Abstract
Persistent infections with human papillomavirus type 16 (HPV16), HPV18, or HPV31 are necessary for the development of cervical cancer, implying that HPVs have evolved immunoevasive mechanisms. Recent global transcriptome analyses indicated that these HPV types downregulate the constitutive expression of interferon (IFN)-stimulated genes (ISGs), but the underlying mechanism is not well understood. Comparative analyses of ISG transcription in keratinocytes with complete HPV16, -18, and -31 genomes revealed that antiviral genes (IFIT1 and MX1), genes involved in IFN signaling (STAT1), proapoptotic genes (TRAIL and XAF1), and pathogen recognition receptors (TLR3, RIG-I, and MDA5) are inhibited to similar extents by HPV16, -18, and -31. The lower expression of pathogen receptors in HPV-positive cells correlated with a greatly impaired induction of IFN-β and also of IFN-λ1, -2, and -3 upon receptor stimulation. IFN-κ is constitutively expressed in normal keratinocytes and is strongly repressed by HPV16, -18, and -31. ISGs downregulated in HPV-positive cells can be reactivated by IFN-κ expression. The viral E6 and E7 oncogenes are sufficient for IFN-κ repression, with E6 being mainly responsible. E6 inhibits IFN-κ transcription independently from binding to PDZ proteins. IFN-κ expression can be activated in only one cell line by E6AP knockdown but can be activated in all tested HPV-positive cells by addition of a DNA methyltransferase inhibitor, suggesting that HPVs modulate DNA methylation. Taken together, these results suggest that carcinogenic HPVs target IFN-κ by different pathways in keratinocytes to inhibit both antiviral ISGs and pathogen recognition receptors, which in turn reduces the expression of inducible IFNs.
INTRODUCTION
Infections with high-risk human papillomavirus (HR-HPV) types such as HPV16, -18, or -31 are a necessary risk factor for the development of invasive cervical cancer (11). A prerequisite for cervical cancer is the establishment of persistent HR-HPV infections, making it likely that HR-HPVs have evolved immunoevasive mechanisms. In support of this idea, genome-wide transcriptome studies of human keratinocyte cell lines harboring HPV18 genomes, HPV31 genomes, or only the E6 and E7 oncogenes of HPV16 have revealed that the expression of interferon (IFN)-stimulated genes (ISGs) is reduced compared to that in HPV-negative keratinocytes (8, 25, 32). This indicated that HR-HPVs interfere with components of the innate immune system.
ISG expression is stimulated when secreted IFN-β or IFN-α binds to the transmembrane IFN-α/β receptor (IFNAR) and activates a signal transduction pathway involving the TYK2 and JAK1 kinases and a transcription factor complex composed of STAT1, STAT2, and IRF9 (5). These IFNs are not present in normal cells but can be strongly induced upon virus infection. Virus components, mainly nucleic acids, are detected by pattern recognition receptors (PRRs), such as transmembrane Toll-like receptors (TLRs), or cytoplasmic receptors, such as RIG-I and MDA5 (6). Cytoplasmic receptors and a subset of TLRs then activate the transcription factors interferon regulatory factor 3 (IRF3) and IRF7, which bind to the IFN-β promoter region and induce IFN-β expression (6). In addition to IFN-α and -β, which can be produced by almost any cell type in the body, tissue-specific interferons that might be involved in HPV pathogenesis have been recently described. The IFN-λ family, which consists of IFN-λ1, -λ2, and -λ3 (also named interleukin-28A, -28B, and -29, respectively), acts predominantly at epithelial surfaces (including keratinocytes), as the IFN-λ receptor displays tissue-specific expression (43). IFN-λs are also induced by pathogen recognition receptors and also stimulate ISG transcription by activating the STAT1/STAT2/IRF9 complex (43).
