School of Medicine, and Director, Center for Adaptation Genetics and Drug Resistance, also at Tufts University School of Medicine. He received his B.A. degree from Williams College and his M.D. from the University of Pennsylvania. He cofounded and remains active in both The Alliance for the Prudent Use of Antibiotics (1981) and Paratek Pharmaceuticals, Inc. (1996). More than 4 decades of studies on the molecular, genetic, and ecologic bases of drug resistance have led to over 250 peer-reviewed publications, authorship of The Antibiotic Paradox, honorary degrees in biology from Wesleyan University (1998) and Des Moines University (2001), ASM's Hoechst-Roussel Award for esteemed research in antimicrobial chemotherapy, and ICS's Hamao Umezawa Memorial Award. Dr. Levy is a Past President of the American Society for Microbiology and a Fellow of the American College of Physicians (ACP), the Infectious Diseases Society of America, the American Academy of Microbiology (AAM), and the American Association for the Advancement of Science. He serves on the National Science Advisory Board for Biosecurity.
Stuart B. Levy is a Board-Certified Internist at Tufts Medical Center, a Professor of Molecular Biology and Microbiology and of Medicine at Tufts University School of Medicine, and Director, Center for Adaptation Genetics and Drug Resistance, also at Tufts University School of Medicine. He received his B.A. degree from Williams College and his M.D. from the University of Pennsylvania. He cofounded and remains active in both The Alliance for the Prudent Use of Antibiotics (1981) and Paratek Pharmaceuticals, Inc. (1996). More than 4 decades of studies on the molecular, genetic, and ecologic bases of drug resistance have led to over 250 peer-reviewed publications, authorship of The Antibiotic Paradox, honorary degrees in biology from Wesleyan University (1998) and Des Moines University (2001), ASM's Hoechst-Roussel Award for esteemed research in antimicrobial chemotherapy, and ICS's Hamao Umezawa Memorial Award. Dr. Levy is a Past President of the American Society for Microbiology and a Fellow of the American College of Physicians (ACP), the Infectious Diseases Society of America, the American Academy of Microbiology (AAM), and the American Association for the Advancement of Science. He serves on the National Science Advisory Board for Biosecurity.
REFERENCES
- 1. Aarestrup F. M. 1999. Association between the consumption of antimicrobial agents in animal husbandry and the occurrence of resistant bacteria among food animals. Int. J. Antimicrob. Agents 12:279–285 [DOI] [PubMed] [Google Scholar]
- 2. Aarestrup F. M., Agerso Y., Gerner-Smidt P., Madsen M., Jensen L. B. 2000. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn. Microbiol. Infect. Dis. 37:127–137 [DOI] [PubMed] [Google Scholar]
- 3. Aarestrup F. M., et al. 1996. Glycopeptide susceptibility among Danish Enterococcus faecium and Enterococcus faecalis isolates of animal and human origin and PCR identification of genes within the VanA cluster. Antimicrob. Agents Chemother. 40:1938–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aarestrup F. M., Bager F., Andersen J. S. 2000. Association between the use of avilamycin for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers: epidemiological study and changes over time. Microb. Drug Resist. 6:71–75 [DOI] [PubMed] [Google Scholar]
- 5. Aarestrup F. M., Carstensen B. 1998. Effect of tylosin used as a growth promoter on the occurrence of macrolide-resistant enterococci and staphylococci in pigs. Microb. Drug Resist. 4:307–312 [DOI] [PubMed] [Google Scholar]
- 6. Aarestrup F. M., Jensen V. F., Emborg H. D., Jacobsen E., Wegener H. C. 2010. Changes in the use of antimicrobials and the effects on productivity of swine farms in Denmark. Am. J. Vet. Res. 71:726–733 [DOI] [PubMed] [Google Scholar]
- 7. Aarestrup F. M., et al. 2001. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45:2054–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Akwar T. H., et al. 2007. Risk factors for antimicrobial resistance among fecal Escherichia coli from residents on forty-three swine farms. Microb. Drug Resist. 13:69–76 [DOI] [PubMed] [Google Scholar]
- 9. Alexander T. W., et al. 2010. Farm-to-fork characterization of Escherichia coli associated with feedlot cattle with a known history of antimicrobial use. Int. J. Food Microbiol. 137:40–48 [DOI] [PubMed] [Google Scholar]
- 10. Alexander T. W., et al. 2008. Effect of subtherapeutic administration of antibiotics on the prevalence of antibiotic-resistant Escherichia coli bacteria in feedlot cattle. Appl. Environ. Microbiol. 74:4405–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alpharma, Inc., Animal Health 2007. Straight talk about antibiotic use in food-producing animals, p. 1–4 In For the record, vol. 6 Alpharma, Inc., Animal Health, Bridgewater, NJ: http://antibiotictruths.com/FTR/FTR_aug07.pdf [Google Scholar]
