Abstract
Sodium gradients in cyanobacteria play an important role in energy storage under photoautotrophic conditions but have not been well studied during autofermentative metabolism under the dark, anoxic conditions widely used to produce precursors to fuels. Here we demonstrate significant stress-induced acceleration of autofermentation of photosynthetically generated carbohydrates (glycogen and sugars) to form excreted organic acids, alcohols, and hydrogen gas by the halophilic, alkalophilic cyanobacterium Arthrospira (Spirulina) maxima CS-328. When suspended in potassium versus sodium phosphate buffers at the start of autofermentation to remove the sodium ion gradient, photoautotrophically grown cells catabolized more intracellular carbohydrates while producing 67% higher yields of hydrogen, acetate, and ethanol (and significant amounts of lactate) as fermentative products. A comparable acceleration of fermentative carbohydrate catabolism occurred upon dissipating the sodium gradient via addition of the sodium-channel blocker quinidine or the sodium-ionophore monensin but not upon dissipating the proton gradient with the proton-ionophore dinitrophenol (DNP). The data demonstrate that intracellular energy is stored via a sodium gradient during autofermentative metabolism and that, when this gradient is blocked, the blockage is compensated by increased energy conversion via carbohydrate catabolism.
INTRODUCTION
Aquatic microbial oxygenic phototrophs (AMOPs) such as green algae, cyanobacteria, and diatoms are being examined as potential feedstocks for renewable energy production (3, 16, 20). While the proven recoverable biomass energy yield per unit area from culturable AMOPs exceeds that from most terrestrial energy crops (16), finding suitable methods for efficient conversion of this biomass to liquid and/or gaseous fuels and bioelectricity is a subject of intensive research (9, 20, 29, 44). One strategy for this conversion is autofermentation of live cells to produce hydrogen gas, ethanol, and/or organic acids in a recurring “cell factory” mode of photosynthesis and autofermentation (39).
Some AMOPs have the capacity to produce cellular energy via autofermentation, which involves the catabolism of photosynthetically produced energy storage compounds, such as soluble sugars and glycogen, under dark, anoxic conditions (39). For this method to become a competitive strategy for fuel production, current limitations with respect to efficiency and low rates of autofermentation of biomass to fuels must be overcome. Strategies to address these issues include boosting the photosynthetic production of the fermentable carbohydrate fraction and its conversion to hydrogen or ethanol, accelerating the rate of autofermentation to make it comparable to that rate seen with the diurnal solar cycle, and suppressing essential biopolymer catabolism.
The filamentous, nondiazotrophic cyanobacterium Arthrospira maxima CS-328 is a promising organism for sunlight-to-biofuel conversion. A. maxima thrives under extreme salt and pH conditions in alkaline (pH 9.5 to 11) soda lakes at high concentrations (0.4 to 1.4 M) of sodium (bi)carbonate. These conditions lead to accumulation of high levels of sugars and glycogen as an osmolyte and an energy reserve, respectively. A. maxima exhibits a high quantum efficiency of charge separation induced by photosystem II (PSII), resulting in a complex with the fastest in vivo water-oxidizing activity of all oxygenic phototrophs examined to date (2). For these and other reasons, including (i) high biomass productivity (23) and a developed technology for mass production (5, 46), (ii) toleration of large changes in osmotic pressure (salt concentration) without lysis (13), and (iii) production of long (∼1-mm) filaments that are often buoyant and amenable to easy harvesting and medium exchange, A. maxima represents an attractive starting point to serve as a cell factory for large-scale hydrogen, ethanol, or organic acid production.
In its natural environment, Arthrospira sp. filaments grow to high cell densities, leading to thick mats at the surface of saline alkaline lakes (23). At night, cells respire oxygen at high rates to generate energy for osmotic balance, thereby regularly encountering microoxic or anoxic conditions. To survive these conditions, it has a well-developed fermentative metabolism. Studies of the yield of autofermentative hydrogen production by A. maxima reveal it to be among the highest reported, and this can be influenced by a range of environmental factors to accelerate the rate to better match the rate seen with the diurnal cycle (1, 11). For example, by the use of strategies such as dietary supplementation of nickel to fully load [NiFe]- hydrogenase (bidirectionally encoded by hox) and a two-stage growth cycle to stimulate metabolic processes under conditions of darkness, thus leading to a reduction in the supply of (bi) carbonate, nitrate removal, and other environmental stresses, substantial increases in hydrogen rate and yield in those studies were reported. Arthrospira spp. have been reported to produce ethanol, acetate, lactate, and formate along with hydrogen gas as autofermentative end products (4, 12, 13).
Arthrospira spp. have a higher energy demand for homeostasis than freshwater AMOPs, owing to their native ecological niche in environments of high alkalinity and ionic strength. They are known to maintain a sodium gradient that is used to import carbonate via an obligate sodium carbonate symporter (6, 10). As expected, they also have a membrane-bound sodium extrusion pump that is dependent on the presence of ATP (6), allowing adaptation to various salt concentrations. High-level extracellular salt conditions do not lead to increases in intracellular sodium or potassium ions but are instead osmotically balanced by an increase in the biosynthesis of low-molecular-weight sugars (osmolytes) (13, 45, 48). Highlighting the importance of sodium ions for proper maintenance of homeostasis, it was previously shown that replacement of sodium with potassium in growth medium caused rapid cell death of Arthrospira platensis under aerobic conditions via cell lysis and that the cell death was accelerated by exposure to light (35).
There are many examples of anoxic nonphotosynthetic bacteria that can couple energy conversion by substrate-level phosphorylation generated during autofermentation of substrates to sodium ion gradients generated by electron transport (see reviews in references 28, 31, and 38). Here we examine whether the sodium ion gradient, which is known to be involved in cellular functions in A. maxima during photoautotrophic growth, can be used to accelerate the rate and yield of autofermentation under dark, anoxic conditions.
MATERIALS AND METHODS
Cell culturing.