In addition, keratinocytes express IFN-κ, which has unusual features as it is constitutively expressed at detectable levels in uninfected cells and also appears to act predominantly in an autocrine manner (7, 27). IFN-κ is distantly related to IFN-α and -β and presumably uses the IFNAR to induce ISG transcription (27), but the role of IFN-κ in keratinocytes is poorly understood. Interestingly, it has been recently shown that IFN-κ expression is inhibited in some HPV16-positive cervical cancer cells by promoter methylation, but it is currently unknown whether this plays a role during the normal HPV replication cycle (37).
The reduction of constitutive ISG transcription in keratinocytes by HR-HPVs might be due to the inhibition of IFN induction and/or interference with IFN signaling by HR-HPVs. Consistent with the first model, it has been reported that the HPV16 E6 protein binds with high affinity to IRF3 and that this prevents IFN-β induction upon Sendai virus infection (38). HPV16 also represses TLR9 expression in keratinocytes, but TLR9 induces IFNs only in plasmacytoid dendritic cells and not in other cell types, making it unlikely to contribute to the reduction of ISG expression in HPV16 E6/E7-positive cells (15, 18, 32). However, HPV18 E6 does not bind to IRF3 and HPV18 E6/E7 is not able to reduce TLR9 levels, indicating that HPV18 has evolved different mechanisms to interfere with ISG expression.
HR-HPVs also modulate the IFN signaling cascade in some experimental systems. HPV16 E7 inhibits the IRF1 and IRF9 transcription factors, which contribute to ISG transcription (2, 33, 34). It has also been demonstrated that HPV18 E6 interferes with the TYK2 kinase, which is required for the activation of ISG transcription (30). In the case of HPV31, it has been suggested that the reduced ISG expression is due to the transcriptional inhibition of STAT1 (8). However, these mechanisms are not sufficient to prevent the antiviral activities of IFN-β against HPV16 and -31, as IFN-β treatment inhibits the growth of cell lines with persistently replicating HPV16 or -31 genomes (9, 21). This might be due to the induction of ISG56/IFIT1, which has been shown to inhibit the replication activity of HPV E1 proteins (41).
A caveat in regard to the above-mentioned transcriptome studies is that not only different HPV types but also different microarray platforms were used. Thus, it remained possible that HR-HPVs regulate different subsets of ISGs by virus-type-specific mechanisms. To resolve these issues, we investigated ISG, pathogen recognition receptor, and IFN expression in cell lines harboring HPV16, -18, or -31 genomes.
We find that HR-HPV16, -18, and -31 regulate an overlapping set of ISGs and that this is due to the inhibition of the constitutive expression of IFN-κ. IFN-κ transcription is mainly repressed by the HPV E6 protein and can be activated in HPV-positive cells by either small interfering RNA (siRNA) knockdown of E6AP, which is dependent on the presence of p53, or by addition of a DNA methyltransferase inhibitor, suggesting that HPVs modulate DNA methylation. The two mechanisms appear to be mutually exclusive. IFN-κ not only regulates antiviral-ISG expression and components of the IFN pathway but also induces the expression of the pathogen recognition receptors TLR3, RIG-I, and MDA5, which in turn control inducible IFN-β and -λ expression in keratinocytes. Taken together, our results suggest that HR-HPVs target IFN-κ, which is a master regulator of ISG expression and inducible IFNs in keratinocytes.
MATERIALS AND METHODS
Cell culture.
