Abstract
The H-NS protein represses the transcription of hundreds of genes in Gram-negative bacteria. Derepression is achieved by a multitude of mechanisms, many of which involve the binding of a protein to DNA at the repressed promoter in a manner that compromises the maintenance of the H-NS–DNA nucleoprotein repression complex. The principal virulence gene promoters in Shigella flexneri, the cause of bacillary dysentery, are repressed by H-NS. VirB, a protein that closely resembles members of the ParB family of plasmid-partitioning proteins, derepresses the operons that encode the main structural components and the effector proteins of the S. flexneri type III secretion system. Bioinformatic analysis suggests that VirB has been co-opted into its current role as an H-NS antagonist in S. flexneri. To test this hypothesis, the potential for VirB to act as a positive regulator of proU, an operon that is repressed by H-NS, was assessed. Although VirB has no known relationship with the osmoregulated proU operon, it could relieve H-NS-mediated repression when the parS-like VirB binding site was placed appropriately upstream of the RpoD-dependent proU promoter. These results reveal the remarkable facility with which novel regulatory circuits can evolve, at least among those promoters that are repressed by H-NS.
INTRODUCTION
Gene regulatory circuits in bacteria are subject to evolution and can be adapted to meet the changing needs of the organisms that harbor them. Similar circuits can differ in detail in even closely related species of bacteria (60) and in eukaryotes (11, 12, 47). Transcription control in bacteria functions along a spectrum of sophistication, and some very effective genetic switches operate through relatively simple mechanisms. Some of the most basic mechanisms involve those that relieve repression that is mediated by the nucleoid-associated protein H-NS (69).
H-NS is a dimeric DNA binding protein with a preference for A+T-rich sequences associated with DNA curvature (7, 14, 21, 30). The protein can bind to a nucleation site and then spread along the DNA through a mechanism that can also involve the formation of DNA-protein-DNA bridges, where each of the DNA binding domains of the dimer can associate with a separate DNA molecule or spatially separate portions of the same DNA molecule (2, 16, 17, 25, 45, 79). The binding and spreading and/or bridging activities lend themselves to transcriptional silencing by either excluding RNA polymerase from a promoter or trapping RNA polymerase at a promoter, preventing transcription initiation (15, 65, 77). The resulting repressive nucleoprotein complex can then be disrupted by a wide variety of mechanisms, many of which rely on DNA binding proteins that target either the same sites in DNA as those of H-NS or adjacent ones such that H-NS fails to maintain its repressive relationship with the target promoter. This can be achieved by the displacement of H-NS or by a remodeling of the H-NS–DNA complex such that transcription can begin (69).
H-NS is associated with the silencing of the transcription of horizontally acquired genes, including virulence genes, in Gram-negative bacterial pathogens (20, 22, 43, 53, 54, 56). It has been suggested that the repression of the expression of genes acquired by lateral transfer avoids any loss of competitive fitness that might arise due to their inappropriate expression and provides time for the new genes to be integrated into the existing gene regulatory networks of the bacterium (20, 22, 53, 54, 69). This integration requires, at least in part, the evolution of a suitable mechanism for the derepression of the new genes. Ideally, this derepression mechanism should respond to environmental cues that are characteristic of the optimal conditions for the expression of the H-NS-repressed genes. For example, in the case of virulence genes, these cues might signal that the bacterium is in close association with a suitable host.
Shigella flexneri is the etiological agent of bacillary dysentery; it is a facultative intracellular pathogen and, as such, establishes a particularly intimate relationship with its human host (66). S. flexneri virulence depends on the expression of a type III secretion system and its associated effector proteins (61). The genes coding for these proteins are located within the so-called entry region, a 31-kbp segment of the S. flexneri 230-kbp virulence plasmid (9, 23). The genes within the entry region are organized into operons whose promoters are regulated by temperature, osmolarity, and pH (9, 26). The DNA of the plasmid is A+T rich, and all of the virulence promoters are silenced by H-NS (3, 9). Shifting the bacterium to a temperature of 37°C at an osmolarity equivalent to that of physiological saline and at a neutral pH derepresses the transcription of the virulence genes (29, 46, 52, 62). The genes are controlled by a regulatory cascade in which the expression of an AraC-like protein called VirF results in the derepression of an intermediate regulatory gene called virB (1, 27, 64, 74, 75). The virB gene is identical to invE in Shigella sonnei (78), and it is thought that the derepression of the virB gene represents a regulatory checkpoint that must be passed prior to a full commitment to the expression of the elaborate type III secretion system and its effector proteins (23). In addition to transcriptional regulation, the expression of the virB gene is also controlled at the level of mRNA stability in an Hfq-dependent manner in response to temperature and osmolarity (49, 50).
The VirB protein is required to derepress the principal promoters governing the expression of the structural genes encoding the type III secretion system and their associated effector proteins. Its activity has been studied in detail at the icsB promoter, where it has been shown to act as an antirepressor rather than a direct activator of RNA polymerase (76). VirB binds to the icsB regulatory region within a segment of DNA that is also targeted by H-NS. There, VirB polymerizes from its initial binding site and wraps the DNA, compromising the stability of the H-NS–DNA repression complex. As the concentration of VirB rises, the H-NS protein becomes displaced, leading to the concomitant upregulation of the icsB promoter (76). The responses of VirB-dependent promoters to this protein are dose dependent; even in the absence of the appropriate environmental signals, the artificial overexpression of VirB will upregulate the transcription of the genes in the VirB regulon (3).
The origin of the VirB-controlled regulatory checkpoint within the S. flexneri virulence cascade has been a subject of speculation. VirB does not resemble other transcription factors but instead shows considerable amino acid sequence identity to members of the ParB/SopB family of plasmid partition proteins (4, 78). The modern S. flexneri plasmid is a complicated mosaic of ancestor plasmids and mobile genetic elements; it has two fully functioning plasmid partition systems, and VirB seems to be a vestige of a third (9, 68). ParB-like proteins usually have a ParA partner protein that is also required for plasmid partitioning; VirB has no known partner of this type (9). Certainly, VirB is not required for the normal segregation of the S. flexneri virulence plasmid, and this has led to the suggestion that VirB may have been redirected to gene regulation from an earlier role in plasmid segregation in either the virulence plasmid or one of its ancestor plasmids (4).
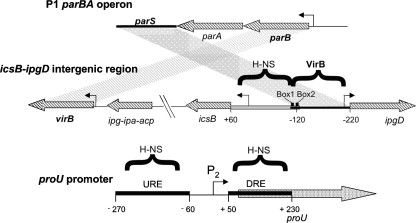
ParB-like proteins bind to a cis-acting DNA sequence called parS (5, 28, 35, 40, 67, 70), and it has been found that the VirB binding site at the icsB promoter retains the features of a parS sequence (73, 76). Normally, such parS elements are located adjacent to the genes coding for the ParA and ParB proteins (35). In the case of virB, the VirB binding site is located at a distance, due to the presence of the icsB operon between them (Fig. 1). This suggests that the VirB-dependent VirB/H-NS regulatory circuit at icsB may have arisen due to a simple local rearrangement of the virulence plasmid (Fig. 1). The essence of the regulatory switch is that one DNA binding protein (VirB) interferes with the repression complex established by a second DNA binding protein (H-NS) by virtue of the location of their respective binding sites. The extremely simple nature of the VirB/H-NS regulatory switch suggests that similar regulatory motifs could arise easily at other genes due to the emergence of suitably placed binding sites. This has already happened on the S. flexneri virulence plasmid, where VirB relieves the H-NS-mediated repression of the icsP gene from a VirB-binding site that has a minimal similarity to the full parS sequence (13). In this study, we sought to bring proU, a known target of H-NS-mediated transcription silencing, under the positive control of VirB, a virulence regulatory protein with no known history of involvement in proU regulation.