Arthrospira (Spirulina) maxima (CS-328) was obtained from the Tasmanian CSIRO Collection of Living Microalgae and was grown at 30°C in batch culture in 4-liter Erlenmeyer flasks or 2.8-liter Fernbach flasks containing 1 liter of modified Zarrouk's medium (6.5 g liter−1 NaHCO3 and 7.0 g liter−1 Na2CO3 Na2CO3 instead of 18.0 g liter−1 NaHCO3, 10 to 30 mM NaNO3, initial pH 9.8) supplemented with 1 μM NiCl as previously described (11). Cultures were illuminated by cool white fluorescent lamps with a light flux of 40 μE m−2 s−1 on a 12-h light-dark cycle. Cells were harvested as early as 10 days after inoculation (cell density, 0.9 g of dry weight [DW]/liter of culture) or remained in batch culture for up to 20 days (1.5 to 2.3 g DW/liter of culture). For some experiments, cells were then transferred to fresh growth medium containing no nitrogen source (30 mM NaNO3 replaced with 30 mM NaCl). Nitrate-free cultures were then incubated under the same light and temperature conditions for an additional 2 to 4 weeks, resulting in chlorosis (partial loss of phycobillins and chlorophyll) and accumulation of higher sugar and glycogen content (1, 4). Cell densities of nitrogen-starved cultures at harvest were between 1.5 and 2.3 g DW/liter of culture, as indicated in the figure legends.
Cell autofermentation.
A. maxima filaments were harvested from liquid cultures by vacuum filtration onto porous filter discs (Pall Life Sciences, Ann Arbor, MI) (5 μm pore size) and resuspended in fermentation buffer (described in Results) at cell densities equivalent to those at harvest (0.9 to 2.3 g of dry weight liter−1). Aliquots (3 ml) of these suspensions were pipetted into small glass vials (10 ml total volume). Vials were subsequently sealed with Teflon-coated rubber septa and covered with aluminum foil for darkness. The headspace in each vial was purged with argon gas to induce microoxic conditions. Residual dissolved oxygen in these samples was rapidly consumed by respiration, as confirmed in independent samples with a Clark-type electrode (data not shown). Vials were then incubated at 30°C under these dark, anoxic conditions for up to 3 days. All biological samples were prepared in triplicate. For time course experiments, 9 biological replicate samples, 3 of which were measured at each time point, were prepared in parallel. We note that the minimal buffer capacity of the fermentation medium was around 50 mM whereas the total concentration of organic acids produced in the experiments presented here did not exceed 2 mM, indicating that the pH was likely stable throughout the fermentative time course.
Fermentation product measurements.
Hydrogen in the headspace above the cultures was measured by withdrawing headspace samples (200 μl) with a gastight syringe (Hamilton) and injection in a calibrated gas chromatograph (Gow-Mac) equipped with a 13× molecular sieve column and argon carrier gas. Following hydrogen measurements, vials were opened and A. maxima cell suspensions were immediately passed through prewashed 0.2-μm-pore-size syringe filters (Whatman, Florham Park, NJ). The resulting cell-free solutions were collected into Eppendorf tubes. An aliquot (540 μl) of the solution was pipetted into a Pyrex nuclear magnetic resonance (NMR) tube (New Era Enterprises, Inc., Vineland, NJ), to which 60 μl of D2O containing 30 μg/ml TSP [3-(trimethylsilyl)propionic-2,2,3,3-d4 acid, sodium salt] (Aldrich) was added, and tubes were mixed by inversion and vortexing. Water-soluble end products were measured by 1H-cryoprobe-assisted nuclear magnetic resonance spectroscopy at 500 MHz as described elsewhere (12).
Total carbohydrate determinations.
Total sugar amounts were determined from filament suspensions based on a modified anthrone method that was developed for whole-cell suspensions of yeast (43) and that has been applied to studies performed with intact filaments of A. maxima (42). Cell suspensions (100 μl) were added to anthrone reagent (Sigma) solution (900 μl) (0.2 g of anthrone per 100 ml of 71% sulfuric acid in water), and the solutions were heated at 100°C for 10 min. The absorbance at 620 nm of the resulting solution was measured in a spectrophotometer. The absorbance was compared against a calibration curve prepared with glucose standards (Sigma) that ranged from 10 to 50 μg of glucose per 100 μl of solution. A calibration curve prepared with bovine glycogen (Sigma) had the same slope as the one prepared with glucose (data not shown), and calibration curves prepared with trehalose and sucrose had slopes that were twice as steep as that of glucose on a molar basis (data not shown). The anthrone method was used to determine carbohydrate content in cells before and after autofermentation, and the difference was taken to represent the total amount of carbohydrate that had been catabolized.
Glycogen determination.
Glycogen content of cells during fermentation was determined by a method similar to that reported by Ernst et al. (17). Briefly, 1 ml of cells was pelleted by centrifugation after 1 to 3 days of fermentation. To each pellet, 200 μl of 48% aqueous KOH was added. Alkaline suspensions were subjected to vortexing and then incubated at 100°C for 1 h. Subsequently, 600 μl of cold (0°C) absolute ethanol was added and the suspension was centrifuged at 13,000 × g. The supernatant was discarded, and the pellet was washed with cold ethanol, dried using air at 70°C, and suspended in 1 ml of 200 mM sodium acetate buffer (pH 4.75) containing 2.7 U of amyloglucosidase and 2.1 U of amylase (Sigma). This solution was incubated at 37°C overnight, after which glucose concentrations were determined by the use of a glucose assay kit that relies on glucose-oxidase, peroxidase, and o-dianisidine (Sigma) and light absorption at 520 nm. The level of glycogen recovery determined by this method with known quantities of bovine glycogen (Sigma) was greater than 95%.
Genome sequencing.
The draft whole-genome shotgun sequencing of the A. maxima was performed by the Joint Genome Institute (U.S. Department of Energy, Walnut Creek, CA). Total genomic DNA was prepared from an axenic culture of A. maxima according to specifications of the Joint Genome Institute. The draft genome was assembled into 129 contigs greater than 2 kb in size and was approximately 6.0 Mb in total. The resulting data were subjected to autoannotation and were deposited in GenBank and are also available at http://genome.jgi-psf.org/draft_microbes/artma/artma.info.html.
Nucleotide sequence accession number.
The draft genome sequence of A. maxima strain CS-328 has been deposited in GenBank (accession no. ABYK00000000).