HPV16-positive keratinocyte cell lines were kindly provided by L. A. Laimins. HPV18- and HPV31-positive cell lines have been previously described (13, 25, 40). CIN612-9E cells were derived from a CIN1 lesion and contain replicating HPV31 genomes (3). Normal human foreskin keratinocytes (NHKs) and HPV-positive keratinocyte cell lines were cocultured with mitomycin C-treated NIH 3T3 J2 fibroblasts in E medium supplemented with 5% fetal bovine serum as described previously (13). To obtain E6/E7-, E6-, or E7-expressing keratinocytes, NHKs were transduced with amphotropic retroviruses and drug selected as previously described (31). HeLa cells were maintained in Dulbecco modified Eagle medium (DMEM) (Invitrogen, Karlsruhe, Germany) supplemented with gentamicin and 10% fetal bovine serum (Thermo Scientific, South Logan, UT) as described previously (13). Cells (2 × 105) were stimulated in 6-well plates by adding 10 μg/ml of poly(I·C) (Sigma-Aldrich, Munich, Germany) dissolved in phosphate-buffered saline (PBS) to the medium and then lysed after 6 h to obtain RNA for quantitative PCR (qPCR) analysis. Cells (2.5 × 105) were treated with 10 μmol/liter 5-aza-2′-deoxycytidine (5-aza-dC) (Alexis Biochemicals, Lörrach, Germany) dissolved in 25% (vol/vol) acetic acid in 6-well plates, and 96 h later cells were lysed to obtain RNA for qPCR analysis.
qPCR.
RNA was isolated using the RNeasy minikit (Qiagen, Hilden, Germany). RNA (200 to 1,000 ng) was reverse transcribed using the QuantiTect reverse transcription kit (Qiagen, Hilden, Germany). Ten to fifty nanograms of cDNA was analyzed in duplicate reactions by qPCR using 0.3 μM gene-specific primers and 1× LightCycler 480 SYBR green I Master (Roche Applied Science, Mannheim, Germany) in a total volume of 20 μl. PCRs were carried out in a Light Cycler 480 (Roche Applied Science, Mannheim, Germany) using a thermal profile of 10 min at 95°C followed by 45 cycles of 10 s at 95°C, 15 s at 55°C, 15 s at 72°C, a melting curve of 10 s at 95°C, 30 s at 60°C, heating to 90°C, and cooling for 30 s at 40°C, and the results were analyzed using the LightCycler 480 software, version 1.5 (Roche Applied Science, Mannheim, Germany). Relative expression levels were calculated as described previously (35) with PGK1 as a reference gene. The following gene-specific primers were designed with Primer 3 plus software (39): PGK1 sense (5′-CTGTGGGGGTATTTGAATGG-3′) and antisense (5′-CTTCCAGGAGCTCCAAACTG-3′), MDA5 sense (5′-ACCAAATACAGGAGCCATGC-3′) and antisense (5′-GCGATTTCCTTCTTTTGCAG-3′), RIG-I sense (5′-CTCCCGGCACAGAAGTGTAT-3′) and antisense (5′-CTTCCTCTGCCTCTGGTTTG-3′), PKR sense (5′-TACGTGTGAGTCCCAAAGCA-3′) and antisense (5′-ATGCCAAACCTCTTGTCCAC-3′), STAT1 sense (5′-CCGTTTTCATGACCTCCTGT-3′) and antisense (5′-GCTCAGTCGGGGAATATT-3′), IFNL1 sense (5′-GAAGCAGTTGCGATTTAGCC-3′) and antisense (5′-GAAGCTCGCTAGCTCCTGTG-3′), IFNL2 sense (5′-CACCCTGCACCATATCCTCT-3′) and antisense (5′-GGAGGGTCAGACACACAGGT-3′), IFNL3 sense (5′-CTGCTGAAGGACTGCAAGTG-3′) and antisense (5′-GAGGATATGGTGCAGGGTGT-3′), TNFSF10 (TRAIL) sense (5′-TTCACAGTGCTCCTGCAGTC-3′) and antisense (5′-ACGGAGTTGCCACTTGACTT-3′), and XAF1 sense (5′-GCCTGCAAGAAACGAAACTC-3′) and antisense (5′-CTTCTGCATGCTCTGCTGAC-3′). Primer pairs for TLR3 (QT00007714), IFIT1 (QT00201012), MX1 (QT00090895), IFNB (QT00203763), IFNK (QT00197512), and CCL20 (QT00012971) were commercially obtained (Qiagen, Hilden, Germany). For the absolute quantification of IFN levels, total cellular DNA was isolated from 16-1 cells by standard methods (40), and standard curves using 100 to 0.001 ng of DNA were generated for IFN-β, IFN-κ, IFN-λ1, and PGK1. Using these standard curves and LightCycler 480 software version 1.5, arbitrary copy numbers for IFN-β, IFN-κ, IFN-λ1, and PGK1 present in 25 ng of cDNA derived from NHKs were obtained. RNA copy numbers were then corrected for the corresponding PGK1 copy number.
