Abstract
Marek's disease virus (MDV) is a highly contagious oncogenic alphaherpesvirus that causes disease that is both a cancer model and a continuing threat to the world's poultry industry. This comprehensive gene expression study analyzes the host response to infection in both resistant and susceptible lines of chickens and inherent expression differences between the two lines following the infection of the host. A novel pathogenicity mechanism, involving the downregulation of genes containing HIC1 transcription factor binding sites as early as 4 days postinfection, was suggested from this analysis. HIC1 drives antitumor mechanisms, suggesting that MDV infection switches off genes involved in antitumor regulation several days before the expression of the MDV oncogene meq. The comparison of the gene expression data to previous QTL data identified several genes as candidates for involvement in resistance to MD. One of these genes, IRG1, was confirmed by single nucleotide polymorphism analysis to be involved in susceptibility. Its precise mechanism remains to be elucidated, although the analysis of gene expression data suggests it has a role in apoptosis. Understanding which genes are involved in susceptibility/resistance to MD and defining the pathological mechanisms of the disease gives us a much greater ability to try to reduce the incidence of this virus, which is costly to the poultry industry in terms of both animal welfare and economics.
INTRODUCTION
Marek's disease (MD) is a major disease affecting poultry health and welfare, with estimated annual global losses of $2 billion (36). MD virus (MDV) is a highly contagious, cell-associated, oncogenic alphaherpesvirus with biological properties more similar to those of gammaherpesviruses, as it is associated with T-cell lymphomas (43). MD continues to be a serious threat to the health and welfare of poultry, because although it has been controlled by vaccination for more than 40 years, there is growing evidence that the intensive use of vaccines is driving the virus to increasing virulence (50), and new, more virulent strains of the virus continue to emerge (12, 16). Infection with virulent strains of serotype 1 MDV causes an early cytolytic infection (3 to 7 days postinfection [dpi]), primarily in B lymphocytes with temporary, often profound, immunosuppression (7) and T-cell activation. Once activated, T lymphocytes themselves become susceptible to infection, which can be lytic, but after about 7 dpi the virus enters latency (45). In susceptible genotypes, a further cytolytic phase may occur about 2 weeks after infection, resulting in a permanent immunosuppression (8). The proliferative/transformation phase leading to lymphoma formation starts around 21 to 28 dpi, although T cells are likely to become transformed much earlier during infection. The infection usually occurs by the respiratory route, and the virus is spread through dander, the feather-follicle epithelium being the only source of enveloped and infectious cell-free MDV and therefore the site of virus shedding.
As the current vaccination practices are thought to be driving the virus to increasing virulence, an alternative means to control MD would be through the selection and breeding of birds with enhanced genetic resistance to MD. Differences in genetic resistance to MD were first reported nearly 80 years ago (2). Genes encoded in the major histocompatibility complex (MHC) locus have long been known to contribute to MD resistance (24, 26) with a rough hierarchy of resistance between haplotypes, with B21 being the most resistant and B19 generally the most susceptible (24). Despite this, other genes also have a strong influence on MD resistance. This has been determined predominantly in studies of the MHC-congenic lines 61 (resistant) and 72 (susceptible), which were developed in East Lansing, MI, which differ in their resistance to a large number of MDV isolates (38). Several potential MD resistance quantitative trait loci (QTL) have been identified from crosses between these lines and others (48, 52, 33, 11, 19), but relatively few actual genes have been associated with the disease. Those which have been identified include growth hormone (GH1) (27, 30), small inducible cytokine subfamily C member 1 (SCYC1) (29), and stem cell antigen 2 (SCA2) (31).
The availability of the chicken genome sequence (21) and commercially available whole-genome microarrays has revolutionized our ability to test candidate genes for disease resistance. In recent years, a small number of microarray experiments have been carried out to determine gene expression changes occurring during the host response to MDV infection and to identify genes involved in disease resistance (29, 40, 41). However, these have been on a much smaller scale and therefore are more limited in their findings.
In this study, we have carried out a comprehensive gene expression study using Affymetrix chicken whole-genome arrays analyzing the host response to infection in both resistant (61) and susceptible (72) lines and inherent expression differences between the two lines. Our assumption is that genes controlling disease resistance will be involved during the initial stages of infection, i.e., during the induced innate response to infection, and we thus have concentrated on responses at 2, 3, and 4 dpi.
The comparison of our gene expression data with QTL previously associated with MDV traits allowed us to identify candidate genes for resistance to MD, which then were fine mapped using single nucleotide polymorphisms (SNPs) to determine potential causative mutations for MD resistance/susceptibility. To our knowledge, this is the first time a large-scale study has been carried out to assess the host innate response to infection, identify SNPs in candidate genes, and validate those disease resistance/susceptibility genes.
MATERIALS AND METHODS
Ethics statement.
All animal work was conducted according to United Kingdom Home Office guidelines.
Experimental animals and design.
Chicks were obtained from specified-pathogen-free (SPF) parent flocks of the White Leghorn inbred lines 61 (MD resistant) and 72 (MD susceptible), maintained at the Institute for Animal Health, Compton, United Kingdom. As SPF chickens, the breeder hens were not vaccinated and were shown to be free of MDV, avian leukosis virus, infectious bursal disease virus, chicken infectious anemia virus, and other poultry pathogens, so the chicks used in these experiments were assumed to be free of maternal antibodies against MDV.
At 2 weeks of age, chicks were selected at random and placed in infected or control groups. Infected birds of both lines were housed in separate cages in the same filtered-air positive-pressure isolation room and injected with 1,000 PFU MDV strain RB1B in chicken kidney cell (CKC) suspension (0.2 ml) by the intraperitoneal route. Similarly, controls were kept together in a separate isolation room and injected with the same number of uninfected CKC (0.2 ml) by the intraperitoneal route. Spleen and thymus samples from nine birds from each group were collected at 2, 3 and 4 dpi.
RNA preparation.
Tissue samples were stabilized in RNAlater (Ambion, Warrington, United Kingdom) and disrupted using a bead mill (Retsch, Haan, Germany) at 20 Hz for 4 min. Total RNA was prepared using the RNeasy minikit (Qiagen, Crawley, United Kingdom) extraction according the manufacturer's protocol (Qiagen, Crawley, United Kingdom) to clean up the RNA further. Concentrations of the RNA samples were calculated by measuring the optical density at 260 nm (OD260) and OD280 on a spectrophotometer (NanoDrop; Thermo Scientific). The quality of the RNA was checked on a bioanalyzer (Agilent Technologies, South Queensferry, United Kingdom).
Microarray hybridization.
Biotinylated fragmented cRNA was hybridized to the Affymetrix chicken genome array. As well as genome-wide coverage of the chicken, this array also contains 689 probe sets for detecting 684 transcripts from 17 avian viruses (http://www.affymetrix.com/support/technical/datasheets/chicken_datasheet.pdf). For each experimental group, three biological replicates, consisting of three pools of three individual samples, were hybridized. Thus, 72 arrays were used in total. Hybridization was performed at 45°C for 16 h in a hybridization oven with constant rotation (60 rpm). The microarrays then were automatically washed and stained with streptavidin-phycoerythrin conjugate (SAPE; Invitrogen, Paisley, United Kingdom) in a Genechip Fluidics station (Affymetrix, Santa Clara, CA). Fluorescence intensities were scanned with a GeneArray scanner 3000 (Affymetrix, Santa Clara, CA). The scanned images were inspected and analyzed using established quality-control measures.
Statistical analysis of gene expression data.
Gene expression data generated from the GeneChip operating software (GCOS) was normalized using the PLIER (probe logarithmic intensity error) method (1) within the Affymetrix Expression Console software package. The normalized data then were analyzed using the limma and FARMS (46) packages within R in Bioconductor. Probes with a false discovery rate (FDR) of <0.05 and a fold change of ≥1.5 were deemed significant.
Functional analysis of differentially expressed gene sets.
The BioMart data-mining tool within the Ensembl database (release 62) (http://www.ensembl.org/biomart/index.html) was used to identify genes lying in areas of the genome known to be associated with QTL for resistance to MD (as defined in Table S1 in the supplemental material). This information then was analyzed in conjunction with the gene expression data to identify potential candidate genes for disease resistance.