RESULTS
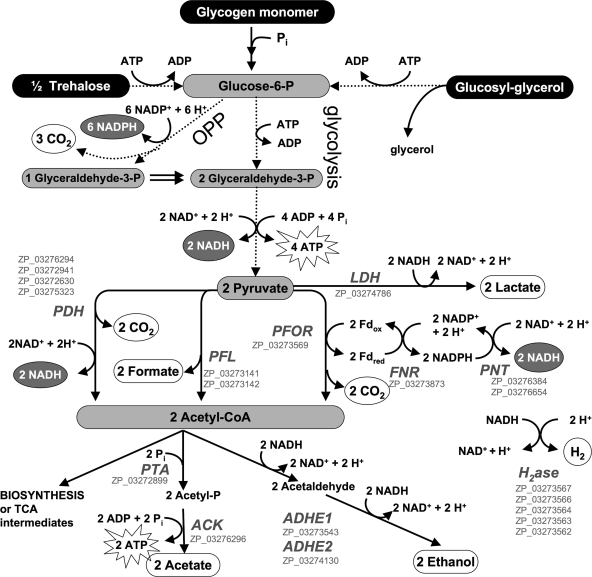
A draft sequence of the genome of A. maxima strain CS-328 was recently determined, and an initial annotation of the data was performed. Given the observed fermentative end products and the genes present in the genome that presumably encode enzymes in glycolysis, the oxidative pentose phosphate (OPP) pathway, and fermentation, a scheme to determine the possible fates of carbohydrates during autofermentation in A. maxima was assembled to serve as a theoretical framework for hypothesis testing (Fig. 1). Intracellular carbohydrates were determined to be dominated by a mixture of osmolytes (glucosyl-glycerol and trehalose) and glycogen, as has been previously reported for Arthrospira species (12, 48). Of specific interest in Fig. 1 are the stoichiometries of fermentative end products determined via examination of the Embden-Meyerhof-Parnas pathway (glycolysis) and the OPP pathway, both of which are active during fermentation in other cyanobacteria (39). During fermentation, one glucose equivalent can yield two lactate, two acetate, or two ethanol molecules via glycolysis, whereas one glucose equivalent yields only one lactate, one acetate, or one ethanol molecule via the OPP pathway. The more extensive oxidation occurring via the OPP pathway produces 6 additional NADPH equivalents per glucose molecule. Control of the fluxes through the OPP pathway and glycolysis provides the basis for redox balancing. A more detailed scheme that illustrates the stoichiometry of the OPP pathway is provided in Fig. S1 in the supplemental material.
Fig. 1.
Catabolism of glucose equivalents derived from previously observed substrates (black ovals) (13), the experimentally observed excreted products (white ovals), and enzymes encoded in the A. maxima genome (gray italic characters) as well as their NCBI reference sequences (gray roman characters). All proteins necessary for these reactions are encoded in the A. maxima draft genome. Multistep reactions are abbreviated (indicated by dotted arrows without enzyme annotation). The OPP pathway abbreviated here was determined based on the idea of full cycling of fructose-6-phosphate. A more detailed scheme to illustrate the stoichiometry of OPP is provided in Fig. S1 in the supplemental material. Enzyme abbreviations: LDH, lactate dehydrogenase; PDH, pyruvate dehydrogenase; PFL, pyruvate formate lyase; PFOR, pyruvate ferredoxin oxidoreductase; FNR, ferredoxin-NADP reductase; PNT, pyridine-NADH transhydrogenase; PTA, phosphotransacetylase; ACK, acetate kinase; ADHE, bifunctional aldehyde alcohol dehydrogenase; H2ase, hydrogenase.
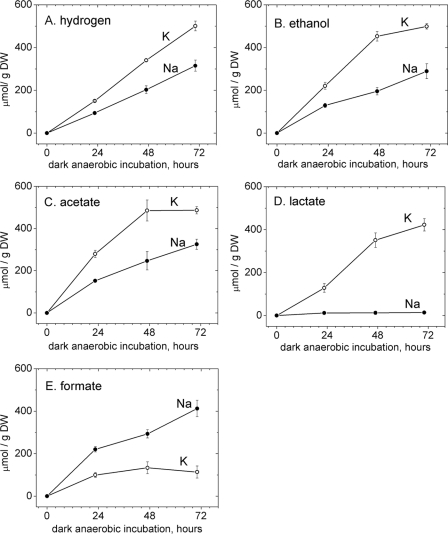
To investigate the influence of the sodium gradient on autofermentation products, filaments from nitrate-free growth medium were resuspended in 50 mM sodium phosphate buffer or 50 mM potassium phosphate buffer and excreted products were monitored by NMR. The replacement of sodium by potassium led to 67% increases in hydrogen, ethanol, and acetate yields (Fig. 2 A to C). Lactate, which was nearly undetectable from filaments suspended in sodium phosphate buffer, was produced in significant amounts from filaments in isotonic potassium phosphate buffer (Fig. 2D). In contrast, formate production from filaments suspended in potassium phosphate buffer was 2- to 4-fold lower than from those in sodium phosphate buffer (Fig. 2E). No other hydrogen-containing autofermentative products were excreted in detectable (>50 μM) amounts in the experiments presented here.
Fig. 2.
Effect of replacement of sodium with potassium on autofermentative product yields of N-starved cells, fermented at pH 7.0. A. maxima filaments were harvested from a culture adapted for 22 days in nitrate-free growth medium at a cell density of 1.5 g DW/liter of culture. Filaments were resuspended at this cell density in 50 mM sodium or potassium phosphate buffer and incubated for up to 3 days under dark, anaerobic conditions.
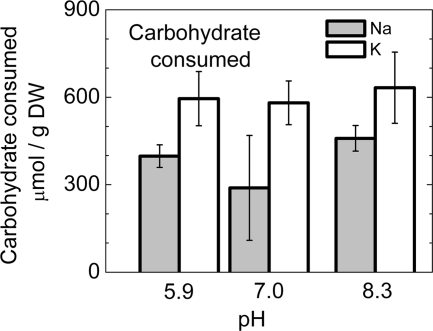
Total intracellular carbohydrate content before and after fermentation was measured using actively growing cultures in complete growth media after 10 days of growth (DW=0.9 g/liter of culture) after they were resuspended in 50 mM buffer at three different pH values (MES [morpholineethanesulfonic acid], pH 5.9; potassium phosphate, pH 7.0; Tricine, pH 8.3) with the addition of either 100 mM sodium chloride or 100 mM potassium chloride. We studied the effects on young cells rather than on N-starved cells (see the results presented in Fig. 2) to demonstrate that the apparent sodium gradient effect was not specific to N-starved cells. Figure 3 shows increased carbohydrate catabolism for cells in the absence of sodium ions. The total volume of carbohydrate degraded after 1 day is about one-quarter to one-third greater in the absence of sodium and is independent of pH levels between 5.9 and 8.3.
Fig. 3.
Carbohydrate consumption of N-replete cells in the presence of 100 mM sodium or potassium chloride in 50 mM fermentation buffer after 1 day of anaerobic incubation in the dark. Cells were grown from inoculation for 10 days to a density of 0.9 g DW/liter of culture and harvested for fermentation studies. The fermentation buffers used were MES (pH 5.9), potassium phosphate (pH 7.0), and Tricine (pH 8.3).
