Abstract
In this study, we examined molecular mechanisms associated with multidrug resistance (MDR) in a collection of Escherichia coli isolates recovered from hospitalized animals in Ireland. PCR and DNA sequencing were used to identify genes associated with resistance. Class 1 integrons were prevalent (94.6%) and contained gene cassettes recognized previously and implicated mainly in resistance to aminoglycosides, β-lactams, and trimethoprim (aadA1, dfrA1-aadA1, dfrA17-aadA5, dfrA12-orfF-aadA2, blaOXA-30-aadA1, aacC1-orf1-orf2-aadA1, dfr7). Class 2 integrons (13.5%) contained the dfrA1-sat1-aadA1 gene array. The most frequently occurring phenotypes included resistance to ampicillin (97.3%), chloramphenicol (75.4%), florfenicol (40.5%), gentamicin (54%), neomycin (43.2%), streptomycin (97.3%), sulfonamide (98.6%), and tetracycline (100%). The associated resistance determinants detected included blaTEM, cat, floR, aadB, aphA1, strA-strB, sul2, and tet(B), respectively. The blaCTX-M-2 gene, encoding an extended-spectrum β-lactamase (ESβL), and blaCMY-2, encoding an AmpC-like enzyme, were identified in 8 and 18 isolates, respectively. The mobility of the resistance genes was demonstrated using conjugation assays with a representative selection of isolates. High-molecular-weight plasmids were found to be responsible for resistance to multiple antimicrobial compounds. The study demonstrated that animal-associated commensal E. coli isolates possess a diverse repertoire of transferable genetic determinants. Emergence of ESβLs and AmpC-like enzymes is particularly significant. To our knowledge, the blaCTX-M-2 gene has not previously been reported in Ireland.
INTRODUCTION
Antimicrobial agents are extensively used in animal therapy, for prophylaxis and metaphylaxis, and in some geographical regions for growth promotion. Many of these drugs belong to the same families of antimicrobial compounds that are used for treating humans (76). Surveillance studies conducted in different countries generally report an increase in the level of resistance in Escherichia coli isolates to major classes of antibiotics used for the treatment of livestock and companion animals (28, 34). It has been suggested that an important factor in the emergence and dissemination of resistance is the selective pressure exerted following antibiotic exposure.
Drug-resistant commensal Escherichia coli isolates may constitute a significant reservoir of antibiotic resistance determinants which can spread to those bacteria pathogenic for animals and/or humans (77). For this reason, the reported emergence of resistance to new drugs such as extended-spectrum β-lactams and fluoroquinolones (FQs) is of particular concern to both animal and public health alike. The potential for transmission of E. coli clones between different animal hosts and humans has been documented previously (26). In addition, transmission of genetic determinants of resistance in vitro and in vivo has been described in several studies (13, 37, 49). Multidrug-resistant (MDR) E. coli strains are often isolated from animals in veterinary hospitals and are frequently associated with opportunistic infections (59). However, currently, detailed knowledge of the resistance mechanisms in bacteria cultured from hospitalized animals is limited.
The dissemination of resistance markers can be attributed to a number of independent genetic events collectively known as horizontal gene transfer (HGT). Most of the genes conferring antibiotic resistance are not specific to one bacterial host. HGT has been attributed to mobile and mobilizable genetic elements such as plasmids, transposons, and integrons since resistance determinants frequently map within these structures. Furthermore, many of the resistance markers persist despite the withdrawal of certain antibiotics from use in therapy (2, 58). The physical linkage of genes, in some cases at least, may explain these epidemiological data (65). Transfer of resistance plasmids between bacteria of veterinary importance, including E. coli, has been demonstrated in vitro and in vivo (14, 49).
Integrons are genetic structures involved in the transmission and expression of the resistance genes located within gene cassettes. Several classes of integrons are recognized on the basis of the sequence of specific recombinases known as integrase, of which class 1 is the most clinically relevant. Their 5′ conserved segment (5′-CS) contains intI1, a gene encoding the integrase along with a promoter region (Pc) required for the expression of the gene cassette. The 3′-CS typically contains a defective quaternary ammonium compound resistance gene denoted qacEΔ1, followed by sul1, conferring sulfonamide resistance. Class 1 integrons have emerged and continue to evolve as a result of site-specific recombination events catalyzed by the intl1-encoding enzyme between the attI1 site of the 5′-CS and the 59-base element located toward the distal end of the gene cassette (21, 52, 55). As integrons are frequently present in MDR bacteria and are associated with conjugative broad-host-range plasmids and transposons, their role in the evolution and dissemination of resistance among different bacterial species and genera is widely recognized (8, 9, 52).
While antibiotic-resistant zoonotic food-borne pathogens constitute an obvious threat to public health, the problem of resistance in other bacteria colonizing animals cannot be ignored. Data on the genetic mechanisms of antibiotic resistance in both food-producing and companion animals in Ireland are limited. This study was undertaken to characterize MDR in a diverse collection of animal E. coli isolates at the molecular level and evaluate the potential risk of resistance determinant(s) transmission.
MATERIALS AND METHODS
Selection of bacterial isolates for the study and resistance profiling.
A collection of 74 E. coli isolates resistant to 3 or more antimicrobial compounds of different classes was cultured from samples from selected animals presenting at the University Veterinary Hospital in Dublin, Ireland, between February and December 2007. These were consecutive MDR clinical isolates processed for routine laboratory testing. Forty-four equine isolates, 17 bovine isolates, 9 porcine isolates, 3 canine isolates, and 1 ovine isolate of E. coli were recovered from a variety of samples, predominantly fecal. Prior to testing, isolates were streaked for purity on MacConkey agar (Oxoid, Basingstoke, England), subcultured to tryptic soy agar (Oxoid), and stored at −80°C on cryoprotectant beads (Technical Service Consultants Ltd., Lancashire, England). Susceptibility testing by disc diffusion was performed, and results were interpreted as recommended by Clinical and Laboratory Standards Institute (CLSI) guidelines (11). Susceptibility testing included a panel of 19 different antimicrobial agents: amikacin (Ak), 30 μg; amoxicillin-clavulanic acid (Amc), 20/10 μg; ampicillin (Amp), 10 μg; cefpirome (Cfp), 30 μg; cefpodoxime (Cpd), 10 μg; ceftiofur (Cfr), 30 μg; cephalothin (Cpl), 30 μg; chloramphenicol (C), 30 μg; ciprofloxacin (Cip), 5 μg; colistin (Ct), 25 μg; florfenicol (Ffc), 30 μg; furazolidone (Fr), 15 μg; gentamicin (Gm), 10 μg; neomycin (Neo), 30 μg; nalidixic acid (Na), 30 μg; streptomycin (S), 10 μg; sulfonamide compound (Su), 300 μg; tetracycline (Te), 30 μg; and trimethoprim (Tmp), 5 μg. The discs were purchased from Oxoid, and E. coli ATCC 25922 was included as a quality control strain.
Phylogenetic grouping.
Phylogenetic groups were determined for each E. coli isolate using an established multiplex PCR targeting chuA, yja, and TSPE4.7 according to the protocol of Clermont et al. (10). The method was previously developed to classify E. coli into 4 phylogenetic groups designated A, B1, B2, and D. Target amplification was performed using the original primer concentrations and cycling conditions. Amplicons generated were separated by conventional 1.7% (wt/vol) agarose gel electrophoresis, stained with 0.1 μg/ml ethidium bromide (Sigma-Aldrich, Ireland) in 0.5× Tris-EDTA-boric acid buffer, and subsequently assigned to one of the lineages (A, B1, B2, or D) using the criteria outlined previously (10).
Detection of antibiotic resistance genes and class 1 and class 2 integrons.