To determine which biological pathways are involved in the responses to viral infection, Pathway Express within the Onto-Tools suite (http://vortex.cs.wayne.edu/projects.htm) was used. Genes differentially expressed during the host response (P < 0.05) were analyzed against a reference background consisting of all genes found to be expressed on the arrays. Annotation was based upon the equivalent human genes.
Genes were clustered by similar expression patterns and analyzed for enriched gene ontology (GO) terms and transcription factor binding sites using the Expander (v4.1.1) software package (http://acgt.cs.tau.ac.il/expander/expander.html). Normalized expression data from control samples were compared to data from infected samples to examine the host response, while expression data from each of the susceptible and resistant lines also were compared. The enrichment of particular GO terms or transcription factor (TF) binding sites within clusters was done by using the TANGO and PRIMA algorithms, respectively, within the Expander package.
The use of the ingenuity pathway analysis (IPA) program (Ingenuity Systems) revealed which canonical pathways are being switched on by MDV infection in the host (with Benjamini-Hochberg multiple testing correction) and allowed us to analyze the gene interaction networks involved in both the host response and the interaction with IRG1.
Genes having an expression profile similar to that of IRG1 were identified with Biolayout Express 3D (update 7) (47). Total raw expression data from the host response comparison were analyzed with a cutoff of P < 0.9. EasyGO (http://bioinformatics.cau.edu.cn/easygo/) then was used to examine enriched gene ontology terms within the genes which grouped together with IRG1.
Quantitative real-time PCR and reverse transcription-PCR (RT-PCR).
Primers and probe were designed using the Primer Express software program (Applied Biosystems, Warrington, United Kingdom) (see Table S2 in the supplemental material). All probes were labeled with the fluorescent reporter dye 5-carboxyfluorescein (FAM) at the 5′ end and the quencher N, N, N, N′-tetramethyl-6-carboxyrhodamine (TAMRA) at the 3′ end.
Real-time quantitative RT-PCR (qRT-PCR) was performed using the reverse transcriptase qPCR Master Mix RT-PCR kit (Applied Biosystems). The amplification and detection of specific products were performed using the ABI 7500 FAST sequence detection system (Applied Biosystems) with the following cycle profile: 1 cycle of 50°C for 2 min, 60°C for 30 min, and 95°C for 5 min, and 40 cycles of 94°C for 20 s and 59°C for 1 min. Normalization was carried out against 28S rRNA, which was used as a housekeeping gene. Fold changes in RNA levels were calculated from mean 40-Ct values by the formula 2(40-Ct in infected line − 40-Ct in control line) or 2(40-Ct in control line 61 − 40-Ct in control line 72).
Sequencing of candidate gene promoters.
Primers were designed around the proximal promoter region (∼700 bp preceding the ATG start codon) of each candidate gene (see Table S2 in the supplemental material). Promoter regions were amplified from 20 ng genomic DNA with 10 pmol of each primer in a final volume of 20 μl using FastStart Taq polymerase (Roche, Welwyn Garden City, United Kingdom). This was used with the supplied reaction buffer containing 200 μM each deoxynucleoside triphosphate (dNTP) and 1× GC-RICH solution (Roche, Welwyn Garden City, United Kingdom). Amplification conditions were 10 min at 95°C; 30 cycles of 30 s at 95°C, 30 s at 55°C or 60°C, and 1 min at 72°C; and 5 min at 72°C. Prior to sequencing, 10 μl of each PCR was treated with 8 U of exonuclease I (New England BioLabs, Hitchin, United Kingdom) and 0.8 U of shrimp alkaline phosphatase (USB, Staufen, Germany) for 15 min at 37°C and 15 min at 80°C to remove primers and dNTPs, respectively. The cleaned-up PCR products were sequenced in both directions with the same primers used for amplification using ABI BigDye Terminator chemistry, version 3.1, and an ABI 3730 sequencer (Applied Biosystems, Foster City, CA).
Backcross population.
A first-generation backcross was generated by crossing birds of line 61 × line 72 F1 generation (6 males) with susceptible line 72 birds (20 females). Members of the resulting backcross progeny were infected at 3 weeks of age with the moderate, classical strain of MDV, HPRS16. Blood samples were taken at 4, 11, and 39 dpi, and viral loads were determined by quantitative PCR. On postmortem, tumor development was recorded as N (neural), V (visceral), NV (both), or negative (−). Death and viral load assessed by PCR were used as QTL traits (the latter shows a high correlation with subsequent tumor development). From the experiment of 85 birds plus parental birds, 47 animals were chosen for mapping on the basis of survival (strongly correlated to viral load PCR) or death with the highest viral load titers.
SNP genotyping.
Thirteen SNPs from the promoter regions of the MD resistance candidate genes were screened in the parent lines 61 and 72 to identify which were fully informative for the mapping study. These SNPs were previously identified from sequencing the promoter regions within a single line 61 bird and a single line 72 bird for the gene of interest. SNPs were selected on the basis of their homozygosity in each line and the divergence of the homozygous allele in the parent lines. Thus, only six SNPs that were fully fixed and divergent between the parent lines were selected for a fully informative analysis. Informative SNPs (see Table S2 in the supplemental material) were PCR amplified using 50 to 100 ng genomic DNA, 200 μM each dNTP, and 400 pmol of each primer, in a total reaction volume of 12.5 μl. Genotypes were generated in the MDV backcross mapping panel using a fragment analysis assay on a Beckman CEQ8000 capillary sequencer. Cycling conditions using touchdown PCR were 95°C for 2 min, 30 s of denaturing, 30 s of annealing (starting at 5°C above the calculated annealing temperature and dropping by 1°C in each cycle), and 2 min of extension at 72°C. A further 25 cycles was performed at the annealing temperature, followed by a final 4-min extension at 72°C. PCR products were purified by incubating with ExoSAP-IT (Amersham, Little Chalfont, United Kingdom) for 45 min, followed by enzyme inactivation at 80°C for 15 min. A Genomelab SNP primer extension kit (Beckman Coulter, High Wycombe, United Kingdom) was used for the SNP assay reactions. Each reaction was carried out using 3.5 μl of cleaned-up PCR product combined with 4 μl SNPStart mastermix (Beckman Coulter, High Wycombe, United Kingdom) and 50 pmol of each SNP assay primer in a 10-μl reaction. Each assay primer in a given multiplex was designed to be of a different length for accurate genotyping during fragment analysis. The duplexing of PCR products in a single reaction mixture used 1 μl of each assay product.
Genetic marker association analysis.
All statistical analyses were conducted in Genstat. Virus counts were analyzed by REML using a split-plot fixed-effect model with time postinfection, sex, genotype, and their two- and three-way interactions. Plots of residuals showed a positive mean variance relationship, and the virus count was transformed to natural logarithms. Time of death was analyzed by analysis of variance (ANOVA) with right censoring and by a proportional hazards model; treatment effects were sex, genotype, and their two-way interaction. Lesion scores were analyzed by chi-square analysis of tables of lesion scores and genotype ignoring sex to maximize the size of subclasses.
Microarray accession number.
Array data have been submitted to Array Express under accession number E-TABM-721.
RESULTS
Host response to MDV infection.
To examine the early host response to infection with MDV, gene expression differences between infected and control birds in the susceptible line were analyzed using an Affymetrix chicken whole-genome microarray at 2, 3, and 4 dpi. Little difference was seen between infected and control birds at 2 dpi, except for the statistically significant downregulation of IgG heavy chain (IgG-H) in infected birds in both the thymus and the spleen. Statistically significant gene expression differences in both spleen and thymus of infected versus control birds were seen at both 3 and 4 dpi, with more genes differentially expressed (DE) at the later time point (1,245 DE genes in spleen and 1,234 DE genes in thymus) (see Table S3 in the supplemental material).