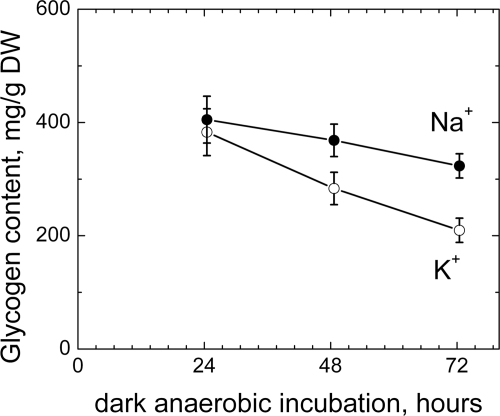
As used to produce the results shown in Fig. 3, the anthrone method measures total intracellular carbohydrate levels in cells; however, it would be useful to know whether glycogen catabolism, specifically, is increased when sodium is removed during fermentation. Figure 4 shows the glycogen content of N-starved cells from N-depleted growth media that were resuspended in 50 mM Tricine (pH 8.3) containing a 100 mM addition of either sodium or potassium chloride. The consumption rate of glycogen increased approximately 2-fold after sodium had been replaced with potassium.
Fig. 4.
Glycogen content of N-starved cells in the presence of sodium or potassium chloride in fermentation buffer (Tricine; pH 8.3). Cells were grown in full media and then subjected to nitrogen starvation under the same growth conditions. The cell density was 2.3 g DW/liter of culture at harvesting, and cells were resuspended in Tricine buffer with the addition of 100 mM sodium or potassium chloride at this cell density.
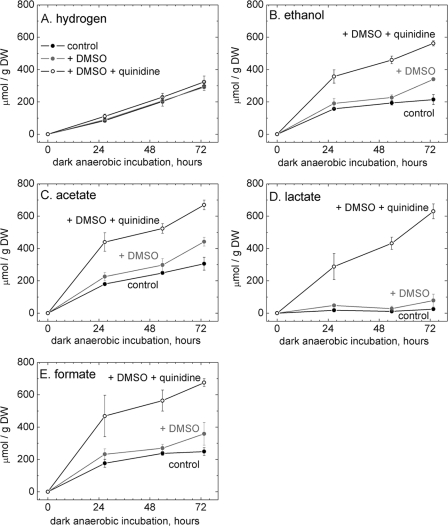
Quinidine is a sodium-specific ion-channel blocker that has often been used in cardiac studies (18) but has been shown to exhibit efficacy in Arthrospira spp. for sodium-dependent (bi)carbonate transport (6, 10). The effect of quinidine on fermentative product yields was tested on N-starved cultures grown as described for Fig. 2 (e.g., DW=2.3 g/liter of culture). Quinidine (100 μM) was delivered to suspensions of A. maxima in 50 mM sodium phosphate buffer containing perdeuterated (d6) dimethyl sulfoxide (d6-DMSO) as a carrier solvent (final d6-DMSO concentration, 1% [vol/vol]). Quinidine addition to suspensions of A. maxima in 50 mM sodium phosphate buffer significantly increased the autofermentative yields of ethanol, acetate, lactate, and formate (Fig. 5). The effect was similar to that of replacement of sodium with potassium on the autofermentative yields of ethanol, acetate, and lactate. The effect of adding only 1% d6-DMSO was insignificant (Fig. 5).
Fig. 5.
Effect of 100 μM quinidine on autofermentative product yields from N-starved cells fermented at pH 7.0. Filaments were harvested from a culture adapted for 28 days in nitrate-free growth medium (cell density, 1.6 g DW/liter of culture). Quinidine was dissolved in d6-DMSO (deuterated) and added to A. maxima cells suspended in 50 mM sodium phosphate buffer (+ DMSO + quinidine) at this cell density. The effect of pure DMSO (+ DMSO) relative to the results seen with the control (no additions) was also tested. In the suspensions to which DMSO was added, the DMSO concentration was 1% (vol/vol).
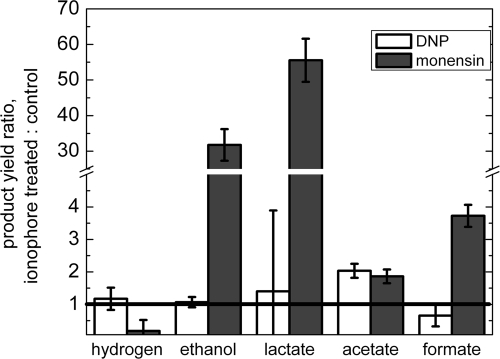
To further test whether the effect of sodium replacement with potassium is a sodium ion-specific effect, we measured the yields of excreted products for cells autofermented in the presence of either dinitrophenol (DNP), a known proton ionophore, or the sodium proton-exchanger monensin (33). The observed products (excreted at or above the detection limit of >50 μM) from these and all other experiments were hydrogen, acetate, ethanol, formate, and lactate. Figure 6 gives the ratios of yields relative to untreated cell results (control with no additions). Within the error ranges, all fermentation product yields were equivalent to those of the control for cells treated with DNP, except for the acetate yield, which showed a 2-fold increase. In contrast, cells treated with monensin exhibited large increases in stimulation of carbon fermentation products: for acetate, 2×; for formate, 4×; and especially for ethanol (30×) and lactate (55×). The hydrogen yield decreased appreciably (5×), as expected based on the considerable consumption of NADH required for the additional lactate and ethanol excretion. An approximate 2-fold stimulation of total carbohydrate consumption occurred for cells treated with monensin relative to control levels (see Fig. S2 in the supplemental material).
Fig. 6.
Effect of the ionophores dinitrophenol (DNP) and monensin on excreted metabolite yields at pH 7.0. Cells in 50 mM sodium phosphate buffer were incubated under conditions of dark anoxia in the presence of 100 μM DNP, 100 μM monensin, or no addition. The ratios of yields relative to untreated cell results are given.
DNP is a poor uncoupler under conditions of alkaline pH and was therefore not tested in fermentation buffers above pH 7.0. However, we did test the effect of another proton uncoupler, 2 μM p-trifluoromethoxycarbonylcyanidephenylhydrazone (FCCP), on N-starved cells fermented at pH 8.3. This treatment led to complete cell lysis (verified by microscopy) in less than 24 h under dark, anoxic conditions.
