Abstract
Salicylate, a nonsteroidal anti-inflammatory compound, has been shown to increase the resistance of Campylobacter to antimicrobials. However, the molecular mechanism underlying salicylate-induced resistance has not yet been established. In this study, we determined how salicylate increases antibiotic resistance and evaluated its impact on the development of fluoroquinolone-resistant Campylobacter mutants. Transcriptional fusion assays, real-time quantitative reverse transcription-PCR (RT-PCR), and immunoblotting assays consistently demonstrated the induction of the CmeABC multidrug efflux pump by salicylate. Electrophoretic mobility shift assays further showed that salicylate inhibits the binding of CmeR (a transcriptional repressor of the TetR family) to the promoter DNA of cmeABC, suggesting that salicylate inhibits the function of CmeR. The presence of salicylate in the culture medium not only decreased the susceptibility of Campylobacter to ciprofloxacin but also resulted in an approximately 70-fold increase in the observed frequency of emergence of fluoroquinolone-resistant mutants under selection with ciprofloxacin. Together, these results indicate that in Campylobacter, salicylate inhibits the binding of CmeR to the promoter DNA and induces expression of cmeABC, resulting in decreased susceptibility to antibiotics and in increased emergence of fluoroquinolone-resistant mutants under selection pressure.
INTRODUCTION
Sodium salicylates are commonly used as nonsteroidal anti-inflammatory drugs (NSAIDs). The acetyl form of salicylate, aspirin, is used widely in medicines and cosmetics. It has been estimated that around 40,000 metric tons of aspirin are consumed each year in the world (32). The main functions of aspirin are to relieve minor aches and pains and to reduce fever. Aspirin also has functions in decreasing the incidence of strokes and heart attacks (3). Salicylic acid and salicylate are the principal metabolites of aspirin (11). Sodium salicylate is also used as an antipyretic, antiphlogistic, and analgesic agent in livestock and poultry (12). In addition, salicylic acid is a common compound in plants and in numerous foods and beverages (13, 33). Therefore, salicylate is available to humans and food-producing animals via multiple sources.
In addition to its effects in mammalian cells, salicylate also alters the susceptibility of bacteria to antibiotics. Growth of several bacterial species in the presence of salicylate induces nonheritable resistance to multiple antibiotics (25). In Escherichia coli, the presence of salicylate increases resistance to multiple antibiotics, including quinolones, cephalosporins, ampicillin, nalidixic acid, tetracycline, and chloramphenicol (25, 27). Salicylate-induced multiple antibiotic resistance in E. coli is mediated by increased transcription of the marRAB operon. Salicylate inhibits the binding of the repressor protein MarR to marO, the operator region of the mar operon, which then leads to overexpression of the transcriptional activator protein MarA (4). MarA modulates the transcription of a number of genes, including decreased expression of OmpF (a porin) and increased expression of the multidrug efflux pump AcrAB-TolC, which results in multiple antibiotic resistance (2). Increased resistance to chloramphenicol and enoxacin in Salmonella enterica serovar Typhimurium is also due to induction of the mar regulon by salicylate (31). In Klebsiella pneumoniae, salicylate-induced antibiotic resistance is due to increased expression of a MarA homologue, RamA, and the reduced production of two porins (5, 7).
Campylobacter is recognized as a leading bacterial cause of food-borne diseases in the United States and other developed countries (30). According to a CDC report, campylobacteriosis is estimated to affect over 0.84 million people every year in the United States (29). Worldwide, Campylobacter infections account for 400 to 500 million cases of diarrhea each year (28). Antibiotic treatment is recommended when the infection by Campylobacter is severe or occurs in immunocompromised patients. However, Campylobacter has become increasingly resistant to antimicrobials (18, 24). Among the known antibiotic resistance mechanisms in Campylobacter, the CmeABC efflux pump is an important player and confers resistance to structurally diverse antibiotics and toxic compounds (17). It has been demonstrated that CmeABC belongs to the RND family of efflux transporters and is regulated by a transcriptional repressor, CmeR, which binds to a specific site in the promoter region of cmeABC (15, 17). Expression of CmeABC is inducible by bile compounds, which interact with the ligand-binding domain of CmeR and prevent binding of CmeR to the cmeABC promoter in Campylobacter jejuni (14, 16). Furthermore, it has been shown that overexpression of CmeABC in Campylobacter significantly increases the frequency of emergence of fluoroquinolone-resistant mutants (35).