Genes of known immune function upregulated during the host response to infection with MDV (i.e., expression levels upregulated in infected compared to control birds) included some which had been identified in previous studies. These included both type I (IFNA) and type II (IFNG) interferons (IFN), some interleukins and interleukin receptors (IL-6, IL13R2A, and IL-18), the proinflammatory chemokine CCLi7 (previously called ah221), genes involved in the interferon response (such as the signaling molecule interferon regulatory factor 1 [IRF1]), inducible nitric oxide synthase (NOS2A), the lymphocyte antigen Ly-6E precursor (SCA2), the quiescence-specific protein P20K, and the proinflammatory protease granzyme A (GZMA) (29, 30, 31, 41).
In addition, several other innate immune function genes were upregulated, including the Toll-like receptors TLR3 and TLR15, the chemokines CCLi2, CCLi3, CCLi6, CCL17, and CCL19, more genes involved in controlling the interferon response (BATF3, IFIH1, IFIT-like ISG12, MX1, STAT1, SOCS1, and SOCS3), the lysozyme gene (LYG2), and the avidin gene (AVD).
In contrast, a subset of genes was downregulated at 3 and 4 dpi with MDV, i.e., expression levels were downregulated in infected birds. In this category, genes included IgG-H (as at 2 dpi), some avian beta-defensins (AvBD1, AvBD2, and AvBD4), the matrix metalloprotein genes MMP2, MMP7, and MMP13, several lectins and collectin genes (CLEC3B, COLEC10, and COLEC12), the chemokine receptor CCR6, the adhesion molecule AMIGO2, and the dendritic cell (DC) surface molecule gene Tim4 (a costimulatory molecule for Th2 T cells). Interestingly, the most highly downregulated genes have no known function in innate immune responses, including type XII collagen (COL12A1) and SLC40A1.
Functional analysis of differentially expressed gene sets.
To determine which biological pathways are altered during the host response to infection with MDV, the microarray data were analyzed using Pathway Express (15). Based upon the KEGG pathways (23), this program displays, pictorially, genes with altered expression patterns (compared to a relevant control) in any given biological pathway. Factors considered by Pathway Express include the magnitude of a gene's expression change and its position and interactions within any given pathway, thus including an impact factor when calculating statistically significant pathways. These pathway diagrams are useful in establishing which gene networks are involved in a particular experimental response. Table 1 shows the pathways which are significantly (FDR-corrected P value of <0.25) affected during the host response to MDV infection. Pathways seen to be involved include cytokine-receptor interaction and the Toll-like receptor and JAK-STAT signaling pathways, which play integral roles during the innate immune response. Figure S1 in the supplemental material shows pictorial examples of the pathways perturbed during the early response to MDV infection. Genes involved in apoptosis were upregulated, while genes involved in tight junction formation and maintenance were downregulated. MDV requires cell-to-cell contact for dispersal throughout the body, so the downregulation of genes involved in tight junctions presumably reflects a mechanism by which the virus promotes such contact. (These diagrams are based on the human pathways and so in some cases are not completely demonstrative of the chicken pathways, i.e., avian-specific genes are not represented.)
Table 1.
Pathway Express analysis of the host response to MDV infection in the spleen
| Rank | Pathway name | Impact factor | No. of input genes/no. of pathway genes | Corrected P value |
|---|---|---|---|---|
| 1 | Leukocyte transendothelial migration | 69.707 | 10/119 | 3.77E−29 |
| 2 | Cell adhesion molecules | 46.582 | 15/134 | 2.80E−19 |
| 3 | Adherens junction | 24.326 | 3/78 | 6.90E−10 |
| 4 | Cytokine-cytokine receptor interaction | 12.714 | 22/263 | 4.13E−05 |
| 5 | Antigen processing and presentation | 10.45 | 8/89 | 3.31E−04 |
| 6 | Complement and coagulation cascades | 10.161 | 10/69 | 4.31E−04 |
| 7 | Systemic lupus erythematosus | 8.129 | 8/144 | 0.002691804 |
| 8 | Jak-STAT signaling pathway | 8.075 | 15/155 | 0.002824351 |
| 9 | Toll-like receptor signaling pathway | 6.922 | 11/102 | 0.007809953 |
| 10 | Graft-vs-host disease | 6.241 | 3/42 | 0.014104792 |
| 11 | Type II diabetes mellitus | 5.744 | 4/45 | 0.021593849 |
| 12 | Long-term depression | 5.653 | 8/75 | 0.023331938 |
| 13 | Extracellular matrix-receptor interaction | 5.197 | 10/84 | 0.034288862 |
| 14 | Epithelial cell signaling in Helicobacter pylori infection | 4.898 | 9/68 | 0.044007875 |
| 15 | Axon guidance | 4.631 | 13/129 | 0.054874147 |
| 16 | Apoptosis | 4.538 | 8/89 | 0.059227662 |
| 17 | Natural killer cell-mediated cytotoxicity | 4.525 | 6/135 | 0.059861797 |
| 18 | p53 signaling pathway | 4.279 | 7/69 | 0.073148525 |
| 19 | Transcription growth factor β signaling pathway | 4.171 | 4/87 | 0.079823773 |
| 20 | Allograft rejection | 4.1 | 2/38 | 0.084520645 |
| 21 | Tight junction | 3.98 | 9/135 | 0.093054484 |
| 22 | ABC transporters | 3.825 | 5/44 | 0.105273951 |
| 23 | Type I diabetes mellitus | 3.79 | 2/44 | 0.108232933 |
| 24 | Gap junction | 3.507 | 7/96 | 0.135150237 |
| 25 | Long-term potentiation | 3.495 | 3/73 | 0.136417624 |
| 26 | PPAR signaling pathway | 3.486 | 7/70 | 0.137375316 |
| 27 | Basal cell carcinoma | 3.456 | 1/55 | 0.140612346 |
| 28 | Pancreatic cancer | 3.306 | 3/72 | 0.157868858 |
| 29 | Neuroactive ligand-receptor interaction | 3.174 | 11/256 | 0.174623126 |
| 30 | Proteasome | 2.969 | 1/48 | 0.203826563 |
| 31 | Amyotrophic lateral sclerosis | 2.891 | 4/56 | 0.216030904 |
| 32 | Vibrio cholerae infection | 2.775 | 6/62 | 0.235369275 |
| 33 | Pathogenic Escherichia coli infection | 2.737 | 4/54 | 0.242024371 |
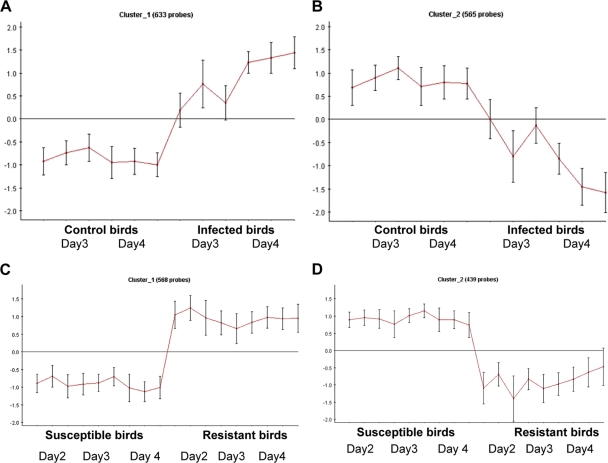
Utilizing the CLICK algorithm within the Expander program (44) allowed us to cluster genes into groups with similar expression profiles, thus identifying genes likely to be involved in the host response to MDV. Figure 1 shows the expression profile of genes either upregulated or downregulated in response to MDV infection compared to levels in age-matched uninfected controls (see Table S3 in the supplemental material for genes involved in this response). It was apparent that similar profiles were present, i.e., some genes were inherently more highly expressed in the resistant line, and some were inherently more highly expressed in the susceptible line (see Table S4 in the supplemental material for genes showing this response).
Fig. 1.
Gene expression clusters generated using the Expander program (http://acgt.cs.tau.ac.il/expander/expander.html). Shown are the expression profiles of genes upregulated during the response to virus (A) and those downregulated (B). When genes expressed in the two different lines were examined, similar profiles were noted: gene expression was inherently higher in the resistant line (C), and genes were more highly expressed in the susceptible line (D).