DISCUSSION
The results demonstrate that interruption of the sodium ion gradient across the cell membrane under dark, anoxic conditions for metabolism causes a large increase in rates of catabolism with respect to intracellular energy reserves (in the form of carbohydrate). This conclusion is supported by the results of all three direct tests we applied: replacement of sodium with potassium in the buffer, addition of the sodium channel blocker quinidine, and addition of the sodium ionophore monensin with sodium present in the fermentation buffer. To our knowledge, no other studies have investigated the role of sodium ion gradients in modulating the autofermentative catabolism of carbohydrates in cyanobacteria under dark, anoxic conditions. However, many other nonphotosynthetic bacteria, especially halophilic bacteria, operate in this way (see reviews in references 28, 31, and 38).
In cyanobacteria, a sodium ion gradient can play a dominant role in solute transport (for example, influx of bicarbonate coupled to sodium influx) under photoautotrophic conditions and pH regulation through sodium-proton antiporters (6, 8, 34, 41). For A. maxima cells, we hypothesize that the sodium gradient is oriented across the cytoplasmic membrane, with high sodium levels in the periplasm of the cell, in analogy to A. platensis cells, in which the intracellular sodium content has been shown to remain unchanged when the sodium concentration in the external medium is increased (45).
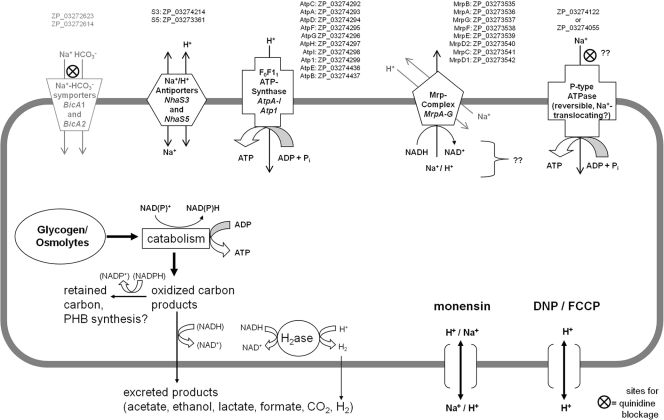
Figure 7 presents a scheme designed to help summarize and explain the observed relationship between fermentative metabolism and ion gradients in A. maxima deduced from our work. It is also based on information from the literature and identification of gene homologues that presumably encode key enzymes involved in sodium and proton ion gradients. However, as membrane energetics in cyanobacteria has been studied predominantly, if not exclusively, under aerobic conditions, we emphasize that this model is provided to generate hypotheses that can be tested in future work.
Fig. 7.
Proposed mechanisms involving sodium and proton gradients and ATP production and hydrolysis in A. maxima. Each circled X indicates a site where quinidine is likely to block the influx of sodium. The ionophore monensin collapses sodium gradients across cell membranes by exchanges with protons. DNP and FCCP are proton ionophores that collapse proton gradients. NCBI reference sequences of putative membrane proteins (possibly) involved in proton-sodium gradient utilization are given from the draft genome sequence of A. maxima. Sodium bicarbonate symporters are likely present but probably do not play a role in catabolism of carbohydrates during autofermentation. That protein representation is therefore shown in gray.
Several insights and questions arise from the observation that catabolic flux can increase as a result of removing the sodium ion gradient. In bacteria in which this coupling has been examined during respiratory and photosynthetic metabolism, this increase is a response that restores the cellular energy charge (CEC), i.e., the ratio of ATP + 0.5 ADP to ATP + ADP + AMP, by substrate-level phosphorylation via glycolysis (Fig. 1). We hypothesize that the ion gradient either contributes directly to ATP generation through a sodium-proton-dependent ATP synthase or is utilized in some other way to power a process (such as solute transport or pH regulation) that, in the absence of a sodium or proton gradient, requires ATP. From Fig. 7, it is apparent that sodium and proton gradients can be exchanged via sodium-proton antiporters in A. maxima.
Ohmori and Ohmori have suggested that A. platensis has the ability to couple the influx of sodium across its cytoplasmic membrane to drive ATP synthesis (30). Those authors showed that, as a result of suspending filaments in the presence of 0.22 M sodium and 20 mM cyclic AMP (cAMP) at alkaline pH, the ATP concentration of the cell increased and was accompanied by a decrease in ADP and AMP. They used the sodium channel blocker amiloride and the sodium ionophore monensin to show that this effect was dependent on the sodium gradient. They also showed that addition of the proton ionophore CCCP (carbonyl cyanide m-chlorophenylhydrazone) did not decrease this effect, indicating that energy exchanges between sodium gradients and ATP but not proton gradients. These results are in full agreement with our various observations of the fermentative rate, which is highly accelerated by replacement of sodium with potassium in fermentation buffers, or by addition of quinidine or monensin, but not by addition of DNP.
We note that a high-resolution structure (2.1 Å) of the c-ring of the FoF1-ATP synthase from A. platensis shows a proton-bound state under conditions in which sodium ions are in a 1,000-fold excess compared to the proton level, suggesting high specificity for protons over sodium for the FoF1-ATP synthase (32). Yet Berry et al. proposed that sodium extrusion occurs via the activity of an ATP-dependent primary sodium pump during photosynthesis in the same organism (6), and a similar conclusion was supported by photosynthetic studies performed with A. maxima (10). Wiangnon et al. characterized isolated plasma membranes from another alkaliphilic, halotolerant cyanobacterium, Aphanothece halophytica, and proposed the presence of a P-type ATPase that extrudes sodium at the expense of ATP hydrolysis (49). The gene for this proposed ATPase has not been sequenced to date (A. Incharoensakdi, personal communication), but two P-type ATPase homologues are present in the A. maxima genome (Fig. 7). We propose that a sodium ion gradient is maintained in equilibrium with the CEC in the cell, either via the activity of an unknown protein (possibly one or both of the two P-type ATPase homologues) or via the activity of a combination of Na+/H+ antiporters and a H+-dependent ATP synthase (such as the FoF1-ATP synthase).