Total genomic DNA was extracted using a Promega Wizard genomic DNA purification kit (Madison, WI) following the manufacturer's protocol. The DNA concentration was measured using a Nanodrop ND-1000 spectrophotometer (Thermoscientific, Wilmington, DE). Detection of antibiotic resistance markers and integron-associated genes was performed using the primers listed in Table 1. The following resistance determinants were investigated by PCR: aac(3)-IV, aadA, aadB, ampC, aphA1, aphA2, blaCARB, blaCMY-2, blaCTX-M, blaOXA, blaSHV, blaTEM, cat1, cmlA, floR, strA-strB, sul1, sul2, sul3, tet(A), tet(B), tet(C), tet(D), tet(E), and tet(G). Previously published PCR methods were employed to determine the presence of conserved integron-associated genes, including intI1 and intI2 (coding for integrases of classes 1 and 2, respectively), qacEΔ1, sul1, and the variable regions of class 1 and 2 integrons (Table 1).
Table 1.
PCR primer characteristics
| Target gene | Primer directiona | Nucleotide sequence (5′-3′) | Annealing temp (°C) | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| intI1 | F | CAG TGG ACA TAA GCC TGT TC | 59 | 160 | Koeleman et al. (31) |
| R | CCC GAG GCA TAG ACT GTA | ||||
| Class 1 gene cassette | F | GGC ATC CAA GCA GCA AGC | 55 | Variable | Lévesque et al. (36) |
| R | AAG CAG ACT TGA CCT GAT | ||||
| qacEΔ1 | F | ATC GCA ATA GTT GGC GAA GT | 53 | 250 | Sandvang et al. (60) |
| R | GAA GCT TTT GCC CAT GAA GC | ||||
| sul1 | F | CGG CGT GGG CTA CCT GAA CG | 66 | 433 | Kerrn et al. (29) |
| R | GCC GAT CGC GTG AAG TTC CG | ||||
| intI2 | F | CAC GGA TAT GCG ACA AAA AGG T | 54 | 788 | Mazel et al. (45) |
| R | GTA GCA AAC GAG TGA CGA AAT G | ||||
| Class 2 gene cassette | F | CGG GAT CCC GGA CGG CAT GCA CGA TTT GTA | 62 | Variable | White et al. (75) |
| R | GAT GCC ATC GCA AGT ACG AG | ||||
| aadA | F | GTG GAT GGC GGC CTG AAG CC | 60 | 525 | Madsen et al. (42) |
| R | AAT GCC CAG TCG GCA GCG | ||||
| strA-strB | F | ATG GTG GAC CCT AAA ACT CT | 60 | 893 | Tamang et al. (66) |
| R | CGT CTA GGA TCG AGA CAA AG | ||||
| aac(3)-IV | F | TGC TGG TCC ACA GCT CCT TC | 60 | 653 | Boerlin et al. (7) |
| R | CGG ATG CAG GAA GAT CAA | ||||
| aphA1 | F | ATG GGC TCG CGA TAA TGT C | 53 | 600 | Maynard et al. (44) |
| R | CTC ACC GAG GCA GTT CCA T | ||||
| aphA2 | F | GAT TGA ACA AGA TGG ATT GC | 53 | 347 | Travis et al. (68) |
| R | CCA TGA TGG ATA CTT TCT CG | ||||
| aadB | F | GAG GAG TTG GAC TAT GGA TT | 53 | 208 | Travis et al. (68) |
| R | CTT CAT CGG CAT AGT AAA A | ||||
| ampC | F | CCC CGC TTA TAG AGC AAC AA | 53 | 634 | Féria et al. (18) |
| R | TCA ATG GTC GAC TTC ACA CC | ||||
| blaCARB | F | CAA GTA CTT TYA AAA CAA TAG C | 50 | 534 | Henriques et al. (24) |
| R | GCTGTAATACTCCKAGCAC | ||||
| blaCMY-2 | F | AAC ACA CTG ATT GCG TCT GAC | 60 | 1,226 | Pérez-Pérez and Hanson (53) |
| R | CTG GGC CTC ATC GTC AGT TA | ||||
| blaCTX | F | CGA TGT GCA GTA CCA GTA A | 60 | 585 | Batchelor et al. (4) |
| R | TTA GTG ACC AGA ATC AGC GG | ||||
| blaCTX-SEQb | F | TTA ATG ATG ACT CAG AGC ATT C | 52 | 902 | Villegas et al. (72) |
| R | GAT ACC TCG CTC CAT TTA TTG | ||||
| blaOXA | F | TAT CTA CAG CAG CGC CAG TG | 53 | 199 | Féria et al. (18) |
| R | CGC ATC AAA TGC CAT AAG TG | ||||
| blaSHV | F | TCA GCG AAA AAC ACC TTG | 53 | 475 | M'Zali et al. (48) |
| R | TCC CGC AGA TAA ATC ACCA | ||||
| blaTEM | F | TAC GAT ACG GGA GGG CTT AC | 53 | 716 | Belaaouaj et al. (5) |
| R | TTC CTG TTT TTG CTC ACC CA | ||||
| blaTEM-SEQb | F | GGG GAG CTC ATA AAA TTC TTG AAG AC | 59 | 1,100 | Spanu et al. (64) |
| R | GGG GGA TCC TTA CCA ATG CTT AAT CA | ||||
| cat | F | AGT TGC TCA ATG TAC CTA TAA CC | 55 | 547 | Van et al. (70) |
| R | TTG TAA TTC ATT AAG CAT TCT GCC | ||||
| cmlA | F | CCG CCA CGG TGT TGT TGT TAT C | 59 | 698 | Keyes et al. (30) |
| R | CAC CTT GCC TGC CCA TCA TTA G | ||||
| floR | F | TAT CTC CCT GTC GTT CCA G | 53 | 399 | Keyes et al. (30) |
| R | AGA ACT CGC CGA TCA ATG | ||||
| sul2 | F | CGG CAT CGT CAA CAT AAC CT | 66 | 721 | Lanz et al. (34) |
| R | TGT GCG GAT GAA GTC AGC TC | ||||
| sul3 | F | CAA CGG AAG TGG GCG TTG TGG A | 66 | 244 | Kozak et al. (32) |
| R | GCT GCA CCA ATT CGC TGA ACG | ||||
| tet(A) | F | GCT ACA TCC TGC TTG CCT TC | 55 | 210 | Ng et al. (51) |
| R | CAT AGA TCG CCG TGA AGA GG | ||||
| tet(B) | F | TTG GTT AGG GGC AAG TTT TG | 55 | 659 | Ng et al. (51) |
| R | GTA ATG GGC CAA TAA CAC CG | ||||
| tet(C) | F | CTT GAG AGC CTT CAA CCC AG | 55 | 418 | Ng et al. (51) |
| R | ATG GTC GTC ATC TAC CTG CC | ||||
| tet(D) | F | AAA CCA TTA CGG CAT TCT GC | 55 | 787 | Ng et al. (51) |
| R | GAC CGG ATA CAC CAT CCA TC | ||||
| tet(E) | F | AAA CCA CAT CCT CCA TAC GC | 55 | 278 | Ng et al. (51) |
| R | AAA TAG GCC ACA ACC GTC AG | ||||
| tet(G) | F | GCT CGG TGG TAT CTC TGC TC | 55 | 468 | Ng et al. (51) |
| R | AGC AAC AGA ATC GGG AAC AC |
F, forward; R, reverse.
Primers blaCTX-SEQ and blaTEM-SEQ were used to PCR amplify blaCTX and blaTEM genes for sequencing, respectively.