- 12. Anderson S. A., Yeaton Woo R. W., Crawford L. M. 2001. Risk assessment of the impact on human health of resistant Campylobacter jejuni from fluoroquinolone use in beef cattle. Food Control 12:13–25 [Google Scholar]
- 13. Angulo F. J., Nargund V. N., Chiller T. C. 2004. Evidence of an association between use of anti-microbial agents in food animals and anti-microbial resistance among bacteria isolated from humans and the human health consequences of such resistance. J. Vet. Med. B Infect. Dis. Vet. Public Health 51:374–379 [DOI] [PubMed] [Google Scholar]
- 14. Anonymous. 2010. Hearing on antibiotic resistance and the use of antibiotics in animal agriculture. House Energy and Commerce Committee, Washington, DC: http://energycommerce.house.gov/hearings/hearingdetail.aspx?NewsID=8001 [Google Scholar]
- 15. Arnold S., Gassner B., Giger T., Zwahlen R. 2004. Banning antimicrobial growth promoters in feedstuffs does not result in increased therapeutic use of antibiotics in medicated feed in pig farming. Pharmacoepidemiol. Drug Saf. 13:323–331 [DOI] [PubMed] [Google Scholar]
- 16. Aubry-Damon H., et al. 2004. Antimicrobial resistance in commensal flora of pig farmers. Emerg. Infect. Dis. 10:873–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bager F., Aarestrup F. M., Madsen M., Wegener H. C. 1999. Glycopeptide resistance in Enterococcus faecium from broilers and pigs following discontinued use of avoparcin. Microb. Drug Resist. 5:53–56 [DOI] [PubMed] [Google Scholar]
- 18. Bager F., Madsen M., Christensen J., Aarestrup F. M. 1997. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prev. Vet. Med. 31:95–112 [DOI] [PubMed] [Google Scholar]
- 19. Bailar J. C., III, Travers K. 2002. Review of assessments of the human health risk associated with the use of antimicrobial agents in agriculture. Clin. Infect. Dis. 34(Suppl. 3):S135–S143 [DOI] [PubMed] [Google Scholar]
- 20. Bartholomew M. J., Vose D. J., Tollefson L. R., Travis C. C. 2005. A linear model for managing the risk of antimicrobial resistance originating in food animals. Risk Anal. 25:99–108 [DOI] [PubMed] [Google Scholar]
- 21. Bartlett J. 15 December 2003. Antibiotics and livestock: control of VRE in Denmark. Medscape Infect. Dis. http://www.medscape.com/viewarticle/465032?rss
- 22. Barza M. D., et al. (ed.). 2002. The need to improve antimicrobial use in agriculture: ecological and human health consequences. Clinical infectious diseases, vol. 34, suppl. 3 Infectious Diseases Society of America, Arlington, VA: [Google Scholar]
- 23. Barza M. D., et al. (ed.). 2002. Policy recommendations. Clin. Infect. Dis. 34:S76–S77 [DOI] [PubMed] [Google Scholar]
- 24. Bates J., Jordens J. Z., Griffiths D. T. 1994. Farm animals as a putative reservoir for vancomycin-resistant enterococcal infection in man. J. Antimicrob. Chemother. 34:507–514 [DOI] [PubMed] [Google Scholar]
- 25. Benbrook C. M. 2002. Antibiotic drug use in U.S. aquaculture. The Northwest Science and Environmental Policy Center, Sandpoint, ID: http://www.healthobservatory.org/library.cfm?RefID=37397 [Google Scholar]
- 26. Berge A. C., Atwill E. R., Sischo W. M. 2005. Animal and farm influences on the dynamics of antibiotic resistance in faecal Escherichia coli in young dairy calves. Prev. Vet. Med. 69:25–38 [DOI] [PubMed] [Google Scholar]
- 27. Berge A. C., Moore D. A., Besser T. E., Sischo W. M. 2009. Targeting therapy to minimize antimicrobial use in preweaned calves: effects on health, growth, and treatment costs. J. Dairy Sci. 92:4707–4714 [DOI] [PubMed] [Google Scholar]
- 28. Bertrand S., et al. 2006. Clonal emergence of extended-spectrum beta-lactamase (CTX-M-2)-producing Salmonella enterica serovar Virchow isolates with reduced susceptibilities to ciprofloxacin among poultry and humans in Belgium and France (2000 to 2003). J. Clin. Microbiol. 44:2897–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boerlin P., Wissing A., Aarestrup F. M., Frey J., Nicolet J. 2001. Antimicrobial growth promoter ban and resistance to macrolides and vancomycin in enterococci from pigs. J. Clin. Microbiol. 39:4193–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Borgen K., Sorum M., Kruse H., Wasteson Y. 2000. Persistence of vancomycin-resistant enterococci (VRE) on Norwegian broiler farms. FEMS Microbiol. Lett. 191:255–258 [DOI] [PubMed] [Google Scholar]
- 31. Brooks J. P., McLaughlin M. R. 2009. Antibiotic resistant bacterial profiles of anaerobic swine lagoon effluent. J. Environ. Qual. 38:2431–2437 [DOI] [PubMed] [Google Scholar]