siRNA treatment.
Cells (2.5 × 105) were transfected with 25 nmol/liter control siRNA (AllStars negative control; Qiagen no. 1027281), siE6AP (human UBE3A [E6AP]; Dharmacon L-005137-00), or sip53 (tumor protein 53 [TP53]; Applied Biosystems/Ambion, S605) in 6-well plates using HiPerfect reagent (Qiagen, Hilden, Germany). At 72 h posttransfection, cells were lysed to obtain whole-cell extracts for immunoblotting or RNA for qPCR analysis.
Transient-transfection experiments.
HeLa or 18-1 cells (3 × 105) were transfected with 1.5 μg of plasmid DNA using FuGENE HD (Promega, Madison, WI). At 72 h posttransfection, cells were lysed to obtain whole-cell extracts for immunoblotting or RNA for qPCR analysis.
Immunoblotting.
Whole-cell extracts were prepared by lysing cell pellets in 30 μl of 4× SDS gel loading buffer (Carl Roth, Karlsruhe, Germany) and then heating to 95°C for 10 min. After brief sonication, extracts were separated by 12% SDS-PAGE and transferred to nitrocellulose membranes. Blocked membranes were incubated with diluted primary antibodies (IFN-κ, Abnova H00056832-MO1, 1:500; α-tubulin, Calbiochem CP06, 1:2,500; E6AP [H-182], Santa Cruz sc-25509, 1:1,000; p53 [DO-I], Santa Cruz sc-126, 1:1,000; and HSP90 [4F10], Santa Cruz sc-69703, 1:2,000) overnight. Bound antibodies were detected with secondary antibodies conjugated to horseradish peroxidase and SuperSignal West Dura reagent (Perbio Science, Bonn, Germany). Chemiluminescent signals were recorded with a FluorSMax Imaging system (Bio-Rad, Munich, Germany).
Recombinant plasmids.
The IFN-κ-coding sequence was amplified by PCR using cDNA from normal keratinocytes. After addition of BglII restriction sites by PCR, the amplicon was cloned into the BglII site of the pSG5 expression vector (Stratagene, Heidelberg, Germany). Retroviral transfer plasmids were constructed as follows. Plasmid pLXSN 31E6/E7 was constructed by inserting the PCR-amplified HPV31 E6/E7-coding sequence between the EcoRI and BamHI sites of pLXSN (Clontech). Plasmid pbabe-neo 18E6/E7 was constructed by inserting the PCR-amplified HPV18 E6/E7-coding sequence into the BamHI sites of pbabe-neo. Plasmid pLXSN 18E6 was constructed by inserting the PCR-amplified HPV18 E6-coding sequence between the EcoRI and XhoI sites of pLXSN. In plasmid pLSXN 18E6 PDZmt, the wild-type PDZ binding domain (ETVQ; amino acids [aa] 155 to 158) was replaced with AAAA by site-directed mutagenesis. In plasmid pLXSN 18E6 fl, the splice donor sequence AGGT (nucleotides [nt] 233 to 236) was mutated to AAGT by site-directed mutagenesis. The HPV18 E7-coding sequence was amplified by PCR and inserted between the XhoI and EcoRI sites of pMSCV-puro (Clontech) to generate plasmid pMSCV-puro 18E7. All cloned inserts were verified by DNA sequencing (GATC, Konstanz, Germany). Plasmid pLXSN 16E6/E7 has been described previously (17). Plasmid pMalc2x-18E6 has been described previously (31). Its derivative pMalc2x-18E6 PDZ mt was generated by PCR using pLSXN 18E6 PDZmt as a template.
