Abstract
When Legionella pneumophila grows on agar plates, it secretes a surfactant that promotes flagellum- and pilus-independent “sliding” motility. We isolated three mutants that were defective for surfactant. The first two had mutations in genes predicted to encode cytoplasmic enzymes involved in lipid metabolism. These genes mapped to two adjacent operons that we designated bbcABCDEF and bbcGHIJK. Backcrossing and complementation confirmed the importance of the bbc genes and suggested that the Legionella surfactant is lipid containing. The third mutant had an insertion in tolC. TolC is the outer membrane part of various trimolecular complexes involved in multidrug efflux and type I protein secretion. Complementation of the tolC mutant restored sliding motility. Mutants defective for an inner membrane partner of TolC also lacked a surfactant, confirming that TolC promotes surfactant secretion. L. pneumophila (lspF) mutants lacking type II protein secretion (T2S) are also impaired for a surfactant. When the tolC and lspF mutants were grown next to each other, the lsp mutant secreted surfactant, suggesting that TolC and T2S conjoin to mediate surfactant secretion, with one being the conduit for surfactant export and the other the exporter of a molecule that is required for induction or maturation of surfactant synthesis/secretion. Although the surfactant was not required for the extracellular growth, intracellular infection, and intrapulmonary survival of L. pneumophila, it exhibited antimicrobial activity toward seven other species of Legionella but not toward various non-Legionella species. These data suggest that the surfactant provides L. pneumophila with a selective advantage over other legionellae in the natural environment.
INTRODUCTION
The aquatic bacterium Legionella pneumophila is the causative agent of Legionnaires' disease, a potentially fatal pneumonia (69). The environmental life cycle of L. pneumophila includes growth, survival, and spread within the planktonic phase, protozoan hosts, and multiorganismal biofilms (58, 91). Human infection occurs after inhalation of L. pneumophila-containing droplets that are generated from a variety of devices, including cooling towers (74). Once the bacteria enter the lung, proliferation occurs inside the resident alveolar macrophages, with lung epithelial cells likely providing an additional replicative niche (64, 69). L. pneumophila secretes many factors, proteinaceous and nonproteinaceous, that permit the bacterium's survival and proliferation in both the environment and the human host. Such factors include the many enzymes and effectors secreted via the type II and type IV protein secretion systems, the iron chelator legiobactin, and a pyomelanin pigment (3, 19, 23, 69).
Previously, we determined that L. pneumophila exhibits surface translocation when it is grown on buffered charcoal yeast extract (BCYE) plates containing 0.5% agar (90). The spreading legionellae appear in an amorphous, lobed pattern that is most manifest at 30°C. When bacteria were spotted onto the BCYE plates, the numbers and sizes of the lobes that formed were variable, ranging from only one or a few main lobes to many large and small lobes. Furthermore, in some cases, the lobes emanate from one side of the inoculation point, whereas in other instances, they spread out in all directions. This translocation is displayed by some, but not all, Legionella species. Because L. pneumophila mutants that lack flagella and/or type IV pili spread on low-percentage agar, this surface translocation is not swarming or twitching motility (90). Importantly, a translucent film appears atop the agar, moving ahead of the spreading legionellae. This film consisted of a surfactant, based on its ability to disperse water droplets and promote surface translocation of nonspreading Escherichia coli (90). L. pneumophila type II protein secretion (T2S) mutants are defective for surface translocation and surfactant. When these mutants are spotted onto the film produced by the wild type, they are able to spread, indicating that T2S, directly or indirectly, promotes the elaboration of a surfactant (90). Thus, L. pneumophila secretes a surfactant which promotes a form of surface translocation that is best described as “sliding motility.”
Biosurfactants (i.e., surfactants made by microbes) are produced by many types of bacteria, including Bacillus subtilis, Pseudomonas aeruginosa, and Serratia marcescens (8, 85). As a group, they display diversity in structure, possessing lipid, sugar, and/or peptide moieties (8). Although biosurfactants have been intensely studied for their role in motility as well as industrial applications (8, 50), their role in the growth, ecology, and virulence of pathogens has, with the exception of the P. aeruginosa rhamnolipid, been minimally addressed (1). The results reported here suggest that the surfactant made by L. pneumophila has lipid characteristics and is secreted by a process that involves TolC, an outer membrane protein that is best known for its role in antibiotic efflux. Although surfactant was not critical for the extra- or intracellular growth of L. pneumophila, it displayed a unique antimicrobial activity against other species of Legionella.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
L. pneumophila 130b, also known as AA100 or Wadsworth, served as our wild-type strain. Additional wild-type bacteria, as well as mutants of 130b that were also examined are listed in Table 1. Legionellae were routinely grown at 37°C in buffered yeast extract (BYE) broth or on BCYE agar containing 1.5% agar (90). When appropriate, bacteria were supplemented with 2.5 μg of gentamicin/ml, 25 μg of kanamycin/ml, or 4.5 μg of chloramphenicol/ml. In some experiments, L. pneumophila was also grown in a deferrated chemically defined medium (CDM) (20). E. coli strain DH5α (Invitrogen, Carlsbad, CA), as a host for recombinant plasmids, was routinely grown in Luria-Bertani medium. Unless otherwise noted, chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Table 1.
Wild-type and mutant strains used in this study
| Species and strain(s)a | Description | Reference or source |
|---|---|---|
| L. pneumophila | ||
| 130b (ATCC BAA-74) | Clinical isolate | 90 |
| NU275 | lspF mutant | 83 |
| NU388, NU393 | lpw_24151 (bbcB) mutants | This study |
| NU389, NU394 | lpw_24111 (bbcI) mutants | This study |
| NU390, NU395 | lpw_07761 (tolC) mutants | This study |
| NU391, NU392 | lpw_23601 mutants | This study |
| NU396, NU397 | lpw_24191 (bbcF) mutants | This study |
| NU400, NU401 | lpw_27401 mutants | This study |
| NU402, NU403 | lpw_07981 mutants | This study |
| lpnE::km strain | lpw_24081 (lpnE) mutant | 70 |
| L. anisa ATCC 35292 | Environmental isolate | 35 |
| L. feeleii ATCC 35072 | Environmental isolate | 42 |
| L. hackeliae ATCC 35250 | Clinical isolate | 13 |
| L. jordanis ATCC 33263 | Environmental isolate | 21 |
| L. moravica ATCC 43877 | Environmental isolate | 94 |
| L. micdadei ATCC 33218 | Clinical isolate | 39 |
| L. wadsworthii ATCC 33877 | Clinical isolate | 28 |
| Bacillus subtilis KY42 | Environmental isolate | 31 |
| Escherichia coli DH5α | Laboratory strain | Invitrogen |
| Klebsiella pneumoniae DMDS 92-08-28a | Environmental isolate | 67 |
| Listeria monocytogenes EGDe | Clinical isolate | 84 |
| Micrococcus luteus | Environmental isolate | 63 |
| Pseudomonas aeruginosa ATCC 7700 | Environmental isolate | 67 |
All L. pneumophila strains listed under 130b are derivatives of strain 130b. ATCC, American Type Culture Collection.
Assays for surface translocation and biosurfactant activity.
Surface translocation and surfactant production were monitored as described before (90). Briefly, Legionella strains were grown, with shaking, in BYE broth at 37°C to early stationary phase, and then 10 μl of culture was spotted onto fresh (i.e., <24-h-old) BCYE plates containing 0.5% agar. In some BCYE plates, we added 5 μg/ml surfactin (Sigma catalog no. S3523), a surfactant produced by B. subtilis (53). The inoculated plates were incubated at 30°C, and growth was observed for the next 14 days. Surface translocation was evident as lobed, wavelike areas of growth emanating from the point of inoculation, and the surfactant was seen as a film advancing ahead of the spreading bacteria. The presence of the surfactant was confirmed by the collapse of a water droplet when it was spotted onto the film-covered area of the plate.
Extracellular growth and effects of a biosurfactant.
To assess the extracellular growth capacity of L. pneumophila, strains were inoculated into BYE broth and then incubated in air, with shaking. Growth was assessed by measuring the optical density at 660 nm (OD660) of cultures, using a DU720 UV-visible light spectrophotometer (Beckman Coulter, Fullerton, CA). To judge the efficiency of plating on solid medium, legionellae were precultured for 3 days on BCYE agar and suspended in phosphate-buffered saline (PBS) at 109 CFU per ml, and then 10-μl aliquots taken from 10-fold serial dilutions in PBS were spotted onto the medium. Growth was recorded after 3 to 5 days of incubation at various temperatures. To ascertain the effect of the L. pneumophila surfactant on the extracellular growth of other bacteria (Table 1), the heterologous bacteria were spotted, as described above, onto standard BCYE containing 1.5% agar, BCYE containing 0.5% agar, and the surfactant film produced by L. pneumophila previously grown on the low-percentage agar medium. After 1 to 3 days at 37°C in an air incubator, the growth of the bacteria in the presence or absence of the surfactant was monitored and recorded.
DNA and protein sequence analysis.
DNA was isolated from L. pneumophila as described before (24). Sequencing was done at the Northwestern Biotech Laboratory, with primers from Integrated DNA Technologies (Coralville, IA). Sequences were analyzed using Lasergene (DNASTAR, Madison, WI). Protein alignments were done using the Clustal method, and predictions for the cellular locations of proteins were done using versions 3.0 of both PSORTb and SignalP (32, 71). BLAST homology searches were done through the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) and Legionella databases at http://genolist.pasteur.fr/LegioList/, http://www.ncbi.nlm.nih.gov/genomeprj/48801, and http://www.ebi.ac.uk/ena/data/view/FR687201.
RT-PCR analysis of L. pneumophila gene expression.
Reverse transcription (RT)-PCR was done as described previously (4, 20). The RNA was isolated from late log and early stationary BYE cultures grown at 30°C using the RNA Stat-60 reagent (IsoTex, Friendswood, TX). Controls in which reverse transcriptase was omitted from the reaction mixtures were included to rule out contributions from contaminating DNA in the DNase-treated samples. The primers used are described in Table 2. Primers 24151rtF and 24151rtR were used to assess transcription of lpw_24151. To assess cotranscription in the lpw_24151-containing operon, four intergenic regions, denoted as regions 1 to 4 in Fig. 3A, were amplified with primers 24141rt and 24151rt2R for region 1, primers 24151rt2F and 24161rtR for region 2, primers 24161rt and 24171rt for region 3, and primers 24191F and 24181rt for region 4. Primers 24111rtF and 24111rtR were used to assess transcription of lpw_24111. To assess cotranscription in the lpw_24111-containing operon, four intergenic regions, denoted as regions 1 to 4 in Fig. 3B, were amplified with primers 24091rt and 24101rt for region 1, primers 24101dsrt and 24111rt2 for region 2, primers 24111rt and 24121rt2 for region 3, and primers 24121rt and 24131rtF for region 4. Primers tolCF3 and TolC-5′-StuI assessed transcription of lpw_07761. To judge cotranscription within the lpw_07761-containing operon, two intergenic regions, denoted as regions 1 and 2 in Fig. 4A, were amplified with primers TolC-3′-F2 and TolC-R2-XbaI for region 1 and primers TolC-3′-F2-XbaI and TolC-5′-R2 for region 2.