All PCRs were performed in a final volume of 25 μl consisting of 2.5 μl 10× PCR buffer (New England BioLabs, Ipswich, MA), 25 pmol of each primer (MWG-Biotech AG, Ebersberg, Germany), deoxynucleoside triphosphates at a concentration of 200 μM (Promega), 1 U Taq DNA polymerase (New England BioLabs), and 50 ng of genomic DNA.
Sequencing of integron gene cassettes and β-lactam resistance genes.
Amplicons produced using degenerate primer pairs targeting variable regions of class 1 and class 2 integrons and PCR-amplified blaCTX and blaTEM genes of isolates displaying cefpirome resistance were purified using a Promega Wizard PCR and a gel purification system and sequenced commercially by Qiagen Sequencing Services (Qiagen, Hilden, Germany). Sequences were initially assembled using DNAStar software (DNAStar Inc., Madison, WI), and similarity searches were carried out using the BLAST program, available at the NCBI BLAST homepage (http://www.ncbi.nlm.nih.gov/BLAST/).
Plasmid profiling.
Plasmid DNA was purified using a QuickGene plasmid kit S (Fuji, Tokyo, Japan) according to the manufacturer's instructions, followed by electrophoresis in a 0.7% agarose gel (SeaKem LE agarose; Lonza Wokingham, Ltd., United Kingdom) as described above. Two E. coli strains, V517 and 39R861, containing multiple reference plasmids were used as controls and size markers (41).
Conjugation experiments and verification.
The ability to transfer antimicrobial resistance determinants was tested by broth mating experiments for 8 isolates, all of which were selected on the basis of their heterogeneous plasmid profiles and resistance genes. Rifampin-resistant, lactose-negative strain E. coli 26R793 served as a recipient in the conjugation assays. In brief, equal volumes of overnight cultures of the donor and recipient strains grown in Luria-Bertani (LB) broth (Difco Laboratories, Becton Dickinson, Sparks, MD) were mixed and incubated at 37°C for 18 h. The transconjugants were selected on MacConkey agar (Oxoid) supplemented with 100 μg/ml rifampin (Sigma-Aldrich) along with either 50 μg/ml ampicillin, 20 μg/ml chloramphenicol, 50 μg/ml nalidixic acid, 50 μg/ml streptomycin, 30 μg/ml tetracycline, or 50 μg/ml trimethoprim (Sigma-Aldrich, Ireland). Presumptive transconjugants (3 colonies from each antibiotic selection) were subjected to further investigation. Susceptibility tests were performed to confirm transfer as described previously, followed by PCR to examine which resistance genes and integrons had been transferred. A selection of the transconjugants exhibiting different resistance patterns was subjected to plasmid extraction.
RESULTS
Phylogenetic classification.
Isolates belonging to phylogenetic group A were found to be the most abundant in the collection (n = 38; 51%). Twenty-eight percent (n = 21) of the isolates were grouped in the B1 lineage. No isolate was identified in B2, the phylogenetic lineage associated with virulent extraintestinal strains. Twenty percent of the isolates (n = 15) were assigned to group D, which is associated with pathogenic bacteria, although less frequently than group B2 (10).
Resistance patterns.
All isolates studied were resistant to several antimicrobial agents, ranging from 5 to 15 of the antimicrobial compounds tested (see Table S1 in the supplemental material). Resistance to tetracycline (n = 74; 100%), trimethoprim (n = 74; 100%), sulfonamides (n = 73; 98.6%), ampicillin (n = 72; 97.3%), and streptomycin (n = 72; 97.3%) was the most common, followed by cephalothin (n = 54; 73%), chloramphenicol (n = 51; 75.4%), the amoxicillin-clavulanic acid combination (n = 42; 56.8%), cefpodoxime (n = 41; 55.4%), nalidixic acid (n = 41; 55.4%), gentamicin (n = 40; 54%), and ciprofloxacin (n = 38; 51.4%). Thirty-two isolates (43.2%) displayed resistance to neomycin, 30 (40.5%) to florfenicol, 30 (40.5%) to ceftiofur, 9 (12.2%) to cefpirome, and 4 (5.4%) to furazolidone. No amikacin or colistin resistance was observed.
Occurrence of resistance determinants.
Genotyping data corresponded with the resistance phenotypes determined, and in most cases of resistance, previously recognized marker genes were identified.
Resistance to streptomycin was mediated mainly by the strA-strB gene pair, present in 60 isolates (81%), and aadA, identified in 57 isolates (77%). Two isolates phenotypically susceptible to streptomycin (isolates 18 and 19; see Table S1 in the supplemental material) both carried strA-strB, and another isolate, isolate 18, also possessed aadA. All of the isolates displaying neomycin resistance (n = 31) harbored the aphA1 determinant. The aphA1 marker was also present in 9 neomycin-susceptible isolates (a total of 40 aphA1-positive isolates in the collection; 54%). Nineteen isolates (25.7%) were positive for aadB, encoding gentamicin resistance. Seventeen of these were phenotypically resistant, while 2 were susceptible to this antibiotic (isolates 5B and 33). The underlying mechanisms of gentamicin resistance were not identified in a further 23 aadB-negative isolates, all of which showed high-level resistance to this compound. Simultaneous carriage of multiple aminoglycoside resistance determinants was common.
Genes encoding various exogenous β-lactamases were identified in all but 1 (isolate 5B) of the 72 isolates that were defined as being resistant to at least one drug within this class. The blaTEM gene was the most prevalent (n = 66; 89.2%), followed by blaCMY-2 (n = 18; 24.3%), blaCARB (n = 11; 14.9%), blaCTX (n = 8; 10.8%), blaSHV (n = 5; 6.8%), and blaOXA (n = 1; 1.35%). Twenty and 9 isolates simultaneously contained 2 and 3 β-lactam-resistance-associated markers, respectively. The β-lactam resistance phenotypes of the isolates that carried blaTEM (the only detectable β-lactamase gene) differed greatly and were as follows: ampicillin resistance only (n = 9); ampicillin and amoxicillin-clavulanate resistance (n = 7); ampicillin and cephalothin resistance (n = 7); ampicillin, amoxicillin-clavulanate, and cephalothin resistance (n = 5); ampicillin, cefpodoxime, and cephalothin resistance (n = 1); ampicillin, amoxicillin-clavulanate, cefpodoxime, and cephalothin resistance (n = 7); and amoxicillin-clavulanate, ampicillin, amoxicillin-clavulanate, cefpirome, cefpodoxime, ceftiofur, and cephalothin resistance (n = 1). In contrast, blaCTX-positive isolates, all of which also carried blaTEM, had identical resistance patterns and were resistant to the full panel of β-lactam compounds tested, except for the amoxicillin-clavulanate combination. Of the 18 blaCMY-2-positive isolates, 14 also carried blaTEM and 9 also carried blaCARB. All were resistant to a range of β-lactams, including ampicillin, amoxicillin-clavulanate, cefpodoxime, and cephalothin, and 15 displayed additional resistance to ceftiofur. The blaSHV genes were always accompanied by blaTEM, and 4 of 5 isolates that were positive for both of these genes were resistant to ampicillin, cefpodoxime, ceftiofur, and cephalothin, while the remaining isolate displayed resistance to ampicillin and amoxicillin-clavulanate. Isolate 29, which was positive for blaOXA, was also determined to be resistant to ampicillin and amoxicillin-clavulanate. β-Lactam-susceptible isolates did not possess any of these markers, as determined by PCR, except for the endogenous ampC.