Previously, it was shown that growth of Campylobacter in the presence of salicylate resulted in a small but statistically significant increase in resistance to ciprofloxacin, tetracycline, and erythromycin (26). Later, Hannula and Hanninen confirmed a salicylate-induced increase in resistance to ciprofloxacin in almost all examined Campylobacter strains (10). These studies indicated that salicylate modulates Campylobacter resistance to antibiotics, but how salicylate influences antibiotic resistance and if it affects the emergence of antibiotic-resistant Campylobacter mutants are unknown. Based on previous findings on salicylate and cmeABC regulation, we hypothesized that salicylate modulates antibiotic resistance in Campylobacter by altering the expression of the CmeABC efflux pump. To examine this hypothesis, we sought to compare the expression levels of cmeABC with and without salicylate, to determine the interaction of salicylate with the CmeR regulator, and to assess the impact of salicylate on the emergence of fluoroquinolone-resistant Campylobacter mutants.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. C. jejuni strains were cultured on Mueller-Hinton (MH) agar or in MH broth at 42°C microaerobically (5% O2, 10% CO2, and 85% N2) in a gas incubator. C. jejuni strains with antimicrobial resistance markers were grown on kanamycin (30 μg/ml) or chloramphenicol (4 μg/ml) when appropriate. All strains were preserved as 30% glycerol stocks at −80°C.
Table 1.
Bacterial plasmids and strains used in this study
| Bacterial strain or plasmid | Description or relevant genotype | Source or reference |
|---|---|---|
| Plasmids | ||
| pMW10 | Shuttle plasmid with promoterless lacZ | 34 |
| pABC11 | pMW10 with the cmeABC promoter sequence cloned in front of lacZ | 1 |
| pMW561 | pMW10 with the promoter of cj0561c inserted upstream of lacZ | 9 |
| C. jejuni strains | ||
| NCTC 11168 | Wild-type C. jejuni | 22 |
| 11168/pABC11 | NCTC 11168 containing pABC11 | 1 |
| W7/pMW561 | 11168W7 containing pMW561 | 9 |
Antimicrobial susceptibility tests.
The MICs of antibiotics against C. jejuni NCTC 11168 were determined using either Campylobacter MIC plates (Trek Diagnostic Systems) or a broth microdilution method as described previously (17). All assays were repeated at least three times.
Bacterial growth assays.
Overnight cultures of C. jejuni NCTC 11168 were diluted 100 times in fresh MH broth. Cultures were grown in 200-μl volumes in 96-well plates and then supplemented with ciprofloxacin (0.125 μg/ml), erythromycin (0.125 μg/ml), novobiocin (16 μg/ml), or tetracycline (0.031 μg/ml), alone or together with salicylate (100 μg/ml). The plate was incubated at 42°C for 20 h in a microaerobic atmosphere, and the optical density at 600 nm was measured by use of a FLUOstar Omega instrument (BMG Labtech, Offenburg, Germany).
β-Galactosidase assay.
To determine if salicylate induced the promoter activity of cmeABC, C. jejuni 11168 containing pABC11 (Table 1) was grown in MH broth or MH broth supplemented with salicylate (100 μg/ml) for 20 h, and the cells were harvested to measure β-galactosidase activity as described in a previous study (1). Since cj0561c is also regulated by CmeR (9), we further analyzed the promoter activity of cj0561c in the presence of salicylate. The promoter fusion construct for cj0561c was described by Guo et al. (9) and is listed in Table 1. All β-galactosidase assays were repeated three times.
Real-time qRT-PCR.
To further assess if the cmeABC operon is subject to induction by salicylate, C. jejuni NCTC 11168 was cultured in MH broth, with or without salicylate, for 20 h. The final concentrations of salicylate in the cultures were 0, 100, and 200 μg/ml. Total RNA was extracted from each of the cultures by use of an RNeasy minikit (Qiagen, Valencia, CA) according to the protocol supplied with the product and was further treated with a Turbo DNA-free kit (Ambion, Austin, TX) to eliminate DNA contamination. Real-time quantitative reverse transcription-PCR (qRT-PCR) analyses were conducted using an iScript one-step RT-PCR kit with SYBR green (Bio-Rad) along with an MyiQ iCycler real-time PCR detection system (Bio-Rad, Hercules, CA) as described previously (16). The primer sets used to detect the transcription levels of cmeB (9) and cmeR (16) were described in previous publications. The 16S rRNA gene was used for normalization. The qRT-PCR experiment was repeated three times, using RNA samples prepared from three independent experiments.
SDS-PAGE and immunoblotting assay.