Table 2.
Primers used in this study
| Primer name | Sequence (5′→3′)a |
|---|---|
| 24151rtF | CCCTGGTTGATGTCGTAAGG |
| 24151rtR | TGTGATCCGCAATCATCAGT |
| 24141rt | GATGCTGATACCATAGGC |
| 24151rtR | TGTGATCCGCAATCATCAGT |
| 24151rtF | GTCCTGATTCCTCTTAAAGCAATG |
| 24161rtR | GGTGAAGAGAATAAGTGCTGTCTGG |
| 24161rt | CAAACGCATCTACTACCCAGGA |
| 24171dsrt | GAAACCCGTCACGTCAATGTT |
| 24191F | GCGTTCACTCGGGTCACC |
| 24181dsrt | CCAGTTGAAAATCCAATCACGG |
| 24111rtF | GGCAGCCCAGCATTCTACTA |
| 24111rtR | AAAGACCTCATCAGGCATGG |
| 24091rt | GGTAGCAGGCTATTCATTGAC |
| 24101rt | GGAGAGTCCTTGCTTCATGG |
| 24101dsrt | TTGGTATCGGTTGGCTGAG |
| 24111rt2 | GCATGGGCAAACCAAAC |
| 24111rt | TGCCAACAATCCTTCCTGAT |
| 2412rt2 | TGACCCACGCAGTTTAGC |
| 24121rt | TTGGAAGTCAAATCGGTTCC |
| 24131rtF | TCATTGGGCTTGATTGATGA |
| tolCF3 | GTGTTTCAACCCATGTGTTTG |
| TolC-5′R-StuI | AGGCCTGTAAAGAGCGGCACGAGC |
| TolC-3′-F2 | GATTAACTCATGGTTAGCAACT |
| TolC-R2-XbaI | AAAAAATCTAGACTTTTCCGCCAGAC |
| TolC-3′-F2-XbaI | AAAAAATCTAGACGCAATCCTTGGTA |
| TolC-5′-R2 | ATGGTATCGTTCTCCAGAGC |
| 23601F | CCCGGGTCTGAGACAAATGAATGAAAGAATGC |
| 23601R | TCTAGATTCGCTTCTATAGTATGGC |
| 27401F | TTATGGTCAAATCACGAACGC |
| 27401R | AGTTGTTTCGCTCCCTGAATA |
| 07981F | CAGGCTCAGGCGAAAGTAG |
| 07981-5′R-HpaI | GTTAACAAATCCTGCTATGAGTTCAGC |
| 07981-3′F-HpaI | GTTAACGACTCGTTATCTCCACAGGTT |
| 07981R | TGGCAACGGCACAATCACC |
| 24191Fcomp | GGTACCGCACCAGTCGATACCCTGTT |
| 24191Rcomp | TCTAGACGCAAGCCTGATTGTTACC |
| 24111Fcomp | GGTACCAAGGAGGATAGACTCGGTTTGTT |
| 24111Rcomp | TCTAGACCTCATACTCGCTATCATCATTG |
| 07981F2comp | GGCTTGTTGTTCTTTACTACC |
| 07981R3comp | GTCGCAGGTATCCACAAAGT |
| tolCF | CGTCTGCCTTGAGTGTTTC |
| tolCR | TCACATTGGATTGACTGCGT |
Restriction sites are underlined.
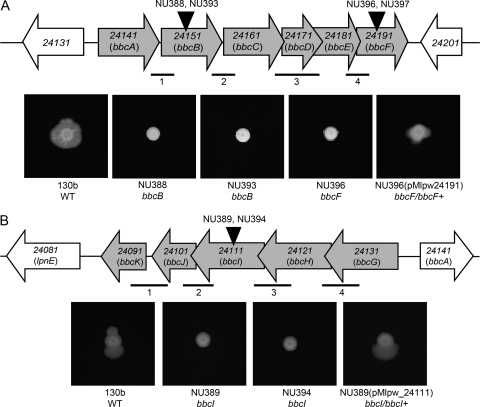
Fig. 3.
L. pneumophila bbcABCDEF and bbcGHIJK operons and their impact on surfactant and surface translocation. (A, top) Depiction of the region of the strain 130b chromosome containing genes lpw_24141 through lpw_24191, i.e., the bbcABCDEF operon. The horizontal gray arrows denote the relative size and orientation of the bbc genes, whereas the horizontal white arrows denote the two flanking genes. The vertical black arrowheads mark the approximate locations of the insertion mutations in mutants NU388, NU393, NU396, and NU397. The thinner horizontal lines below the gene map signify the approximate size and location of the four intergenic transcripts identified by RT-PCR analysis. (A, bottom) Surface translocation and surfactant phenotypes of bbcB and bbcF mutants. WT 130b, bbcB surfactant mutants NU388 and NU393, bbcF surfactant mutant NU396, and complemented mutant NU396(pMlpw24191) were grown on BCYE plates containing 0.5% agar at 30°C for 7 days. The bbcF mutant NU397 also consistently lacked surface translocation and surfactant (data not shown). (B, top) Depiction of the region of the strain 130b chromosome containing genes lpw_24131 through lpw_24091, i.e., the bbcGHIJK operon. The black arrowhead marks the location of the insertion in mutants NU389 and NU394, and the thinner horizontal lines signify the four intergenic transcripts identified by RT-PCR analysis. (B, bottom) Surface translocation and surfactant phenotypes of L. pneumophila bbcI mutants. WT 130b, bbcI surfactant mutants NU389 and NU394, and complemented mutant NU389(pMlpw_24111) were grown on BCYE plates containing 0.5% agar at 30°C for 7 days. The images of bacterial growth presented in panels A and B are representative of those obtained from at least three independent experiments.
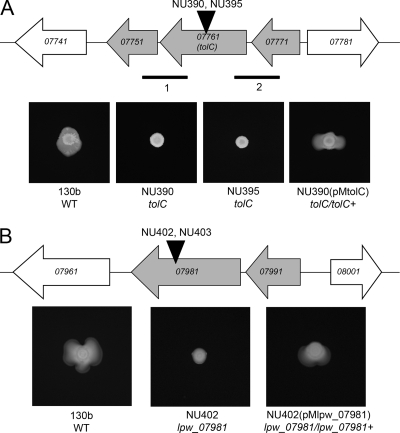
Fig. 4.
L. pneumophila tolC and lpw_07981 and their impact on surfactant and surface translocation. (A, top) Depiction of the region of the 130b chromosome containing genes lpw_07771, lpw_07761, and lpw_07751; i.e., the tolC operon. The horizontal gray arrows denote the size and orientation of the tolC operon, and the white arrows denote the two flanking genes. The vertical black arrowheads mark the locations of the insertion mutations in mutants NU390 and NU395. The thinner horizontal lines below the gene map signify the size and location of the two intergenic transcripts identified by RT-PCR. (A, bottom) Surface translocation and surfactant phenotypes of tolC mutants. WT 130b, tolC mutants NU390 and NU395, and complemented mutant NU390(pMtolC) were grown on BCYE plates containing 0.5% agar and 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 30°C for 7 days. (B, top) Depiction of the region of the 130b chromosome containing genes lpw_07991 and lpw_07981. The black arrowhead marks the locations of the insertions in lpw_07981 mutants NU402 and NU403. In the 130b database, the lpw_07981 ORF is also (erroneously) denoted lpw_07971. (B, bottom) Surface translocation and surfactant phenotypes of lpw_07981 mutants. WT 130b, lpw_07981 mutant NU402, and complemented mutant NU402(pMlpw_07981) were grown on BCYE plates containing 0.5% agar at 30°C for 7 days. The lpw_07981 mutant NU403 also consistently lacked surface translocation and surfactant (data not shown). The images presented in panels A and B are representative of those obtained from at least three independent experiments.
Construction and screening of an L. pneumophila mutant library.
Strain 130b was electroporated with mini-Tn10-containing pCDP05, and transposon-containing transformants were selected on BCYE agar with kanamycin (76). Individual mutants were inoculated into the wells of a 96-well, nontreated, polystyrene microtiter plate containing 200 μl of BYE and then grown at 37°C with shaking. Following 14 to 16 h of culturing, 5-μl aliquots from the wells were spotted onto BCYE plates containing 0.5% agar and incubated at 30°C. Over the next 3 to 5 days, the inocula were examined for the presence of surface translocation and surfactant. For mutants displaying impaired translocation and surfactant, the location of the mini-Tn10 insertion was ascertained by inverse PCR and subsequent sequence determination as previously described (19, 60). To confirm that the mutant phenotypes were due to transposon insertions, we backcrossed the Tn10-linked mutations into the chromosome of wild-type strain 130b. DNA from the transposon-containing mutants was treated with SacI, and then the digested DNA was mixed with competent 130b, giving a final DNA concentration of 4 μg per ml. DNA transformation proceeded as previously described (90), with the mixture incubating for 72 h at 30°C with shaking (i.e., 100 rpm). Transformants were selected by plating bacteria on kanamycin-containing BCYE agar.
Additional mutant constructions and gene complementation.
To obtain mutants specifically lacking lpw_23601, a 7-kb region containing the gene was amplified from 130b DNA using primers 23601F and 23601R (see Table 2 for these and all other primers). The resultant fragment was then cloned into pGEM-T Easy (Promega, Madison, WI) to give pG23601. Plasmid pG23601 was then digested with NruI, producing a 2-kb deletion in the lpw_23601 gene, and ligated to a kanamycin resistance (Kmr) cassette derived from pMB2190 (83), giving pG23601K1. Mutated lpw_23601 was introduced into strain 130b by transformation (90). Verification of the mutant genotype was carried out by PCR, using primers 23601F and 23601R. A similar allelic exchange procedure was used to introduce mutations into the lpw_24191 and lpw_27401 genes of strain 130b, with primers 24191F and 24191Rcomp being used to construct the lpw_24191 mutant and primers 27401F and 27401R being used to make the lpw_27401 mutant. In order to mutate lpw_07981, the 5′ and 3′ ends of the gene were separately amplified from 130b DNA by using the primer pairs 07981F/07981-5′R-HpaI and 07981-3′F-HpaI/07981R, respectively. Each of the generated fragments was ligated into pGEM-T Easy, and then the two resulting plasmids were digested with HpaI and SacI. Finally, a trimolecular ligation was done by placing the Kmr-containing cassette between the beginning of lpw_07981 and the end of the gene. The plasmid thus obtained (i.e., pGEM-lpw_07981-Kan) carried a 2.7-kb deletion in the central lpw_07981 coding region. Introduction and confirmation of the mutation in strain 130b was achieved as noted above for the other mutants. For each of the targeted genes, two independent mutants were obtained by performing separate transformations of the mutagenizing plasmid. Genetic complementation of each of the mutants was achieved by PCR amplifying the corresponding intact gene, ligating the resulting fragment into pGEM-T Easy, and then following a unique restriction digestion, moving the gene into pMMB2002 as described previously (20). The primer pairs used for complementation were 24191Fcomp and 24191Rcomp to create pMlpw24191 for analysis of the lpw_24191 mutant, 24111Fcomp and 24111Rcomp to make pMlpw_24111 for analysis of the lpw_24111 mutant, and 07981F2comp and 07981R3comp to make pM07981 for analysis of the lpw_07981 mutant. For the lpw_07761 mutant, a 3-kb PCR fragment of tolC generated using primers tolCF and tolCR was digested with HindIII-XmnI, and the resulting fragment was cloned into vector pMMB2002 to generate pMtolC.