Mechanisms of chloramphenicol resistance were identified for 47 of the 51 resistant isolates. The cat gene, encoding chloramphenicol acetyltransferase, was identified in 29 strains (21.5%), whereas the prevalence of cmlA conferring nonenzymatic resistance was significantly lower (n = 12; 8.9%). The chloramphenicol-florfenicol transporter floR was detected in 28 (20.7%) isolates, all of which were coresistant to florfenicol, as expected. This gene was frequently detected along with cat (n = 10), cmlA (n = 10), or both genes (n = 1). In addition, 2 isolates (isolates 21 and CL59) classified florfenicol resistant did not carry floR but were positive by PCR for the cat gene. No associated resistance genes were detected in the 23 isolates susceptible to phenicols.
Resistance to sulfonamides was mediated by the sul2 gene in 66 (89.2%) and the sul1 gene in 56 (75.7%) of the 73 sulfonamide-resistant isolates. Forty-five isolates possessed both sul1 and sul2. A single isolate (denoted isolate 17; see Table S1 in the supplemental material) possessed the sul3 marker. A single sulfonamide-susceptible isolate did not carry any of the known sul genes.
Tetracycline resistance, observed in all isolates of this collection, was mediated predominantly by tet(B), identified in 37 (50%) isolates, or tet(A), identified in 31 isolates (41.9%), while 3 isolates (4%) were found to possess tet(D). Six of these (8.1%) possessed both tet(A) and tet(B). Tetracycline resistance genes were not identified in the further 9 isolates classified to be resistant to this antibiotic.
Sequence analysis of selected β-lactamase genes.
Nucleotide sequence analysis revealed that all 8 cefpirome-resistant isolates that were positive for blaTEM and blaCTX carried identical variants of these genes and included blaTEM-1, encoding a classical narrow-spectrum β-lactamase conferring ampicillin resistance, and blaCTX-M-2, conferring the extended-spectrum-β-lactam resistance phenotype.
Integrons and gene cassettes.
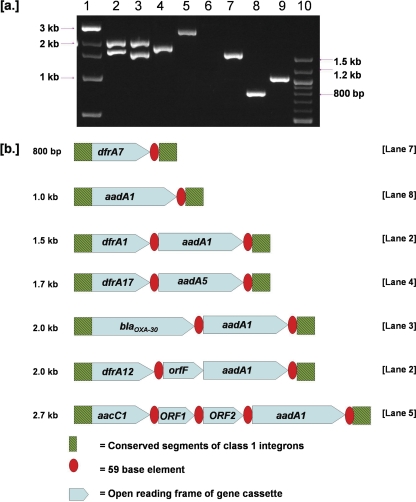
Seventy isolates (94.6%) were determined to possess the class 1 integrase gene (intI1), and most of these (n = 56; 76%) also carried both the qacEΔ1 and sul1 genes, located within the 3′-CS. PCR amplification with consensus primers targeting the regions flanking the gene cassettes yielded 11 different gene cassettes. Integrase I (intI1)-positive isolates possessed none (n = 9; 12.2%), 1 (n = 53; 71.6%), 2 (n = 4; 5.4%), or 3 (n = 4; 5.4%) gene cassettes. The following amplicons and their corresponding sizes were identified: 800 bp (n = 1; 1.4%), 1 kb (n = 17; 23%), 1.3 kb (n = 4; 5.4%), 1.5 kb (n = 45; 60.8%), 1.7 kb (n = 5; 6.8%), 2 kb (n = 5; 6.8%), and 2.7 kb (n = 6; 8.1%). A representative of each of these gene cassettes was subjected to DNA sequencing. A summary of the sequencing data is provided in Table 2, and a genetic map is shown in Fig. 1.
Table 2.
Integron-associated gene cassettes identified in MDR E. coli isolates recovered from animals presenting at the University Veterinary Hospital
| Integron class | Approximate size (kb) | Gene cassette array |
|---|---|---|
| 1 | 0.8 | dfr7 |
| 1 | 1.0 | aadA1 |
| 1 | 1.3 | aadA1 |
| 1 | 1.5 | dfrA1-aadA1 |
| 1 | 1.7 | dfrA17-aadA5 |
| 1 | 2.0 | dfrA12-orfF-aadA2 |
| 1 | 2.0 | blaOXA-30-aadA1 |
| 1 | 2.7 | aacC1-orf1-orf2- aadA1 |
| 2 | 2.6 | dfrA1-sat1-aadA1 |
Fig. 1.
(a) Representation of the sequenced gene cassettes amplified with primers targeting conserved segments of the class 1 integrons after electrophoretical separation in 1.7% agarose gel. Lane 1, 1-kb DNA size marker (New England BioLabs); lane 2, E. coli 18; lane 3, E. coli 29; lane 4, E. coli 28; lane 5, E. coli 21; lane 6, empty; lane 7, E. coli 16; lane 8, E. coli 2; lane 9, E. coli CL45; lane 10, 100-bp DNA size marker. (b) Schematic representation of the organization of the sequenced gene cassettes of the class 1 integrons. Arrowheads represent the direction of the transcription. Integron-associated conserved features are indicated.
The 800-bp amplicon contained a single open reading frame (ORF) that was 99% similar to a previously reported dfr7 gene cassette (GenBank accession no. X58425). The aadA1 gene, encoding an aminoglycoside-modifying enzyme, was identified within the variable regions of both the 1- and 1.3-kb cassettes (99% similarity to a sequence from GenBank with accession no. EF078697). Variable regions of 1.5 kb contained an array of the dfrA1 and aadA1 genes, associated with trimethoprim and streptomycin resistance, respectively (99% similarity with the sequence with GenBank accession no. GQ293501). The variable region of 1.7 kb contained a dfrA17 marker conferring resistance to trimethoprim and the aminoglycoside acetyltransferase aadA5 gene conferring resistance to streptomycin-spectinomycin and exhibited 100% similarity to the sequence with GenBank accession no. GU055937. Sequence analysis of the 2-kb amplicon revealed that it contained a dfrA12-orfF-aadA2 gene arrangement with 99% similarity to the sequence with GenBank accession no. HM043573. Another amplicon of approximately 2 kb from isolate 29 (see Table S1 in the supplemental material) was positive for blaOXA and found to contain this gene located within the gene cassette with the classical head-to-tail blaOXA-30-aadA1 arrangement. These genes were identical to a sequence previously deposited in GenBank (accession no. F543148). The aacC1 gene (encoding a 3-N-aminoglycoside acetyltransferase, involved in gentamicin resistance), orf1, orf2, and the aadA1 gene array were identified within the 2.7-kb variable region (99% similarity to the sequence with GenBank accession no. AF550679).
Ten isolates (13.5%) possessed a class 2 integrase gene along with a 2.6-kb variable region containing a dfrA1-sat1-aadA1 gene cassette which exhibited 100% nucleotide sequence similarity to a sequence deposited in the GenBank database under accession no. AB188272.
Analysis of plasmid content.
Overall, 35 diverse plasmid profiles were observed in the strain collection (data not shown). Nineteen isolates carried single plasmids and 17 possessed 2 plasmids, while 13 and 5 isolates carried 3 and 4 plasmids, respectively. No plasmids were detected in 20 (27%) of the 74 isolates. Large plasmids with sizes equal to or greater than 50 kb were observed in 40 isolates (54%).
Conjugal transfer of resistance markers.
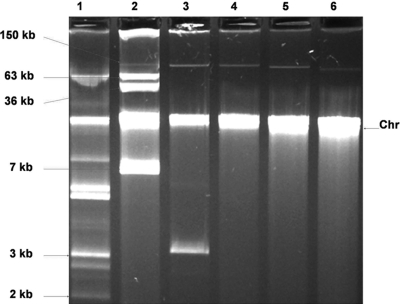
Eight isolates were selected and tested for their ability to transfer one or more of the resistance markers to a susceptible E. coli recipient (Table 3). Resistance to between 1 and 5 antimicrobial classes was transferred, and multiresistance transfer occurred frequently. One isolate (E. coli CL42; see Table S1 in the supplemental material) could transfer its entire resistance repertoire (resistance to 12 antimicrobial compounds). Relevant genes matching the phenotypic resistance profiles were also identified (Table 3). Plasmid profiling revealed that transconjugants principally acquired high-molecular-weight plasmids of approximately 100 kb or larger (Fig. 2).