Our data do not prove that the sodium ion gradient is in a direct exchange with the CEC, although this is the simplest interpretation and has been shown to be the case in other bacteria. Instead, it could be used to directly facilitate energy-dependent processes, such as solute transport or intracellular pH regulation. Physiological evidence of the presence of a sodium-bicarbonate symporter that can be blocked by quinidine has been previously reported for A. platensis and A. maxima (6, 10). In Synechocystis sp. PCC 6803, this enzyme is encoded by the gene designated sbtA (37). We found no homologues of this gene in the A. maxima genome. However, marine cyanobacteria such as Synechococcus sp. PCC 7002 also have an additional sodium-bicarbonate symport protein that is encoded by the gene designated bicA (34); two genes homologous to bicA are present in the A. maxima genome (Fig. 7). It is not expected that bicarbonate uptake would be important during dark anaerobiosis in A. maxima, but it is a likely site for quinidine blockage. Under the conditions presented in the manuscript, the only energy needs for transport might involve importing bicarbonate or exporting fermentative end products, as there are no other components (i.e., no added trace metals or macronutrients such as nitrate) in the fermentation buffers. Because bicarbonate would not seem to be needed during dark anaerobisis (carbon fixation is not needed, although the possibility of such fixation is not precluded), the most likely explanation in accordance with this hypothesis is that cells use energy to export fermentative end products to allow catabolism to continue, using energy from the sodium gradient. In the absence of a sodium gradient, ATP would be used to export these products.
Sodium is necessary for maintaining pH homeostasis, presumably with the assistance of sodium-proton antiporters in A. platensis (35). In alkaliphilic heterotrophic bacteria, it has been shown that the removal of sodium is coupled to a loss of regulation of the intracellular pH and a concomitant shift to that of the medium (24, 25). Two sodium-proton antiporters are present in the genome of A. maxima (Fig. 7). They are well understood in Synechocystis sp. PCC 6803 (8, 21). These antiporters couple the influx of a proton with the extrusion of a sodium ion. We note here, however, that the increase in carbohydrate consumption in cells suspended in buffers containing potassium rather than sodium does not appear to be pH dependent (Fig. 3). At all pH values between 5.9 and 8.3 that were tested, cells consumed the same amount of carbohydrate in 2 days when suspended in buffers in which all sodium has been replaced with potassium. If the sodium ion gradient were used solely for pH maintenance, one would expect the difference in carbohydrate consumption between buffers containing sodium and buffers containing only potassium to be higher at pH values further from neutral pH (e.g., a larger increase in carbohydrate consumption upon replacement of sodium with potassium at pH 5.9 and/or 8.3 than at pH 7.0). No such trend was observed, and the collapse of a proton gradient alone at pH 7.0 with addition of DNP did not result in substantial increases in lactate and ethanol production at pH 7.0 (Fig. 6). This result was observed in experiments involving replacement of quinidine, monensin, or sodium with potassium.
Another insight worth noting is that, because addition of DNP does not significantly stimulate fermentative product formation (Fig. 6), a sodium ion gradient formed under fermentative conditions at pH 7.0 does not have an electrical component. Instead, the sodium ion gradient observed is strictly due to a sodium concentration gradient across the membrane at that tested pH level. Otherwise, the addition of an ionophore that allowed protons, but not sodium ions, to diffuse across the cell membrane would significantly stimulate fermentative product formation.
Thus, we return to the conclusion that the sodium gradient is in equilibrium with the CEC and that, in the absence of the sodium gradient, carbohydrate catabolism accelerates and deepens to compensate for a lower CEC. This raises another interesting question: how is the sodium gradient generated under anaerobic conditions in the first place? We note that the replacement of sodium with potassium or the addition of quinidine led to increased rates of fermentative product formation from N-starved cells and that the process continued during the 3-day window measured (Fig. 3 and 4). Hence, the effect is not transient.
The coupling of decarboxylation reactions with sodium extrusion across cell membranes has been described for many anoxic nonphotosynthetic bacteria. For example, many species have been found to maintain a sodium gradient across the cell membrane by decarboxylating oxalate to form formate and CO2, decarboxylating malonate to form acetate and CO2, or decarboxylating malate to form lactate and CO2 (15). Other possible mechanisms for sodium translocation known in other microbes could involve Na+-translocating methyltransferases (known only in archaea to date) (14), Na+-translocating ferredoxin-NAD+ reductases (so-called Rnf complexes, first discovered in Rhodobacter capsulatus) (36), Na+-translocating NADH-quinone oxidoreductases (Nrq complexes) (40, 50), or an Na+-pyrophosphatase (27). However, our search of the A. maxima draft genome sequence provided no homologues to any of those genes known to be involved in sodium extrusion.
In contrast, all of the genes that encode a known Mrp complex are, in fact, present in the genome of A. maxima. One of the genes of this complex, mrpA, was identified as playing a role in Na+ resistance in the cyanobacterium Anabaena sp. PCC7120 (7). This complex was first identified in Bacillus halodurans C125 (26). Later, Ito et al. showed that the Mrp system supports anoxic resistance by expressing it in Escherichia coli (22). That result indicates that the Mrp complex plays a role in extruding sodium under anoxic conditions. The complex also has subunits similar in primary sequence to NADH:ubiquinone oxidoreductase (19), suggesting the involvement of pyridine nucleotides and redox poise of fermenting cells in this context. However, it is unclear what the terminal electron acceptor might be under anoxic conditions, if this were the case. Interestingly, transcription of the genes for this complex is upregulated not only by high alkalinity in the cyanobacterium Synechocystis sp. PCC 6803 (41) but also by inorganic carbon limitation (47). It would be interesting to compare the characteristics of transcription of genes that encode the Mrp complex in A. maxima under anoxic conditions with and without sodium and to contextualize these findings as more information regarding the function and mechanism of Mrp complexes becomes available from studies performed with other organisms.
We measured total fluorescence yield due to NADH plus NADPH [NAD(P)H] in fermenting whole cells of A. maxima in both sodium and potassium phosphate buffer at pH 7.0 and saw clear differences in the total reduced pyridine nucleotide pools (see Fig. S3 in the supplemental material). While there was effectively no change in the levels of (reduced) NAD(P)H pools over 20 h of autofermentation in sodium phosphate buffer (pH 7.0) relative to that seen at the start of fermentation (at time 0), cells autofermented in potassium phosphate buffer showed a continual decline in NAD(P)H pools to levels below our detection limit. These data allow us to form the conclusion that the Na+ gradient is involved in maintaining the pyridine nucleotide redox poise such that, when Na+ is replaced by K+, it is compensated by a large drop in redox poise. The H2 data reveal that, as the fluorescence yield of NADH plus NADPH drops, there is a lag in flux through H2 until about 9 h have passed, after which there is a parallel increase in the rate of H2 production.