- 32. Butaye P., Devriese L. A., Haesebrouck F. 2003. Antimicrobial growth promoters used in animal feed: effects of less well known antibiotics on gram-positive bacteria. Clin. Microbiol. Rev. 16:175–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bywater R., et al. 2004. A European survey of antimicrobial susceptibility among zoonotic and commensal bacteria isolated from food-producing animals. J. Antimicrob. Chemother. 54:744–754 [DOI] [PubMed] [Google Scholar]
- 34. Bywater R. J. 2005. Identification and surveillance of antimicrobial resistance dissemination in animal production. Poult. Sci. 84:644–648 [DOI] [PubMed] [Google Scholar]
- 35. Cabello F. C. 2004. Antibiotics and aquaculture in Chile: implications for human and animal health. Rev. Med. Chil. 132:1001–1006 [DOI] [PubMed] [Google Scholar]
- 36. Cabello F. C. 2006. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ. Microbiol. 8:1137–1144 [DOI] [PubMed] [Google Scholar]
- 37. Campagnolo E. R., et al. 2002. Antimicrobial residues in animal waste and water resources proximal to large-scale swine and poultry feeding operations. Sci. Total Environ. 299:89–95 [DOI] [PubMed] [Google Scholar]
- 38. Carnevale R. A. 2005. Antimicrobial use in food animals and human health. Med. Mal. Infect. 35:105–106 [DOI] [PubMed] [Google Scholar]
- 39. Castanon J. I. 2007. History of the use of antibiotic as growth promoters in European poultry feeds. Poult. Sci. 86:2466–2471 [DOI] [PubMed] [Google Scholar]
- 40. CDC 2010. Get smart: know when antibiotics work. Centers for Disease Control, Atlanta, GA: www.cdc.gov/Features/GetSmart [Google Scholar]
- 41. Chapin A., Rule A., Gibson K., Buckley T., Schwab K. 2005. Airborne multidrug-resistant bacteria isolated from a concentrated swine feeding operation. Environ. Health Perspect. 113:137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chaslus-Dancla E., Pohl P., Meurisse M., Marin M., Lafont J. P. 1991. High genetic homology between plasmids of human and animal origins conferring resistance to the aminoglycosides gentamicin and apramycin. Antimicrob. Agents Chemother. 35:590–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chee-Sanford J. C., Aminov R. I., Krapac I. J., Garrigues-Jeanjean N., Mackie R. I. 2001. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67:1494–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chiller T. M., Barrett T., Angulo F. J. 2004. CDC studies incorrectly summarized in ‘critical review.’ J. Antimicrob. Chemother. 54:275–276 [DOI] [PubMed] [Google Scholar]
- 45. Costa P. M., Bica A., Vaz-Pires P., Bernardo F. 2010. Changes in antimicrobial resistance among faecal enterococci isolated from growing broilers prophylactically medicated with three commercial antimicrobials. Prev. Vet. Med. 93:71–76 [DOI] [PubMed] [Google Scholar]
- 46. Cox L. A., Jr., Popken D. A. 2010. Assessing potential human health hazards and benefits from subtherapeutic antibiotics in the United States: tetracyclines as a case study. Risk Anal. 30:432–457 [DOI] [PubMed] [Google Scholar]
- 47. Cox L. A., Jr., Popken D. A. 2004. Quantifying human health risks from virginiamycin used in chickens. Risk Anal. 24:271–288 [DOI] [PubMed] [Google Scholar]
- 48. Cox L. A., Jr., Ricci P. F. 2008. Causal regulations vs. political will: why human zoonotic infections increase despite precautionary bans on animal antibiotics. Environ. Int. 34:459–475 [DOI] [PubMed] [Google Scholar]
- 49. DANMAP 2008. Consumption of antimicrobial agents and the occurrence of antimicrobial resistance in bacteria from food animals, foods and humans in Denmark. DANMAP, Denmark: http://www.danmap.org/pdfFiles/Danmap_2008.pdf [Google Scholar]
- 50. de Boer E., et al. 2009. Prevalence of methicillin-resistant Staphylococcus aureus in meat. Int. J. Food Microbiol. 134:52–56 [DOI] [PubMed] [Google Scholar]
- 51. de Neeling A. J., et al. 2007. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet. Microbiol. 122:366–372 [DOI] [PubMed] [Google Scholar]
- 52. Diarra M. S., et al. 2007. Impact of feed supplementation with antimicrobial agents on growth performance of broiler chickens, Clostridium perfringens and enterococcus counts, and antibiotic resistance phenotypes and distribution of antimicrobial resistance determinants in Escherichia coli isolates. Appl. Environ. Microbiol. 73:6566–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dibner J. J., Richards J. D. 2005. Antibiotic growth promoters in agriculture: history and mode of action. Poult. Sci. 84:634–643 [DOI] [PubMed] [Google Scholar]
- 54. Dolliver H., Kumar K., Gupta S. 2007. Sulfamethazine uptake by plants from manure-amended soil. J. Environ. Qual. 36:1224–1230 [DOI] [PubMed] [Google Scholar]