Infection assays.
To test the ability of strains to grow intracellularly, Acanthamoeba castellanii and Hartmannella vermiformis amoebae, U937 macrophages, and A549 and WI-26 lung epithelial cells were infected as described previously (20, 64). The production of cytokines by infected U937 cells was also ascertained as described before (64). To assess the ability of L. pneumophila to grow within the lung, strains were inoculated into the tracheas of 6- to 8-week-old A/J mice (Jackson Laboratory, Bar Harbor, ME), and then at various times postinoculation, the numbers of CFU in the lungs were determined by plating bacteria on BCYE agar (20).
Assays for PHB content, secreted enzymes, and sensitivity to antibiotics.
In order to measure the level of poly-3-hydroxybutyrate (PHB) in L. pneumophila, strains were grown at 37°C in broth and then stained with the lipophilic dye Nile red as previously described (47). Following various periods of incubation in deferrated CDM broth, bacteria were collected by centrifugation, resuspended to an OD660 of 0.3, and then fixed with 1% formaldehyde. After being washed and resuspended in PBS, the cells were finally stained with Nile red (at 25 μM) for 60 min. Fluorescence was recorded at a λexcitation of 545 nm and λemission of 600 nm and blanked against a sample of unstained bacteria. The presence of secreted proteins in L. pneumophila culture supernatants was confirmed by assaying for lipase and phosphatase activities (5, 6). Antibiotic and detergent sensitivity assays were performed as described previously (79, 81). Briefly, strains were grown at 37°C in BYE broth to early stationary phase and then diluted in fresh BYE to 5 × 105 CFU per ml. In triplicate wells of a microtiter polystyrene tray, 50 μl of the bacterial suspension was added to an equal volume of BYE containing various concentrations of SDS or erythromycin. Following incubation at 37°C for 3 days, the lowest concentration of detergent or antibiotic resulting in no visible growth was deemed to be the MIC for that compound for that strain.
RESULTS
Isolation of L. pneumophila mutants defective for surfactant production.
As a first approach to identifying L. pneumophila genes that promote surfactant production, we examined the genome database of strain 130b (87) for a gene(s) that is predicted to encode a protein with sequence similarity to a bacterial enzyme known to be involved in the synthesis of a surfactant. The open reading frame (ORF) lpw_23601 was predicted to encode a product sharing 30% amino acid (aa) identity (E value = 2 × 10−61) with serrawettin W1 synthetase; serrawettin is a surfactant produced by S. marcescens (59). Using allelic exchange, we isolated two mutants lacking lpw_23601 but found that both produced normal levels of a surfactant and surface translocation (see Fig. S1 in the supplemental material). As a next approach, we screened a population of Tn10-mutagenized 130b for strains lacking the ability to elaborate a surface film and/or undergo surface translocation when spotted on BCYE plates containing 0.5% agar. Out of the 1,536 mutants screened, three (NU388, NU389, and NU390) reproducibly displayed the desired phenotype. Each mutant lacked both film production and surface translocation (Fig. 1), confirming that surfactant spreading and surface spreading are linked phenotypes in Legionella. A similar result was obtained when we reexamined our T2S (lspF) mutant of strain 130b (Fig. 1). However, NU388, NU389, and NU390 did not appear to be T2S mutants, because they exhibited a yellow coloration when grown on 0.5% agar BCYE plates whereas lsp mutants have a bluish-gray appearance (82). Also, they were not defective for secreted enzyme activities that are dependent upon T2S (data not shown). In theory, the inability of NU388, NU389, and NU390 to undergo surface translocation could be due to the loss of a surfactant or a cell-associated defect such as loss of a motility organelle. Differentiating between these possibilities, each of the new mutants recovered the ability to spread when spotted onto the film made by parental 130b (Fig. 2). Mutant bacteria spread in the direction away from the central area of wild-type growth, suggesting that they were moving into areas with sufficient surfactant production or away from areas of nutrient depletion. Together, these data suggest that NU388, NU389, and NU390 are defective for the synthesis and/or secretion of a surfactant.
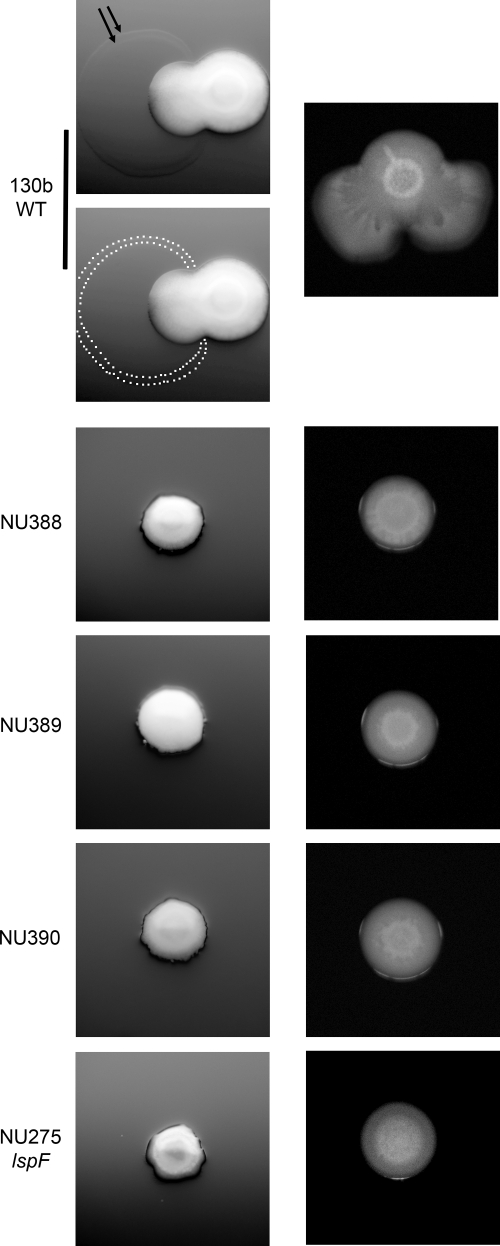
Fig. 1.
L. pneumophila mutants that lack surfactant and sliding motility. Wild-type (WT) strain 130b, mini-Tn10-containing mutants NU388, NU389, and NU390, and type II secretion (lspF) mutant NU275 were grown on BCYE plates containing 0.5% agar at 30°C. Imaged after 4 days, the left panel depicts, in the case of the WT but not the mutants, the early stage of surface translocation and a surfactant film front displaying two delineating rings. To aid in visualization of the surfactant film, two images are presented for the WT, with the upper one showing the delineating rings marked by two black arrows and the lower one having white dots mark the boundary of the film. Imaged after 10 days, the right panel shows the progression of surface translocation by the WT and its continued absence for the mutants. The images presented are representative of those observed for at least 12 experiments.
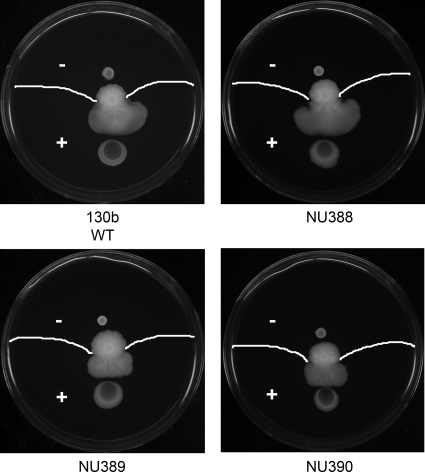
Fig. 2.
Surface translocation by L. pneumophila mutants when spotted onto film produced by wild-type legionellae. WT 130b was inoculated onto the center of a BCYE plate containing 0.5% agar at 30°C. After 10 days, when a layer of surface film was clearly evident beyond the spreading legionellae, a 10-μl aliquot of the WT, mutant NU388, mutant NU389, and mutant NU390 was spotted onto the agar surface either in the surfactant-covered area (“+”) or at a point outside of it but still equidistant from the center of WT growth (“−” area). After 3 days of further incubation, images were taken in order to visualize any differences in spreading. The white lines show the film front. The images are representative of three independent experiments.
Identification of L. pneumophila genes required for biosurfactant biosynthesis.
To discern the genetic basis for the observed losses of surfactant, we used inverse PCR and sequence analysis to identify the location of the Tn10 insertions. Mutant NU388 had its insertion at nucleotide (nt) 757 in the 1,746-bp gene designated lpw_24151 in the 130b genome database (87). The lpw_24151 gene was predicted to be part of a six-gene operon (Fig. 3A). The operon structure was confirmed by RT-PCR (data not shown), identifying transcripts spanning each intergenic region (Fig. 3A). Transcription of lpw_24151 was detected in the wild type grown in BYE broth at both 30°C and 37°C (data not shown). Using allelic exchange, we isolated a new mutant (NU393) specifically defective for lpw_24151. This mutant, like NU388, was impaired for both the production of film and surface translocation (Fig. 3A), confirming that NU388's loss of a surfactant was due to mutation of lpw_24151 and not an unlinked spontaneous mutation. When an insertion mutation was introduced into lpw_24191, the last gene in the lpw_24151 operon (Fig. 3A), the resulting mutants (NU396 and NU397) displayed the simultaneous loss of a surfactant and surface translocation (Fig. 3A). The introduction of lpw_24191 on a plasmid (i.e., pMlpw_24191) into NU396 reversed the mutant phenotype (Fig. 3A). Thus, these data indicate that lpw_24191 and most likely the entire lpw_24151-containing operon are required for the production of a surfactant by strain 130b. Utilizing the L. pneumophila databases (18, 22, 26, 34), we determined that the operon containing lpw_24141 through lpw_24191 (lpw_24141-lpw_24191) is largely conserved in the five other sequenced strains of L. pneumophila (Table 3). Bioinformatics analysis indicated that all of the proteins encoded within the lpw_24141-lpw_24191 operon are cytoplasmic proteins. We next used BLAST to begin to gain insight into the activities or functions associated with the proteins encoded by the lpw_24141-lpw_24191 operon. Although the protein encoded by lpw_24171 did not share sequence similarity to any known or hypothetical non-Legionella protein in the database, each of the five other predicted proteins shared significant similarity with cytoplasmic enzymes involved in lipid metabolism (Table 4) (36, 49, 57). Lpw_24191, for example, had a predicted ketoacyl-acyl carrier protein synthase III domain, which, in proteins from other bacteria, is known to initiate elongation in fatty acid biosynthesis (57). Based upon our mutational and bioinformatics analyses, we designated the lpw_24141-containing operon bbcABCDEF for Legionella biosurfactant biosynthesis cluster genes A through F (Fig. 3A). We posited that the surfactant made by L. pneumophila has a lipid component and that the enzymes encoded by bbcABCDEF help mediate the production of that lipid component.