In order to determine if salicylate alters cmeABC expression at the protein level, C. jejuni 11168 was grown overnight in MH broth containing 0, 100, or 200 μg/ml of salicylate. Bacterial cells were harvested from the cultures, and whole-cell proteins were analyzed by SDS-PAGE and Western blotting, using anti-CmeA, anti-CmeB, anti-CmeC, anti-Cj0561c, and anti-Momp antibodies as described previously (9, 17).
EMSA.
To examine if salicylate inhibits CmeR binding to the promoter DNA of cmeABC, an electrophoretic mobility shift assay (EMSA) was performed using an EMSA kit (Invitrogen, Carlsbad, CA). Briefly, primers GSF and GSR1 (15) were used to amplify the 170-bp promoter region of cmeABC. The purified PCR products were incubated in 30-ng aliquots with 120 ng of purified recombinant CmeR (rCmeR) carrying the C69S and C166S changes (15, 21) in 10 μl of binding buffer containing 250 mM KCl, 0.1 mM dithiothreitol, 0.1 mM EDTA, and 10 mM Tris. The reaction mixtures were supplemented with salicylate (10 and 100 μg), taurocholate (10 and 100 μg), or ampicillin (10 and 100 μg) and then incubated for 20 min at room temperature. The reaction mixtures were then subjected to electrophoresis on a nondenaturing 6% (wt/vol) polyacrylamide gel in 0.5× TBE (44 mM Tris, 44 mM boric acid, 1 mM EDTA [pH 8.0]) at 200 V for 45 min. The gel was stained in 1× TBE containing 1× SYBR gold nucleic acid staining solution for 20 min. After washing of the gel in 150 ml of distilled H2O for 10 s, the DNA bands were visualized under UV light at 254 nm.
Detection of spontaneous fluoroquinolone-resistant mutants.
The emergence of spontaneous fluoroquinolone-resistant Campylobacter mutants in the presence or absence of salicylate was detected as described previously (35). Briefly, strain NCTC 11168 was grown on MH agar plates for 20 h under microaerobic conditions. The cells were collected and suspended in 1 ml MH broth. The total CFU in the concentrated culture was determined by serial dilution and plating on MH plates. Equal amounts of bacterial concentrate were plated onto salicylate-free and salicylate-containing MH plates that contained three different concentrations of ciprofloxacin. The concentrations of ciprofloxacin used for enumerating the mutants were 0.625, 1.25, and 4 μg/ml, which are 5-, 10-, and 16-fold higher, respectively, than the MIC for wild-type NCTC 11168. The frequency of emergence of fluoroquinolone-resistant mutants was calculated as the ratio of the CFU on ciprofloxacin-containing MH agar plates to the CFU of the inoculated culture concentrate. This experiment was repeated three times. The significance of differences in the frequencies of emergence of fluoroquinolone-resistant mutants between salicylate-treated cultures and nontreated cultures was determined by Student's t test.
RESULTS
Salicylate increases Campylobacter resistance to several antibiotics.
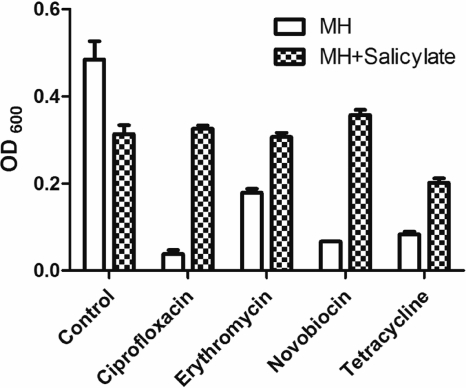
To verify if salicylate affects the antimicrobial susceptibility of Campylobacter, we measured the MICs of different antibiotics in the absence and presence of salicylate (100 μg/ml). Growth of C. jejuni NCTC 11168 with salicylate resulted in a moderate (2-fold) but reproducible increase in the MIC of ciprofloxacin in multiple experiments. This result is consistent with previous findings reported by others (10, 26). In this study, salicylate did not affect the MICs of other examined antimicrobials, including azithromycin, gentamicin, florfenicol, nalidixic acid, clindamycin, rifampin, cefotaxime, and streptomycin. However, the presence of salicylate increased the growth rate of strain 11168 in the presence of various antibiotics, including ciprofloxacin, erythromycin, novobiocin, and tetracycline, at their corresponding MICs (Fig. 1). Together, these results confirmed that salicylate decreased the susceptibility of Campylobacter to antibiotics. It should be pointed out that the enhanced resistance induced by salicylate was not inheritable. After removing salicylate from the medium, the MIC of ciprofloxacin for strain 11168 returned to the baseline level (data not shown).
Fig. 1.