Table 3.
Summary of phenotypic and genotypic characterization of transconjugants obtained in broth mating assays with MDR E. coli isolates from animals presenting at the University Veterinary Hospitala
| Strain no./origin | Resistance profile of donor | Resistance transferred to E. coli recipient/resistance gene(s) identified in transconjugantsd | Plasmid(s) acquired (kb) |
|---|---|---|---|
| 11/equine | AmcAmpCCfrCipCpdCplGmNaSSuTeTmp | AmcAmpCfrCpdCpl/blaCMY-2 | ∼100 |
| AmcAmpCfrCpdCplGmS/intI1-1-kb GCb(1)-qacEΔ1-aadA-blaCMY-2 | ∼100 | ||
| AmcAmpCfrCpdCplSSuTmp/intI1-1.5-kb GC(1)-qacEΔ1-sul1- blaCMY-2-blaTEM-strA-strB-sul2 | 2×c ∼100 | ||
| 15/equine | AmcAmpCCfrCipCpdCplFfcNNaSSuTeTmp | AmpGmNS/aphA1-blaTEM-strA-strB | ∼100 |
| AmpGmNSSuTeTmp/intI1-1.5-kb GC(1)-qacEΔ1-sul1-aadA- aphA1-blaTEM-strA-strB-sul2-tet(A) | ∼7 | ||
| 31/canine | AmcAmpCCipFfcNaSSuTeTmp | CFfcSSuTe/floR-strA-strB--sul2-tet(B) | ∼150 |
| 32/canine | AmcAmpCCipFfcNaSSuTeTmp | AmcAmp/blaTEM | ∼100 |
| CFfcSSuTe/floR-strA-strB-sul2-tet(B) | 2× ∼100 | ||
| CL42/equine | AmpCCfpCfrCpdCplGmNSSuTeTmp | AmpCCfpCfrCpdCplGmNSSuTeTmp/intI1-1.5-kb GC(1)-qacEΔ1-sul1-aadA-aphA1-blaCTX-M-2-blaTEM-cat-strA-strB-sul2-tet(B) | ∼150 |
| AmpCCfpCfrCpdCplGmNSuTeTmp/intI1-1.5-kb GC(1)-qacEΔ1-sul1-aadA-aphA1-blaCTX-M-2-cat-tet(B) | |||
| CL62/equine | AmcAmpCCfrCpdCplFfcGmNSSuTeTmp | AmpNSSuTe/aphA1-blaTEM-strA-strB-sul2-tet(A) | ∼150 |
Two independent mating-out experiments were performed. Resistance transfer was not observed with E. coli isolates 6 and 29.
GC, gene cassette.
2×, the presence of two plasmids of equal approximate size.
(1) indicates the presence of a class 1 integron type.
Fig. 2.
Agarose gel image showing plasmid profiles of E. coli 31 and its transconjugants. Chr, chromosomal DNA. Lane 1, E. coli 39R861; lane 2, E. coli V517; lane 3, E. coli 31 (AmcAmpCCipFfcNaSSuTeTmp resistance profile); lanes 4, 5, and 6, E. coli 31-derived transconjugants selected on chloramphenicol, streptomycin, and tetracycline, respectively, all with the CFfcSSuTe resistance profile.
DISCUSSION
In veterinary hospitals, the ongoing usage of antimicrobial compounds for treatment of animals increases the selective pressure for emergence of MDR organisms and dissemination of resistance (16, 50). Antimicrobial agents frequently used in referral veterinary hospitals include broad-spectrum-activity drugs, such as β-lactams and new cephalosporins, as well as fluoroquinolones. Thus, hospitalized animals may constitute an important reservoir of antimicrobial resistance (20).
Of the 74 E. coli isolates investigated in this study, the majority (80%) belonged to phylogroups A and B1 and are therefore classified commensal bacteria, but a number of strains, within group D (20%), were classified potentially pathogenic. Bacteria of classes A and B1 are rarely reported to possess virulence genes (10). However, although typing of isolates from humans showed a good correlation of phylogenetic group with pathogenicity (10), animal-associated extraintestinal pathogenic E. coli (ExPEC) can be phylogenetically distinct. Thus, caution should be exercised when defining such bacteria as commensals (19, 22). Furthermore, commensal E. coli strains can cause extraintestinal disease when predisposing factors for infection are present (57). Therefore, although most of the isolates investigated in this study may be phylogenetically classified commensal organisms, they are not representative of commensal strains from healthy animals in the community.
The presence of class 1 integrons, associated with resistance to trimethoprim, streptomycin, and sulfonamide, is in agreement with previous investigations. The variable gene cassette regions characterized in this study were previously reported and are widely disseminated among the Enterobacteriaceae (28, 40). In addition, identical class 2 integrons, associated with Tn7 and Tn7-like transposable elements, have been identified previously (3, 40, 73). The absence of the 3′ conserved structure (containing qacEΔ1-sul1) in several isolates has been reported on occasions by others, and such genetic elements are sometimes associated with the sul3 gene (63).
Molecular investigations of the underlying aminoglycoside resistance mechanisms revealed that the strA-strB gene pair was the most prevalent (81%) among the determinants identified, followed by aadA (77%) and the less frequent aphA1 (54%) and aadB (25.7%) markers. This observation is consistent with earlier reports showing that these genes are common in isolates resistant to streptomycin and/or other aminoglycoside drugs (65). The fact that aphA1 was present in 9 isolates phenotypically susceptible to neomycin and, likewise, the presence of strA-strB and/or aadA in streptomycin-susceptible isolates could be explained by the existence of defective genes or reduced expression of these markers, required to confer the phenotype.
The most commonly detected β-lactamase gene in this study was blaTEM, occurring in 66 of 74 strains. The prevalence of blaTEM is in good agreement with previous reports that found this marker to be widely disseminated among the Enterobacteriaceae from veterinary sources (38). The diversity of the β-lactam resistance phenotype among blaTEM-positive isolates from the current strain collection may be due to the presence of altered gene variants in some isolates or the overexpression of endogenous, chromosomally encoded AmpC enzyme (27). TEM enzymes, including ESβLs, are typically inhibited by clavulanic acid. Nonetheless, we demonstrated the transfer of ampicillin and amoxicillin-clavulanate resistance from a canine isolate (E. coli isolate 32) by conjugation (Table 3) that was directly linked with the presence of blaTEM, indicating that an inhibitor-resistant enzyme variant was present. Inhibitor-resistant TEM enzymes (IRT) have been reported worldwide (5, 43).
The shv determinant was detected in a smaller number of isolates: a canine isolate (denoted isolate 31 in Table S1 in the supplemental material) was resistant to ampicillin and amoxicillin-clavulanate, along with 4 equine fecal isolates (denoted isolates CL50, CL61, CL64, and CL78). It is possible that variants of these enzymes in this isolate collection may have arisen following different levels of selective pressure. Moreover, due to the concomitant presence of blaTEM genes, it is difficult to speculate as to the contribution of each of the genes to this resistance phenotype. Both TEM and SHV enzymes belong to the class A family of β-lactamases and are ubiquitous among E. coli isolates, being frequently associated with therapeutic failure (43, 69).