- 55. Donabedian S. M., et al. 2003. Molecular characterization of gentamicin-resistant enterococci in the United States: evidence of spread from animals to humans through food. J. Clin. Microbiol. 41:1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55a. Dumonceaux T. J., Hill J. E., Hemmingsen S. M., Van Kessel A. G. 2006. Characterization of intestinal microbiota and response to dietary virginiamycin supplementation in the broiler chicken. Appl. Environ. Microbiol. 72:2815–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Duran G. M., Marshall D. L. 2005. Ready-to-eat shrimp as an international vehicle of antibiotic-resistant bacteria. J. Food Prot. 68:2395–2401 [DOI] [PubMed] [Google Scholar]
- 57. Dutil L., et al. 2010. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis. 16:48–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ekkelenkamp M. B., Sekkat M., Carpaij N., Troelstra A., Bonten M. J. 2006. Endocarditis due to meticillin-resistant Staphylococcus aureus originating from pigs. Ned. Tijdschr. Geneeskd. 150:2442–2447 [PubMed] [Google Scholar]
- 59. Emborg H. D., et al. 2003. Relations between the occurrence of resistance to antimicrobial growth promoters among Enterococcus faecium isolated from broilers and broiler meat. Int. J. Food Microbiol. 84:273–284 [DOI] [PubMed] [Google Scholar]
- 60. Emborg H. D., et al. 2007. Tetracycline consumption and occurrence of tetracycline resistance in Salmonella typhimurium phage types from Danish pigs. Microb. Drug Resist. 13:289–294 [DOI] [PubMed] [Google Scholar]
- 61. Endtz H. P., et al. 1991. Quinolone resistance in campylobacter isolated from man and poultry following the introduction of fluoroquinolones in veterinary medicine. J. Antimicrob. Chemother. 27:199–208 [DOI] [PubMed] [Google Scholar]
- 62. Ervik A., Thorsen B., Eriksen V., Lunestad B. T., Samuelsen O. B. 1994. Impact of administering antibacterial agents on wild fish and blue mussels Mytilus edulis in the vicinity of fish farms. Dis. Aquat. Org. 18:45–51 [Google Scholar]
- 63. Fairchild A. S., et al. 2005. Effects of orally administered tetracycline on the intestinal community structure of chickens and on tet determinant carriage by commensal bacteria and Campylobacter jejuni. Appl. Environ. Microbiol. 71:5865–5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. FAO 2007. The state of world fisheries and aquaculture 2006. Fisheries and Aquaculture Department, Food and Agriculture Organisation of the United Nations, Rome, Italy: http://www.fao.org/docrep/009/a0699e/A0699E00.htm [Google Scholar]
- 65. FDA 2000. FDA Task Force on Antimicrobial Resistance: key recommendations and report, Washington, DC. FDA, Washington, DC: http://www.fda.gov/downloads/ForConsumers/ConsumerUpdates/UCM143458.pdf [Google Scholar]
- 66. FDA 2010. The judicious use of medically important antimicrobial drugs in food-producing animals. Draft guidance 209. Center for Veterinary Medicine, FDA, Washington, DC: http://www.fda.gov/downloads/AnimalVeterinary/GuidanceComplianceEnforcement/GuidanceforIndustry/UCM216936.pdf [Google Scholar]
- 67. FDA 2009. Summary report on antimicrobials sold or distributed for use in food-producing animals. www.fda.gov/downloads/ForIndustry/UserFees/AnimalDrugUserFeeActADUFA/UCM231851.pdf
- 68. Ferber D. 2002. Livestock feed ban preserves drug's power. Science 295:27–28 [DOI] [PubMed] [Google Scholar]
- 69. Fey P. D., et al. 2000. Ceftriaxone-resistant salmonella infection acquired by a child from cattle. N. Engl. J. Med. 342:1242–1249 [DOI] [PubMed] [Google Scholar]
- 70. Franklin A. 1999. Current status of antibiotic resistance in animal production. Acta Vet. Scand. Suppl. 92:23–28 [PubMed] [Google Scholar]
- 71. Fuhrman J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541–548 [DOI] [PubMed] [Google Scholar]
- 72. Gibbs S. G., et al. 2006. Isolation of antibiotic-resistant bacteria from the air plume downwind of a swine confined or concentrated animal feeding operation. Environ. Health Perspect. 114:1032–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gilbert J. M., White D. G., McDermott P. F. 2007. The US national antimicrobial resistance monitoring system. Future Microbiol. 2:493–500 [DOI] [PubMed] [Google Scholar]
- 74. Gorbach S. L. 2001. Antimicrobial use in animal feed—time to stop. N. Engl. J. Med. 345:1202–1203 [DOI] [PubMed] [Google Scholar]
- 75. Graslund S., Holmstrom K., Wahlstrom A. 2003. A field survey of chemicals and biological products used in shrimp farming. Mar. Pollut. Bull. 46:81–90 [DOI] [PubMed] [Google Scholar]