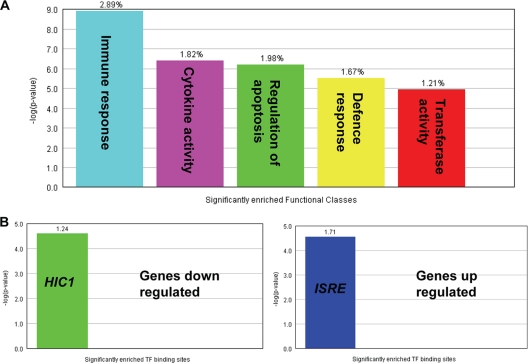
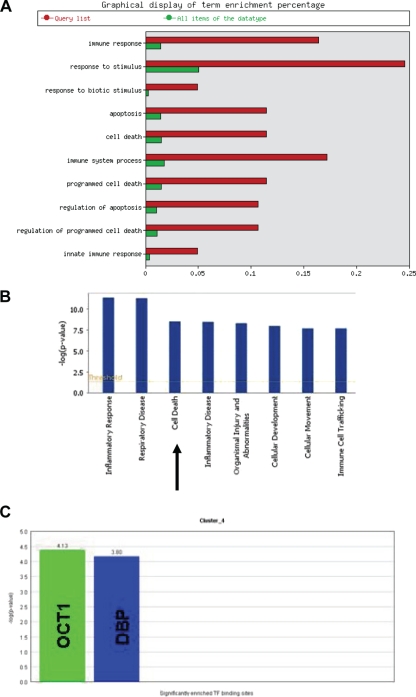
To gain insight into the function of the genes activated during the host response, the Expander program was also used to analyze the gene ontology (GO) functional annotations of the differentially expressed genes. Figure 2 A shows the biological process terms which were significantly enriched in the genes differentially expressed during the host response to infection. As might be expected, these include immune response, cytokine activity, and regulation of apoptosis. How these genes are transcriptionally regulated also can be determined using Expander software. Potential or known transcription factor binding sites present in the differentially expressed genes which are significantly overrepresented can be identified. Figure 2B shows that genes upregulated during the host response had a higher proportion of ISRE (interferon-stimulated response element) sites than would be represented by chance in a random sample of genes. The frequency ratio (actual frequency of sites in a gene set divided by the frequency of the background) is 1.71, i.e., nearly twice as many ISRE sites than would be expected by chance (P < 0.0001).
Fig. 2.
Overrepresentation analysis using the Expander program. (A) The GO biological processes which are significantly enriched during the host response to infection. The frequency of genes of a functional class within the examined set is described as a percentage of the total. (B) Transcription factor binding sites present in differentially expressed genes which are significantly overrepresented in up- and downregulated genes during the host response to MDV infection. The frequency ratio (frequency of the set divided by the frequency of the background) is shown.
HIC1 (hypermethylated in cancer 1) binding sites were significantly (P < 0.0001) enriched (frequency ratio of 1.24) in the promoters of genes which were repressed during the host response. HIC1 acts as a tumor suppressor (17, 49).
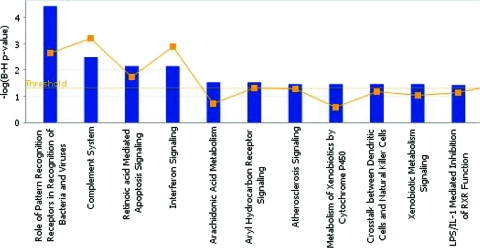
Complementing the findings from Pathway Express, ingenuity pathway analysis (IPA) allowed us to identify which biological networks and pathways are most highly involved during the host response to MDV infection. The most significant pathways are represented in Fig. 3. It can be clearly seen that processes involved in innate immunity and signaling constitute the majority of pathways highlighted. Receptor recognition of PAMPs, the interferon response, apoptosis signaling, and the involvement of various immune cell types (DC, natural killer cells, etc.) all can be seen to play an important role in MDV infection.
Fig. 3.
Ingenuity pathway analysis of the host response to MDV infection. Shown are the most highly represented canonical pathways as revealed after IPA of genes differentially expressed during the host response to MDV (in the spleen). The line represents the ratio of the number of genes represented within each pathway to the total number of genes in the pathway.
Inherent differences between susceptible and resistant lines.
Resistance to MD inherent between the lines could be due to a number of mechanisms. For example, line 61 could simply express certain genes, such as those involved in key innate immune responses, at a constitutively higher level than that of line 72 and thus mount a stronger innate response upon infection, limiting viral replication and disease. Alternatively, following infection, line 61 could upregulate the expression of key immune function genes to a greater degree than line 72, thus mounting a stronger induced immune response. Either or both mechanisms could contribute to MD resistance.
Gene expression differences were identified between the two lines (395 DE genes in spleen and 177 DE genes in thymus) (see Table S4 in the supplemental material) in tissues from the control, uninfected birds, including the following genes, which were more highly expressed in line 61 than line 72, that are known to be involved in the innate immune response: DNAJC3, DDT, NMU, GSTO1, VIP, HPS5, MMP7, FGFR3, HSCB, E2F4, SFTPA2, and GNG12. There are therefore genes whose constitutive expression is different between susceptible and resistant birds, even before infection occurs.
Following infection, differences in gene expression levels also were seen between the two lines (539 DE genes in spleen and 156 DE genes in thymus) (see Table S5 in the supplemental material). Immune genes more highly expressed in the resistant line included IgG-H, AMIGO2, MMP13, and CLEC3B, while genes more highly expressed in the susceptible line included AVD, IRG1, HSP25, ART1, IL-18, NOS2A, CXCL13, CCLi2, MX1, SOCS1, and IL-6. There are therefore also immune genes whose expression is differentially upregulated following infection.
MDV QTL candidate genes.
Many different regions of the chicken genome have been identified as being QTL involved in MDV infection (see Table S1 in the supplemental material), although very few actual genes have been shown to have a causative role in the disease. Genes underlying the known QTL (based on Ensembl data, release 62) were compared to the lists of differentially expressed genes from these experiments, and there was a degree of overlap (see Tables S3 and S5 in the supplemental material). Between 26 and 30% of the differentially expressed genes are located under a previously reported QTL for MD resistance and therefore are potential candidate genes for controlling the resistance phenotype.
Confirmatory real-time quantitative RT-PCR.
The differential expression of 17 genes (Table 2), chosen as potential candidates for involvement in disease resistance, was verified by qRT-PCR (see Fig. S2 in the supplemental material). These genes showed differential expression between the susceptible and resistant lines of birds (either inherently or following infection), and 10 of the genes chosen also lie within a known MD QTL region (Table 2).
Table 2.
Candidate genes for Marek's disease resistance
| Gene | Description | Analysisa | Fold change | Tissueb | Comment | Accession no. |
|
|---|---|---|---|---|---|---|---|
| GenBank | Ensembl | ||||||
| IgG-H | IgG H-chain (HC36) D-J-C | D | 10–22 | S and T | X07174 | ||
| AMIGO2 | Adhesion molecule with Ig-like domain 2 | D | 5 | S | XM_416052 | ENSGALG00000009739 | |
| MMP13 | Matrix metalloproteinase-13 | D | 3 | S | XM_001235209 | ENSGALG00000017183 | |
| CLEC3B | Tetranectin | D | 3 | S | NM_204666 | ENSGALG00000011883 | |
| AVD | Avidin precursor | D | 38 | S | Between 2 QTL | NM_205320 | ENSGALG00000023622 |
| IFNA | Alpha interferon | D | 28 | T | Between 2 QTL | NM_205427 | ENSGALG00000013245 |
| MMP9 | Matrix metalloproteinase-9 | D | 10 | T | NM_204667 | ENSGALG00000006992 | |
| IL-18 | Interleukin-18 precursor | D | 5 | S | NM_204608 | ENSGALG00000007874 | |
| TLR15 | Toll-like receptor 15 | D | 3 | S | NM_001037835 | ENSGALG00000008166 | |
| MX1 | Interferon-induced GTP-binding protein Mx. | D | 3–4 | S | Under QTL (MD2) | NM_204609 | ENSGALG00000016142 |
| IL-6 | Mature chil-6 | D | 2–4 | S | Near MD3 | NM_204628 | ENSGALG00000010915 |
| FN1 | Fibronectin | D | 3 | S | Under QTL (MD10) | XM_421868 | ENSGALG00000003578 |
| IRG1 | Immunoresponsive protein 1 | D | 14 | S | Quite near MD2 | NM_001030821 | ENSGALG00000016919 |
| DDT | d-Dopachrome tautomerase | I | 79–110 | S and T | Under QTL (MD24) | NM_001030667 | ENSGALG00000006350 |
| DNAJC3 | DnaJ homolog subfamily C member 3 | I | 14–27 | S and T | Under QTL (MD2) | NM_001008437 | ENSGALG00000016894 |
| SFTPA2 | Lung lectin precursor | I | 17 | S | Under QTL (MD22) | NM_001039166 | ENSGALG00000002496 |
| GNG12 | Guanine nucleotide-binding protein | I | 9 | S | Under QTL (MD23) | XM_001234702 | ENSGALG00000020596 |
D, differential response between the lines upon infection; I, inherently different between control birds of the resistant and susceptible lines.