Effects of salicylate (100 μg/ml) on the growth of C. jejuni 11168 in the presence of various antibiotics, including ciprofloxacin (0.125 μg/ml), erythromycin (0.125 μg/ml), novobiocin (16 μg/ml), and tetracycline (0.031 μg/ml). The bars represent the means and standard deviations for triplicate samples from a single representative experiment.
Salicylate induces the expression of cmeABC in Campylobacter.
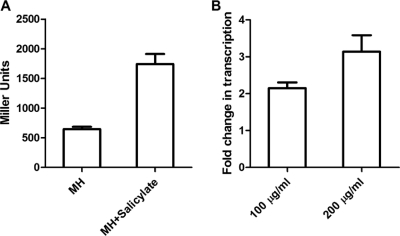
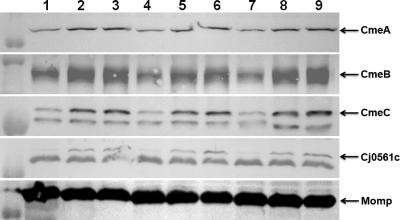
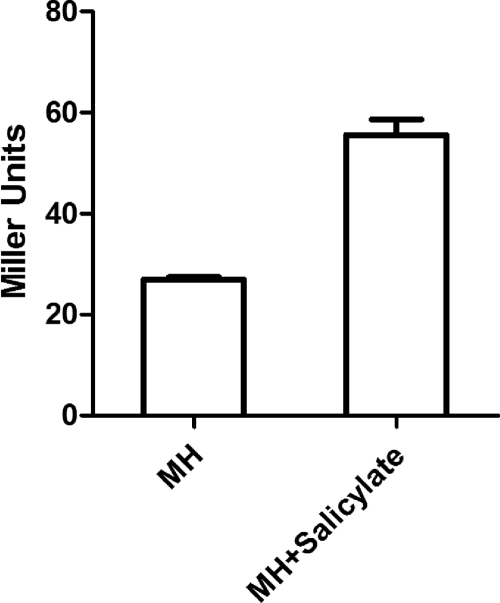
To determine if salicylate induces the expression of cmeABC, we measured the β-galactosidase activity of strain 11168/pABC11 grown in the presence or absence of salicylate. Compared to the basal level of transcription in MH broth, addition of salicylate (100 μg/ml) to the culture resulted in a 3-fold increase in the expression of cmeABC (Fig. 2A). We further examined the levels of the cmeB transcript in Campylobacter cultures grown with different concentrations of salicylate (0, 100, and 200 μg/ml), using real-time qRT-PCR. As shown in Fig. 2B, salicylate induced the transcription of cmeB 2- to 3-fold, in a dose-dependent manner. An immunoblotting assay using anti-CmeABC antibodies further confirmed the induction of CmeABC by salicylate (Fig. 3). According to densitometric analysis (data not shown), the amounts of CmeABC proteins increased 1.5- to 3-fold in the presence of salicylate compared to the baseline control levels (in MH broth). The major outer membrane protein (MOMP) band, which was used as an internal control, did not show any changes among the samples (Fig. 3). The protein data and the transcriptional results all indicated that salicylate is an inducer of CmeABC.
Fig. 2.
Induction of the cmeABC operon in C. jejuni 11168 by salicylate. (A) Transcriptional fusion (β-galactosidase assay) demonstrating the increased expression of cmeABC in the presence of salicylate (100 μg/ml). The bars represent the means and standard deviations for triplicate samples from a single representative experiment. (B) qRT-PCR results showing the increased transcript levels of cmeB with salicylate (100 and 200 μg/ml). Bacterial cells grown in MH broth alone were used as the control for baseline expression. The bars represent the means and standard deviations for three independent experiments.
Fig. 3.
Immunoblotting analysis of CmeA, CmeB, CmeC, Cj0561c, and MOMP production in NCTC 11168 grown with 0 (lanes 1, 4, and 7), 100 (lanes 2, 5, and 8), or 200 (lanes 3, 6, and 9) μg/ml of salicylate. Whole-cell proteins were used for immunoblotting with specific antibodies against the indicated proteins. The position of each protein is indicated by an arrow. MOMP was used as an internal control.
Since expression of CmeABC is controlled by CmeR (15), we further determined if salicylate affected the transcription of cmeR by using qRT-PCR. Results from three independent experiments did not reveal any significant changes in the transcription level of cmeR in the presence of salicylate (data not shown), indicating that salicylate did not alter the expression of cmeR. Thus, the enhanced expression of cmeABC by salicylate is unlikely to be due to a change in cmeR transcription.