Table 3.
Surfactant biosynthetic operons in six sequenced L. pneumophila strains
| Strain 130b ORF (gene) | Gene annotation using indicated database: |
||||
|---|---|---|---|---|---|
| Alcoyb | Corby | Lens | Paris | Philadelphia | |
| lpw_24141 (bbcA) | lpa_03202 | lpc_1695 | lpl2153 | lpp2180 | lpg2228 |
| lpw_24151 (bbcB) | lpa_03203 | lpc_1696 | lpl2154 | lpp2181 | lpg2229 |
| lpw_24161 (bbcC) | lpa_03204 | lpc_1697 | lpl2155 | lpp2182 | lpg2230 |
| lpw_24171 (bbcD) | lpa_03205 | lpc_1698 | lpl2156 | lpp2183 | —c |
| lpw_24181 (bbcE) | lpa_03207 | lpc_1699 | lpl2157 | lpp2184 | lpg2231 |
| lpw_24191 (bbcF) | lpa_03209 | lpc_1700 | lpl2158 | lpp2185 | lpg2232 |
| lpw_24131 (bbcG) | lpa_03201 | lpc_1694 | lpl2152 | lpp2178 | lpg2227 |
| lpw_24121 (bbcH) | lpa_03200 | lpc_1693 | lpl2151 | lpp2177 | lpg2226 |
| lpw_24111 (bbcI) | lpa_03198 | lpc_1692 | lpl2150 | lpp2176 | lpg2225 |
| lpw_24101 (bbcJ)a | lpa_03196 | lpc_1691 | lpl2149 | lpp2175 | lpg2223 |
| lpw_24091 (bbcK)a | lpa_03196 | lpc_1691 | lpl2149 | lpp2175 | lpg2223 |
The genome databases are slightly divergent at this junction, such that bbcJ and bbcK are annotated as two open reading frames in strain 130b but as a single open reading frame in the other strains.
In the Alcoy database, there are no genes annotated as lpa_03206, lpa_03208, and lpa_03199.
—, the Philadelphia-1 annotation does not include this gene.
Table 4.
Conserved domains in surfactant biosynthetic proteins of L. pneumophila
| Strain 130b ORF | Gene | Conserved domain in the predicted proteina | E value |
|---|---|---|---|
| lpw_24141 | bbcA | Ketoacyl-acyl carrier protein synthase III | 2 × 10−39 |
| lpw_24151 | bbcB | Acyl-CoA synthetase | 5 × 10−120 |
| lpw_24161 | bbcC | Acyl-CoA/AMP-acid ligase II | 9 × 10−54 |
| lpw_24171 | bbcD | None | |
| lpw_24181 | bbcE | 3-Ketoacyl-acyl carrier protein reductase | 4 × 10−30 |
| lpw_24191 | bbcF | Ketoacyl-acyl carrier protein synthase III | 8 × 10−71 |
| lpw_24131 | bbcG | Carboxyltransferase of acetyl-CoA carboxylase | 5 × 10−96 |
| lpw_24121 | bbcH | Acyl-CoA dehydrogenase | 1 × 10−52 |
| lpw_24111 | bbcI | GH3 domain | 7 × 10−90 |
| lpw_24101 | bbcJ | None | |
| lpw_24091 | bbcK | None |
Protein sequences from the 130b genome (http://www.ebi.ac.uk/ena/data/view/FR687201) were analyzed using the conserved domain database at the NCBI. CoA, coenzyme A.
Mutant NU389 contained its Tn10 insertion at nt 723 in the 1,530-bp gene denoted lpw_24111. The lpw_24111 ORF was predicted to be part of an operon, and this arrangement was confirmed by RT-PCR analysis (data not shown), which identified transcripts spanning four intergenic regions (Fig. 3B). Transcripts from lpw_24111 were detected in 130b bacteria grown in BYE broth at both 30°C and 37°C (data not shown). When we backcrossed the mutation in lpw_24111, the new mutant (NU394) lacked the ability to both produce a film and spread over the agar (Fig. 3B). These data indicated that the loss of the surfactant that we had seen for NU389 was due to mutation in lpw_24111 and not a spontaneous second-site mutation. Genetic complementation of NU389 with a plasmid copy of lpw_24111 (Fig. 3B) confirmed that lpw_24111 promotes the production of a surfactant. Examination of the genome database revealed that the first three genes in the lpw_24111-containing operon (i.e., lpw_24131, lpw_24121, and lpw_24111) are conserved within the five other sequenced strains of L. pneumophila (Table 3). Bioinformatics analysis indicated that most of the proteins encoded within the lpw_24091-lpw_24131 operon are cytoplasmic proteins, with the exception being Lpw_24121, an inner membrane protein. BLAST analysis and examination of the conserved domain database indicated that Lpw_24091 and Lpw_24101 encoded by the last two genes in the 130b operon did not share sequence similarity to any known or hypothetical non-Legionella protein in the database. However, Lpw_24111 and the two remaining proteins from the 130b locus had similarity with enzymes involved in lipid metabolism (Table 4) (25, 36). These findings support our hypothesis that the surfactant of L. pneumophila has a lipid component. Interestingly, this second operon identified by our screen mapped immediately upstream but transcribed in the direction opposite from bbcABCDE (compare Fig. 3A and B). Thus, we designated the lpw_24111-containing operon bbcGHIJK (Fig. 3B). Although all of the predicted activities associated with the bbc loci relate to lipid metabolism, those from bbcABCDE were distinct from those of bbcGHIJK.
Incidentally, the gene (lpw_24081) downstream of the bbcGHIJK operon is lpnE (Fig. 3B), which encodes a protein that is important for bacterial entry into host cells (70). An lpnE mutant had no alteration in surfactant or surface translocation (data not shown).
Outer membrane protein TolC promotes the secretion of L. pneumophila biosurfactant.
The third mutant (NU390) isolated in our screen had its Tn10 insertion mutation in lpw_07761, which encodes the outer membrane protein TolC. TolC has been very well studied in a variety of Gram-negative bacteria and is best known for its key role in antibiotic efflux (54, 96). Previous mutational analysis of L. pneumophila strain Lens determined that TolC also promotes antibiotic and detergent efflux by Legionella (30), and we confirmed this when we compared our tolC mutant NU390 to parental 130b for sensitivity to erythromycin and SDS (data not shown). In strain 130b, tolC (i.e., lpw_07761) was predicted to be the second gene in a three-gene operon (Fig. 4A), with the upstream gene lpw_07751 predicted to encode a protein with some similarity to nucleotide synthesis enzymes and the downstream gene lpw_07771 predicted to encode protein-l-isoaspartate-O-methyltransferase. This operon structure was confirmed by using RT-PCR (data not shown) to amplify the junction between lpw_07761 and lpw_07771 and the region between lpw_07761 and lpw_07751 (Fig. 4A). TolC transcript was detected in cultures at both 37°C and 30°C (data not shown). When we backcrossed the mutation in tolC, the newly derived mutant (NU395) lacked the ability to both produce a film and spread over agar (Fig. 4A), implying that the loss of the surfactant that we had seen for NU390 was due to mutation in tolC and not a spontaneous second-site mutation. Complementation of NU390 with a plasmid copy of tolC (Fig. 4A) confirmed that TolC is required for the elaboration of a surfactant by L. pneumophila. Given the well-known outer membrane location of TolC, we hypothesized that our tolC mutants were defective for surfactant secretion and not impaired for biosynthesis.
In other systems studied, TolC forms a trimolecular complex with multiple pairs of inner membrane and membrane fusion (adaptor) proteins in order to mediate efflux of different types of substrates (10, 72). The inner membrane anchor protein and its associated adaptor protein are often referred to as the “translocase.” Therefore, in order to gain further support for the role of TolC-mediated efflux in surfactant secretion, we scanned the L. pneumophila database for putative TolC partners and then mutated the corresponding genes in strain 130b. The operon containing lpw_07981 and lpw_07991 (Fig. 4B) encoded a protein (Lpw_07981) with sequence relatedness to the E. coli O157:H7 inner membrane protein AcrB (i.e., 32% aa identity and 52% aa similarity, with an E value of 2 × 10−141) and a protein (Lpw_07991) showing sequence relatedness to the E. coli membrane fusion (adaptor) protein AcrA (i.e., 22% aa identity and 38% aa similarity, with an E value of 5 × 10−11) (10). Two strains (NU402 and NU403) containing a mutation in lpw_07981 were made. As would be predicted, both mutants showed increased sensitivity to erythromycin and SDS, similar to the tolC mutant (data not shown). More significantly, NU402 and NU403 were impaired for both surfactant and surface translocation (Fig. 4B). Complementation of NU402 with a plasmid copy of lpw_07981 (Fig. 4B) confirmed that the targeted AcrB-like protein is required for elaboration of the surfactant by L. pneumophila. In contrast to these results, mutants NU400 and NU401, defective for lpw_27401, a gene predicted to encode an adaptor protein similar to E. coli OqxA (37% aa identity and 53% aa similarity, with an E value of 3 × 10−51) (37), exhibited a surfactant phenotype that was similar to that of the wild type (see Fig. S2 in the supplemental material). Based upon the combination of the results obtained from analyzing a TolC mutant (and its complement) and an AcrB-like mutant (and its complement), we hypothesized that a TolC-containing membrane complex is required for secretion of the L. pneumophila surfactant. To our knowledge, these data represent the first genetic proof for the role of TolC and its partners in surfactant secretion.