Fig. 1.
Comparison of the parBA operon from bacteriophage P1, the icsB-ipgD intergenic region from S. flexneri, and the proU promoter from E. coli. The parS centromere-like site is located downstream of the parBA operon in bacteriophage P1 (top). The P1 parS region corresponds to the VirB binding region in the S. flexneri virulence plasmid, and the P1 parB gene corresponds to the S. flexneri virB gene (middle) (3). The gray parallelogram denotes DNA sequence homology between parS and the S. flexneri VirB binding site; the striped parallelogram illustrates homology between the parB and virB genes. DNA sequences bound by the H-NS protein or by the VirB protein are indicated by horizontal brackets. The positions of the essential box 1/box 2 VirB binding and nucleation sites are shown. The regulatory region of proU is illustrated (bottom), together with the locations of the proU P2 promoter and the URE and DRE H-NS binding sites (gray boxes). The angled arrows represent promoters; the diagram is not drawn to scale.
The proU operon encodes an uptake system for the osmoprotectant glycine-betaine, and it is induced at the level of transcription in response to hyperosmotic stress (32). The operon is transcribed from a σ70-dependent promoter (called P2) that is flanked by two binding sites for H-NS (Fig. 1). These binding sites are the downstream regulatory element (DRE), lying within proV, the first gene of the operon, and the upstream regulatory element (URE), located on the other side of P2 (Fig. 1) (31, 33, 44, 58, 59, 71, 72). This arrangement is reminiscent of other H-NS-repressed genes, where the promoter is flanked by H-NS binding sites that may lend themselves to the formation of a bridged nucleoprotein transcription complex (15, 19, 76).
MATERIALS AND METHODS
Bacterial strains and growth media.
All bacterial strains were derivatives of Escherichia coli K-12 or S. flexneri 2a 2457T and are listed in Table 1. It should be noted that the open reading frames of the hns genes of E. coli K-12 and S. flexneri are identical (36). Bacteria were grown in LB broth (172 mM NaCl) or low-salt medium (LO; LB broth without NaCl) at 37°C. Antibiotics were used at the following concentrations: carbenicillin at 50 μg/ml, chloramphenicol at 25 μg/ml, kanamycin at 50 μg/ml, and tetracycline at 15 μg/ml.
Table 1.
Bacterial strains
| Strain | Relevant characteristic(s) | Reference or source |
|---|---|---|
| S. flexneri 2a 2457T | ||
| BS184 | mxiC::mudI1734 Kmr | 46 |
| BS185 | BS184 hns::Tn10 Tcr | 46 |
| CJD1018 | BS184 virB::pMEP151 | 63 |
| E. coli K-12 | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lacΔFΔproAB lacIqZΔM15 Tn10 (Tcr) | Stratagene |
| MG1655 | F− ΔilvG rfb-50 rph-1 | Laboratory stocks |
Plasmid construction and genetic manipulations.
For flow cytometric analysis, the proU promoter region of E. coli K-12 derivative MG1655 was amplified as a 500-bp fragment using oligonucleotide primers proU fw −270.NotI and proU rev +230.XbaI (see Table S1 in the supplemental material). The PCR fragment was digested by using the restriction enzymes NotI and XbaI, and the digested fragment was ligated into NotI- and XbaI-digested plasmid pZep08. The structure of the resulting construct was verified by DNA sequencing, and the construct was designated pZep-proU-1 (Table 2). The VirB binding site from icsB was amplified as a 160-bp fragment by PCR using the primer pair icsB.fw and icsB.rev, which were NotI tagged (Table S1). The PCR fragment was digested by using the restriction enzyme NotI, and the digested fragment was ligated into NotI-digested plasmid pZep-proU-1. The structure of the resulting construct was verified by DNA sequencing, and the construct was designated either pZep-proU-2, if the binding site was inserted in the reverse orientation, or pZep-proU-3, if the site was inserted in the forward orientation. Plasmids pZep-proU-4 to pZep-proU-7 were constructed in a similar manner. Control plasmids (denoted by the letter M) were also constructed, which included the 160-bp icsB region with mutations in the essential box 1/box 2 as previously described (76).
Table 2.
Plasmids
| Plasmid | Relevant characteristic(s) | Reference |
|---|---|---|
| pZep08 | GFP reporter plasmid; Apr Cmr | 34 |
| pZep-proU-1 | Positions −270–+230 of PproU cloned into pZep08 | This study |
| pZep-proU-2 | Positions −240–−80 of PicsB cloned into pZep-proU-1 in the reverse orientation | This study |
| pZep-proU-3 | Positions −240–−80 of PicsB cloned into pZep-proU-1 in the forward orientation | This study |
| pZep-proU-4 | Positions −150–+230 of the PproU promoter cloned into pZep08 | This study |
| pZep-proU-5 | Positions −240–−80 of PicsB cloned into pZep-proU-4 in the reverse orientation | This study |
| pZep-proU-6 | Positions −60–+230 of the PproU promoter cloned into pZep08 | This study |
| pZep-proU-7 | Positions −240–−80 of PicsB cloned into pZep-proU-8 in the reverse orientation | This study |
| pZep-proU-8 | Positions −240–−80 of PicsB cloned into pZep-proU-6 in the reverse orientation with the URE upstream | This study |
| pZep-proU-9 | 210-bp yaeT coding region cloned into pZep-proU-6 | This study |
| pZep-proU-10 | Positions −240–−80 of PicsB cloned into pZep-proU-9 in the reverse orientation | This study |
The VirB binding site from icsB was inserted between the URE and the DRE at position −60, resulting in pZep-proU-8, by site-directed mutagenesis of the proU promoter to include a unique FseI restriction site using primer set proU SDM FseI fw and proU SDM FseI rev. The FseI-tagged icsB PCR product was then digested and inserted at this position. Site-directed mutagenesis was performed by using the QuikChange (Stratagene) kit according to the manufacturer's guidelines. The replacement of the URE with 180 bp of the yaeT coding region was performed by PCR amplification of yaeT with primer set yaeT fw.SmaI and yaeT rev.NotI and the insertion of this amplicon into pZep-proU-6 to create pZep-proU-9. The NotI-tagged icsB PCR product was then digested and inserted into NotI-digested pZep-proU-9, creating pZep-proU-10.
Mutagenesis by lambda red recombination.
All knockout mutations and chromosome integrations were made by homologous recombination using the λ red recombination system described previously by Datsenko and Wanner (18). The λ red recombination system facilitates integration into the chromosome of PCR products containing 40 bp of DNA sequence homology at both the 5′ and 3′ ends of the region of the chromosome where the PCR product is to be integrated. To generate a proU::tet mutant, the tetRA resistance cassette was PCR amplified with primer set tet.40 bp proU fw and tet.40 bp proU rev (see Table S1 in the supplemental material) and spin column purified by using a HiYield PCR DNA fragment extraction kit (RBC Bioscience). It was then transformed into electrocompetent S. flexneri BS184 cells containing the red helper plasmid as previously described (18). The structure of the proU::tet lesion was confirmed by PCR and DNA sequencing (GATC Biotech) and then used as a template for the insertion of the modified icsB-proU-gfp-cat promoters using primer set icsB.fw.40 bp proU and cat.rev.40 bp proU or icsB.rev.40 bp proU and cat.rev.40 bp proU.