- 76. Grave K., Jensen V. F., Odensvik K., Wierup M., Bangen M. 2006. Usage of veterinary therapeutic antimicrobials in Denmark, Norway and Sweden following termination of antimicrobial growth promoter use. Prev. Vet. Med. 75:123–132 [DOI] [PubMed] [Google Scholar]
- 77. Grave K., Kaldhusdal M. C., Kruse H., Harr L. M., Flatlandsmo K. 2004. What has happened in Norway after the ban of avoparcin? Consumption of antimicrobials by poultry. Prev. Vet. Med. 62:59–72 [DOI] [PubMed] [Google Scholar]
- 78. Graveland H., et al. 2010. Methicillin resistant Staphylococcus aureus ST398 in veal calf farming: human MRSA carriage related with animal antimicrobial usage and farm hygiene. PLoS One 5:e10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gupta A., et al. 2004. Antimicrobial resistance among Campylobacter strains, United States, 1997-2001. Emerg. Infect. Dis. 10:1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hammerum A. M., Fussing V., Aarestrup F. M., Wegener H. C. 2000. Characterization of vancomycin-resistant and vancomycin-susceptible Enterococcus faecium isolates from humans, chickens and pigs by RiboPrinting and pulsed-field gel electrophoresis. J. Antimicrob. Chemother. 45:677–680 [DOI] [PubMed] [Google Scholar]
- 81. Hamscher G., Pawelzick H. T., Sczesny S., Nau H., Hartung J. 2003. Antibiotics in dust originating from a pig-fattening farm: a new source of health hazard for farmers? Environ. Health Perspect. 111:1590–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hanselman B. A., et al. 2006. Methicillin-resistant Staphylococcus aureus colonization in veterinary personnel. Emerg. Infect. Dis. 12:1933–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hershberger E., et al. 2005. Epidemiology of antimicrobial resistance in enterococci of animal origin. J. Antimicrob. Chemother. 55:127–130 [DOI] [PubMed] [Google Scholar]
- 84. Heuer O. E., et al. 2009. Human health consequences of use of antimicrobial agents in aquaculture. Clin. Infect. Dis. 49:1248–1253 [DOI] [PubMed] [Google Scholar]
- 85. Heuer O. E., Pedersen K., Jensen L. B., Madsen M., Olsen J. E. 2002. Persistence of vancomycin-resistant enterococci (VRE) in broiler houses after the avoparcin ban. Microb. Drug Resist. 8:355–361 [DOI] [PubMed] [Google Scholar]
- 86. Ho P. L., et al. 2010. Genetic identity of aminoglycoside-resistance genes in Escherichia coli isolates from human and animal sources. J. Med. Microbiol. 59:702–707 [DOI] [PubMed] [Google Scholar]
- 87. Holmberg S. D., Osterholm M. T., Senger K. A., Cohen M. L. 1984. Drug-resistant Salmonella from animals fed antimicrobials. N. Engl. J. Med. 311:617–622 [DOI] [PubMed] [Google Scholar]
- 88. Holmberg S. D., Wells J. G., Cohen M. L. 1984. Animal-to-man transmission of antimicrobial-resistant Salmonella: investigations of U.S. outbreaks, 1971–1983. Science 225:833–835 [DOI] [PubMed] [Google Scholar]
- 89. Howells C. H., Joynson D. H. 1975. Possible role of animal feeding-stuffs in spread of antibiotic-resistant intestinal coliforms. Lancet i:156–157 [DOI] [PubMed] [Google Scholar]
- 90. Hummel R., Tschape H., Witte W. 1986. Spread of plasmid-mediated nourseothricin resistance due to antibiotic use in animal husbandry. J. Basic Microbiol. 26:461–466 [DOI] [PubMed] [Google Scholar]
- 91. Hurd H. S., Malladi S. 2008. A stochastic assessment of the public health risks of the use of macrolide antibiotics in food animals. Risk Anal. 28:695–710 [DOI] [PubMed] [Google Scholar]
- 92. Inglis G. D., et al. 2005. Effects of subtherapeutic administration of antimicrobial agents to beef cattle on the prevalence of antimicrobial resistance in Campylobacter jejuni and Campylobacter hyointestinalis. Appl. Environ. Microbiol. 71:3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Jackson C. R., Fedorka-Cray P. J., Barrett J. B., Ladely S. R. 2004. Effects of tylosin use on erythromycin resistance in enterococci isolated from swine. Appl. Environ. Microbiol. 70:4205–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Johnson J. R., et al. 2006. Similarity between human and chicken Escherichia coli isolates in relation to ciprofloxacin resistance status. J. Infect. Dis. 194:71–78 [DOI] [PubMed] [Google Scholar]
- 95. Johnson J. R., et al. 2007. Antimicrobial drug-resistant Escherichia coli from humans and poultry products, Minnesota and Wisconsin, 2002–2004. Emerg. Infect. Dis. 13:838–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Jones F. T., Ricke S. C. 2003. Observations on the history of the development of antimicrobials and their use in poultry feeds. Poult. Sci. 82:613–617 [DOI] [PubMed] [Google Scholar]