Salicylate interferes with CmeR binding to the cmeABC promoter.
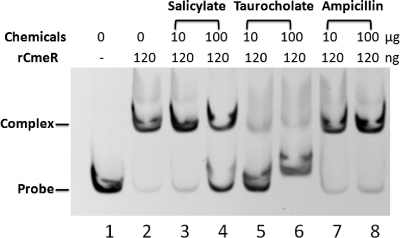
Since salicylate induced expression of CmeABC but did not alter the transcription of cmeR, we hypothesized that salicylate might affect the function of CmeR by reducing its affinity for the promoter DNA. To test this hypothesis, we determined the effect of salicylate on the binding of CmeR to the cmeABC promoter by using EMSA. The results showed that salicylate produced a dose-dependent inhibition of CmeR binding to the cmeABC promoter DNA (Fig. 4). Taurocholate is a known inducer of cmeABC and was used as a control for inhibition of CmeR binding. Although the inhibition by salicylate was not as strong as that by taurocholate, the inhibitory effect of salicylate on CmeR binding was clearly evident (Fig. 4). In contrast, ampicillin, which is not an inducer of cmeABC, did not inhibit the binding of CmeR to the cmeABC promoter DNA (Fig. 4).
Fig. 4.
Effects of salicylate, taurocholate, and ampicillin on the formation of CmeR-DNA complexes as determined by EMSA. Each reaction mixture contained 30 ng cmeABC promoter DNA and 120 ng rCmeR (except for that in lane 1). The amounts of chemicals used in the assay are indicated at the top.
Salicylate induces the expression of Cj0561c.
In addition to the control of cmeABC expression, CmeR also functions as a repressor of Cj0561c (a putative periplasmic protein) in Campylobacter by specifically binding to the cj0561c promoter (9). Since salicylate inhibited the function of CmeR, we suspected that it might also induce the expression of Cj0561c in Campylobacter. To examine this possibility, we evaluated the promoter activity of cj0561c by using a transcriptional fusion construct (pMW561) (Table 1). The β-galactosidase activity of strain W7/pMW561 grown in MH broth supplemented with 100 μg/ml of salicylate increased 2.5-fold compared with that for growth in MH broth without salicylate (Fig. 5). An immunoblotting assay using anti-Cj0561c antibodies further confirmed the induction of Cj0561c by salicylate (Fig. 3). This induction result for Cj0561c is consistent with the result that salicylate interferes with the function of CmeR in Campylobacter.
Fig. 5.
Induction of cj0561c in C. jejuni 11168 by salicylate (100 μg/ml), as determined by transcriptional fusion (β-galactosidase assay). The bars represent the means and standard deviations for triplicate samples from a single representative experiment.
Salicylate increases the frequency of emergence of fluoroquinolone-resistant Campylobacter mutants.
Since salicylate induced the expression of CmeABC, we further examined if the presence of salicylate modulates the emergence of fluoroquinolone-resistant Campylobacter. The results are shown in Table 2. When 0.625 or 1.25 μg/ml ciprofloxacin was used in the selection plates for enumeration of mutants, C. jejuni 11168 exhibited similar frequencies of emergence of fluoroquinolone-resistant mutants, regardless of the presence of salicylate in the growth medium (∼10−6; P ≥ 0.05). However, at a ciprofloxacin concentration of 4 μg/ml, incubation of strain 11168 with salicylate resulted in a 70-fold increase in the frequency of emergence of ciprofloxacin-resistant mutants, and the difference was statistically significant (P < 0.05). These findings indicate that salicylate increases the frequency of emergence of fluoroquinolone-resistant mutants on plates with a high concentration of ciprofloxacin.
Table 2.
Frequencies of emergence of fluoroquinolone-resistant C. jejuni mutants under different selection pressures
| Ciprofloxacin level (μg/ml) | Frequency of mutant emergencea |
|
|---|---|---|
| No salicylate | Salicylate (100 μg/ml) | |
| 0.625 | 3.33 × 10−6 ± 1.58 × 10−6 | 4.60 × 10−6 ± 2.70 × 10−6 |
| 1.25 | 3.77 × 10−6 ± 0.74 × 10−6 | 4.16 × 10−6 ± 2.96 × 10−6 |
| 4 | 4.24 × 10−8 ± 1.42 × 10−8 | 3.23 × 10−6 ± 1.55 × 10−6* |
Data are means ± standard deviations for three independent experiments. *, the difference from the no-salicylate control is statistically significant (P = 0.024).