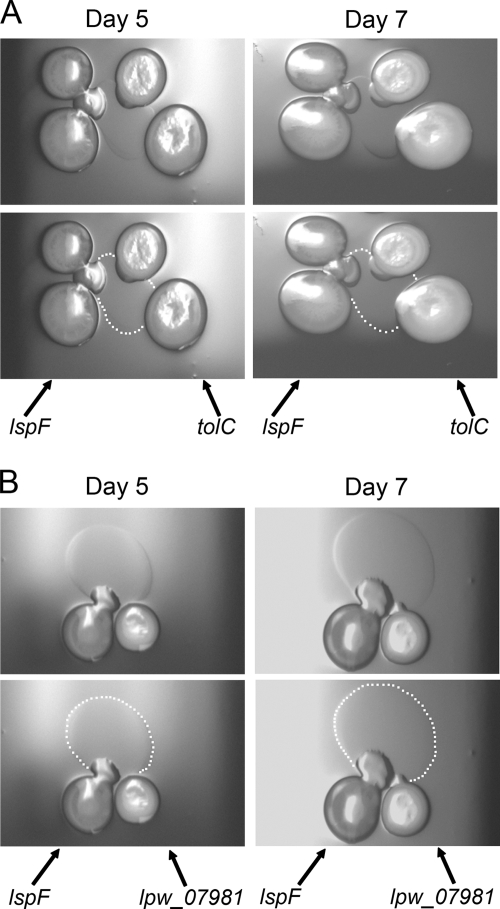
In considering the different ways in which TolC might promote surfactant secretion, it was important to revisit a key result from our previous study, namely, that an intact T2S system is also required for elaboration of the L. pneumophila surfactant (90). Therefore, we posited that the TolC and T2S systems work together to mediate secretion, with one directly exporting the surfactant and the other exporting a signaling molecule and/or activating enzyme. To test this hypothesis, we spotted an lspF mutant and a tolC mutant next to each other on the low-percentage-agar BCYE plates and then monitored bacterial growth, surface translocation, and surfactant film production over time (Fig. 5A). After 4 to 5 days of growth, the lspF mutant showed surface translocation and surfactant film emerged from the mutant's spreading lobe. By 7 days, the expanding film resulted in spreading from the tolC mutant spots. A similar result was obtained when we spotted the lspF mutant next to a mutant lacking the presumptive AcrB-like, inner membrane partner of TolC (Fig. 5B). Taken together, these results indicate that the lspF mutant is able to secrete a surfactant when exposed to a diffusible factor (that is still) made by mutants lacking a TolC complex. These data also imply that the lack of surfactant secretion that we had observed for the T2S and TolC mutants was most likely due to the specific loss of mediators of secretion and not due solely to indirect effects on global gene expression and/or growth rate when lsp, tolC, or lpw_07981 is mutated. Because it was the lspF mutant and not the TolC mutant that was able to now produce a surfactant, it would appear that a TolC complex is the conduit for surfactant secretion and that the T2S apparatus mediates the export of a signaling or activating factor.
Fig. 5.
Surfactant secretion by an L. pneumophila lspF mutant when grown in the presence of an L. pneumophila tolC or lpw_07981 mutant. (A) As indicated by the arrows, two inocula of the lspF mutant NU275 and two inocula of the tolC mutant NU390 were spotted next to each other on a BCYE plate containing 0.5% agar. After incubation at 30°C for 5 days or 7 days, bacterial growth and surface translocation were imaged (top row). To help with visualization of the surfactant that was produced by the sliding lspF mutant, white dots have been added to mark the boundary of the spreading film (bottom row). The surfactant film was also evident when single spots of the two mutants were placed next to each other (data not shown). (B) As indicated, lspF mutant NU275 and lpw_07981 mutant NU402 were spotted next to each other on low-agar BCYE plates, and then after incubation at 30°C for 5 or 7 days, bacterial growth, surface translocation, and surfactant production were imaged. As in panel A, the bottom images depict the boundary of the film produced by the lspF mutant with white dots. The results depicted in panels A and B are representative of those obtained from three independent experiments.
Effect of a surfactant on L. pneumophila growth.
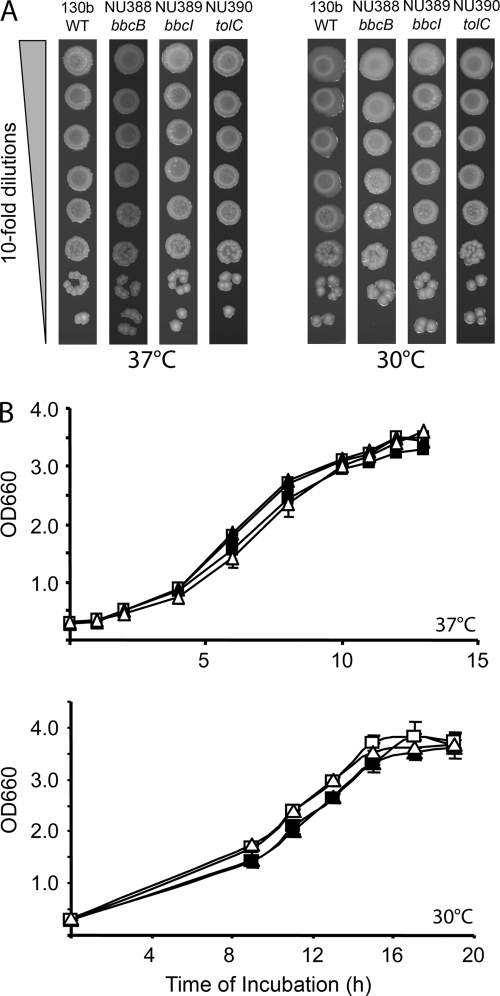
Utilizing our new mutants, we next sought to determine the importance of surfactant for L. pneumophila growth. The bbcB mutant NU388, bbcI mutant NU389, and tolC mutant NU390 had an efficiency of plating on standard BCYE agar that was similar to that of wild-type 130b, whether the plates were incubated at 30°C or 37°C (Fig. 6A). In a similar vein, NU388, NU389, and NU390 grew like the wild type did in standard BYE broth (Fig. 6B). Previous studies had established that the lspF T2S mutant also grows normally on standard BCYE agar and in BYE broth at 30°C and 37°C (90). These data indicate that these mutants do not have a generalized growth defect and that surfactant is not required for optimal extracellular growth by L. pneumophila at least under standard lab conditions.
Fig. 6.
Extracellular growth of the L. pneumophila wild type and bbcB, bbcI, and tolC mutants. (A) Growth of legionellae on BCYE agar. We spotted dilutions of WT 130b, bbcB mutant NU388, bbcI mutant NU389, and tolC mutant NU390 onto standard BCYE agar and incubated the plates at various temperatures. Bacterial growth was then imaged at 3 days for the 37°C plates and 4 days for the 30°C plates. (B) Growth of legionellae in BYE broth. WT 130b (▪), bbcB mutant NU388 (□), bbcI mutant NU389 (▴), and tolC mutant NU390 (▵) were inoculated into BYE broth and then incubated at 37°C or 30°C. At various times postinoculation, the extent of growth was monitored spectrophotometrically. The data points represent the means and standard deviations for triplicate cultures. The results presented in both panels A and B are representative of three independent experiments.
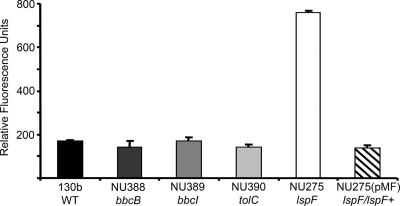
In some cases, the enzymes involved in surfactant synthesis are also required for the synthesis of other metabolites; e.g., P. aeruginosa RhlG is needed for synthesis of rhamnolipids and poly-β-hydroxyalkanoate lipid storage molecules (16). L. pneumophila produces poly-3-hydroxybutyrate (PHB) storage molecules when grown in bacteriological medium, but the basis of its synthesis is only partly known (7, 47). Given this and the fact that BbcE belongs to the same FabG-like family as RhlG, we considered the possibility that our surfactant mutants might lack PHB. However, the bbc and tolC mutants accumulated PHB levels similar to those of the wild type (Fig. 7). On the other hand, the T2S mutant NU275, but not its complemented derivative (83), had higher levels of PHB (Fig. 7), indicating that in L. pneumophila a T2S-dependent factor unrelated to the surfactant is critical for maintaining normal levels of PHB. To our knowledge, these data are the first link between T2S and PHB. Since T2S is not known to directly influence factors in the cytoplasm, which is where enzymes involved in PHB synthesis and utilization would reside (7), a T2S-dependent enzyme may be indirectly influencing PHB metabolism.
Fig. 7.
PHB in the L. pneumophila wild type and bbcB, bbcI, tolC, and lspF mutants. WT 130b, bbcB mutant NU388, bbcI mutant NU389, tolC mutant NU390, lspF mutant NU275, and complemented NU275(pMF) were grown in BYE to early log phase. The cells were then subcultured into CDM and grown at 37°C. After staining with Nile red, fluorescence was measured using a spectrophotometer blanked with unstained bacteria. Data are the means and standard deviations for three replicates obtained from duplicate cultures. The levels of PHB exhibited by the NU275 mutant were significantly increased compared to those for the other strains (P < 0.05, Student's t test). The results shown here are representative of at least four independent experiments.
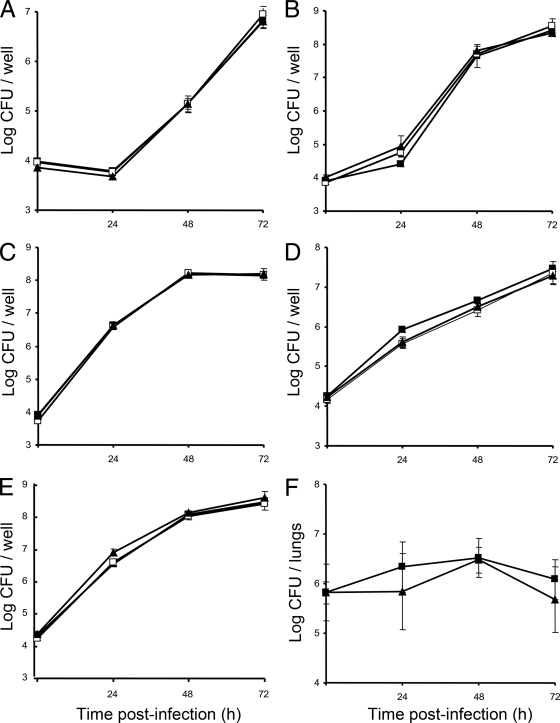
When the bbcB and bbcI mutants were tested for their relative ability to infect and replicate in A. castellanii and H. vermiformis, no defect was observed at a coculture incubation temperature of 35°C (Fig. 8A and B) or 30°C (data not shown). The surfactant biosynthesis mutants also grew in the way that the wild type did when tested in the U937 macrophage cell line, the A549 type II lung epithelial cell line, and the WI-26 type I lung epithelial cell line (Fig. 8C to E). Recently, we defined the cytokine output produced when U937 cells are infected with L. pneumophila 130b (64). Since some biosurfactants can influence host cytokine responses (33, 86), we examined the levels of interleukin-6 (IL-6) in supernatants from U937 cell cultures infected with the bbc mutants but did not find an altered cytokine profile (data not shown). Lastly, the bbcI mutant NU389 was tested for replication in the lungs of A/J mice and found to grow and persist in a manner similar to that of the wild type (Fig. 8F). Together, these data indicate that the surfactant linked to the bbc loci is not required for optimal intracellular infection or pathogenicity.