β-Galactosidase assay.
The transcription of the mxiC::lacZ fusion was measured in cultures of the wild type and hns and virB mutant strains grown overnight in LO or LB broth at 37°C. The β-galactosidase activity was measured as described previously by Miller (48).
Flow cytometry analysis.
The bacterial culture to be assayed was harvested and immediately fixed in 4% formaldehyde in phosphate-buffered saline (PBS) and then stored at 4°C in the dark. Before analysis, samples were diluted to a concentration of approximately 106 bacteria per ml and then analyzed with an Epics-XL flow cytometer (Beckman Coulter). Approximately 10,000 bacteria per sample were assayed, and the results were expressed as the mean channel fluorescence after analysis with EXPO-XL software. Each sample was measured in duplicate, and the mean values were determined from the results of at least three independent experiments.
Immunodetection of the VirB protein.
Total protein extracts were separated through a 12% SDS-PAGE gel. The separated proteins were electroblotted onto a nitrocellulose membrane using the Bio-Rad Miniprotean II system for 1 h at 80 V. Nitrocellulose membranes were stained with Ponceau stain (0.2% Ponceau dye, 3% trichloroacetic acid) to check the efficiency of transfer before being blocked overnight with 5% dried skimmed milk in phosphate-buffered saline (PBS). The detection of the VirB protein was preformed by use of PBS containing 1% dried skimmed milk with primary polyclonal anti-VirB antiserum (1:500) and a secondary goat anti-rabbit horseradish peroxidase-conjugated antiserum (1:10,000). Membranes were developed by using the chemiluminescent Pierce West Pico Super Signal kit.
Real-time quantitative PCR.
Total RNA was isolated from cultures by using the SV Total RNA Isolation system (Promega), and purity and quality were assessed by electrophoresis in 1% agarose (1× Tris-acetate-EDTA [TAE]). Residual DNA contamination was removed by treatment with DNase I (Ambion DNA-free kit). cDNA templates were synthesized by the random priming of 1 μg of RNA in a 20-μl reaction mixture using the GoScript reverse transcription system (Promega). Quantitative PCR (qPCR) primers are listed in Table S1 in the supplemental material. PCRs were carried out in duplicate with primer sets on an ABI 7500 sequence detection system (Applied Biosystems) using FastStart SYBR green master mix with ROX (Roche). Standard curves were generated for each primer set using five serial 10-fold dilutions of chromosomal DNA and were included in every run.
RESULTS
Creation of VirB dependence at the proU promoter.
The proU operon is transcribed from a promoter (P2) whose transcription start site is located approximately 60 nucleotides upstream of the initiation codon of the first structural gene (proV) and which is recognized by the σ70-RNA polymerase holoenzyme (E σ70). A second promoter, P1, located 250 nucleotides upstream of proV, is σS (RpoS) dependent, but its physiological significance is uncertain, since the deletion of P1 or the elimination of rpoS expression does not affect proU transcription in vivo (32). Furthermore, quantitative real-time PCR (qRT-PCR) showed that under the conditions used in this study, the transcription of proU is driven by P2 and not P1 (see Fig. S1 in the supplemental material).
The proU promoter region contains two regulatory domains, the upstream regulatory element (URE) and the downstream regulatory element (DRE), both of which are known H-NS binding sites (13). These high-affinity binding sites contain identical matches to the consensus sequence for H-NS binding (Fig. 1). Under conditions of low osmolarity, H-NS is bound in these regions and represses transcription; upon the addition of salt, the proU promoter is derepressed as H-NS is displaced, allowing transcription to proceed; full expression occurs at 300 mM NaCl (32). Transcription silencing at proU may involve a repressive complex consisting of a DNA–H-NS–DNA bridge at the URE and the DRE (7). However, the DRE alone is sufficient to maintain significant repression under low-salt conditions, where it inhibits the formation of an open transcription complex (7, 51). In this study, the previously characterized binding site for VirB at the icsB promoter was introduced into the promoter region of proU at different locations, and the synthetic promoters were monitored for VirB-dependent derepression under normally repressive conditions.
The VirB binding site at its native location at icsB is found upstream of the promoter, bordering the region that is bound by H-NS (76). To mimic this arrangement, the VirB binding site, including the essential box 1/box 2 motif, was inserted immediately upstream of the URE at the proU promoter. Transcription was monitored by using a gfp fusion, where levels of the green fluorescent protein (Gfp) reflected proU promoter activity (Fig. 2). The expression of proU-gfp was monitored in wild-type, virB mutant, and hns mutant backgrounds under growth conditions that were normally nonpermissive for proU transcription, i.e., LB broth containing 0 M NaCl. Control constructs that completely lacked a VirB binding site were also monitored to determine the level of native proU expression under nonpermissive conditions. Further controls that included constructs with a modified VirB binding site with base pair substitutions in the essential box 1/box 2 motif were also monitored to examine proU expression in constructs where the antagonist cannot bind. Functional or mutated VirB binding sites were introduced at position −270 with respect to the transcription start site. Insertions were made in either a forward or a reverse orientation so as to address the possibility that VirB binding site directionality might influence derepression (Fig. 2A to C). The gfp gene was placed under the control of each promoter on a plasmid to monitor proU transcription. All of these experiments were performed with S. flexneri.
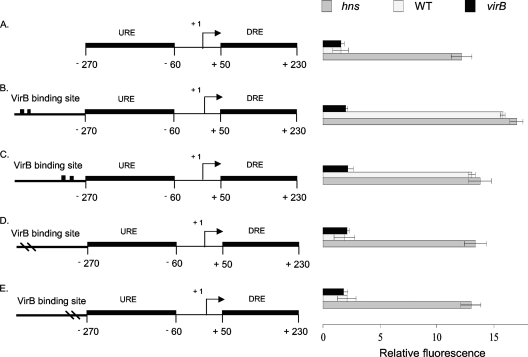
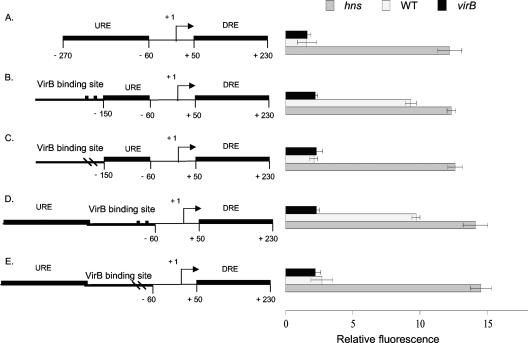
Fig. 2.
Derepression of the proU promoter by VirB. Summaries of the structures of various derivatives of the E. coli proU promoter region are presented at the left; levels of expression of the proU-gfp transcriptional reporter fusion are given at the right (diagrams not to scale). The native proU promoter is shown (A); proU incorporating functional VirB binding and nucleation sites (denoted by two black boxes) is shown in the forward (B) and reverse (C) orientations, and proU with inactivated VirB binding sites (denoted by two dashes in the VirB site) is shown in the forward (D) and reverse (E) orientations (drawings not to scale). The constructs were assessed in S. flexneri for proU-gfp expression in a virB mutant (black bars), the wild type (WT) (white bars), and the hns mutant (gray bars) under conditions normally repressive of proU transcription.