The presence of blaCTX-M-2 in a number of isolates from horses is interesting, considering the global distribution of these genes. CTX-M enzymes are currently regarded as the predominant ESβL type of animal origin, while they have also been associated with human isolates in Europe since the late 1990s (39). In Ireland, blaCTX-M-mediated extended-spectrum β-lactam resistance is widespread, as demonstrated by a recent nationwide survey of Enterobacteriaceae isolates recovered from clinical cases in humans over an 11-year period (46). CTX-M group 1 followed by CTX-M group 9, represented by CTX-M-15 and CTX-M-14, respectively, are the most frequently reported examples (46). In our study, the transmissibility of blaCTX-M-2 was demonstrated following conjugation, and this observation was consistent with our current knowledge of CTX-M enzymes, which are typically carried by multiresistance plasmids (56).
We identified the blaCMY-2 gene in 16 isolates (2 bovine, 1 canine, and 13 equine). Consistent with the genotype, all of these isolates displayed amoxicillin-clavulanate resistance and, and in most cases, were resistant to cefpodoxime and ceftiofur. To our knowledge, neither blaCMY-2 nor blaCTX-M-2 has previously been reported in animals in Ireland. A growing incidence of blaCMY-2 in recent years has been linked by some investigators to the extensive use of ceftiofur (25, 67), an expanded-spectrum veterinary cephalosporin, while others have suggested no significant impact of selection pressure on the frequency and transfer of the blaCMY-2 genes (13). Two recent studies investigating the mechanisms of β-lactam resistance in horses identified blaCTX-M-1, blaCMY-2, blaTEM-1, and blaSHV-1 β-lactamase-encoding genes (1, 73). However, to our knowledge, the blaCTX-M-2 gene has never been reported in bacteria of equine origin.
Despite the fact that chloramphenicol is banned by European Union legislation from use in food-producing animals, resistance to this antimicrobial remains prevalent (61). The cat gene, responsible for enzymatic resistance, was found to be predominant among our isolates. Furthermore, conjugal transfer of cat-mediated chloramphenicol resistance associated with a high-molecular-weight plasmid was demonstrated for one equine isolate. The resulting transconjugants were also resistant to a wide array of β-lactams, aminoglycosides, sulfonamides, and tetracycline (Table 3). These data support the findings in other studies that reported chloramphenicol acetyltransferases to be frequent causes of resistance to this antimicrobial compound (47). The occurrence of cmlA, encoding a specific chloramphenicol transporter, was significantly lower in the collection (n = 12; 9%), and its transfer by conjugation was not observed. As expected, florfenicol resistance occurred concomitantly with resistance to chloramphenicol and could be linked directly to the presence of floR (28 of 30 Ffc-resistant strains) or cat (2 isolates). Florfenicol, a fluorinated derivative of chloramphenicol, is a strictly controlled veterinary drug that is licensed for use in respiratory infections in cattle and pigs (61). The floR gene is a common genetic determinant responsible for florfenicol resistance in Gram-negative bacteria, including animal-derived E. coli strains (12, 15, 30). In our study, floR-mediated florfenicol resistance was transferred by conjugation along with plasmids of approximately 100 to 150 kb from one equine and two canine isolates. Previous reports showed that the floR determinant was either chromosomally located or resided on large conjugative plasmids (6, 12, 15, 74).
Common sulfonamide resistance in the collection could be attributed to the presence of the sul1 and sul2 genes. Our PCR data showed that of 74 strains investigated, 56 tested positive for sul1, 66 tested positive for sul2, and only 1 carried sul3. Transfer of both sul1 and sul2 along with other resistance genes was observed. Examination of the transconjugants revealed that sulfonamide resistance was associated with large self-transmissible plasmids of approximately 100 to 150 kb and, in one case, a plasmid of 7 kb, all of which conferred a multidrug resistance phenotype (see Table 3 for details). This is in agreement with earlier studies that reported sul1 and sul2 to be common and occurring at similar frequencies among bacteria from the Enterobacteriaceae family (32, 54). The suI1 gene is usually plasmid encoded and strongly associated with Tn21-like class 1 integrons, wherein it constitutes part of the 3′ conserved structure (21, 55). Thus, it is to be expected that sul1 would be cotransferred with genetic markers such as intI1 and qacEΔ1. The sul2 marker is usually located on small plasmids of the IncQ family and also on plasmids represented by pBP1 (71, 78). Enne and coworkers, in their study, demonstrated the transfer of a sul2 determinant along with its association with large multiresistance plasmids in a collection of bacteria originating from the United Kingdom (17). In a more recent study, the occurrence of sul3 was found to be limited mainly to porcine E. coli isolates (32). Similarly, in our study, this genetic determinant was identified in only a single porcine isolate.
Of the 6 tetracycline resistance genes analyzed by PCR, only tet(A) and tet(B) were detected in the collection, with 42% of strains carrying tet(A) and 50% being positive for tet(B). Six of these isolates possessed both determinants. Transfer of both tet(A) and tet(B) was subsequently demonstrated in mating assays with selected strains. These data supported the findings of others with respect to the prevalence of the tet genes. Both determinants were previously found to be particularly abundant in tetracycline-resistant E. coli isolates cultured from animals. However, their distribution varied in different animal species and in different geographical regions (23, 33, 35, 62).
In summary, this report showed that the occurrence of integrons among MDR E. coli isolates from hospitalized animals in Ireland is high (94.6%). Their contribution to resistance appears to be directed against older antimicrobial compounds, including streptomycin, trimethoprim, and sulfonamides. MDR phenotypes, including resistance to newer drugs, are transferable and in some cases directly linked to the presence of large conjugative plasmids carrying multiple resistance determinants and integrons. This observation can help to explain the coresistance to several unrelated drug classes that was detected in these isolates and, importantly, emphasizes the dangers of coselection. Colonization of hospitalized horses with multiresistant ESβL and/or AmpC-like enzymes producing E. coli is particularly challenging, given the importance of these drugs and the potential for their dissemination to the environment and individuals such as veterinarians and others who have direct contact with these animals. Identification of resistance genes allied to the investigation of their transfer potential among bacteria of animal origin provides valuable information regarding this important resistance gene pool and the dynamics of resistance gene exchange. Further studies are required to establish the true nature of the animal resistance reservoir.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Department of Agriculture, Fisheries and Food of Ireland (Research Stimulus Fund 06-364).
We thank Jørgen Leisner (Department of Veterinary Pathobiology, Faculty of Life Sciences, University of Copenhagen) for the kind gift of bacterial strains E. coli V517 and E. coli 39R861 and Vivi Miriagou (Laboratory of Bacteriology, Hellenic Pasteur Institute, Athens, Greece) for E. coli 26R793 (Rifr, lac negative).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 19 August 2011.