Fig. 8.
Intracellular and intrapulmonary growth of the L. pneumophila wild type and bbcB and bbcI mutants. (A to E) H. vermiformis (A), A. castellanii (B), U937 macrophages (C), A549 type II epithelial cells (D), and WI-26 type I epithelial cells (E) were infected with WT 130b (▪), bbcB mutant NU388 (□), and bbcI mutant NU389 (▴), and then at the indicated times, the numbers of CFU in the infected monolayers were determined by plating. Data are the means and standard deviations for four infected wells. Each panel is representative of three independent experiments. (F) WT 130b (▪) and bbcI mutant NU389 (▴) were inoculated into the lungs of A/J mice, and then after 24, 48, and 72 h, the numbers of CFU in total lung homogenates were determined by plating. Data are the means and standard deviations for five infected animals. While there might appear to be no growth and reduced recovery of the mutant at 24 h, this result was not statistically significant and was not obtained in the repeat experiment (data not shown).
The L. pneumophila surfactant impacts the growth of other legionellae.
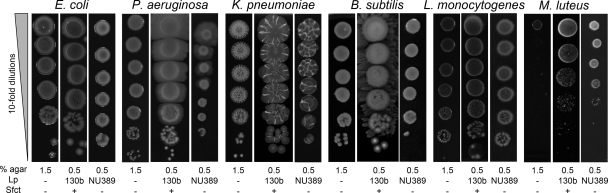
Because some biosurfactants possess antimicrobial activity (8), we determined whether the surfactant produced by L. pneumophila 130b influenced the growth of heterologous bacteria. Wild-type 130b and surfactant mutant NU389 had no effect on the efficiency of plating (i.e., numbers of CFU) of Gram-negative E. coli, Klebsiella pneumoniae, and P. aeruginosa as well as Gram-positive B. subtilis and Listeria monocytogenes (Fig. 9), indicating that the L. pneumophila surfactant does not affect the growth of these microbes. As might be expected, the L. pneumophila surfactant did promote spreading of some of these bacteria; e.g., B. subtilis (Fig. 9) and surfactants produced by other bacteria promoted spreading of L. pneumophila (see Fig. S4 in the supplemental material). In contrast to these results, L. pneumophila greatly stimulated the growth of Gram-positive Micrococcus luteus (Fig. 9). However, this effect was not dependent on the surfactant, because the bbcI mutant promoted growth just as the wild type did.
Fig. 9.
Effect of the L. pneumophila surfactant on heterologous bacteria. As indicated, we spotted 10-fold dilutions of E. coli, P. aeruginosa, K. pneumoniae, B. subtilis, L. monocytogenes, and M. luteus onto BCYE agar plates that were free of L. pneumophila (Lp −), contained a nearby row of WT L. pneumophila that had grown and made a surfactant film (Lp 130b), or contained a row of a bbcI mutant that had grown but not produced a surfactant (Lp NU389). After incubation at 37°C for another day, the growth of the heterologous bacteria in the absence (Sfct −) or presence (Sfct +) of a surfactant was imaged. The results depicted here were observed in at least three independent experiments. On BCYE agar not containing L. pneumophila, the efficiencies of plating for these bacteria were the same whether the medium contained 0.5% agar or 1.5% agar, although plating on low-percentage-agar BCYE did result in greater spreading for those bacteria, such as P. aeruginosa, that are known to make a surfactant and/or undergo surface translocation (see Fig. S3 in the supplemental material).
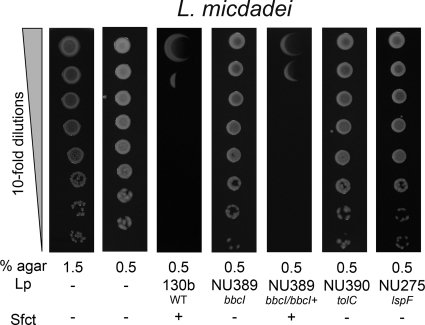
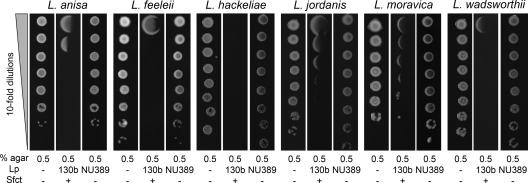
We had previously determined that surfactant production levels vary among the different Legionella species (90). Most notably, Legionella micdadei lacks film production and surface translocation when spotted onto BCYE agar containing a low percentage of agar (90). Therefore, we next examined the impact of L. pneumophila and its surfactant on the growth of nonsliding L. micdadei. In marked contrast to the observations made with nonlegionellae, the growth of L. micdadei was deleteriously affected by the presence of L. pneumophila (Fig. 10). Compared to its behavior when plated alone on standard BCYE agar or BCYE containing 0.5% agar, the growth of L. micdadei was reduced ca. 106-fold when spotted on the film produced by nearby strain 130b (Fig. 10). However, when L. micdadei was spotted next to the surfactant-lacking bbcI mutant NU2388, there was no reduction in CFU (Fig. 10), indicating that this impaired growth was due, directly or indirectly, to the surfactant. In support of this conclusion, a similar result was obtained when we spotted L. micdadei next to bbcB mutant NU388, tolC mutant NU390, and lspF mutant NU275 (Fig. 10 and data not shown). Further confirmation was obtained when the reduction in L. micdadei growth was observed after the strain was spotted adjacent to a complemented bbcI mutant (Fig. 10). Given these results, we next assessed the effect of the L. pneumophila surfactant on the growth of six additional species of Legionella. As we had observed for L. micdadei, L. anisa, L. feeleii, L. hackeliae, L. jordanis, L. moravica, and L. wadsworthii were all impaired by the presence of wild-type 130b but not the bbcI surfactant mutant (Fig. 11). Taken together, these data indicate that the surfactant made by L. pneumophila has an antagonistic or antibacterial activity which may be specific against species of Legionella. An antagonism such as this, surfactant or otherwise, has not been reported before.
Fig. 10.
Effect of the L. pneumophila surfactant on the growth of L. micdadei. For the two leftmost panels, we spotted dilutions of L. micdadei onto BCYE plates containing either 1.5% agar or 0.5% agar but no nearby L. pneumophila (Lp −). For the next five panels, we spotted L. micdadei onto 0.5% agar BCYE containing a nearby row of WT L. pneumophila that had grown and made a surfactant film (Lp 130b; third panel), a row of a bbcI mutant that had grown but not produced a surfactant (Lp NU389; fourth panel), a row of the complemented bbcI mutant that had restored surfactant production [Lp NU389(pMlpw_24111); fifth panel], a row of a tolC mutant that had grown but not made a surfactant (Lp NU390; sixth panel), or a row of an lspF mutant that had grown but lacked a surfactant (Lp NU275; last panel). After incubation at 37°C for another 3 days, growth of L. micdadei in the absence (Sfct −) or presence (Sfct +) of a surfactant was imaged. The results depicted here were observed in at least three independent experiments.
Fig. 11.
Effect of the L. pneumophila surfactant on the growth of L. anisa, L. feeleii, L. hackeliae, L. jordanis, L. moravica, and L. wadsworthii. For the leftmost panel, we spotted dilutions of the indicated strains onto BCYE plates containing 0.5% agar but no nearby L. pneumophila (Lp −). For the next two panels, we spotted the bacteria onto 0.5% agar BCYE containing either a nearby row of WT L. pneumophila that had grown and made a surfactant film (Lp 130b; second panel) or a row of a bbcI mutant that had grown but not produced a surfactant (Lp NU389; third panel). After incubation at 37°C for another 3 days, growth of the legionellae in the absence (Sfct −) or presence (Sfct +) of a surfactant was imaged. The results depicted here were observed in at least three independent experiments.
DISCUSSION
Surfactants are amphipathic molecules that reduce surface and interfacial tension (66, 95). Hence, the various bacteria and fungi that make surfactants are usually capable of spreading across an agar surface (90). In our previous survey of wild-type Legionella that included 10 strains of L. pneumophila and 12 non-pneumophila strains, the expression of the surfactant correlated with surface translocation (90). That all L. pneumophila surfactant mutants, including the five (bbcB, bbcF, bbcI, tolC, and lpw_07981) currently isolated and the four (lspDE, lspF, lspG, and pilD) from our past study (90), lack both a surfactant and surface translocation indicates that the surfactant made by L. pneumophila is promoting translocation. We have still not uncovered a role for flagella, pili, or any other motility organelle in the L. pneumophila surface translocation described here. Thus, we continue to use the term sliding as a way of denoting the surface translocation exhibited by L. pneumophila; sliding is a passive form of motility produced by the expansive forces in a growing culture in combination with the reduced friction between a cell and a substrate that is mediated by a surfactant (40, 48, 50). Other bacteria that exhibit sliding include species of Acinetobacter, Alcaligenes, Bacillus, Mycobacterium, Pseudomonas, Serratia, Stenotrophomonas, and Vibrio (2, 29, 38, 40, 44, 68, 78).
Surfactants are represented by a wide variety of molecular structures, including fatty acids, neutral lipids, phospholipids, glycolipids, glycopeptidolipids, polysaccharides, lipopeptides, and lipoproteins (2, 9, 11, 66, 73, 95). Based upon the genes within the bbcABCDEF and bbcGHIJK operons, it is probable that the L. pneumophila surfactant contains a lipid structure and is being synthesized, at least in part, by a polyketide synthase (PKS) (92). More specifically, our data are most compatible with the involvement of a type II PKS (41, 52). Indeed, the bbcABCDEF locus is predicted to encode monofunctional enzymes and provide three of the four enzymes that typically constitute a minimal type II PKS. BbcA and BbcF would represent two ketosynthases, whereas the gene directly downstream of bbcF (lpw_24201 in Fig. 3) would be the acyl carrier protein. The fourth typical enzyme, an acyltransferase, might be encoded by one of the more novel genes in the locus (e.g., bbcD) or encoded elsewhere in the chromosome, perhaps as part of a fatty acid biosynthetic pathway (52). The identification of the bbcI gene permits one further prediction, that the surfactant is a lipopeptide or lipoprotein. As a GH3-like enzyme (89), BbcI is predicted to conjugate an amino acid, from a peptide or protein, to an acyl substrate. In the future, the structural determination of a purified surfactant, when combined with the genetic information obtained here, should reveal much of the mechanism of synthesis for this new Legionella surfactant.