S, spleen; T, thymus.
Genetic association of genetic variants in candidate genes with MD.
The 5′ flanking region (assumed to contain the proximal promoter) of 16 of the candidate genes (IL-6 primers failed to generate sequence in line 72) were sequenced from DNA from several individuals from lines 61 and 72 to identify SNPs between the two lines. Nine of these genes exhibited polymorphisms between the lines. Six of these contained SNPs which result in changes to known transcription factor binding sites, as determined using the TRANSFAC database (32) (Table 3; also see Fig. S3 and S4 in the supplemental material). SNPs from five of these genes (MMP13, SFTPA2, IRG1, AMIGO2, and DDT) were genotyped in a backcross experiment, specifically in progeny of a (61× 72)×72 backcross, to assess allele segregation (the SNPs found within the MMP9 promoter region were not fixed in the lines). The backcross progeny had been challenged with MDV and scored postinfection for several different phenotypes: viral load at 4, 11, and 39 dpi, tumor formation, and survival. Forty-seven animals were used for the mapping analysis, being the extremes of the experimental population as either survivors or those that died with the highest viral titers.
Table 3.
SNPs found in transcription factor binding sites in the promoter regions of six candidate genes
| MDV strain | No. of SNPs between lines | No. of SNPs in TF binding sites | Comment(s) |
|---|---|---|---|
| MMP13 | 12 | 3 | Line 6 has an HMF1 site; line 7 has GATA4 and Hand1:E47 sites |
| SFTPA2 | 8 | 3 | Line 6 has MRF2 and FAC1 sites; line 7 has IPF1 and TBP sites |
| IRG1 | 7 | 2 | Line 7 has extra Arnt and PPARγ:RXRα sites |
| AMIGO2 | 5 | 2 | Line 6 has extra BRCA1:USF2 and MEIS1B:HOXA9 sites |
| DDT | 4 | 2 | Kid3 site created (line 7); TCF11 site instead of PAX5, which is in line 6 |
| MMP9 | 4 | 2 | Line 6 has extra NF-1 and PPARalpha sites; line 7 has an extra Pax5 site |
The means, standard errors, and medians from the raw data for virus counts (per spleen) by SNP genotype for each gene for the top/tail data set are given in Table 4. There were no statistically significant differences between the two genotypes for AMIGO2, DDT, MMP13, or SFTPA2. There was a significant (P < 0.05) effect for IRG1 genotypes, and these were further investigated after genotyping the remaining 38 middle-ranking birds of the backcross. The raw data from the full data set are summarized by IRG1 genotype in Table 4. Interaction effects were not significant, and the final model included main effects for time postinfection (p.i.), sex, and IRG1 genotype. There was a significant effect of time p.i. (P < 0.001) and SNP genotype for IRG1 (P = 0.033), whereas differences between the sexes were not significant. The marginal means for each time point postinfection were 9.23, 10.79, and 10.82 (sed 0.125), respectively, for 4, 11, and 39 dpi (back-transformed medians from viral DNA counts were 10,199, 48,533, and 50,011); means for IRG1 SNP AG and GG were 10.41 and 10.15 (sed 0.126) (back-transformed medians from viral DNA counts were 33,190 and 25,591); means for female and male chicks were 10.29 and 10.27 (sed 0.126) (back-transformed medians from viral DNA counts were 29,437 and 28,854). Both analyses by censored ANOVA (survivors were killed 150 dpi) and proportional hazard models of survival time p.i. showed no statistical differences between sexes or genotypes. The mean (± standard errors of the means)-adjusted times to death p.i. for SNP genotypes AG and GG were 81 ± 6.9 and 84 ± 6.9 days, respectively. The pattern of death p.i. was similar for both genotypes (proportional hazard results not shown). There were no statistical differences in lesion scores between genotypes. However, comparing the types of lesions found suggests that the homozygote genotype preferentially targets the neural system, and the heterozygote targets the viscera (P = 0.029). This helps explain the lower viral loads seen in homozygous birds, because the IRG1 homozygote alleles confer resistance to virus uptake and thus provide protection from visceral lymphoma formation.
Table 4.
Raw data for virus countsa
| Gene | SNP | Virus count (means ± SEM) at dpi: |
||
|---|---|---|---|---|
| 4 | 11 | 39 | ||
| AMIGO2 | TC | 15,336 ± 3,126 (9,775) | 55,763 ± 5,710 (57,075) | 65,321 ± 6,109 (73,387) |
| CC | 13,002 ± 2,104 (12,215) | 54,677 ± 3,908 (53,420) | 53,739 ± 6,542 (52,344) | |
| DDT | CT | 10,452 ± 1,327 (10,062) | 56,160 ± 4,782 (53,132) | 62,411 ± 5,938 (70,093) |
| TT | 19,537 ± 3,877 (14,048) | 53,864 ± 4,952 (55,989) | 55,208 ± 7,046 (59,029) | |
| MMP13 | CG | 16,467 ± 3,386 (12,072) | 58,376 ± 5,280 (59,370) | 66,909 ± 6,334 (71,740) |
| GG | 11,871 ± 1,537 (12,014) | 52,218 ± 4,502 (51,932) | 52,862 ± 6,213 (56,159) | |
| SFTPA2 | TC | 14,158 ± 2,832 (11,734) | 57,632 ± 5,681 (51,714) | 54,323 ± 6,242 (57,716) |
| CC | 14,176 ± 2,532 (12,206) | 53,454 ± 4,348 (55,989) | 63,048 ± 6,347 (64,016) | |
| IRG1 | AG | 17,154 ± 2,793 (12,748) | 62,757 ± 4,091 (61,230) | 65,134 ± 5,771 (66,335) |
| GG | 9,857 ± 1,794 (6,497) | 45,072 ± 5,230 (45,072) | 51,413 ± 6,986 (55,215) | |
| IRG1 all | AG | 16,415 ± 2,546 (12,349) | 59,843 ± 3,303 (55,989) | 66,813 ± 4,449 (67,500) |
| GG | 12,948 ± 2,064 (7,974) | 47,563 ± 3,276 (50,954) | 61,017 ± 5,223 (63,878) | |
Raw data, with median values in parentheses, are for backcrossed chickens classified by different SNP genotypes for five genes in the top-and-tail-selected data set and for the complete set of data for IRG1 (IRG1 all).
The fact that a backcross was utilized assumes that the susceptible allele is codominant, because if it was dominant no difference between the two genotypes would be seen. The lack of effect of the other candidate genes therefore may not mean that they are not involved in resistance.
Potential role of IRG1 in MD susceptibility.
Immunoresponsive gene 1 (IRG1) is an orthologue of the bacterial methylcitrate dehydratase gene (9) and is induced by lipopolysaccharide (LPS) (28). Our expression data show that this gene is more highly expressed in the susceptible line following infection with MDV, indicating a candidate susceptibility locus. When the promoter regions of this gene were sequenced in lines 61 and 72 and analyzed by TRANSFAC, seven SNPs were identified, two of which resulted in changes to known transcription factor binding sites. Compared to line 61 (MD resistant), line 72 (MD susceptible) has extra ARNT (aryl hydrocarbon receptor nuclear translocator) and PPARγ (peroxisome proliferator-activated receptor gamma) binding sites (see Fig. S4 in the supplemental material). The SNP which was analyzed and found to segregate with susceptibility to MD is that which creates an ARNT binding site in line 72. ARNT is required for the activity of the dioxin receptor and for the ligand-binding subunit to translocate from the cytosol to the nucleus after ligand binding. The complex then initiates the transcription of genes involved in the activation of polycyclic aromatic hydrocarbon (PAH) procarcinogens. The role, if any, of PAH procarcinogens in MD pathology has never been evaluated, although it is interesting that increased IRG1 expression, in a line of chickens susceptible to infection with a tumor-causing herpesvirus, correlates with an SNP that creates a binding site in the IRG1 promoter for a transcription factor involved in the induction of carcinogens in another system.