REFERENCES
- 1. Ahmed M. O., Clegg P. D., Williams N. J., Baptiste K. E., Bennett M. 2010. Antimicrobial resistance in equine faecal Escherichia coli isolates from North West England. Ann. Clin. Microbiol. Antimicrob. 7:9–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bager F., Aarestrup F. M., Madsen M., Wegener H. C. 1999. Glycopeptide resistance in Enterococcus faecium from broilers and pigs following discontinued use of avoparcin. Microb. Drug Resist. 5:53–56 [DOI] [PubMed] [Google Scholar]
- 3. Barlow R. S., Gobius K. S. 2006. Diverse class 2 integrons in bacteria from beef cattle sources. J. Antimicrob. Chemother. 58:1133–1138 [DOI] [PubMed] [Google Scholar]
- 4. Batchelor M., et al. 2005. BlaCTX-M genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob. Agents Chemother. 49:1319–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belaaouaj A., et al. 1994. Nucleotide sequences of the genes coding for TEM-like β-lactamases IRT-1 and IRT-2 (formerly called TRI-1 and TRI-2). FEMS Microbiol. Lett. 120:75–80 [DOI] [PubMed] [Google Scholar]
- 6. Blickwede M., Schwarz S. 2004. Molecular analysis of florfenicol-resistant Escherichia coli isolates from pigs. J. Antimicrob. Chemother. 53:58–64 [DOI] [PubMed] [Google Scholar]
- 7. Boerlin P., et al. 2005. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl. Environ. Microbiol. 71:6753–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carattoli A. 2001. Importance of integrons in the diffusion of resistance. Vet. Res. 32:243–259 [DOI] [PubMed] [Google Scholar]
- 9. Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clermont O., Bonacorsi S., Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clinical Laboratory Standards Institute 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 3rd ed. CLSI document M31-A3 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 12. Cloeckaert A., et al. 2000. Plasmid-mediated florfenicol resistance by the floR gene in Escherichia coli isolated from cattle. Antimicrob. Agents Chemother. 44:2858–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daniels J. B., et al. 2009. Role of ceftiofur in selection and dissemination of blaCMY-2-mediated cephalosporin resistance in Salmonella enterica and commensal Escherichia coli isolates from cattle. Appl. Environ. Microbiol. 75:3648–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dhanarani T. S., et al. 2009. Study on acquisition of bacterial antibiotic resistance determinants in poultry litter. Poult. Sci. 88:1381–1387 [DOI] [PubMed] [Google Scholar]
- 15. Doublet B., et al. 2002. Molecular analysis of chromosomally florfenicol resistant Escherichia coli isolates from France and Germany. J. Antimicrob. Chemother. 49:49–54 [DOI] [PubMed] [Google Scholar]
- 16. Dunowska M., Morley P. S., Traub-Dargatz J. L., Hyatt D. R., Dargatz D. A. 2006. Impact of hospitalization and antimicrobial drug administration on antimicrobial susceptibility patterns of commensal Escherichia coli isolated from the feces of horses. J. Am. Vet. Med. Assoc. 228:1909–1917 [DOI] [PubMed] [Google Scholar]
- 17. Enne V. I., Livermore D. M., Stephens P., Hall L. M. 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325–1328 [DOI] [PubMed] [Google Scholar]
- 18. Féria C., Ferreira E., Correia J. D., Gonçalves J., Caniça M. 2002. Patterns and mechanisms of resistance to β-lactams and β-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. J. Antimicrob. Chemother. 49:77–85 [DOI] [PubMed] [Google Scholar]
- 19. Ghanbarpour R., Oswald E. 2010. Phylogenetic distribution of virulence genes in Escherichia coli isolated from bovine mastitis in Iran. Res. Vet. Sci. 88:6–10 [DOI] [PubMed] [Google Scholar]
- 20. Guardabassi L., Schwarz S., Lloyd D. H. 2004. Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 54:321–332 [DOI] [PubMed] [Google Scholar]
- 21. Hall R. M., Collis C. M. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593–600 [DOI] [PubMed] [Google Scholar]
- 22. Hancock V., Nielsen E. M., Krag L., Engberg J., Klemm P. 2009. Comparative analysis of antibiotic resistance and phylogenetic group patterns in human and porcine urinary tract infectious Escherichia coli. APMIS 117:786–790 [DOI] [PubMed] [Google Scholar]
- 23. Hartman A. B., Essiet I. I., Isenbarger D. W., Lindler L. E. 2003. Epidemiology of tetracycline resistance determinants in Shigella spp. and enteroinvasive Escherichia coli: characterization and dissemination of tet(A)-1. J. Clin. Microbiol. 41:1023–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Henriques I. S., Fonseca F., Alves A., Saavedra M. J., Correia A. 2006. Occurrence and diversity of integrons and beta-lactamase genes among ampicillin-resistant isolates from estuarine waters. Res. Microbiol. 157:938–947 [DOI] [PubMed] [Google Scholar]
- 25. Jiang X., Yang H., Dettman B., Doyle M. P. 2006. Analysis of fecal microbial flora for antibiotic resistance in ceftiofur-treated calves. Foodborne Pathog. Dis. 3:355–365 [DOI] [PubMed] [Google Scholar]
- 26. Johnson J. R., Clabots C., Kuskowski M. A. 2008. Multiple-host sharing, long-term persistence, and virulence of Escherichia coli clones from human and animal household members. J. Clin. Microbiol. 46:4078–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jørgensen R. L., Nielsen J. B., Friis-Møller A., Fjeldsøe-Nielsen H., Schønning K. 2010. Prevalence and molecular characterisation of clinical isolates of Escherichia coli expressing an AmpC phenotype. J. Antimicrob. Chemother. 65:460–464 [DOI] [PubMed] [Google Scholar]
- 28. Kadlec K., Schwarz S. 2008. Analysis and distribution of class 1 and class 2 integrons and associated gene cassettes among Escherichia coli isolates from swine, horses, cats and dogs collected in the BfT-GermVet monitoring study. J. Antimicrob. Chemother. 62:469–473 [DOI] [PubMed] [Google Scholar]
- 29. Kerrn M. B., Klemmensen T., Frimodt-Møller N., Espersen F. 2002. Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. J. Antimicrob. Chemother. 50:513–516 [DOI] [PubMed] [Google Scholar]
- 30. Keyes K., et al. 2000. Detection of florfenicol resistance genes in Escherichia coli from sick chickens. Antimicrob. Agents Chemother. 44:421–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koeleman J. G., Stoof J., Van Der Bijl M. W., Vandenbroucke-Grauls C. M., Savelkoul P. H. 2001. Identification of epidemic strains of Acinetobacter baumannii by integrase gene PCR. J. Clin. Microbiol. 39:8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kozak G. K., et al. 2009. Distribution of sulfonamide resistance genes in Escherichia coli and Salmonella from swine and chickens at abattoirs in Ontario and Quebec, Canada. Appl. Environ. Microbiol. 75:5999–6001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumai Y., et al. 2005. Characterisation of multidrug-resistance phenotypes and genotypes of Escherichia coli strains isolated from swine from an abattoir in Osaka, Japan. Epidemiol. Infect. 133:59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lanz R., Kuhnert P., Boerlin P. 2003. Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet. Microbiol. 91:73–84 [DOI] [PubMed] [Google Scholar]
- 35. Lee C., Langlois B. E., Dawson K. A. 1993. Detection of tetracycline resistance determinants in pig isolates from three herds with different histories of antimicrobial agent exposure. Appl. Environ. Microbiol. 59:1467–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lévesque C., Piché L., Larose C., Roy P. H. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Levy S. B., Fitzgerald G. B., Macone A. B. 1976. Spread of antibiotic resistance plasmids from chicken to chicken and from chicken to man. Nature 260:40–42 [DOI] [PubMed] [Google Scholar]
- 38. Li X. Z., Mehrotra M., Ghimire S., Adewoye L. 2007. Beta-lactam resistance and beta-lactamases in bacteria of animal origin. Vet. Microbiol. 121:197–214 [DOI] [PubMed] [Google Scholar]
- 39. Livermore D. M., et al. 2007. CTX-M: changing the face of ESΒLs in Europe. J. Antimicrob. Chemother. 59:165–174 [DOI] [PubMed] [Google Scholar]