DISCUSSION
The results from this study revealed that salicylate-mediated decreases in the susceptibility of Campylobacter to antibiotics are due at least partially to induction of expression of the CmeABC efflux pump. This conclusion is based on multiple pieces of experimental evidence derived from transcriptional fusion assays (Fig. 2A), real-time qPCRs (Fig. 2B), and immunoblotting assays (Fig. 3). We further showed that salicylate inhibits the binding of CmeR to its target promoters (Fig. 4), leading to enhanced expression of CmeABC and Cj0561c in the presence of salicylate (Fig. 2, 3, and 5). Together, these results provide a molecular basis for salicylate-induced resistance to antibiotics in Campylobacter.
The CmeABC efflux system plays an important role in resistance to antibiotics and toxic compounds in Campylobacter (17), and the expression of this efflux pump is modulated by CmeR (15). Previous studies demonstrated that overexpression of CmeABC results in modest increases in the MICs of antibiotics, including ciprofloxacin, erythromycin, novobiocin, tetracycline, cefotaxime, and fusidic acid (15, 16). In this study, we found that salicylate caused a small but reproducible increase in the MIC of ciprofloxacin and facilitated the growth of Campylobacter in the presence of inhibitory concentrations of antibiotics (Fig. 1). This finding is consistent with the results from previous studies using salicylate (10, 26).
CmeR is a pleiotropic regulator and functions as a transcriptional repressor of cmeABC and cj0561c (9, 15). Cj0561c is a periplasmic protein and is tightly controlled by CmeR due to the presence of two CmeR-binding sites in the promoter sequence of Cj0561c (9). Although the exact function of Cj0561c is unknown, a previous study showed that it contributes to the in vivo fitness of Campylobacter in chickens (9). In this study, we found that salicylate induced the expression of both CmeABC and Cj0561c. This induction can be explained by the fact that salicylate inhibited the binding of CmeR to promoter DNA, as shown by EMSA (Fig. 4). Compared with that by bile salts, which are strong inhibitors of CmeR binding (16), the inhibition by salicylate was relatively weak but was visually apparent (Fig. 4). This finding suggests that salicylate releases the repression of CmeR on cmeABC and cj0561c, leading to increased expression of the two genes.
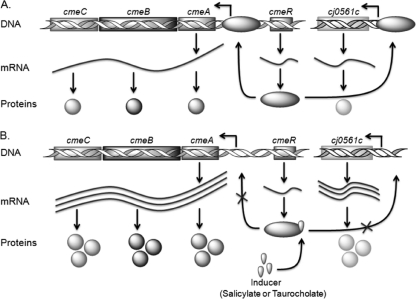
Salicylate inhibits the function of CmeR, not the expression level of this regulatory protein. This conclusion is based on the fact that qRT-PCR did not detect altered transcription of cmeR in the presence of salicylate (data not shown). How salicylate modulates the function of CmeR is unclear at present, but it is known that CmeR has a DNA-binding motif in the N-terminal region and a flexible ligand-binding pocket in the C-terminal region (8). It is possible that salicylate interacts with the ligand-binding pocket and triggers a conformational change in the DNA-binding domain, preventing CmeR binding to promoter DNA. This possibility remains to be examined in future studies. Based on results from this study and previously known information on CmeR regulation, we present a model that depicts the induction mechanisms of salicylate in Campylobacter (Fig. 6). The binding of salicylate to CmeR appears to be reversible, since removal of salicylate from the culture medium restored cmeABC expression to the basal level (data not shown).
Fig. 6.
Diagram depicting the molecular basis of salicylate-mediated induction of CmeABC and Cj0561c. (A) Baseline expression of the genes in the absence of salicylate. Transcription of cmeABC and cj0561c is at a low level due to inhibition by CmeR. (B) Induction of the genes by salicylate. When salicylate is present, it inhibits the binding of CmeR and ameliorates the repression on cmeABC and cj0561c, leading to overexpression of CmeABC and Cj0561c.
Salicylate not only increases antibiotic resistance but also promotes the emergence of spontaneous fluoroquinolone-resistant Campylobacter mutants under selection pressure. In Campylobacter, fluoroquinolone resistance is mediated by target modification (GyrA mutations) and by the efflux function of CmeABC (6, 20, 23). A single point mutation in the quinolone resistance-determining region of gyrA DNA is sufficient to significantly increase the resistance of Campylobacter to fluoroquinolone antimicrobials (6, 18, 19, 24). A T86I substitution in GyrA confers high-level resistance to fluoroquinolones, while T86K, A70T, and D90N substitutions are associated with moderate resistance to fluoroquinolones (18, 24). It is important that CmeABC functions synergistically with GyrA mutations in conferring fluoroquinolone resistance and that without CmeABC, GyrA mutants are unable to maintain the resistance phenotype (18, 35). Thus, the expression level of CmeABC affects the frequency of emergence of fluoroquinolone-resistant mutants in Campylobacter (35). In this study, we showed that addition of salicylate to the culture medium resulted in a 70-fold increase in the frequency of emergence of ciprofloxacin-resistant mutants at a higher concentration of the antibiotic (4 μg/ml) (Table 2). This result is consistent with our previous finding for a cmeR mutant in which CmeABC was overexpressed, for which the frequency of emergence of fluoroquinolone-resistant mutants increased significantly (35).