Based on currently available literature, homology comparisons, and measurements of reduced pyridine nucleotide pools in whole cells, we tentatively identify the Mrp complex as the Na+-extruding complex possibly responsible for sodium gradient generation under anoxic conditions. This complex may possibly function with the assistance of NADH (Fig. 7; see also Fig. S3 in the supplemental material). This suggestion is made without direct proof and hence is given for the purpose of future hypothesis testing.
In any case, it is noted that a significant increase in carbohydrate catabolism can be observed when sodium ion gradients are inaccessible to cells, thus allowing faster mobilization of substrates to fermentative products. For biofuel applications, such a removal of sodium gradients could be used to accelerate fermentative product formation to better match the diurnal cycle of the cells. This effect should be independent of and potentially additive to other manipulations, such as pathway engineering, to direct fermentative products toward products of greater desirability.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by grants from the U.S. Air Force Office of Scientific Research (MURI-FA9550-05-1-0365 to G.C.D. and D.A.B.) and the DOE-GTL (DE-FG02-07ER64488 to G.C.D.), the DOE-JGI (project DOEM_782004 to D.A.B.) for genome sequencing, the Deutsche Forschungsgemeinschaft, through Sfb498 (to O.L.), and the Cluster of Excellence UniCat (to O.L.).
D.C. thanks PinChing Maness and Kate Brown, both in the Biosciences Center at the National Renewable Energy Laboratory, for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 2 September 2011.
REFERENCES
- 1. Ananyev G., Carrieri D., Dismukes G. C. 2008. Optimizing metabolic capacity and flux through environmental cues to maximize hydrogen production by cyanobacterium Arthrospira maxima. Appl. Environ. Microbiol. 74:6102–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ananyev G., Dismukes G. C. 2005. How fast can Photosystem II split water? Kinetic performance at high and low frequencies. Photosynth Res. 84:355–365 [DOI] [PubMed] [Google Scholar]
- 3. Angermayr S. A., Hellingwerf K. J., Lindblad P., Teixeira de Mattos M. J. 2009. Energy biotechnology with cyanobacteria. Curr. Opin. Microbiol. 20:257–263 [DOI] [PubMed] [Google Scholar]
- 4. Aoyama K., Uemura I., Miyake J., Asada Y. 1997. Fermentative metabolism to produce hydrogen gas and organic compounds in a cyanobacterium, Spirulina platensis. J. Ferment. Bioeng. 83:17–20 [Google Scholar]
- 5. Belay A. 1997. Mass culture of Spirulina outdoors—the Earthrise Farms experience, p. 131–158 In Vonshak A. (ed.), Spirulina platensis (Arthrospira): physiology, cell-biology and biotechnology. Taylor & Francis Inc., Bristol, PA [Google Scholar]
- 6. Berry S., Bolychevtseva Y. V., Rogner M., Karapetyan N. V. 2003. Photosynthetic and respiratory electron transport in the alkaliphilic cyanobacterium Arthrospira (Spirulina) platensis. Photosynth. Res. 78:67–76 [DOI] [PubMed] [Google Scholar]
- 7. Blanco-Rivero A., Leganes F., Fernandez-Valiente E., Calle P., Fernandez-Pinas F. 2005. mrpA, a gene with roles in resistance to Na+ and adaptation to alkaline pH in the cyanobacterium Anabaena sp PCC7120. Microbiology 151:1671–1682 [DOI] [PubMed] [Google Scholar]
- 8. Buck D. P., Smith G. D. 1995. Evidence for a Na+/H+ electrogenic antiporter in an alkaliphilic cyanobacterium Synechocystis. FEMS Microbiol. Lett. 128:315–320 [Google Scholar]
- 9. Burrows E. H., Chaplen F. W. R., Ely R. L. 2008. Optimization of media nutrient composition for increased photofermentative hydrogen production by Synechocystis sp. PCC 6803. Int. J. Hydrogen Energy 33:6092–6099 [Google Scholar]
- 10. Carrieri D., Ananyev G., Brown T., Dismukes G. C. 2007. In vivo bicarbonate requirement for water oxidation by Photosystem II in the hypercarbonate-requiring cyanobacterium Arthrospira maxima. J. Inorg. Biochem. 101:1865–1874 [DOI] [PubMed] [Google Scholar]
- 11. Carrieri D., Ananyev G., Garcia Costas A. M., Bryant D. A., Dismukes G. C. 2008. Renewable hydrogen production by cyanobacteria: nickel requirements for optimal hydrogenase activity. Int. J. Hydrogen Energy 33:2014–2022 [Google Scholar]
- 12. Carrieri D., et al. 2009. Identification and quantification of water-soluble metabolites by cryoprobe-assisted nuclear magnetic resonance spectroscopy applied to microbial fermentation. Magn. Reson. Chem. 47:S138–S146 [DOI] [PubMed] [Google Scholar]
- 13. Carrieri D., et al. 2010. Boosting autofermentation rates and product yields with sodium stress cycling: application to production of renewable fuels by cyanobacteria. Appl. Environ. Microbiol. 76:6455–6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deppenmeier U., Lienard T., Gottschalk G. 1999. Novel reactions involved in energy conservation by methanogenic archaea. FEBS Lett. 457:291–297 [DOI] [PubMed] [Google Scholar]
- 15. Dimroth P., Schink B. 1998. Energy conservation in the decarboxylation of dicarboxylic acids by fermenting bacteria. Arch. Microbiol. 170:69–77 [DOI] [PubMed] [Google Scholar]
- 16. Dismukes G. C., Carrieri D., Bennette N., Ananyev G. M., Posewitz M. C. 2008. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 19:235–240 [DOI] [PubMed] [Google Scholar]
- 17. Ernst A., Kirschenlohr H., Diez J., Boger P. 1984. Glycogen content and nitrogenase activity in Anabaena variabilis. Arch. Microbiol. 140:120–125 [Google Scholar]
- 18. Hill R. J., Duff H. J., Sheldon R. S. 1988. Determinants of stereospecific binding of type I antiarrhythmic drugs to cardiac sodium channels. Mol. Pharmacol. 34:659–663 [PubMed] [Google Scholar]
- 19. Hiramatsu T., Kodama K., Kuroda T., Mizushima T., Tsuchiya T. 1998. A putative multisubunit Na+/H+ antiporter from Staphylococcus aureus. J. Bacteriol. 180:6642–6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu Q., et al. 2008. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54:621–639 [DOI] [PubMed] [Google Scholar]