The expression of proU-gfp in the control construct, either completely lacking the binding site for VirB or containing the version with a mutated box 1/box 2 motif, was repressed, and repression was maintained regardless of the presence or absence of the VirB protein (Fig. 2A, D, and E). Repression was alleviated only when the hns gene was inactivated. However, the modified proU promoters that contained a functional binding site for VirB were derepressed in the presence of a functioning hns gene (Fig. 2B and C). These proU-gfp expression levels were similar to those seen for the hns mutant, suggesting that the upregulation of the proU promoter involved a relief of H-NS-mediated repression. This derepression of proU in low concentrations of salt was contingent on the presence of the VirB protein. The ability of VirB to derepress the promoter was not influenced by the orientation of the VirB binding site (Fig. 2B and C).
To address the possibility that plasmid-associated artifacts might have influenced the results, the proU-gfp fusion constructs with the various forms of the VirB binding site were integrated into the chromosome at the native proU locus. The results obtained with these strains were similar to those obtained from the plasmid-based experiments (data not shown).
VirB-dependent derepression is conditional on binding-site location.
The VirB binding site was inserted into different locations upstream of the proU promoter to investigate the importance of its position relative to the URE and the DRE, the cis-acting negative regulatory elements that are bound by H-NS. The DRE found downstream of the proU promoter is known to be imperative for the maintenance of proU repression by H-NS. In contrast, the URE has been shown to be less essential for H-NS-mediated repression under low-osmolarity growth conditions (44, 51). To assess the significance of the URE for the VirB-mediated derepression mechanism, two new proU promoter constructs were made, one with the VirB binding site inserted into the URE and the other with the site located between the URE and the DRE (Fig. 3).
Fig. 3.
Partial derepression of the proU promoter by VirB. Summaries of the structures of various derivatives of the E. coli proU promoter region are presented at the left; levels of expression of the proU-gfp transcriptional reporter fusion are given at the right (diagrams not to scale). Shown are the native proU promoter (A), proU with a functional VirB binding site located at position −150 (B) or with an inactivated binding site for VirB (C), and proU with a functional binding site for VirB at position −60 with the URE upstream (D) or with an inactivated binding site for VirB (E). All constructs were assessed in S. flexneri for proU-gfp expression in a virB mutant (black bars), the wild type (white bars), and the hns mutant (gray bars) under conditions normally repressive of proU transcription.
The URE and VirB-dependent derepression of proU.
The insertion of the VirB binding site at position −150 with respect to the proU transcription start site interrupted the URE at approximately its midpoint (Fig. 3A to C). Insertions were made at this position of the native VirB binding site (Fig. 3B) and of the site with the box 1/box 2 mutations (Fig. 3C). The mutated site was unable to support the derepression of the proU-gfp fusion in the presence of VirB, while the presence of the native binding site resulted in only partial VirB-dependent depression. In both cases, derepression was assessed by comparison with the level of proU-gfp expression seen for the hns knockout mutant (Fig. 3).
The VirB binding site was next inserted at position −60, placing it between the intact URE and DRE elements. This insertion also had the effect of displacing the URE further upstream from the transcription start site; its promoter-proximal boundary was now at position −160 rather than position −60 (Fig. 3). Insertions were made at this position of both the intact binding site (Fig. 3D) and the mutant derivative with the box 1/box 2 base pair substitutions (Fig. 3E). As before, the mutant binding site could not support the VirB-dependent derepression of proU-gfp transcription, whereas the intact binding site did (Fig. 3B and C). However, the level of derepression achieved was not as great as that seen for the hns-deficient strain. Taken together, these data indicated that the successful antagonism of proU repression was affected by both the presence and the location of an intact URE.
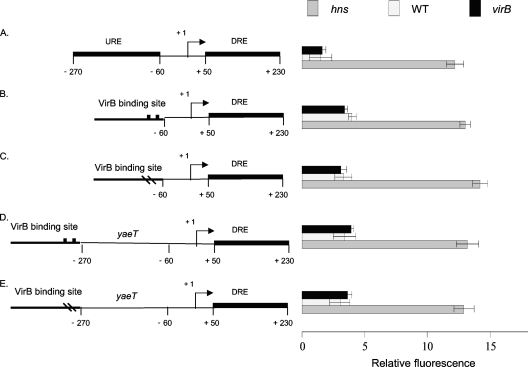
Removal of the URE abolishes VirB-dependent derepression of proU.
The proU promoter region was modified so that the URE was removed completely and replaced by the VirB binding site (Fig. 4A and B). A control construct was produced in which the altered form of the VirB binding site containing the box 1/box 2 mutations had replaced the URE (Fig. 4C). Neither of these constructs supported the VirB-dependent derepression of the proU-gfp fusion, although the fusion was derepressed when introduced into an hns mutant strain (Fig. 4).
Fig. 4.
The URE and VirB-dependent proU derepression. Summaries of the structures of various derivatives of the E. coli proU promoter region are presented at the left; levels of expression of the proU-gfp transcriptional reporter fusion are given at the right (diagrams not to scale). Shown are the native proU promoter (A), proU with a functional VirB binding site at position −60 (B) or a mutated VirB binding site at the same position (C), and proU with a functional VirB binding site at position −270 with the URE removed (D) or with an inactivated VirB binding site at the same position (E). All constructs were assessed in S. flexneri for proU-gfp expression in a virB mutant (black bars), the wild type (white bars), and the hns mutant (gray bars) under conditions normally repressive of proU transcription.
Full VirB-dependent depression of proU transcription had been achieved when the VirB binding site was located immediately upstream of the URE (Fig. 2B and C). In this construct, the promoter-proximal boundary of the binding site was at position −270. The constructs where the URE had been replaced by the VirB binding site (Fig. 4B and C) were modified by the insertion of a DNA sequence from the yaeT gene, which is known not to bind the H-NS protein (30). This insertion had the effect of moving the promoter-proximal boundary of the VirB binding site to position −270 with respect to the transcription start site, restoring the distance (but not the H-NS binding activity) associated with the presence of the URE (Fig. 4D and E). The presence of an intact VirB binding site did not result in the VirB-dependent derepression of proU-gfp transcription in the absence of the URE, despite the placement of the binding site at a location from where it was effective when the URE was present (compare Fig. 2C and 4D).
Neither set of constructs in which the URE was removed showed an alleviation of repression compared to the hns mutant. This result is consistent with the hypothesis that while the URE and the DRE may act synergistically, the DRE alone can maintain significant repression (51).
VirB protein levels are sufficient for proU derepression under conditions of low osmolarity.
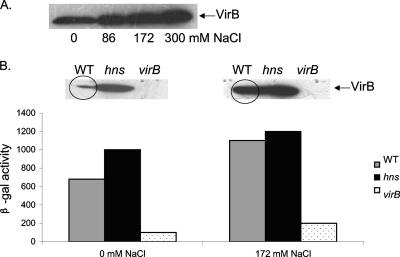
The VirB protein is known to be present at reduced levels under the low-osmolarity growth conditions of these experiments (63). To confirm this, VirB protein levels in bacteria exposed to increasing concentrations of NaCl were monitored. The results showed a clear increase in protein levels with increasing osmolarity (Fig. 5A). Western blots were used to monitor variations in VirB protein concentrations in wild-type bacteria and an hns mutant grown under 0 mM NaCl conditions or in standard LB broth with 172 mM NaCl. As expected, VirB protein levels increased in the hns mutant (the virB promoter is repressed by H-NS), and no VirB was detected in a virB-null mutant (Fig. 5B). Despite levels of VirB being lower in wild-type cells under conditions of low osmolarity, the proU expression data showed that there was still a sufficient quantity of VirB to exert an effect on transcription. To verify this, the expression of a lacZ reporter fusion to mxiC, a VirB-regulated gene encoding a structural component of the type III secretion system, was analyzed. β-Galactosidase activities were assessed at both osmolarities in the wild-type, virB, and hns strains. The results showed that although VirB protein levels were depleted at 0 M NaCl compared to those found with the standard concentration of NaCl (172 mM), there was a sufficient amount of protein to derepress mxiC transcription, in agreement with the data obtained with the proU promoter constructs (Fig. 5B).