For Campylobacter, it is known that different GyrA mutations confer different levels of resistance, and the measured frequencies of emergence of resistant mutants vary with the concentration of ciprofloxacin in the plates (35). The presence of salicylate in the medium did not alter the frequencies of emergence of fluoroquinolone-resistant mutants (∼10−6) when the concentrations of ciprofloxacin in the selection plates were 5 times (0.625 μg/ml) and 10 times (1.25 μg/ml) higher than the MIC (Table 2). This result can be explained by the facts that the basal expression of CmeABC is sufficient and that overexpression of this efflux pump is not required for GyrA mutants to survive at low selection pressure (0.625 and 1.25 μg/ml) (35). In contrast, with 4 μg/ml of ciprofloxacin, those GyrA mutants with lower MICs would require overexpression of cmeABC to survive the selection pressure, resulting in a difference in the numbers of detected mutants with and without salicylate. These results indicate that salicylate does not affect the spontaneous mutation rate but facilitates the emergence of fluoroquinolone-resistant mutants under antibiotic selection by inducing the expression of cmeABC. Similar findings were obtained in a previous study in which mutation of cmeR (overexpressed CmeABC) led to increased emergence of ciprofloxacin-resistant mutants for selection with ciprofloxacin at 4 μg/ml but not at 0.625 and 1.25 μg/ml (35).
In summary, this study identified the molecular mechanism underlying salicylate-induced antibiotic resistance in Campylobacter and revealed that salicylate promotes the emergence of fluoroquinolone-resistant Campylobacter mutants under selection pressure. Although these findings were made under laboratory conditions, they might be applicable to the ecological niches occupied by C. jejuni under natural conditions. Considering the common presence of salicylate in plant and food as well as its widespread use in veterinary and human medicine, it is possible that C. jejuni is exposed to salicylate in animal reservoirs and in the human host. Such exposure could conceivably influence the development of antibiotic resistance in this pathogenic organism. Those treating Campylobacter infections with fluoroquinolones should consider this possibility and avoid the simultaneous use of salicylate-containing medicine or salicylate-rich nutrients.
ACKNOWLEDGMENTS
We thank Orhan Sahin for critical readings of the manuscript.
This work was supported by NIH grant RO1DK063008 and by National Research Initiative Competitive Grants Program grant 2007-35201-18278 from the USDA National Institute of Food and Agriculture.
Footnotes
Published ahead of print on 5 August 2011.
REFERENCES
- 1. Akiba M., Lin J., Barton Y. W., Zhang Q. 2006. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J. Antimicrob. Chemother. 57:52–60 [DOI] [PubMed] [Google Scholar]
- 2. Alekshun M. N., Levy S. B. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baigent C., et al. 2009. Aspirin in the primary and secondary prevention of vascular disease: collaborative meta-analysis of individual participant data from randomised trials. Lancet 373:1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohen S. P., Levy S. B., Foulds J., Rosner J. L. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol. 175:7856–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Domenico P., Hopkins T., Cunha B. A. 1990. The effect of sodium salicylate on antibiotic susceptibility and synergy in Klebsiella pneumoniae. J. Antimicrob. Chemother. 26:343–351 [DOI] [PubMed] [Google Scholar]
- 6. Ge B., McDermott P. F., White D. G., Meng J. 2005. Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 49:3347–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. George A. M., Hall R. M., Stokes H. W. 1995. Multidrug resistance in Klebsiella pneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology 141:1909–1920 [DOI] [PubMed] [Google Scholar]
- 8. Gu R., et al. 2007. Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni. J. Mol. Biol. 372:583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo B., et al. 2008. CmeR functions as a pleiotropic regulator and is required for optimal colonization of Campylobacter jejuni in vivo. J. Bacteriol. 190:1879–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hannula M., Hanninen M. L. 2008. Effect of putative efflux pump inhibitors and inducers on the antimicrobial susceptibility of Campylobacter jejuni and Campylobacter coli. J. Med. Microbiol. 57:851–855 [DOI] [PubMed] [Google Scholar]