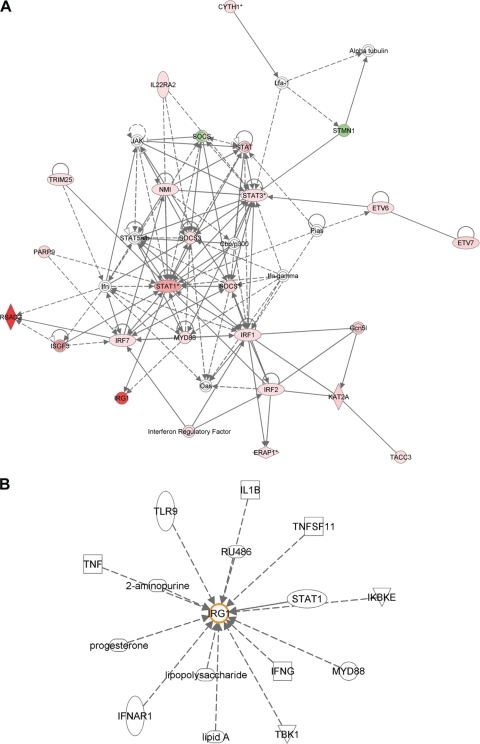
The analysis of the host response to MDV in spleen at 4 dpi using IPA enables us to see the biological interaction network involving IRG1. Figure 4A shows how IRG1 is involved in the inflammatory response via the actions of MyD88 (myeloid differentiation primary response gene 88) and STAT1 (signal transducer and activator of transcription 1). IPA also allows us to examine all of the previously identified interactions in which IRG1 is thought to be involved (Fig. 4B). Toll-like receptor 9, tumor necrosis factor, gamma interferon, interleukin-1B, and bacterial lipopolysaccharide are just some of the molecules which are known to be involved with the currently unidentified role of IRG1.
Fig. 4.
Network analysis using the IPA program. (A) Interaction network representing genes involved in the inflammatory response, showing genes upregulated in response to MDV infection (red) and genes downregulated (green). IRG1 is seen to be highly upregulated in this analysis. (B) Genes currently known to interact with IRG1.
To investigate the possible function of IRG1, we analyzed the expression profile of this gene during the host response to MDV infection and compared it to other genes with similar profiles. Using Biolayout Express 3D (47), we identified all genes (179) from this analysis having similar expression profiles and that cluster with IRG1. This cluster of genes was then analyzed using programs such as EasyGO and IPA. Many of the genes which cluster together with IRG1 are shown to be involved in apoptosis or the regulation of apoptosis (Fig. 5A and B). Based on these findings and the mitochondrial localization of IRG1 in mice, we hypothesize that the product of this gene also plays a part in the apoptosis pathway.
Fig. 5.
Analysis of the IRG1-containing cluster of genes identified using Biolayout Express 3D software. (A) EasyGO analysis shows a significant enrichment of genes involved in apoptosis and the regulation of apoptosis. (B) IPA analysis also reveals cell death to be a highly featured function of the genes in this cluster. (C) A subcluster identified using the Expander program is seen to be highly overrepresented (more than 4-fold) by genes which contain a binding site for the transcription factor OCT1 (octamer-binding transcription factor 1). Genes with binding sites for DBP (albumin D box-binding protein) also are seen to be enriched. DBP is thought to be involved in the modulation of circadian rhythms.
To understand how IRG1 is regulated, we used the Expander program to cluster the genes with similar expression profiles (generated using Biolayout Express 3D software). This generated four subclusters. These subclusters were examined for the enrichment of transcription factor binding sites, and one was found to be highly overrepresented (more than four times the level expected by chance) by genes containing binding sites for the transcription factor OCT1 (also called POU2F1) (Fig. 5C). OCT1 binds to the octamer motif (5′-ATTTGCAT-3′) and activates the promoters of the genes, such as some small nuclear RNAs (snRNA), histone H2B, and immunoglobulins. OCT1 modulates transcription transactivation by NR3C1, AR, and PGR. It is interesting that during another herpesvirus infection, human herpes simplex virus (HSV), OCT1 forms a multiprotein-DNA complex with the viral transactivator protein VP16 and HCFC1, thereby enabling the transcription of the viral immediate-early genes.
DISCUSSION
The early host response to MDV infection.
To further characterize the chicken innate immune response to infection with MDV, spleen and thymus samples from 2, 3, and 4 dpi were analyzed. Infected and control birds of the susceptible line were compared, with the hypothesis being that these birds had the most viral replication, which therefore was most likely to trigger stronger, but not necessarily more effective, immune responses. Responses in the spleen and thymus, as measured at the transcriptional level using the whole-genome microarrays, were detectable only at significant levels after 2 dpi (when the only difference in expression level between infected and control birds was for IgG-H, which was decreased following infection). MDV is normally detectable in most secondary lymphoid organs, including the spleen, from 2 to 7 dpi, peaking at 4 dpi (3). The lack of response at 2 dpi in this experiment possibly reflects the low level of virus in the spleen at this time point. By 3 dpi, an induced innate response has been established (22), and many proinflammatory cytokines and chemokines were activated. The expression of genes involved in apoptosis, immune-related signaling pathways (e.g., the TLR and Jak-Stat pathways), transcription factors, and receptors was upregulated. Genes involved in tissue remodelling were downregulated, including some involved in leukocyte transendothelial migration, and in the formation and maintenance of both adherens and tight junctions. As expected, there was a higher transcriptional response in the spleen than in the thymus, the former being the primary target organ for MDV.
The expression of certain TLRs also was increased during the host response, specifically TLR3, TLR15, TLR1B, and TLR4 in the spleen, and the same four TLRs plus TLR21 in the thymus. The modulation of TLR expression following the recognition of a pathogen-associated molecular pattern (PAMP) is poorly understood in the chicken. It is interesting that MDV, presumably recognized through its CpG motifs in its double-stranded DNA genome by TLR21 (which recognizes CpG in the chicken [5, 25], rather than TLR9, which is absent), upregulates the expression of TLRs that recognize double-stranded RNA (TLR3) and bacterial cell surface components (TLR1B and TLR4). TLR15 is an avian-specific receptor and is of unknown function, although it is upregulated in response to Salmonella infection (20) and has been predicted to recognize a pathogen-surface PAMP (37).
In mammals, the chemokine receptor CCR6 is normally found on immature DCs, Th17 cells, and unactivated memory T cells. It is also downregulated on activated T cells, which would fit with its downregulation in the spleen and thymus at 4 dpi with MDV, as at this stage of infection T cells are becoming activated and in turn infected with MDV. It also could be explained in part by the maturation of DCs in response to the MDV infection, as mature DCs in both mammals (39) and chickens (51) downregulate CCR6. The expression of the DC-expressed costimulatory molecule for Th2 cells, Tim4, also was downregulated, suggesting, as one would expect, the maturation of DCs to drive an antiviral Th1 response. This is in contrast to recent reports suggesting that MDV infection biases the immune response toward a Th2 (18) or Treg (6) response, either of which would be advantageous to the virus.
The analysis of potential or known transcription factor binding sites in the differentially expressed genes that were statistically significantly overrepresented yielded both expected and unexpected results. Genes whose expression was upregulated during the host response had a high proportion of ISRE sites in their promoters. This was to be expected, as MDV infection induces type I IFNs (18, 22), and downstream signaling events from the IFN-α receptor, used by all type I IFNs, lead to the induction of the expression of IFN-stimulated genes which contain ISRE sites.