- 21. Inaba M., Sakamoto A., Murata N. 2001. Functional expression in Escherichia coli of low-affinity and high-affinity Na+(Li+)/H+ antiporters of Synechocystis. J. Bacteriol. 183:1376–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ito M., Guffanti A. A., Krulwich T. A. 2001. Mrp-dependent Na+/H+ antiporters of Bacillus exhibit characteristics that are unanticipated for completely secondary active transporters. FEBS Lett. 496:117–120 [DOI] [PubMed] [Google Scholar]
- 23. Kebede E., Ahlgren G. 1996. Optimum growth conditions and light utilization efficiency of Spirulina platensis (equals Arthrospira fusiformis) (Cyanophyta) from Lake Chitu, Ethiopia. Hydrobiologia 332:99–109 [Google Scholar]
- 24. Kitada M., Guffanti A. A., Krulwich T. A. 1982. Bioenergetic properties and viability of alkalophilic Bacillus firmus RAB as a function of pH and Na+ contents of the incubation medium. J. Bacteriol. 152:96–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krulwich T. A., Guffanti A. A. 1989. Alkalophilic bacteria. Annu. Rev. Microbiol. 43:435–463 [DOI] [PubMed] [Google Scholar]
- 26. Kudo T., Hino M., Kitada M., Horikoshi K. 1990. DNA sequences required for the alkalophily of Bacillus sp. strain C-125 are located close together on its chromosomal DNA. J. Bacteriol. 172:7282–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malinen A. M., Belogurov G. A., Baykov A. A., Lahti R. 2007. Na+-pyrophosphatase: a novel primary sodium pump. Biochemistry 46:8872–8878 [DOI] [PubMed] [Google Scholar]
- 28. Mesbah N. M., Wiegel J. 2008. Life at extreme limits: the anaerobic halophilic alkalithermophiles. Ann. N. Y. Acad. Sci. 1125:44–57 [DOI] [PubMed] [Google Scholar]
- 29. Nguyen M. T., Choi S. P., Lee J., Lee J. H., Sim S. J. 2009. Hydrothermal acid pretreatment of Chlamydomonas reinhardtii biomass for ethanol production. J. Microbiol. Biotechnol. 19:161–166 [DOI] [PubMed] [Google Scholar]
- 30. Ohmori K., Ohmori M. 2002. cAMP stimulates Na+-dependent ATP formation in the alkalophylic cyanobacterium Spirulina platensis. Microbes Environ. 17:144–147 [Google Scholar]
- 31. Ollivier B., Caumette P., Garcia J. L., Mah R. A. 1994. Anaerobic bacteria from hypersaline environments. Microbiol. Rev. 58:27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pogoryelov D., Yildiz O., Faraldo-Gomez J. D., Meier T. 2009. High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat. Struct. Mol. Biol. 16:1068–1073 [DOI] [PubMed] [Google Scholar]
- 33. Pressman B. C. 1976. Biological applications of ionophores. Annu. Rev. Biochem. 45:501–530 [DOI] [PubMed] [Google Scholar]
- 34. Price G. D., Woodger F. J., Badger M. R., Howitt S. M., Tucker L. 2004. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 101:18228–18233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schlesinger P., Belkin S., Boussiba S. 1996. Sodium deprivation under alkaline conditions causes rapid death of the filamentous cyanobacterium Spirulina platensis. J. Phycol. 32:608–613 [Google Scholar]
- 36. Schmehl M., et al. 1993. Identification of a new class of nitrogen-fixation genes in Rhodobacter capsulatus—a putative membrane complex involved in electron-transport to nitrogenase. Mol. Gen. Genet. 241:602–615 [DOI] [PubMed] [Google Scholar]
- 37. Shibata M., et al. 2002. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria—function and phylogenetic analysis. J. Biol. Chem. 277:18658–18664 [DOI] [PubMed] [Google Scholar]
- 38. Speelmans G., Poolman B., Konings W. N. 1995. Na+ as coupling ion in energy transduction in extremophilic Bacteria and Archaea. World J. Microb. Biotechnol. 11:58–70 [DOI] [PubMed] [Google Scholar]
- 39. Stal L. J., Moezelaar R. 1997. Fermentation in cyanobacteria. FEMS Microbiol. Rev. 21:179–211 [Google Scholar]
- 40. Steuber J. 2001. Na+ translocation by bacterial NADH:quinone oxidoreductases: an extension to the complex-I family of primary redox pumps. Biochim. Biophys. Acta 1505:45–56 [DOI] [PubMed] [Google Scholar]
- 41. Summerfield T. C., Sherman L. A. 2008. Global transcriptional response of the alkali-tolerant cyanobacterium Synechocystis sp. strain PCC 6803 to a pH 10 environment. Appl. Environ. Microbiol. 74:5276–5284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tadros M. G., MacElroy R. D. 1988. Characterization of Spriulina biomass for CELSS diet potential. NASA-CR-185329, NASA contractor NCC 2-501. Alabama A&M University, Normal, AL [Google Scholar]
- 43. Trevelyan W. E., Harrison J. S. 1952. Studies on yeast metabolism. 1. Fractionation and microdetermination of cell carbohydrates. Biochem. J. 50:298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Velasquez-Orta S. B., Curtis T. P., Logan B. E. 2009. Energy from algae using microbial fuel cells. Biotechnol. Bioeng. 103:1068–1076 [DOI] [PubMed] [Google Scholar]
- 45. Vonshak A., Guy R., Guy M. 1988. The response of the filamentous cyanobacterium Spirulina platensis to salt stress. Arch. Microbiol. 150:417–420 [Google Scholar]
- 46. Vonshak A., Richmond A. 1988. Mass production of the blue-green-alga Spirulina—an overview. Biomass 15:233–247 [Google Scholar]
- 47. Wang H. L., Postier B. L., Burnap R. L. 2004. Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 279:5739–5751 [DOI] [PubMed] [Google Scholar]
- 48. Warr S. R. C., Reed R. H., Chudek J. A., Foster R., Stewart W. D. P. 1985. Osmotic adjustment in Spirulina platensis. Planta 163:424–429 [DOI] [PubMed] [Google Scholar]
- 49. Wiangnon K., Raksajit W., Incharoensakdi A. 2007. Presence of a Na-stimulated P-type ATPase in the plasma membrane of the alkaliphilic halotolerant cyanobacterium Aphanothece halophytica. FEMS Microbiol. Lett. 270:139–145 [DOI] [PubMed] [Google Scholar]
- 50. Zhou W., et al. 1999. Sequencing and preliminary characterization of the Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio harveyi. Biochemistry 38:16246–16252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.