- 11. Hartog E., Menashe O., Kler E., Yaron S. 2010. Salicylate reduces the antimicrobial activity of ciprofloxacin against extracellular Salmonella enterica serovar Typhimurium, but not against Salmonella in macrophages. J. Antimicrob. Chemother. 65:888–896 [DOI] [PubMed] [Google Scholar]
- 12. Huff G. R., Huff W. E., Balog J. M., Rath N. C., Izard R. S. 2004. The effects of water supplementation with vitamin E and sodium salicylate (Uni-Sol) on the resistance of turkeys to Escherichia coli respiratory infection. Avian Dis. 48:324–331 [DOI] [PubMed] [Google Scholar]
- 13. Janssen P. L., Hollman P. C., Venema D. P., van Staveren W. A., Katan M. B. 1996. Salicylates in foods. Nutr. Rev. 54:357–359 [DOI] [PubMed] [Google Scholar]
- 14. Lei H. T., et al. 2011. Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni. Protein Sci. 20:712–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin J., Akiba M., Sahin O., Zhang Q. 2005. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob. Agents Chemother. 49:1067–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin J., et al. 2005. Bile salts modulate expression of the CmeABC multidrug efflux pump in Campylobacter jejuni. J. Bacteriol. 187:7417–7424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin J., Michel L. O., Zhang Q. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46:2124–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luangtongkum T., et al. 2009. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 4:189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luo N., et al. 2005. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. U. S. A. 102:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luo N., Sahin O., Lin J., Michel L. O., Zhang Q. 2003. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob. Agents Chemother. 47:390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oakland M., Jeon B., Sahin O., Shen Z., Zhang Q. 2011. Functional characterization of a lipoprotein-encoding operon in Campylobacter jejuni. PLoS One 6:e20084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parkhill J., et al. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668 [DOI] [PubMed] [Google Scholar]
- 23. Payot S., et al. 2004. Relative contribution of target gene mutation and efflux to fluoroquinolone and erythromycin resistance, in French poultry and pig isolates of Campylobacter coli. Int. J. Antimicrob. Agents 23:468–472 [DOI] [PubMed] [Google Scholar]
- 24. Payot S., et al. 2006. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 8:1967–1971 [DOI] [PubMed] [Google Scholar]
- 25. Price C. T., Lee I. R., Gustafson J. E. 2000. The effects of salicylate on bacteria. Int. J. Biochem. Cell Biol. 32:1029–1043 [DOI] [PubMed] [Google Scholar]
- 26. Randall L. P., et al. 2003. Prevalence of multiple antibiotic resistance in 443 Campylobacter spp. isolated from humans and animals. J. Antimicrob. Chemother. 52:507–510 [DOI] [PubMed] [Google Scholar]
- 27. Rosner J. L. 1985. Nonheritable resistance to chloramphenicol and other antibiotics induced by salicylates and other chemotactic repellents in Escherichia coli K-12. Proc. Natl. Acad. Sci. U. S. A. 82:8771–8774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ruiz-Palacios G. M. 2007. The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin. Infect. Dis. 44:701–703 [DOI] [PubMed] [Google Scholar]
- 29. Scallan E., et al. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Slutsker L., Altekruse S. F., Swerdlow D. L. 1998. Foodborne diseases. Emerging pathogens and trends. Infect. Dis. Clin. North Am. 12:199–216 [DOI] [PubMed] [Google Scholar]
- 31. Sulavik M. C., Dazer M., Miller P. F. 1997. The Salmonella typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J. Bacteriol. 179:1857–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Warner T. D., Mitchell J. A. 2002. Cyclooxygenase-3 (COX-3): filling in the gaps toward a COX continuum? Proc. Natl. Acad. Sci. U. S. A. 99:13371–13373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wood A., Baxter G., Thies F., Kyle J., Duthie G. 2011. A systematic review of salicylates in foods: estimated daily intake of a Scottish population. Mol. Nutr. Food Res. 55(Suppl. 1):S7–S14 [DOI] [PubMed] [Google Scholar]
- 34. Wosten M. M., Boeve M., Koot M. G., van Nuenen A. C., A. van der Zeijst B. 1998. Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180:594–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yan M., Sahin O., Lin J., Zhang Q. 2006. Role of the CmeABC efflux pump in the emergence of fluoroquinolone-resistant Campylobacter under selection pressure. J. Antimicrob. Chemother. 58:1154–1159 [DOI] [PubMed] [Google Scholar]