Fig. 5.
VirB protein levels are reduced under low-salt conditions but are still sufficient to derepress promoters. (A) Western blot analysis of VirB protein levels in S. flexneri cells grown with increasing amounts of salt. (B) Western blot analysis of VirB levels coupled with expression data from an mxiC-lacZ fusion (a gene expressed from a VirB-dependent promoter) in wild-type, hns, and virB bacteria grown in the presence of 0 mM or 172 mM NaCl. The VirB protein band in the wild-type samples is circled to highlight the increase in the VirB concentration in bacteria growing in medium containing 172 mM NaCl.
The modified proU promoter constructs retain osmosensitivity.
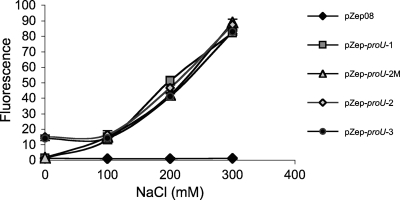
To ensure that the icsB-proU hybrid promoters used in this study retained the osmotic sensitivity that is characteristic of proU, the hybrid promoters were tested for activity under conditions of increasing osmolarity. Both the native proU promoter and those derivatives that had been modified to accept a binding site for the VirB protein showed strong sensitivity to increasing osmolarity, showing that the underlying proU promoter function had not been altered by the introduction of DNA containing the S. flexneri icsB VirB binding site (Fig. 6).
Fig. 6.
Osmoregulation of modified proU promoters. The expression of Gfp was measured in S. flexneri cells growing in increasing concentrations of NaCl from pZep08 (promoterless gfp), pZep-proU-1 (native proU promoter), pZep-proU-2 M (proU with a mutated VirB binding site at position −270), pZep-proU-2 (proU with a functional VirB binding site at position −270 in the reverse orientation), and pZep-proU-3 (proU with a functional VirB binding site at position −270 in the forward orientation). The data are the averages of data from three measurements, and the error bars represent the standard deviations.
DISCUSSION
Gene regulatory mechanisms in bacteria cover a spectrum of sophistication, and among the most intensively studied are those that control transcription initiation (8). The repression and activation of RNA polymerase activity at the initial stages of the transcription process can be achieved by a wide variety of mechanisms. Among the most basic are likely to be mechanisms that rely chiefly on the modulation of DNA structure. For example, the ultrasimple bacterium Mycoplasma genitalium has just one sigma factor and is thought to regulate much of its transcription through changes in DNA supercoiling (24, 80). However, most regulatory mechanisms involve roles for DNA binding proteins, especially proteins whose expression and/or activity is responsive to environmental signals. The distribution in the genome of the DNA sequence motifs that these proteins recognize and bind determines the extent of their regulatory influence (37, 42). Genes can acquire and lose regulatory protein binding sites through mutation, and so they can join and leave regulons as part of the process of regulatory evolution. Regulatory proteins with relatively astringent requirements for DNA binding can bring large numbers of genes under their control, and this is certainly so in the case of the nucleoid-associated protein H-NS (14, 20, 22, 38, 43, 53, 54, 57).
The H-NS protein has a regulatory role throughout the virulence gene cascade in S. flexneri, and its transcription repression activity is opposed there in many cases by the VirB protein (3). This is a particularly interesting case because VirB shares many properties with plasmid-partitioning proteins of the ParB/SopF family, including amino acid sequence identity and common requirements for the makeup of its binding sites in DNA (1, 73, 76). The suggestion that the S. flexneri virulence regulon has co-opted VirB from an earlier role in plasmid maintenance raises interesting questions about the plasticity of gene regulatory circuits and their capacity for evolution.
The existing information on the regulatory activity of VirB suggests that it operates via a very simple mechanism: the concentration of the VirB protein in the cell is controlled in response to environmental signals, and once a threshold concentration is reached, it can displace the H-NS repressor from its target gene promoters (3, 76). Displacement is most likely to be achieved due to the mutually incompatible effects of H-NS and VirB on the local DNA structure at their overlapping binding sites. VirB acts simply as an antirepressor that makes the H-NS–DNA repression complex untenable; it does not act to recruit or to activate RNA polymerase (76).
The simple nature of the VirB regulatory mechanism makes it an interesting subject for artificial-evolution experiments involving genes that are repressed by H-NS but have no known VirB dependency. The proU operon fits this description and is a particularly attractive subject for investigation because, unlike the S. flexneri virulence operons, proU is not regulated in response to temperature (7).
We have modified the proU promoter to make it responsive to the VirB protein. The experiments described in this report were carried out under low-osmolarity growth conditions, where the proU promoter is subject to full transcription repression, at least part of which is due to the binding of the H-NS protein (58). Similar results were obtained when the modified proU promoter was studied in recombinant plasmids or on the chromosome at the native location of the proU operon, increasing our confidence that the data reflect intrinsic properties of the promoter-regulator relationship and not artifacts of the genomic position.
The placement of the VirB binding site immediately adjacent to, and upstream of, the URE brought the proU promoter under the positive control of the VirB protein. This showed that the VirB binding-site combination influences proU transcription from a position that is at least 270 bp upstream of the transcription start site (Fig. 2). At this range, it is highly unlikely that the VirB protein is acting via protein-protein contact with RNA polymerase bound to the proU promoter. It is more likely that VirB is acting as an antirepressor by antagonizing H-NS-mediated promoter repression, just as it does at its native targets in the S. flexneri virulence gene regulatory cascade. The effect on proU promoter activity was the same regardless of the orientation of the sequence of the VirB binding site (Fig. 2D and E). This is consistent with the ability of this same sequence to regulate positively and simultaneously the divergently oriented promoters of the icsB and the ipgD operons on the S. flexneri large virulence plasmid (26, 39). There, VirB-DNA interactions resulted in increased hypersensitivity to DNase I cleavage, a pattern that is consistent with DNA wrapping by the VirB protein. This form of DNA remodeling by the VirB protein is incompatible with the maintenance of an H-NS–DNA nucleoprotein complex both in the same and in the immediately adjacent DNAs (76).
While the placement of the VirB binding site immediately adjacent to the URE facilitated the derepression of the proU promoter by VirB, the placement of the same DNA sequence immediately adjacent to a truncated URE facilitated only partial derepression (Fig. 3B). When the intact URE was relocated to a position on the promoter-distal side of the VirB binding site, the efficacy of VirB-mediated transcriptional derepression was reduced (Fig. 3D). Moreover, the complete removal of the URE from next to the VirB binding site resulted in a complete loss of VirB-dependent derepression (Fig. 4B). That the effects seen were not due to the distance of the VirB binding site from the promoter was shown by experiments in which the location of the binding site at position −270 was preserved by the insertion of a 210-bp segment of DNA known not to contain H-NS binding sites (Fig. 4D). Here, in the absence of the URE, the proU promoter remained repressed unless the H-NS protein was removed from the cell by a mutation of the hns gene. These data suggest that the URE plays an important role in the VirB-dependent derepression mechanism.