- 97. Kaszanyitzky E. J., Tenk M., Ghidan A., Fehervari G. Y., Papp M. 2007. Antimicrobial susceptibility of enterococci strains isolated from slaughter animals on the data of Hungarian resistance monitoring system from 2001 to 2004. Int. J. Food Microbiol. 115:119–123 [DOI] [PubMed] [Google Scholar]
- 98. Katsunuma Y., et al. 2007. Associations between the use of antimicrobial agents for growth promotion and the occurrence of antimicrobial-resistant Escherichia coli and enterococci in the feces of livestock and livestock farmers in Japan. J. Gen. Appl. Microbiol. 53:273–279 [DOI] [PubMed] [Google Scholar]
- 99. Kelly L., et al. 2004. Animal growth promoters: to ban or not to ban? A risk assessment approach. Int. J. Antimicrob. Agents 24:205–212 [DOI] [PubMed] [Google Scholar]
- 100. Khachatryan A. R., Besser T. E., Hancock D. D., Call D. R. 2006. Use of a nonmedicated dietary supplement correlates with increased prevalence of streptomycin-sulfa-tetracycline-resistant Escherichia coli on a dairy farm. Appl. Environ. Microbiol. 72:4583–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Khanna T., Friendship R., Dewey C., Weese J. S. 2008. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet. Microbiol. 128:298–303 [DOI] [PubMed] [Google Scholar]
- 102. Klare I., et al. 1999. Decreased incidence of VanA-type vancomycin-resistant enterococci isolated from poultry meat and from fecal samples of humans in the community after discontinuation of avoparcin usage in animal husbandry. Microb. Drug Resist. 5:45–52 [DOI] [PubMed] [Google Scholar]
- 103. Kriebel D., et al. 2001. The precautionary principle in environmental science. Environ. Health Perspect. 109:871–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kruse H., Sorum H. 1994. Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl. Environ. Microbiol. 60:4015–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kumar K., Gupta S. C., Chander Y., Singh A. K. 2005. Antibiotic use in agriculture and its impact on the terrestrial environment. Adv. Agron. 87:1–54 [Google Scholar]
- 106. Labbate M., Case R. J., Stokes H. W. 2009. The integron/gene cassette system: an active player in bacterial adaptation. Methods Mol. Biol. 532:103–125 [DOI] [PubMed] [Google Scholar]
- 107. Lauderdale T. L., et al. 2007. Effect of banning vancomycin analogue avoparcin on vancomycin-resistant enterococci in chicken farms in Taiwan. Environ. Microbiol. 9:819–823 [DOI] [PubMed] [Google Scholar]
- 108. Lee J. H. 2003. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 69:6489–6494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Levy S. B. 2002. The antibiotic paradox: how the misuse of antibiotics destroys their curative powers, 2nd ed. Perseus Publishing, Cambridge, MA [Google Scholar]
- 110. Levy S. B. 1998. The challenge of antibiotic resistance. Sci. Am. 278:46–53 [DOI] [PubMed] [Google Scholar]
- 111. Levy S. B., FitzGerald G. B., Macone A. B. 1976. Changes in intestinal flora of farm personnel after introduction of a tetracycline-supplemented feed on a farm. N. Engl. J. Med. 295:583–588 [DOI] [PubMed] [Google Scholar]
- 112. Levy S. B., FitzGerald G. B., Macone A. B. 1976. Spread of antibiotic-resistant plasmids from chicken to chicken and from chicken to man. Nature 260:40–42 [DOI] [PubMed] [Google Scholar]
- 113. Levy S. B., Marshall B. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10:S122–S129 [DOI] [PubMed] [Google Scholar]
- 114. Lewis H. C., et al. 2008. Pigs as source of methicillin-resistant Staphylococcus aureus CC398 infections in humans, Denmark. Emerg. Infect. Dis. 14:1383–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Luangtongkum T., et al. 2006. Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of Campylobacter spp. in poultry. Appl. Environ. Microbiol. 72:3600–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Marshall B. M., Ochieng D. J., Levy S. B. 2009. Commensals: underappreciated reservoirs of resistance. Microbe 4:231–238 [Google Scholar]
- 117. Mellon M., Benbrook C., Benbrook K. L. 2001. Hogging it: estimates of antimicrobial abuse in livestock. USC Publications, Cambridge, MA [Google Scholar]
- 118. Molbak K., et al. 1999. An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype Typhimurium DT104. N. Engl. J. Med. 341:1420–1425 [DOI] [PubMed] [Google Scholar]
- 119. Noble W. C., Virani Z., Cree R. G. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol. Lett. 72:195–198 [DOI] [PubMed] [Google Scholar]
- 120. Normanno G., et al. 2007. Methicillin-resistant Staphylococcus aureus (MRSA) in foods of animal origin product in Italy. Int. J. Food Microbiol. 117:219–222 [DOI] [PubMed] [Google Scholar]
- 121. Pantosti A., Del Grosso M., Tagliabue S., Macri A., Caprioli A. 1999. Decrease of vancomycin-resistant enterococci in poultry meat after avoparcin ban. Lancet 354:741–742 [DOI] [PubMed] [Google Scholar]