Among the other Legionella species that we had previously found to produce a surfactant and undergo sliding (90), L. longbeachae is the only one for which a published genome database is available (17, 55). L. longbeachae has homologs of bbcABCDEF and bbcGHI, i.e., llo2601 to llo2609 in strain NSW150 and llb_2731 to llb_2739 in D-4968. These data suggest that the surfactant produced by L. longbeachae is the same as the one made by L. pneumophila. The ability of different species within the same genus to produce identical or similar surfactants has been observed before; e.g., several species of Pseudomonas produce rhamnolipids (1). Complete genome databases are not (yet) available for any other Legionella species, with the exception of the obligate intracellular parasite L. drancourtii (65). Therefore, we cannot immediately say whether those species that do not produce a surfactant lack bbc genes.
The data presented here are both the first documentation of a link between surfactant secretion and TolC and the first indication that a TolC complex may act as the conduit for surfactant secretion. However, several past studies which examined putative TolC partners in other bacteria have suggested that such a connection could exist. For example, an AcrB-like protein known as SwrC is needed for secretion of surfactin by B. subtilis (51), and the loss of a putative TolC partner results in the reduced production of putisolvin by Pseudomonas putida and arthrofactin by Pseudomonas sp. strain MIS38 (27, 61). That TolC may act as the conduit for the export of a surfactant is not unreasonable for two more reasons; first, TolC complexes are known to mediate the export of a wide variety of substrates, ranging from small antibiotics to proteins (10), and second, surfactants are usually small, amphipathic molecules that should be compatible with the TolC outer membrane channel. In pondering the mechanism by which T2S conjoins with TolC to promote surfactant secretion by L. pneumophila, several scenarios are possible. In the first case, T2S promotes the secretion of a “regulatory” molecule that provides a necessary signal to the cell for when to make and/or secrete the surfactant. In the second scenario, T2S mediates the secretion of a factor (e.g., an enzyme) that is needed for the maturation or activation of the surfactant after it exits the cell envelope. In the third case, the T2S system secretes a factor that is necessary for the ability of TolC to export the surfactant. That secreted factors can influence the export of a surfactant is supported by observations in the literature; e.g., quorum sensing has been linked to regulation of surfactant production in B. subtilis, P. aeruginosa, and Serratia liquefaciens (62, 75, 77). Since a connection between TolC and T2S has not been reported previously, further study of the L. pneumophila surfactant should yield novel insight into the mechanism and regulation of Gram-negative secretion. By continuing to screen our mutant library for strains that lack a surfactant and sliding, we should be able to uncover additional factors, such as a hypothetical signal or modifying enzyme, that provide the link between TolC and T2S as it relates to surfactant secretion.
Although we did not find a required role for the surfactant in the extracellular growth, intracellular infection, or intrapulmonary survival of L. pneumophila, we did determine that the substance is antagonistic toward other Legionella species. This inhibitory effect most likely represents a direct antimicrobial activity, since there are numerous past studies ascribing antimicrobial properties to biosurfactants, including those that are lipoproteins (77). Additionally, though fewer, there are other surfactants that are known to act within a genus; e.g., surfactin is a lipoprotein that is produced by B. subtilis and has activity against Bacillus cereus (45). It is also formally possible that the L. pneumophila surfactant impaired the growth of the other legionellae by obstructing nutrient acquisition or somehow negatively affecting the microenvironment. Whatever its precise mode(s) of action, the surfactant may help strains of L. pneumophila gain a selective advantage over other Legionella species that it encounters in natural habitats. Besides providing an antimicrobial activity, the surfactant could foster the environmental survival of L. pneumophila in several other ways. For example, L. pneumophila is known to exist in multiorganismal biofilms (91), and based upon work in other microbial systems, surfactants help bacteria to modulate their biofilm architectures as well as to promote their ability to detach from and/or disperse biofilms (12, 46, 56). Moreover, surfactants are known to mediate solubilization, uptake, and utilization of hydrophobic compounds, solubilization of quorum-sensing molecules, solubilization of outer membrane vesicles, and binding of heavy metals and prevention of their toxicity (2, 14, 15, 43, 80, 88, 93). Thus, another direction for future research will focus on more fully defining the role of the surfactant in L. pneumophila ecology, particularly given that environmental legionellae are directly linked to human disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank past and present members of the Cianciotto lab for their helpful comments and suggestions. We also thank Nancy Freitag for sending us strains of Micrococcus and Bacillus and Barry Fields for strains of Pseudomonas and Klebsiella. We gratefully acknowledge Emily Yip for designing tolC primers and performing some of the antibiotic testing and Kessler McCoy-Simandle for help with the cytokine analysis and animal infections.
This study was funded by NIH grant AI043987 awarded to N.P.C.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 2 September 2011.
REFERENCES
- 1. Abdel-Mawgoud A. M., Lepine F., Deziel E. 2010. Rhamnolipids: diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 86:1323–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agusti G., Astola O., Rodriguez-Guell E., Julian E., Luquin M. 2008. Surface spreading motility shown by a group of phylogenetically related, rapidly growing pigmented mycobacteria suggests that motility is a common property of mycobacterial species but is restricted to smooth colonies. J. Bacteriol. 190:6894–6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allard K. A., et al. 2009. Purification of legiobactin and the importance of this siderophore in lung infection by Legionella pneumophila. Infect. Immun. 77:2887–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allard K. A., Viswanathan V. K., Cianciotto N. P. 2006. lbtA and lbtB are required for production of the Legionella pneumophila siderophore legiobactin. J. Bacteriol. 188:1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aragon V., Kurtz S., Cianciotto N. P. 2001. Legionella pneumophila major acid phosphatase and its role in intracellular infection. Infect. Immun. 69:177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aragon V., Rossier O., Cianciotto N. P. 2002. Legionella pneumophila genes that encode lipase and phospholipase C activities. Microbiology 148:2223–2231 [DOI] [PubMed] [Google Scholar]
- 7. Aurass P., et al. 2009. bdhA-patD operon as a virulence determinant, revealed by a novel large-scale approach for identification of Legionella pneumophila mutants defective for amoeba infection. Appl. Environ. Microbiol. 75:4506–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Banat I. M., et al. 2010. Microbial biosurfactants production, applications and future potential. Appl. Microbiol. Biotechnol. 87:427–444 [DOI] [PubMed] [Google Scholar]
- 9. Berti A. D., Greve N. J., Christensen Q. H., Thomas M. G. 2007. Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 189:6312–6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blair J. M., Piddock L. J. 2009. Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr. Opin. Microbiol. 12:512–519 [DOI] [PubMed] [Google Scholar]
- 11. Bodour A. A., et al. 2004. Structure and characterization of flavolipids, a novel class of biosurfactants produced by Flavobacterium sp. strain MTN11. Appl. Environ. Microbiol. 70:114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boles B. R., Thoendel M., Singh P. K. 2005. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 57:1210–1223 [DOI] [PubMed] [Google Scholar]
- 13. Brenner D. J., et al. 1985. Ten new species of Legionella. Int. J. Syst. Bacteriol. 35:50–59 [Google Scholar]
- 14. Cajal Y., Rogers J., Berg O. G., Jain M. K. 1996. Intermembrane molecular contacts by polymyxin B mediate exchange of phospholipids. Biochemistry 35:299–308 [DOI] [PubMed] [Google Scholar]
- 15. Calfee M. W., Shelton J. G., McCubrey J. A., Pesci E. C. 2005. Solubility and bioactivity of the Pseudomonas quinolone signal are increased by a Pseudomonas aeruginosa-produced surfactant. Infect. Immun. 73:878–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Campos-Garcia J., et al. 1998. The Pseudomonas aeruginosa rhlG gene encodes an NADPH-dependent β-ketoacyl reductase which is specifically involved in rhamnolipid synthesis. J. Bacteriol. 180:4442–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cazalet C., et al. 2010. Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires' disease. PLoS Genet. 6:e1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cazalet C., et al. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165–1173 [DOI] [PubMed] [Google Scholar]
- 19. Chatfield C. H., Cianciotto N. P. 2007. The secreted pyomelanin pigment of Legionella pneumophila confers ferric reductase activity. Infect. Immun. 75:4062–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chatfield C. H., Mulhern B. J., Burnside D. M., Cianciotto N. P. 2011. Legionella pneumophila LbtU acts as a novel, TonB-independent receptor for the legiobactin siderophore. J. Bacteriol. 193:1563–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cherry W. B., et al. 1982. Legionella jordanis: a new species of Legionella isolated from water and sewage. J. Clin. Microbiol. 15:290–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chien M., et al. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966–1968 [DOI] [PubMed] [Google Scholar]
- 23. Cianciotto N. P. 2009. Many substrates and functions of type II protein secretion: lessons learned from Legionella pneumophila. Future Microbiol. 4:797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cianciotto N. P., Fields B. S. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. U. S. A. 89:5188–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cronan J. E., Jr., Waldrop G. L. 2002. Multi-subunit acetyl-CoA carboxylases. Prog. Lipid Res. 41:407–435 [DOI] [PubMed] [Google Scholar]
- 26. D'Auria G., Jimenez-Hernandez N., Peris-Bondia F., Moya A., Latorre A. 2010. Legionella pneumophila pangenome reveals strain-specific virulence factors. BMC Genomics 11:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dubern J. F., Coppoolse E. R., Stiekema W. J., Bloemberg G. V. 2008. Genetic and functional characterization of the gene cluster directing the biosynthesis of putisolvin I and II in Pseudomonas putida strain PCL1445. Microbiology 154:2070–2083 [DOI] [PubMed] [Google Scholar]
- 28. Edelstein P. H., et al. 1982. Legionella wadsworthii species nova: a cause of human pneumonia. Ann. Intern. Med. 97:809–813 [DOI] [PubMed] [Google Scholar]
- 29. Fall R., Kearns D. B., Nguyen T. 2006. A defined medium to investigate sliding motility in a Bacillus subtilis flagella-less mutant. BMC Microbiol. 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ferhat M., et al. 2009. The TolC protein of Legionella pneumophila plays a major role in multi-drug resistance and the early steps of host invasion. PLoS One 4:e7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Freitag N. E., Youngman P., Portnoy D. A. 1992. Transcriptional activation of the Listeria monocytogenes hemolysin gene in Bacillus subtilis. J. Bacteriol. 174:1293–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gardy J. L., et al. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617–623 [DOI] [PubMed] [Google Scholar]