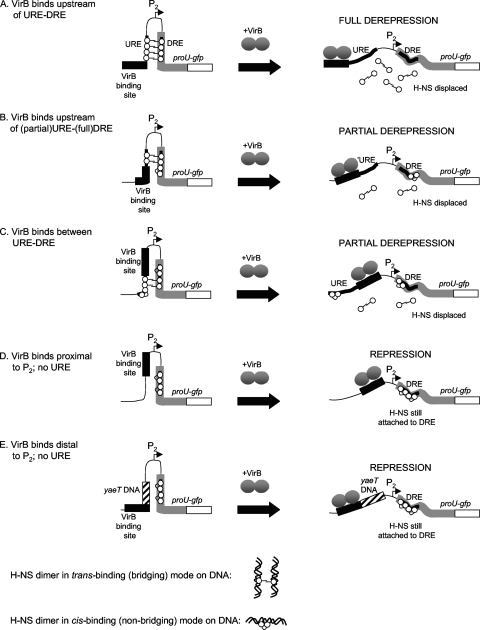
The model that is most likely to account for the VirB-mediated derepression of the proU-gfp fusion under normally nonpermissive growth conditions is one that assumes that both the URE and the DRE participate in an H-NS-dependent transcription repression complex (Fig. 7). The established ability of the VirB protein to act as an antirepressor in S. flexneri through the disruption of H-NS–DNA complexes can be applied to this URE–H-NS–DRE complex, particularly if the ability of H-NS to form DNA-protein-DNA bridges is taken into account (16, 17, 45, 55). Assuming that the URE and the DRE are bridged by H-NS dimers to form a structure that is inhibitory to proU transcription, the binding of the VirB protein to its site located immediately adjacent to the URE with the associated remodeling of the nucleoprotein complex due to the VirB wrapping of DNA might be expected to reduce the stability of the repression complex (Fig. 7A). The proposed displacement of H-NS from the bridged structure would be consistent with the upregulation of the proU-gfp fusion to the levels seen with the hns mutant.
Fig. 7.
VirB-dependent derepression of the proU promoter. The model is based on the differential sensitivity of the known trans-binding (i.e., bridging) and cis-binding (nonbridging) modes of DNA binding exhibited by the H-NS protein to antagonism by the VirB protein. Full derepression is achieved when the URE and the DRE are intact and the binding site for the VirB protein is located immediately upstream of the URE (A). This mimics the situation in icsB, a natural target of VirB in S. flexneri. Here, VirB acts to antagonize DNA–H–NS-VirB bridging. If the URE is truncated (B), the degree of bridging is reduced, allowing H-NS to adopt a nonbridging binding mode over part of the DRE, imposing a partial barrier to full transcriptional derepression. When the VirB binding site is located between the URE and the DRE (C), derepression is also inefficient. The removal of the URE (D) abrogates bridging by H-NS, and the resulting cis-binding mode over the DRE produces a partially effective barrier to proU derepression. Similarly, the displacement of the virB binding site to position −270 but with a non-H-NS-binding DNA sequence (yaeT) in place of the URE also permits the development of the cis-bound H-NS complex at the DRE that is partially effective at preventing proU derepression.
The placement of the VirB binding site between the URE and the DRE or adjacent to a truncated URE also resulted in the upregulation of the proU-gfp fusion under nonpermissive conditions but not to the same level as that seen for the hns mutant. This indicates that residual H-NS-mediated repression activity remained when the VirB protein exerted its antirepressive activity from these sites. Perhaps, the disruption of the URE–H-NS–DRE bridge by the alteration of the URE resulted in some H-NS molecules adopting a cis-binding mode at the DRE rather than a trans-binding mode involving URE–H-NS–DRE bridging, with the H-NS–DRE complex exerting some impediment to transcription (Fig. 7B).
In all cases where VirB derepressed the proU promoter, the VirB binding site was located immediately adjacent to a DNA sequence that is known to bind H-NS. The removal of the URE abolished the VirB-dependent derepression of proU. In the absence of the URE, repression would be maintained by the DRE alone. The binding of VirB at position −60 or −270 without the URE present might place the antagonist at too great a distance for it to displace the repressive complex formed by H-NS binding at the DRE (Fig. 7C and D).
The results reported in this study demonstrate the relative ease with which a novel genetic switch can evolve based on H-NS displacement with a heterologous DNA binding protein. Previous attempts at altering the regulation of the proU promoter have relied on more sophisticated mechanisms where a dedicated transcription factor (TyrR) is used to recruit RNA polymerase to the proU promoter in response to the cognate environmental signals detected by that protein (33). In that case, the TyrR binding site must be placed at the promoter, taking into account spatial constraints that might interfere with physical interactions between the transcription factor and RNA polymerase (33). This is in contrast to the simpler antirepression mechanism that has been exploited in the case of VirB. The creation of the VirB-dependent proU derepression system also differs from a previously reported redesign of the bgl promoter, where the Lac or lambda repressor was recruited as a positive regulator. There, the binding sites for the heterologous DNA binding proteins were inserted at a series of different locations within the bgl equivalent of the URE, disrupting its ability to act as a repressive element (10). This is likely to have created direct competition between the Lac or lambda repressor and H-NS for access to bgl DNA, whereas the VirB protein acts to “pull the rug” from under H-NS at the immediately adjacent proU URE by its DNA-wrapping activity by analogy with its antirepressor mechanism at icsB in S. flexneri (76).
The adaptability of VirB as an antirepressor is reflected in the number of promoters that this protein regulates on the S. flexneri virulence plasmid. Those binding sites that have been studied have shown similarity to the minimal parS element that is bound by ParB-like proteins (6, 35, 41). This is a portion of the full-length parS-like motif that is found between the icsB and ipgD promoters and is approximately equivalent to the region at the box 1/box 2 motif.
It should be noted that the modified proU constructs all retained osmoregulation and that expression was highly induced under conditions of osmotic upshock, comparable to that of the original wild-type promoter (Fig. 5). The VirB dependency added an extra level of regulation to the promoter without altering its native expression pattern. Also, the titer of the VirB protein in the cell was approximately equivalent to, or perhaps slightly below, that of wild-type S. flexneri, as virB is downregulated under low-salt conditions (62, 63). Western blot analysis confirmed a lower concentration of VirB in the cell than in bacteria growing under conditions of standard osmolarity; however, monitoring of the expression of a VirB-dependent mxiC::lacZ fusion on the S. flexneri virulence plasmid demonstrated that the concentration of VirB present was sufficient to exert its normal effect on transcription (Fig. 6). The constructs were not subjected to an overexpression of the VirB protein, but instead, natural levels of the protein were used to effect derepression. Therefore, although artificial constructs were tested here, this system did mimic its native equivalent in that physiologically relevant conditions were used and proU osmoregulation remained intact. This serves to support the proposal that an event of the type engineered here could evolve by natural means.
We have demonstrated that the insertion of a VirB binding site can confer VirB-mediated derepression to an H-NS-repressed promoter that is unrelated to the normal VirB regulon. This is consistent with the hypothesis that the VirB binding site may once have been located in the vicinity of its gene (virB), by analogy with the juxtaposition of its parB homologue and its cis-acting parS site (Fig. 1). The insertion of the icsB-ipg-ipa-acp operon would have disconnected the VirB site from the virB gene while simultaneously placing this site at a position in the icsB promoter region, whereby it can act to displace H-NS, creating a new role for the VirB protein. The experimental results obtained in this study also illustrate that fine-tuning would be required for this regulatory network innovation to be successful: the correct positioning of the binding sites for the antagonist and repressor is essential for efficient derepression to occur. This implies that such a repositioning of protein binding sites in DNA accompanied by the co-option of cognate regulatory proteins are events that can occur routinely in bacteria. The creation of new regulatory circuits may afford bacteria the potential to explore and/or exploit novel environmental niches. It is clear that studies of H-NS antagonism have much to teach us about the evolution of regulatory switches and highlight pitfalls that may arise from the facile assignment of unique biological properties to DNA binding proteins.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Dorman laboratory for valuable discussions.