- 122. Perreten V. 2005. Resistance in the food chain and in bacteria from animals: relevance to human infections, p. 446–464 In White D. G., Alekshun M. N., McDermott P. F. (ed.), Frontiers in antimicrobial resistance: a tribute to Stuart B. Levy. ASM Press, Washington, DC [Google Scholar]
- 123. Petersen A., Andersen J. S., Kaewmak T., Somsiri T., Dalsgaard A. 2002. Impact of integrated fish farming on antimicrobial resistance in a pond environment. Appl. Environ. Microbiol. 68:6036–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Phillips I., et al. 2004. Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J. Antimicrob. Chemother. 53:28–52 [DOI] [PubMed] [Google Scholar]
- 125. Post V., Recchia G. D., Hall R. M. 2007. Detection of gene cassettes in Tn402-like class 1 integrons. Antimicrob. Agents Chemother. 51:3467–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Price L. B., et al. 2007. Elevated risk of carrying gentamicin-resistant Escherichia coli among U.S. poultry workers. Environ. Health Perspect. 115:1738–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Rhodes G., et al. 2000. Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant tet(A). Appl. Environ. Microbiol. 66:3883–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Sanchez R., et al. 1994. Evolution of susceptibilities of Campylobacter spp. to quinolones and macrolides. Antimicrob. Agents Chemother. 38:1879–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Sapkota A. R. 2006. Antibiotic resistance genes in drug resistant Enterococcus spp and Streptococcus spp recovered from indoor air of a concentrated swine-feeding operation. Lett. Appl. Microbiol. 43:534–540 [DOI] [PubMed] [Google Scholar]
- 130. Schroeder C. M., et al. 2003. Isolation of antimicrobial-resistant Escherichia coli from retail meats purchased in Greater Washington, DC, U.S.A. Int. J. Food Microbiol. 85:197–202 [DOI] [PubMed] [Google Scholar]
- 131. Sharfstein J. M. 14 July 2010. Testimony. FDA, Silver Spring, MD: http://democrats.energycommerce.house.gov/documents/20100714/Sharfstein.Testimony.07.14.2010.pdf [Google Scholar]
- 132. Sharma R., et al. 2008. Diversity and distribution of commensal fecal Escherichia coli bacteria in beef cattle administered selected subtherapeutic antimicrobials in a feedlot setting. Appl. Environ. Microbiol. 74:6178–6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Shotwell T. K. (ed.). 2009. Superbugs: E. coli, Salmonella, Staphylococcus and more!, 1st ed. Biontogeny Publications, Bridgeport, TX [Google Scholar]
- 134. Smith D. L., Harris A. D., Johnson J. A., Silbergeld E. K., Morris J. G., Jr 2002. Animal antibiotic use has an early but important impact on the emergence of antibiotic resistance in human commensal bacteria. Proc. Natl. Acad. Sci. U. S. A. 99:6434–6439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Smith D. L., et al. 2003. Assessing risks for a pre-emergent pathogen: virginiamycin use and the emergence of streptogramin resistance in Enterococcus faecium. Lancet Infect. Dis. 3:241–249 [DOI] [PubMed] [Google Scholar]
- 136. Smith H. W., Crabb W. E. 1957. The effect of the continuous administration of diets containing low levels of tetracyclines on the incidence of drug-resistant Bacterium coli in the faeces of pigs and chickens: the sensitivity of the Bact. coli to other chemotherapeutic agents. Vet. Rec. 69:24–30 [Google Scholar]
- 137. Smith K. E., et al. 1999. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992-1998. N. Engl. J. Med. 340:1525–1532 [DOI] [PubMed] [Google Scholar]
- 138. Smith T. C., et al. 2009. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in Midwestern U.S. swine and swine workers. PLoS One 4:e4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Sorensen T. L., et al. 2001. Transient intestinal carriage after ingestion of antibiotic-resistant Enterococcus faecium from chicken and pork. N. Engl. J. Med. 345:1161–1166 [DOI] [PubMed] [Google Scholar]
- 140. Sorum H. 1999. Antibiotic resistance in aquaculture. Acta Vet. Scand. Suppl. 92:29–36 [PubMed] [Google Scholar]
- 141. Sorum H. 2006. Antimicrobial drug resistance in fish pathogens, p. 213–238 In Aarestrup F. M. (ed.), Antimicrobial resistance in bacteria of animal origin. ASM Press, Washington, DC [Google Scholar]
- 142. Stokestad E. L. R., Jukes T. H. 1950. Further observations on the “animal protein factor.” Proc. Soc. Exp. Biol. Med. 73:523–528 [Google Scholar]
- 143. Summers A. O. 2002. Generally overlooked fundamentals of bacterial genetics and ecology. Clin. Infect. Dis. 34(Suppl. 3):S85–S92 [DOI] [PubMed] [Google Scholar]