The overrepresentation of genes with HIC1 binding sites in their promoters among those genes whose expression was repressed during the host response was unexpected and suggests a new pathological mechanism for MDV. HIC1 acts as a tumor suppressor in mammals acting at the intersection of regulatory loops involving p53-dependent and E2F-1-dependent cell survival, growth control, and stress responses (17, 49). HIC1 transcriptional repressor function occurs by the recruitment of CtBP1 through the conserved PXDLSXK/R motif, and we have shown previously that the interaction of the principal MDV oncogene meq with CtBP is crucial for MD oncogenicity (4). The meq oncogene is expressed later in infection, peaking at 3 weeks p.i., but as early as 4 dpi MDV infection causes the downregulation of genes whose expression would be upregulated by the tumor suppressor HIC1. In other words, MDV infection is blocking an antitumor mechanism long before the MDV oncogene is expressed. The precise mechanism by which MDV infection could be driving the suppression of this antitumor response remains to be determined.
MD resistance genes.
The analysis of samples from MD-susceptible and -resistant birds, with and without infection with MDV, using whole-genome microarrays has identified genes which either show different inherent levels of gene expression (without infection) between the lines or are transcribed differently following infection, thus potentially eliciting differing host responses. By analyzing the list of genes differentially expressed between the susceptible and resistant lines (either inherently or following infection), 16 genes were identified as potential candidates for involvement in disease resistance. We sequenced the promoters of these genes from multiple individuals from both lines, seven of which contained promoter SNPs resulting in changes to transcription factor binding sites. The allelic segregation of six of these was tested in a backcross MDV experiment, and one, IRG1 (immunoresponsive gene 1), which is more highly expressed in the susceptible line, showed a significant effect.
Interestingly, higher viral load was associated with the heterozygote genotype at the IRG1 locus (61/72). This is in contrast to what one might have expected, given that 72/72 birds should be more susceptible to MDV infection. However, we must bear in mind that this is the analysis of one locus within a polygenic system. Resistance to MD is not a simple Mendelian trait but is highly complex and involves at least 14 separate quantitative trait loci (33, 52). In addition, epistatic interactions between different QTL influence MDV viremia (10). Such genetic interactions may also modify the effect of the IRG1 alleles found in this study. The exact nature of the polymorphism within the IRG1 promoter (whether it confers susceptibility to MD infection or whether it is a resistance locus that we have highlighted within a susceptible background) and the function of the IRG1 protein itself will obviously require further functional investigation.
Potential function of IRG1.
There is little literature on the potential role of IRG1 in the immune response in biomedical model species. It is 1 of at least 35 genes that constitute the IFN-γ/tumor necrosis factor alpha (TNF-α)-triggered effector program in innate immunity in mice (13), with its expression being highly upregulated by several proinflammatory cytokines and TLR agonists. Moreover, in the same study it was shown to localize subcellularly with mitochondria.
The analysis of other genes with an expression profile similar to that of IRG1 indicates that IRG1 has a role in the apoptosis pathway. Apoptosis is often induced in virally infected cells and is the likely cause of MDV-induced cell death (42). Herpesviruses can both trigger and block apoptosis, the former during productive infections (as would be the case in this study) and the latter during latency or transformation (14). The apoptosis of CD4+ CD8+ T cells in the thymus during lytic infection with MDV has been reported (34), as has that of CD4+ T cells in peripheral blood at 14 to 21 dpi (35). However, it is not clear in either case that the apoptotic cells were infected with MDV. Schat (42) hypothesized that the prolonged cytolytic infection associated with very virulent plus strains of MDV result from those strains having been selected for decreased antiapoptotic or increased apoptotic ability during virus replication in vaccinated chickens. In other words, increased apoptosis would lead to prolonged and/or increased viral replication and hence increased pathogenesis. If IRG1 has a role in apoptosis, one would hypothesize that it is proapoptotic, as its expression is increased in the susceptible line compared to that of the resistant line. Further functional studies obviously need to be carried out to determine the true role of this protein in susceptibility to Marek's disease.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Biotechnology and Biological Sciences Research Council (grant numbers BB/D010705/1 and BB/D013704/1).
We thank Alison Downing (ARK-Genomics, Roslin, United Kingdom) for excellent technical assistance with the Affymetrix microarray experiments, Caroline McCorquodale and Daphne Mouzaki (Roslin) for invaluable statistical support, and the staff of the EAH (IAH) for their excellent care of the animals used in these experiments.
J.S. performed the arrays, analyzed the results, and wrote the manuscript; J.-R.S. carried out challenge experiments, prepared RNA, measured viral load, and performed quantitative RT-PCR; I.R.P. sequenced the promoter regions of the candidate genes; P.M.H. performed genotype analysis; N.S. and M.F. carried out the genotyping; V.N. supplied the challenge virus, was the Home Office license holder, and reviewed the manuscript; and D.W.B. and P.K. conceived and supervised the project and wrote the manuscript.
We declare no conflicts of interest and no competing financial interests.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 24 August 2011.
REFERENCES
- 1. Affymetrix 2005. Guide to probe logarithmic intensity error (PLIER) estimation. Affymetrix I, Santa Clara, CA [Google Scholar]
- 2. Asmundson V. S., Biely J. 1932. Inheritance of resistance to fowl paralysis (neurolymphomatosis gallinarum). I. Differences in susceptibility. Can. J. Res. 6:171–176 [DOI] [PubMed] [Google Scholar]
- 3. Baigent S. J., Davison F. 2004. Marek's disease virus: biology and life cycle, p. 62–77 In Davison F., Nair V. (ed.), Marek's disease: an evolving problem. Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 4. Brown A. C., et al. 2006. Interaction of MEQ protein and C-terminal-binding protein is critical for induction of lymphomas by Marek's disease virus. Proc. Natl. Acad. Sci. U. S. A. 103:1687–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brownlie R., et al. 2009. Chicken TLR21 acts as a functional homologue to mammalian TLR9 in the recognition of CpG oligodeoxynucleotides. Mol. Immunol. 46:3163–3170 [DOI] [PubMed] [Google Scholar]
- 6. Buza J. J., Burgess S. C. 2007. Modeling the proteome of a Marek's disease transformed cell line: a natural animal model for CD30 overexpressing lymphomas. Proteomics 7:1316–1326 [DOI] [PubMed] [Google Scholar]
- 7. Calnek B. W. 1986. Marek's disease-a model for herpesvirus oncology. Crit. Rev. Microbiol. 12:293–320 [DOI] [PubMed] [Google Scholar]
- 8. Calnek B. W. 2001. Pathogenesis of Marek's disease virus infection. Curr. Top. Microbiol. Immunol. 255:25–55 [DOI] [PubMed] [Google Scholar]
- 9. Chen B., Zhang D., Pollard J. W. 2003. Progesterone regulation of the mammalian ortholog of methylcitrate dehydratase (immune response gene 1) in the uterine epithelium during implantation through the protein kinase C pathway. Mol. Endocrinol. 17:2340–2354 [DOI] [PubMed] [Google Scholar]
- 10. Cheng H. H., Zhang Y., Muir W. M. 2007. Evidence for widespread epistatic interactions influencing Marek's disease virus viremia levels in chicken. Cytogenet. Genome Res. 117:313–318 [DOI] [PubMed] [Google Scholar]
- 11. Cheng H. H., et al. 2008. Using integrative genomics to elucidate genetic resistance to Marek's disease in chickens. Dev. Biol. (Basel) 132:365–372 [DOI] [PubMed] [Google Scholar]