This work was supported by a grant from Science Foundation Ireland.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 26 August 2011.
REFERENCES
- 1. Adler B., et al. 1989. A dual transcriptional activation system for the 230-kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol. Microbiol. 3:627–635 [DOI] [PubMed] [Google Scholar]
- 2. Amit R., Oppenheim A. B., Stavans J. 2003. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys. J. 84:2467–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beloin C., Dorman C. J. 2003. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol. Microbiol. 47:825–838 [DOI] [PubMed] [Google Scholar]
- 4. Beloin C., McKenna S., Dorman C. J. 2002. Molecular dissection of VirB, a key regulator of the virulence cascade of Shigella flexneri. J. Biol. Chem. 277:15333–15344 [DOI] [PubMed] [Google Scholar]
- 5. Bignall C., Thomas C. M. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91:1–34 [DOI] [PubMed] [Google Scholar]
- 6. Bouet J.-Y., Funnell B. E. 1991. P1 ParA interacts with the P1 partition complex at parS and an ATP-ADP switch controls ParA activities. EMBO J. 18:1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bouffartigues E., Buckle M., Baudet C., Travers A., Rimsky S. 2007. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 14:441–448 [DOI] [PubMed] [Google Scholar]
- 8. Browning D. F., Busby S. J. W. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57–65 [DOI] [PubMed] [Google Scholar]
- 9. Buchrieser C., et al. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760–771 [DOI] [PubMed] [Google Scholar]
- 10. Caramel A., Schnetz K. 1998. Lac and λ repressors relieve silencing of the Escherichia coli bgl promoter. Activation by alteration of a repressing nucleoprotein complex. J. Mol. Biol. 284:876–883 [DOI] [PubMed] [Google Scholar]
- 11. Carroll S. B. 2005. Evolution at two levels: on genes and form. PLoS Biol. 3:e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carroll S. B. 2008. Evo-Devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134:25–36 [DOI] [PubMed] [Google Scholar]
- 13. Castellanos M. I., et al. 2009. VirB alleviates H-NS repression of the icsP promoter in Shigella flexneri from sites more than one kilobase upstream of the transcription start site. J. Bacteriol. 191:4047–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corcoran C. P., Dorman C. J. 2010. H-NS silences gfp, the green fluorescent protein gene: gfpTCD is a genetically remastered gfp gene with reduced susceptibility to H-NS-mediated transcription silencing and with enhanced translation. J. Bacteriol. 192:4790–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dame R. T., Wyman C., Wurm R., Wagner R., Goosen N. 2002. Structural basis for H-NS-mediated trapping of RNA polymerase in the initiation complex at the rrnB P1. J. Biol. Chem. 277:2146–2150 [DOI] [PubMed] [Google Scholar]
- 16. Dame R. T., et al. 2005. DNA bridging: a property shared among H-NS-like proteins. J. Bacteriol. 187:1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dame R. T., Noom M. C., Wuite G. J. 2006. Bacterial chromatin organisation by H-NS protein unravelled using dual DNA manipulation. Nature 444:387–390 [DOI] [PubMed] [Google Scholar]
- 18. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De la Cruz M. A., Merino E., Oropeza R., Téllez J., Calva E. 2009. The DNA static curvature has a role in the regulation of the ompS1 porin gene in Salmonella enterica serovar Typhi. Microbiology 155:2127–2136 [DOI] [PubMed] [Google Scholar]
- 20. Dillon S. C., et al. 2010. Genome-wide analysis of the H-NS and Sfh regulatory networks in Salmonella Typhimurium identifies a plasmid-encoded transcription silencing mechanism. Mol. Microbiol. 76:1250–1265 [DOI] [PubMed] [Google Scholar]
- 21. Dorman C. J. 2004. H-NS, a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391–400 [DOI] [PubMed] [Google Scholar]
- 22. Dorman C. J. 2007. H-NS, the genome sentinel. Nat. Rev. Microbiol. 5:157–161 [DOI] [PubMed] [Google Scholar]
- 23. Dorman C. J. 2009. The virulence plasmids of Shigella flexneri, p. 151–170 In Schwartz E. (ed.), Microbial megaplasmids. Springer, Heidelberg, Germany [Google Scholar]
- 24. Dorman C. J. 2011. Regulation of transcription by DNA supercoiling in Mycobacterium genitalium: global control in the smallest known self-replicating genome. Mol. Microbiol. 81:302–304 [DOI] [PubMed] [Google Scholar]
- 25. Dorman C. J., Kane K. A. 2009. DNA bridging and antibridging: a role for bacterial nucleoid-associated proteins in regulating the expression of laterally acquired genes. FEMS Microbiol. Rev. 33:587–592 [DOI] [PubMed] [Google Scholar]
- 26. Dorman C. J., Porter M. E. 1998. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 29:677–684 [DOI] [PubMed] [Google Scholar]
- 27. Dorman C. J., McKenna S., Beloin C. 2001. Regulation of virulence gene expression in Shigella flexneri, a facultative intracellular pathogen. Int. J. Med. Microbiol. 291:89–96 [DOI] [PubMed] [Google Scholar]
- 28. Ebersbach G., Gerdes K. 2005. Plasmid segregation mechanisms. Annu. Rev. Genet. 39:453–479 [DOI] [PubMed] [Google Scholar]
- 29. Falconi M., Colonna B., Prosseda G., Micheli G., Gualerzi C. O. 1998. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 17:7033–7043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fang F. C., Rimsky S. 2008. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 11:113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fletcher S. A., Csonka L. N. 1995. Fine-structure deletion analysis of the transcriptional silencer of the proU operon of Salmonella typhimurium. J. Bacteriol. 177:4508–4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gowrishankar J., Manna D. 1996. How is osmotic regulation of transcription of the Escherichia coli proU operon achieved? A review and a model. Genetica 97:363–378 [DOI] [PubMed] [Google Scholar]
- 33. Gowrishankar J., Pittard A. J. 1998. Superimposition of TyrR protein-mediated regulation on osmoresponsive transcription of Escherichia coli proU in vivo. J. Bacteriol. 180:6743–6748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hautefort I., Proenca M. J., Hinton J. C. 2003. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl. Environ. Microbiol. 69:7480–7491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hayes F., Barillá D. 2010. Extrachromosomal components of the nucleoid: recent developments in deciphering the molecular basis of plasmid segregation, p. 49–70 In Dame R. T., Dorman C. J. (ed.), Bacterial chromatin. Springer Press, Dordrecht, Netherlands [Google Scholar]
- 36. Hromockyj A. E., Tucker S. C., Maurelli A. T. 1992. Temperature regulation of Shigella virulence: identification of the repressor gene virR, an analogue of hns, and partial complementation by tyrosyl transfer RNA (tRNA1Tyr). Mol. Microbiol. 6:2113–2124 [DOI] [PubMed] [Google Scholar]
- 37. Janga S. C., Collado-Vides J. 2007. Structure and evolution of gene regulatory networks in microbial genomes. Res. Microbiol. 158:787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kahramanoglou C., et al. 2011. Direct and indirect effects of H-NS and FIS on global gene expression control in Escherichia coli. Nucleic Acids Res. 39:2073–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Le Gall T., et al. 2005. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology 151:951–962 [DOI] [PubMed] [Google Scholar]