- 33. Gerstel U., Czapp M., Bartels J., Schroder J. M. 2009. Rhamnolipid-induced shedding of flagellin from Pseudomonas aeruginosa provokes hBD-2 and IL-8 response in human keratinocytes. Cell. Microbiol. 11:842–853 [DOI] [PubMed] [Google Scholar]
- 34. Glockner G., et al. 2008. Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int. J. Med. Microbiol. 298:411–428 [DOI] [PubMed] [Google Scholar]
- 35. Gorman G. W., et al. 1985. Legionella anisa: a new species of Legionella isolated from potable waters and a cooling tower. Appl. Environ. Microbiol. 49:305–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gulick A. M. 2009. Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4:811–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hansen L. H., Sorensen S. J., Jorgensen H. S., Jensen L. B. 2005. The prevalence of the OqxAB multidrug efflux pump amongst olaquindox-resistant Escherichia coli in pigs. Microb. Drug Resist. 11:378–382 [DOI] [PubMed] [Google Scholar]
- 38. Harshey R. M. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57:249–273 [DOI] [PubMed] [Google Scholar]
- 39. Hebert G. A., Steigerwalt A. G., Brenner D. J. 1980. Legionella micdadei species nova: classification of a third species of Legionella associated with human pneumonia. Curr. Microbiol. 3:255–257 [Google Scholar]
- 40. Henrichsen J. 1972. Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36:478–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hertweck C. 2009. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 48:4688–4716 [DOI] [PubMed] [Google Scholar]
- 42. Herwaldt L. A., et al. 1984. A new Legionella species, Legionella feeleii species nova, causes Pontiac Fever in an automobile plant. Ann. Intern. Med. 100:333–338 [DOI] [PubMed] [Google Scholar]
- 43. Hsueh Y. H., Somers E. B., Lereclus D., Wong A. C. 2006. Biofilm formation by Bacillus cereus is influenced by PlcR, a pleiotropic regulator. Appl. Environ. Microbiol. 72:5089–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang T. P., Lee Wong A. C. 2007. Extracellular fatty acids facilitate flagella-independent translocation by Stenotrophomonas maltophilia. Res. Microbiol. 158:702–711 [DOI] [PubMed] [Google Scholar]
- 45. Huang X., et al. 2007. Optimization of inactivation of endospores of Bacillus cereus by antimicrobial lipopeptides from Bacillus subtilis fmbj strains using a response surface method. Appl. Microbiol. Biotechnol. 74:454–461 [DOI] [PubMed] [Google Scholar]
- 46. Irie Y., O'Toole A. G., Yuk M. H. 2005. Pseudomonas aeruginosa rhamnolipids disperse Bordetella bronchiseptica biofilms. FEMS Microbiol. Lett. 250:237–243 [DOI] [PubMed] [Google Scholar]
- 47. James B. W., Mauchline W. S., Dennis P. J., Keevil C. W., Wait R. 1999. Poly-3-hydroxybutyrate in Legionella pneumophila, an energy source for survival in low-nutrient environments. Appl. Environ. Microbiol. 65:822–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jarrell K. F., McBride M. J. 2008. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 6:466–476 [DOI] [PubMed] [Google Scholar]
- 49. Kavanagh K. L., Jornvall H., Persson B., Oppermann U. 2008. The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 65:3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kearns D. B. 2010. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8:634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kearns D. B., Chu F., Rudner R., Losick R. 2004. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol. Microbiol. 52:357–369 [DOI] [PubMed] [Google Scholar]
- 52. Khosla C., Gokhale R. S., Jacobsen J. R., Cane D. E. 1999. Tolerance and specificity of polyketide synthases. Annu. Rev. Biochem. 68:219–253 [DOI] [PubMed] [Google Scholar]
- 53. Kinsinger R. F., Shirk M. C., Fall R. 2003. Rapid surface motility in Bacillus subtilis is dependent on extracellular surfactin and potassium ion. J. Bacteriol. 185:5627–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Koronakis V., Eswaran J., Hughes C. 2004. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 73:467–489 [DOI] [PubMed] [Google Scholar]
- 55. Kozak N. A., et al. 2010. Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968. J. Bacteriol. 192:1030–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kuiper I., et al. 2004. Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol. Microbiol. 51:97–113 [DOI] [PubMed] [Google Scholar]
- 57. Lai C. Y., Cronan J. E. 2003. Beta-ketoacyl-acyl carrier protein synthase III (FabH) is essential for bacterial fatty acid synthesis. J. Biol. Chem. 278:51494–51503 [DOI] [PubMed] [Google Scholar]
- 58. Lau H. Y., Ashbolt N. J. 2009. The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water. J. Appl. Microbiol. 107:368–378 [DOI] [PubMed] [Google Scholar]
- 59. Li H., Tanikawa T., Sato Y., Nakagawa Y., Matsuyama T. 2005. Serratia marcescens gene required for surfactant serrawettin W1 production encodes putative aminolipid synthetase belonging to nonribosomal peptide synthetase family. Microbiol. Immunol. 49:303–310 [DOI] [PubMed] [Google Scholar]
- 60. Liles M. R., Viswanathan V. K., Cianciotto N. P. 1998. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect. Immun. 66:1776–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lim S. P., Roongsawang N., Washio K., Morikawa M. 2009. Flexible exportation mechanisms of arthrofactin in Pseudomonas sp. MIS38. J. Appl. Microbiol. 107:157–166 [DOI] [PubMed] [Google Scholar]
- 62. Lindum P. W., et al. 1998. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:6384–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mansfield B. E., Dionne M. S., Schneider D. S., Freitag N. E. 2003. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell. Microbiol. 5:901–911 [DOI] [PubMed] [Google Scholar]
- 64. McCoy-Simandle K., et al. 2011. Legionella pneumophila type II secretion dampens the cytokine response of infected macrophages and epithelia. Infect. Immun. 79:1984–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Moliner C., Raoult D., Fournier P. E. 2009. Evidence that the intra-amoebal Legionella drancourtii acquired a sterol reductase gene from eukaryotes. BMC Res. Notes 2:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mukherjee S., Das P., Sen R. 2006. Towards commercial production of microbial surfactants. Trends Biotechnol. 24:509–515 [DOI] [PubMed] [Google Scholar]
- 67. Murga R., et al. 2001. Role of biofilms in the survival of Legionella pneumophila in a model potable-water system. Microbiology 147:3121–3126 [DOI] [PubMed] [Google Scholar]
- 68. Murray T. S., Kazmierczak B. I. 2008. Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J. Bacteriol. 190:2700–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Newton H. J., Ang D. K., van Driel I. R., Hartland E. L. 2010. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin. Microbiol. Rev. 23:274–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Newton H. J., et al. 2007. Sel1 repeat protein LpnE is a Legionella pneumophila virulence determinant that influences vacuolar trafficking. Infect. Immun. 75:5575–5585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nielsen H., Engelbrecht J., Brunak S., von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1–6 [DOI] [PubMed] [Google Scholar]
- 72. Nikaido H., Takatsuka Y. 2009. Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta 1794:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nitschke M., Costa S. G., Contiero J. 2005. Rhamnolipid surfactants: an update on the general aspects of these remarkable biomolecules. Biotechnol. Prog. 21:1593–1600 [DOI] [PubMed] [Google Scholar]
- 74. Pagnier I., Merchat M., La Scola B. 2009. Potentially pathogenic amoeba-associated microorganisms in cooling towers and their control. Future Microbiol. 4:615–629 [DOI] [PubMed] [Google Scholar]
- 75. Pearson J. P., Pesci E. C., Iglewski B. H. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pope C. D., Dhand L., Cianciotto N. P. 1994. Random mutagenesis of Legionella pneumophila with mini-Tn10. FEMS Microbiol. Lett. 124:107–111 [DOI] [PubMed] [Google Scholar]
- 77. Raaijmakers J. M., De Bruijn I., Nybroe O., Ongena M. 2010. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 34:1037–1062 [DOI] [PubMed] [Google Scholar]
- 78. Recht J., Kolter R. 2001. Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacterium smegmatis. J. Bacteriol. 183:5718–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Robey M., O'Connell W., Cianciotto N. P. 2001. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect. Immun. 69:4276–4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ron E. Z., Rosenberg E. 2001. Natural roles of biosurfactants. Environ. Microbiol. 3:229–236 [DOI] [PubMed] [Google Scholar]
- 81. Rossier O., Cianciotto N. P. 2005. The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect. Immun. 73:2020–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rossier O., Cianciotto N. P. 2001. Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect. Immun. 69:2092–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rossier O., Starkenburg S., Cianciotto N. P. 2004. Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires' disease pneumonia. Infect. Immun. 72:310–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Salerno J. A., Waltenbaugh C., Cianciotto N. P. 2001. Ethanol consumption and the susceptibility of mice to Listeria monocytogenes infection. Alcohol Clin. Exp. Res. 25:464–472 [PubMed] [Google Scholar]
- 85. Satpute S. K., Banat I. M., Dhakephalkar P. K., Banpurkar A. G., Chopade B. A. 2010. Biosurfactants, bioemulsifiers and exopolysaccharides from marine microorganisms. Biotechnol. Adv. 28:436–450 [DOI] [PubMed] [Google Scholar]
- 86. Schorey J. S., Sweet L. 2008. The mycobacterial glycopeptidolipids: structure, function, and their role in pathogenesis. Glycobiology 18:832–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schroeder G. N., et al. 2010. The genome of Legionella pneumophila strain 130b contains a unique combination of type IV secretion systems and encodes novel Dot/Icm secretion system effector proteins. J. Bacteriol. 192:6001–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Soberon-Chavez G., Lepine F., Deziel E. 2005. Production of rhamnolipids by Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 68:718–725 [DOI] [PubMed] [Google Scholar]
- 89. Staswick P. E., et al. 2005. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17:616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Stewart C. R., Rossier O., Cianciotto N. P. 2009. Surface translocation by Legionella pneumophila: a form of sliding motility that is dependent upon type II protein secretion. J. Bacteriol. 191:1537–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Taylor M., Ross K., Bentham R. 2009. Legionella, protozoa, and biofilms: interactions within complex microbial systems. Microb. Ecol. 58:538–547 [DOI] [PubMed] [Google Scholar]
- 92. Tsai S. C., Ames B. D. 2009. Structural enzymology of polyketide synthases. Methods Enzymol. 459:17–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Verstraeten N., et al. 2008. Living on a surface: swarming and biofilm formation. Trends Microbiol. 16:496–506 [DOI] [PubMed] [Google Scholar]
- 94. Wilkinson H. W., et al. 1988. Legionella moravica sp. nov. and Legionella brunensis sp. nov. isolated from cooling-tower water. Ann. Inst. Pasteur Microbiol. 139:393–402 [DOI] [PubMed] [Google Scholar]
- 95. Youssef N. H., et al. 2004. Comparison of methods to detect biosurfactant production by diverse microorganisms. J. Microbiol. Methods 56:339–347 [DOI] [PubMed] [Google Scholar]
- 96. Zgurskaya H. I. 2009. Multicomponent drug efflux complexes: architecture and mechanism of assembly. Future Microbiol. 4:919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.