- 12. Davison F., Nair V. 2005. Use of Marek's disease vaccines: could they be driving the virus to increasing virulence? Expert Rev. Vaccines 4:77–88 [DOI] [PubMed] [Google Scholar]
- 13. Degrandi D., Hoffman R., Beuter-Gunia C., Pfeffer K. 2009. The proinflammatory cytokine-induced IRG1 protein associates with mitochondria. J. Interferon Cyt. Res. 29:55–68 [DOI] [PubMed] [Google Scholar]
- 14. Derfuss T., Meinl E. 2002. Herpesviral proteins regulating apoptosis. Curr. Top. Microbiol. Immunol. 269:257–272 [DOI] [PubMed] [Google Scholar]
- 15. Draghici S., et al. 2007. A systems biology approach for pathway level analysis. Genome Res. 17:1537–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gimeno I. M. 2008. Marek's disease vaccines: a solution for today but a worry for tomorrow? Vaccine 26(Suppl. 3):C31–C41 [DOI] [PubMed] [Google Scholar]
- 17. Guerardel C., et al. 2001. Identification in the human candidate tumor suppressor gene HIC-1 of a new major alternative TATA-less promoter positively regulated by p53. J. Biol. Chem. 276:3078–3089 [DOI] [PubMed] [Google Scholar]
- 18. Heidari M., Zhang H. M., Sharif S. 2008. Marek's disease virus induces Th-2 activity during cytolytic infection. Viral Immunol. 21:203–214 [DOI] [PubMed] [Google Scholar]
- 19. Heifetz E. M., et al. 2009. Mapping QTL affecting resistance to Marek's disease in an F6 advanced intercross population of commercial layer chickens. BMC Genomics 10:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Higgs R., et al. 2006. Induction of a novel chicken Toll-like receptor following Salmonella enterica serovar Typhimurium infection. Infect. Immun. 74:1692–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. International Chicken Genome Sequencing Consortium 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716 (Erratum, 433:77, 2005.) [DOI] [PubMed] [Google Scholar]
- 22. Kaiser P., Underwood G., Davison F. 2003. Differential cytokine responses following Marek's disease virus infection of chickens differing in resistance to Marek's disease. J. Virol. 77:762–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanehisa M., Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaufman J. 2000. The simple chicken major histocompatibility complex: life and death in the face of pathogens and vaccines. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:1077–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keestra A. M., de Zoete M. R., Bouwman L. I., van Putten J. P. 2010. Chicken TLR21 is an innate CpG DNA receptor distinct from mammalian TLR9. J. Immunol. 185:460–467 [DOI] [PubMed] [Google Scholar]
- 26. Koch M., et al. 2007. Structures of an MHC class I molecule from B21 chickens illustrate promiscuous peptide binding. Immunity 27:885–899 [DOI] [PubMed] [Google Scholar]
- 27. Kuhnlein U., et al. 1997. DNA polymorphisms in the chicken growth hormone gene: response to selection for disease resistance and association with egg production. Anim. Genet. 28:116–123 [DOI] [PubMed] [Google Scholar]
- 28. Lee C. G., Jenkins N. A., Gilbert D. J., Copeland N. G., O'Brien W. E. 1995. Cloning and analysis of gene regulation of a novel LPS-inducible cDNA. Immunogenetics 41:263–270 [DOI] [PubMed] [Google Scholar]
- 29. Liu H. C., Cheng H. H., Tirunagaru V., Sofer L., Burnside J. 2001. A strategy to identify positional candidate genes conferring Marek's disease resistance by integrating DNA microarrays and genetic mapping. Anim. Genet. 32:351–359 [DOI] [PubMed] [Google Scholar]
- 30. Liu H. C., Kung H. J., Fulton J. E., Morgan R. W., Cheng H. H. 2001. Growth hormone interacts with the Marek's disease virus SORF2 protein and is associated with disease resistance in chicken. Proc. Natl. Acad. Sci. U. S. A. 98:9203–9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu H. C., Niikura M., Fulton J. E., Cheng H. H. 2003. Identification of chicken lymphocyte antigen 6 complex, locus E (LY6E, alias SCA2) as a putative Marek's disease resistance gene via a virus-host protein interaction screen. Cytogenet. Genome Res. 102:304–308 [DOI] [PubMed] [Google Scholar]
- 32. Matys V., et al. 2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34:D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McElroy J. P., et al. 2005. Microsatellite markers associated with resistance to Marek's disease in commercial layer chickens. Poult. Sci. 84:1678–1688 [DOI] [PubMed] [Google Scholar]
- 34. Morimura T., et al. 1996. Apoptosis and CD8-down-regulation in the thymus of chickens infected with Marek's disease virus. Arch. Virol. 141:2243–2249 [DOI] [PubMed] [Google Scholar]
- 35. Morimura T., Hattori M., Ohashi K., Sugimoto C., Onuma M. 1995. Immunomodulation of peripheral T cells in chickens infected with Marek's disease virus: involvement in immunosuppression. J. Gen. Virol. 76:2979–2985 [DOI] [PubMed] [Google Scholar]
- 36. Morrow C., Fehler F. 2004. Marek's disease: a worldwide problem, p. 49–61 In Davison F., Nair V. (ed.), Marek's disease: an evolving problem. Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 37. Nerren J. R., He H., Genovese K., Kogut M. H. 2010. Expression of the avian-specific toll-like receptor 15 in chicken heterophils is mediated by gram-negative and gram-positive bacteria, but not TLR agonists. Vet. Immunol. Immunopathol. 136:151–156 [DOI] [PubMed] [Google Scholar]
- 38. Pazderka F., Longenecker B. M., Law G. R. J., Stone H. A., Ruth R. F. 1975. Histocompatability of chicken populations selected for resistance to Marek's disease. Immunogenetics 2:93–100 [Google Scholar]
- 39. Sallusto F., et al. 1998. Rapid and coordinated switch in chemokine receptor expression during dendritic cell maturation. Eur. J. Immunol. 28:2760–2769 [DOI] [PubMed] [Google Scholar]
- 40. Sarson A. J., Abdul-Careem M. F., Zhou H., Sharif S. 2006. Transcriptional analysis of host responses to Marek's disease viral infection. Viral Immunol. 19:747–758 [DOI] [PubMed] [Google Scholar]
- 41. Sarson A. J., Parvizi P., Lepp D., Quinton M., Sharif S. 2008. Transcriptional analysis of host responses to Marek's disease virus infection in genetically resistant and susceptible chickens. Anim. Genet. 39:232–240 [DOI] [PubMed] [Google Scholar]
- 42. Schat K. A. 2004. Marek's disease immunosuppression, p. 142–155 In Davison F., Nair V. (ed.), Marek's disease: an evolving problem. Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 43. Schat K. A., Xing Z. 2000. Specific and nonspecific immune responses to Marek's disease virus. Dev. Comp. Immunol. 24:201–221 [DOI] [PubMed] [Google Scholar]
- 44. Sharan R., Maron-Katzm A., Shamir R. 2003. CLICK and EXPANDER: a system for clustering and visualizing gene expression data. Bioinformatics 19:1787–1799 [DOI] [PubMed] [Google Scholar]
- 45. Shek W. R., Calnek B. W., Schat K. A., Chen C. H. 1983. Characterization of Marek's disease virus-infected lymphocytes: discrimination between cytolytically and latently infected cells. J. Natl. Cancer Inst. 70:485–491 [PubMed] [Google Scholar]
- 46. Talloen W., et al. 2007. I/NI calls for the exclusion of non-informative genes: a highly effective filtering tool for microarray data. Bioinformatics 23:2897–2902 [DOI] [PubMed] [Google Scholar]
- 47. Theocharidis A., van Dongen S., Enright A. J., Freeman T. C. 2009. Network visualization and analysis of gene expression data using BioLayout Express (3D). Nat. Protoc. 4:1535–1550 [DOI] [PubMed] [Google Scholar]
- 48. Vallejo R. L., et al. 1998. Genetic mapping of quantitative trait loci affecting susceptibility to Marek's disease virus induced tumors in F2 intercross chickens. Genetics 148:349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wales M. M., et al. 1995. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat. Med. 1:570–577 [DOI] [PubMed] [Google Scholar]
- 50. Witter R. L. 1997. Increased virulence of Marek's disease virus field isolates. Avian Dis. 41:149–163 [PubMed] [Google Scholar]
- 51. Wu Z., Hu T., Rothwell L., Young J. R., Kaiser P. 2011. Chicken CCR6 and CCR7 are markers for immature and mature dendritic cells respectively. Dev. Comp. Immunol. 35:563–567 [DOI] [PubMed] [Google Scholar]
- 52. Yonash N., Bacon L. D., Witter R. L., Cheng H. H. 1999. High resolution mapping and identification of new quantitative trait loci (QTL) affecting susceptibility to Marek's disease. Anim. Genet. 30:126–135 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.