- 144. Swann M. 1969. Report of the Joint Committee on the Use of Antibiotics in Animal Husbandry and Veterinary Medicine. Her Majesty's Stationery Office, London, United Kingdom [Google Scholar]
- 145. Tacket C. O., Dominguez L. B., Fisher H. J., Cohen M. L. 1985. An outbreak of multiple-drug-resistant Salmonella enteritis from raw milk. JAMA 253:2058–2060 [PubMed] [Google Scholar]
- 146. Tschape H. 1994. The spread of plasmids as a function of bacterial adaptability. FEMS Microbiol. Ecol. 15:23–31 [Google Scholar]
- 147. van Belkum A., et al. 2008. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg. Infect. Dis. 14:479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. van den Bogaard A. E., Bruinsma N., Stobberingh E. E. 2000. The effect of banning avoparcin on VRE carriage in The Netherlands. J. Antimicrob. Chemother. 46:146–148 [DOI] [PubMed] [Google Scholar]
- 149. van den Bogaard A. E., Willems R., London N., Top J., Stobberingh E. E. 2002. Antibiotic resistance of faecal enterococci in poultry, poultry farmers and poultry slaughterers. J. Antimicrob. Chemother. 49:497–505 [DOI] [PubMed] [Google Scholar]
- 150. van Leeuwen W. J., Guinee P. A., Voogd C. E., van Klingeren B. 1986. Resistance to antibiotics in Salmonella. Tijdschr. Diergeneeskd. 111:9–13 [PubMed] [Google Scholar]
- 151. Van Loo I. H. 2007. Emergence of methicillin-resistant Staphylococcus aureus of animal origins in humans. Emerg. Infect. Dis. 13:1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. van Rijen M. M., Van Keulen P. H., Kluytmans J. A. 2008. Increase in a Dutch hospital of methicillin-resistant Staphylococcus aureus related to animal farming. Clin. Infect. Dis. 46:261–263 [DOI] [PubMed] [Google Scholar]
- 153. Varga C., et al. 2009. Associations between reported on-farm antimicrobial use practices and observed antimicrobial resistance in generic fecal Escherichia coli isolated from Alberta finishing swine farms. Prev. Vet. Med. 88:185–192 [DOI] [PubMed] [Google Scholar]
- 154. Varga C., Rajic A., McFall M. E., Reid-Smith R. J., McEwen S. A. 2009. Associations among antimicrobial use and antimicrobial resistance of Salmonella spp. isolates from 60 Alberta finishing swine farms. Foodborne Pathog. Dis. 6:23–31 [DOI] [PubMed] [Google Scholar]
- 155. Voss A., Loeffen F., Bakker J., Klaassen C., Wulf M. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11:1965–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Weber J. T., et al. 1994. Epidemic cholera in Ecuador: multidrug-resistance and transmission by water and seafood. Epidemiol. Infect. 112:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156a. Wegener H. C. 2003. Ending the use of antimicrobial growth promoters is making a difference. ASM News 69:443–448 [Google Scholar]
- 157. WHO June 2006. Antimicrobial use in aquaculture and antimicrobial resistance. Report of a joint FAO/OIE/WHO expert consultation on antimicrobial use in aquaculture and antimicrobial resistance. WHO, Geneva, Switzerland: ftp://ftp.fao.org/ag/agn/food/aquaculture_rep_13_16june2006.pdf [Google Scholar]
- 158. WHO 2002. Impacts of antimicrobial growth promoter termination in Denmark. Department of Communicable Diseases, World Health Organization, Foulum, Denmark: http://whqlibdoc.who.int/hq/2003/WHO_CDS_CPE_ZFK_2003.1.pdf [Google Scholar]
- 159. WHO October 1997. The medical impact of the use of antimicrobials in food animals: report of a WHO meeting. World Health Organization, Geneva, Switzerland: http://whqlibdoc.who.int/hq/1997/WHO_EMC_ZOO_97.4.pdf [Google Scholar]
- 160. Wierup M. 2001. The Swedish experience of the 1986 year ban of antimicrobial growth promoters, with special reference to animal health, disease prevention, productivity, and usage of antimicrobials. Microb. Drug Resist. 7:183–190 [DOI] [PubMed] [Google Scholar]
- 161. Winokur P. L., Vonstein D. L., Hoffman L. J., Uhlenhopp E. K., Doern G. V. 2001. Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Witte W. 2000. Selective pressure by antibiotic use in livestock. Int. J. Antimicrob. Agents 16(Suppl. 1):S19–S24 [DOI] [PubMed] [Google Scholar]
- 163. Witte W., Strommenger B., Stanek C., Cuny C. 2007. Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg. Infect. Dis. 13:255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Zhang X. Y., Ding L. J., Fan M. Z. 2009. Resistance patterns and detection of aac(3)-IV gene in apramycin-resistant Escherichia coli isolated from farm animals and farm workers in northeastern of China. Res. Vet. Sci. 87:449–454 [DOI] [PubMed] [Google Scholar]
- 165. Zhao S., et al. 2003. Characterization of Salmonella enterica serotype Newport isolated from humans and food animals. J. Clin. Microbiol. 41:5366–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]