- 40. Leonard T. A., Møller-Jensen J., Löwe J. 2005. Towards understanding the molecular basis of bacterial DNA segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Livny J., Yamaichi Y., Waldor M. K. 2007. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J. Bacteriol. 189:8693–8703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lozada-Chávez I., Angarica V. E., Collado-Vides J., Contreras-Moreira B. 2008. The role of DNA binding specificity in the evolution of bacterial regulatory networks. J. Mol. Biol. 379:627–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lucchini S., et al. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lucht J. M., Dersch P., Kempf B., Bremer E. 1994. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J. Biol. Chem. 269:6578–6586 [PubMed] [Google Scholar]
- 45. Luijsterburg M. S., White M. F., van Driel R., Dame R. T. 2008. The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem. Mol. Biol. 43:393–418 [DOI] [PubMed] [Google Scholar]
- 46. Maurelli A. T., Blackmon B., Curtiss R., III 1984. Temperature-dependent expression of virulence genes in Shigella species. Infect. Immun. 43:195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McLean C. Y., et al. 2011. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471:216–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miller J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 49. Mitobe J., Morita-Ishihara T., Ishihama A., Watanabe H. 2008. Involvement of RNA-binding protein Hfq in the post-transcriptional regulation of invE gene expression in Shigella sonnei. J. Biol. Chem. 283:5738–5747 [DOI] [PubMed] [Google Scholar]
- 50. Mitobe J., Morita-Ishihara T., Ishihama A., Watanabe H. 2009. Involvement of RNA-binding protein Hfq in the osmotic-response regulation of invE gene expression in Shigella sonnei. BMC Microbiol. 9:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nagarajavel V., Madhusudan S., Dole S., Rahmouni A. R., Schnetz K. 2007. Repression by binding of H-NS within the transcription unit. J. Biol. Chem. 282:23622–23630 [DOI] [PubMed] [Google Scholar]
- 52. Nakayama S., Watanabe H. 1998. Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J. Bacteriol. 180:3522–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Navarre W. W., et al. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238 [DOI] [PubMed] [Google Scholar]
- 54. Navarre W. W., McClelland M., Libby S. J., Fang F. C. 2007. Silencing of xenogeneic DNA by H-NS—facilitation of lateral DNA transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 21:1456–1471 [DOI] [PubMed] [Google Scholar]
- 55. Noom M. C., Navarre W. W., Oshima T., Wuite G. J., Dame R. T. 2007. H-NS promotes looped domain formation in the bacterial chromosome. Curr. Biol. 17:R913–R914 [DOI] [PubMed] [Google Scholar]
- 56. O'Byrne C. P., Dorman C. J. 1994. Transcription of the Salmonella typhimurium spv virulence locus is regulated negatively by the nucleoid-associated protein H-NS. FEMS Microbiol. Lett. 121:99–105 [DOI] [PubMed] [Google Scholar]
- 57. Oshima T., Ishikawa S., Kurokawa K., Aiba H., Ogasawara N. 2006. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 13:141–153 [DOI] [PubMed] [Google Scholar]
- 58. Overdier D. G., Csonka L. N. 1992. A transcriptional silencer downstream of the promoter in the osmotically controlled proU operon of Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 89:3140–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Owen-Hughes T. A., et al. 1992. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell 71:255–265 [DOI] [PubMed] [Google Scholar]
- 60. Perez J. C., Groisman E. A. 2009. Evolution of transcriptional regulatory circuits in bacteria. Cell 138:233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Phalipon A., Sansonetti P. J. 2007. Shigella's ways of manipulating the host intestinal innate and adaptive immune system: a toolbox for survival? Immunol. Cell Biol. 85:119–129 [DOI] [PubMed] [Google Scholar]
- 62. Porter M. E., Dorman C. J. 1994. A role for H-NS in the thermo-osmotic regulation of virulence gene expression in Shigella flexneri. J. Bacteriol. 176:4187–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Porter M. E., Dorman C. J. 1997. Virulence gene deletion frequency is increased in Shigella flexneri following conjugation, transduction, and transformation. FEMS Microbiol. Lett. 147:163–172 [DOI] [PubMed] [Google Scholar]
- 64. Porter M. E., Dorman C. J. 2002. In vivo DNA binding and oligomerization properties of the Shigella flexneri AraC-like transcriptional regulator VirF as identified by random and site-specific mutagenesis. J. Bacteriol. 184:531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schröder O., Wagner R. 2000. The bacterial DNA-binding protein H-NS represses ribosomal RNA transcription by trapping RNA polymerase in the initiation complex. J. Mol. Biol. 298:737–748 [DOI] [PubMed] [Google Scholar]
- 66. Schroeder G. N., Hilbi H. 2008. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin. Microbiol. Rev. 21:134–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schumacher M. A. 2007. Structure of plasmid segregation proteins. Curr. Opin. Struct. Biol. 17:103–109 [DOI] [PubMed] [Google Scholar]
- 68. Sergueev K., Dabrazhynetskaya A., Austin S. 2005. Plasmid partition system of the P1par family from the pWR100 virulence plasmid of Shigella flexneri. J. Bacteriol. 187:3369–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Stoebel D. M., Free A., Dorman C. J. 2008. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiol. 154:2533–2545 [DOI] [PubMed] [Google Scholar]
- 70. Surtees J. A., Funnell B. E. 2003. Plasmid and chromosome traffic control: how ParA and ParB drive partition. Curr. Top. Dev. Biol. 56:145–180 [DOI] [PubMed] [Google Scholar]
- 71. Tanaka K., Muramatsu S., Yamada H., Mizuno T. 1991. Systematic characterization of curved DNA segments randomly cloned from Escherichia coli and their functional significance. Mol. Gen. Genet. 226:367–376 [DOI] [PubMed] [Google Scholar]
- 72. Tanaka K., Ueguchi C., Mizuno T. 1994. Importance of stereospecific positioning of the upstream cis-acting DNA element containing a curved DNA structure for the functioning of the Escherichia coli proV promoter. Biosci. Biotechnol. Biochem. 58:1097–1101 [DOI] [PubMed] [Google Scholar]
- 73. Taniya T., et al. 2003. Determination of the InvE binding site required for expression of IpaB of the Shigella sonnei virulence plasmid: involvement of a ParB boxA-like sequence. J. Bacteriol. 185:5158–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tobe T., et al. 1991. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large virulence plasmid. Mol. Microbiol. 5:887–893 [DOI] [PubMed] [Google Scholar]
- 75. Tobe T., Yoshikawa M., Mizuno T., Sasakawa C. 1993. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by VirF and repression by H-NS. J. Bacteriol. 175:6142–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Turner E. C., Dorman C. J. 2007. H-NS antagonism in Shigella flexneri by VirB, a virulence gene transcription regulator that is closely related to plasmid partition factors. J. Bacteriol. 189:3403–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Walthers D., et al. 2011. Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J. Biol. Chem. 286:1895–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Watanabe H., Arakawa E., Ito K., Kato J., Nakamura A. 1990. Genetic analysis of an invasion region by use of a Tn3-lac transposon and identification of a second positive regulator gene, invE, for cell invasion of Shigella sonnei: significant homology of invE with ParB of plasmid P1. J. Bacteriol. 172:619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wiggins P. A., Dame R. T., Noom M. C., Wuite G. J. 2009. Protein-mediated molecular bridging: a key mechanism in biopolymer organization. Biophys. J. 97:1997–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhang W., Baseman J. B. 2011. Transcriptional response of Mycoplasma genitalium to osmotic stress. Microbiology 157:548–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.