- 40. Machado E., et al. 2005. Integron content of extended-spectrum-beta-lactamase-producing Escherichia coli strains over 12 years in a single hospital in Madrid, Spain. Antimicrob. Agents Chemother. 49:1823–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Macrina F. L., Kopecko D. J., Jones K. R., Ayers D. J., McCowen S. M. 1978. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid 1:417–420 [DOI] [PubMed] [Google Scholar]
- 42. Madsen L., Aarestrup F. M., Olsen J. E. 2000. Characterisation of streptomycin resistance determinants in Danish isolates of Salmonella Typhimurium. Vet. Microbiol. 75:73–82 [DOI] [PubMed] [Google Scholar]
- 43. Matagne A., Lamotte-Brasseur J., Frère J. M. 1998. Catalytic properties of class A beta-lactamases: efficiency and diversity. Biochem. J. 330:581–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maynard C., et al. 2003. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149:K91 isolates obtained over a 23-year period from pigs. Antimicrob. Agents Chemother. 47:3214–3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mazel D., Dychinco B., Webb V. A., Davies J. 2000. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44:1568–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morris D., et al. 2009. CTX-M enzymes are the predominant extended-spectrum beta-lactamases produced by Enterobacteriaceae in Ireland. J. Antimicrob. Chemother. 64:864–866 [DOI] [PubMed] [Google Scholar]
- 47. Murray I. A., Shaw W. V. 1997. O-Acetyltransferases for chloramphenicol and other natural products. Antimicrob. Agents Chemother. 41:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. M'Zali F. H., Gascoyne-Binzi D. M., Heritage J., Hawkey P. M. 1996. Detection of mutations conferring extended-spectrum activity on SHV β-lactamase using polymerase chain reaction single strand conformational polymorphism (PCR-SSCP). J. Antimicrob. Chemother. 37:797–802 [DOI] [PubMed] [Google Scholar]
- 49. Nagachinta S., Chen J. 2008. Transfer of class 1 integron-mediated antibiotic resistance genes from Shiga toxin-producing Escherichia coli to a susceptible E. coli K-12 strain in storm water and bovine feces. Appl. Environ. Microbiol. 74:5063–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nam H. M., et al. 2010. Prevalence of antimicrobial resistance in fecal Escherichia coli isolates from stray pet dogs and hospitalized pet dogs in Korea. Microb. Drug Resist. 16:75–79 [DOI] [PubMed] [Google Scholar]
- 51. Ng L. K., Martin I., Alfa M., Mulvey M. 2001. Multiplex PCR for the detection of tetracycline-resistant genes. Mol. Cell. Probes 15:209–215 [DOI] [PubMed] [Google Scholar]
- 52. Partridge S. R., Tsafnat G., Coiera E., Iredell J. R. 2009. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33:757–784 [DOI] [PubMed] [Google Scholar]
- 53. Pérez-Pérez F. J., Hanson N. D. 2002. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rådström P., Swedberg G., Sköld O. 1991. Genetic analyses of sulfonamide resistance and its dissemination in gram-negative bacteria illustrate new aspects of R plasmid evolution. Antimicrob. Agents Chemother. 35:1840–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Recchia G. D., Hall R. M. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015–3027 [DOI] [PubMed] [Google Scholar]
- 56. Rossolini G. M., D'Andrea M. M., Mugnaioli C. 2008. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 1(Suppl.):33–41 [DOI] [PubMed] [Google Scholar]
- 57. Russo T. A., Johnson J. R. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5:449–456 [DOI] [PubMed] [Google Scholar]
- 58. Salyers A. A., Amabile-Cuevas C. F. 1997. Why are antibiotic resistance genes so resistant to elimination? Antimicrob. Agents Chemother. 41:2321–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sanchez S., et al. 2002. Characterization of multidrug-resistant Escherichia coli isolates associated with nosocomial infections in dogs. J. Clin. Microbiol. 40:3586–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sandvang D., Aarestrup F. M., Jensen L. B. 1997. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 157:177–181 [DOI] [PubMed] [Google Scholar]
- 61. Schwarz S., Kehrenberg C., Doublet B., Cloeckaert A. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28:519–542 [DOI] [PubMed] [Google Scholar]
- 62. Sengeløv G., Halling-Sørensen B., Aarestrup F. M. 2003. Susceptibility of Escherichia coli and Enterococcus faecium isolated from pigs and broiler chickens to tetracycline degradation products and distribution of tetracycline resistance determinants in E. coli from food animals. Vet. Microbiol. 95:91–101 [DOI] [PubMed] [Google Scholar]
- 63. Soufi L., et al. 2009. Prevalence and diversity of integrons and associated resistance genes in Escherichia coli isolates from poultry meat in Tunisia. Foodborne Pathog. Dis. 6:1067–1073 [DOI] [PubMed] [Google Scholar]
- 64. Spanu T., et al. 2002. Occurrence of extended-Spectrum β-lactamases in members of the family Enterobacteriaceae in Italy: implications for resistance to β-lactams and other antimicrobial drugs. Antimicrob. Agents Chemother. 46:196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sunde M., Norström M. 2006. The prevalence of, associations between and conjugal transfer of antibiotic resistance genes in Escherichia coli isolated from Norwegian meat and meat products. J. Antimicrob. Chemother. 58:741–747 [DOI] [PubMed] [Google Scholar]
- 66. Tamang M. D., et al. 2007. Emergence of multidrug-resistant Salmonella enterica serovar Typhi associated with a class 1 integron carrying the dfrA7 gene cassette in Nepal. Int. J. Antimicrob. Agents 30:330–335 [DOI] [PubMed] [Google Scholar]
- 67. Tragesser L. A., Wittum T. E., Funk J. A., Winokur P. L., Rajala-Schultz P. J. 2006. Association between ceftiofur use and isolation of Escherichia coli with reduced susceptibility to ceftriaxone from fecal samples of dairy cows. Am. J. Vet. Res. 67:1696–1700 [DOI] [PubMed] [Google Scholar]
- 68. Travis R. M., et al. 2006. Chloramphenicol and kanamycin resistance among porcine Escherichia coli in Ontario. J. Antimicrob. Chemother. 58:173–177 [DOI] [PubMed] [Google Scholar]
- 69. Tzouvelekis L. S., Bonomo R. A. 1999. SHV-type beta-lactamases. Curr. Pharm. Des. 5:847–864 [PubMed] [Google Scholar]
- 70. Van T. T., Chin J., Chapman T., Tran L. T., Coloe P. J. 2008. Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int. J. Food Microbiol. 124:217–223 [DOI] [PubMed] [Google Scholar]
- 71. Van Treeck U. F., Schmidt F., Wiedemann B. 1981. Molecular nature of a streptomycin and sulfonamide resistance plasmid (pBPI) prevalent in clinical Escherichia coli strains and integration of an ampicillin resistance transposon (TnA). Antimicrob. Agents Chemother. 19:371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Villegas M. V., et al. 2004. CTX-M-12 β-lactamase in a Klebsiella pneumoniae clinical isolate in Colombia. Antimicrob. Agents Chemother. 48:629–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vo A. T., van Duijkeren E., Fluit A. C., Gaastra W. 2007. Characteristics of extended-spectrum cephalosporin-resistant Escherichia coli and Klebsiella pneumoniae isolates from horses. Vet. Microbiol. 124:248–255 [DOI] [PubMed] [Google Scholar]
- 74. White D. G., et al. 2000. Characterization of chloramphenicol and florfenicol resistance in Escherichia coli associated with bovine diarrhea. J. Clin. Microbiol. 38:4593–4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. White P. A., McIver C. J., Rawlinson W. D. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Witte W. 1998. Medical consequences of antibiotic use in agriculture. Science 279:996–997 [DOI] [PubMed] [Google Scholar]
- 77. Wright G. D. 2007. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5:175–186 [DOI] [PubMed] [Google Scholar]
- 78. Yau S., Liu X., Djordjevic S. P., Hall R. M. 2010. RSF1010-like plasmids in Australian Salmonella enterica serovar Typhimurium and origin of their sul2-strA-strB antibiotic resistance gene cluster. Microb. Drug Resist. 16:249–252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.