MBP pulldown assays.
Expression, purification, and interaction assays using maltose binding protein (MBP) or MBP-18E6 fusion proteins were carried out as previously described (31). Briefly, affinity-purified MBP, MBP-18E6, or MBP-18E6PDZmt protein was incubated with whole-cell extract derived from 3 × 107 C33a cells lysed in 130 mM NaCl, 50 mM Tris (pH 8.0), 1 mM EDTA, 1% (vol/vol) Igepal CA-630, 1 mM dithiothreitol (DTT). and protease inhibitors for 2 h at 4°C. After extensive washing, the retained proteins were eluted with 50 mM Tris (pH 8.0), 12.5 mM MgCl2, 0.1% (vol/vol) Igepal CA-630, 0.1% DTT, 20 mM maltose, and protease inhibitors and analyzed by immunoblotting using anti-MBP conjugated to horseradish peroxidase (HRP) (NEB, E8038), anti-hDlg (BD Bioscience, no. 610874), anti-E6AP, or anti-scribble (Abcam, ab6049).
RESULTS
ISG and pathogen receptor expression is inhibited by HR-HPVs.
To investigate whether the HR-HPV16, -18, and -31 regulate similar sets of ISGs, two human keratinocyte cell lines each harboring HPV16, HPV18, or HPV31 genomes (16-1, 16-2, 18-1, 18-2, 31-1, and 31-2) were used to compare the expression levels of selected genes with that in normal human keratinocytes from several different donors (4, 13, 25, 40). Based on the published transcriptome studies and functional data for HR-HPVs, the expression levels of the ISGs IFIT1, MX1, PKR, and STAT1 were investigated (8, 19, 25, 32, 41). Furthermore, we included TNFSF10/TRAIL and XAF1, as their expression was reduced in HPV18 transcriptome studies and both genes can be induced by IFN-β and are involved in the regulation of apoptosis (25, 29). In addition, the expression of the pathogen receptors TLR3, RIG-I, and MDA5 was analyzed, as their expression was reduced by HPV18 in transcriptome studies and they are involved in the induction of IFNs (20, 24, 25, 45). The relative expression of these genes was determined by quantitative reverse transcription-PCR (qRT-PCR) using PGK1 as a reference gene. This revealed that the expression of IFIT1, MX1, and STAT1 was downregulated in all HR-HPV-positive cell lines to slightly varying extents (Table 1). Similarly, the expression of TRAIL and XAF1 was reduced in all HR-HPV-positive cell lines (Table 1). PKR gene expression was reduced in all cell lines with the exception of the 18-2 cell line (Table 1). Also, the expression of the pathogen receptors TLR3, RIG-I, and MDA5 was consistently reduced in all HR-HPV-positive cell lines (Table 1). This indicated that all HR-HPV-positive cells display reduced levels of different ISGs and certain pathogen receptors, and no obvious differences between the three different HPV types were apparent.
Table 1.
Differential expression of ISGs in HPV-positive cells and normal keratinocytes
| Gene | Relative expression level (mean ± SEM)a in cell line: |
|||||
|---|---|---|---|---|---|---|
| 16-1 | 16-2 | 18-1 | 18-2 | 31-1 | 31-2 | |
| IFIT1 | 0.25 ± 0.06 | 0.25 ± 0.05 | 0.11 ± 0.03 | 0.46 ± 0.13 | 0.25 ± 0.13 | 0.38 ± 0.08 |
| MX1 | 0.36 ± 0.10 | 0.46 ± 0.14 | 0.15 ± 0.06 | 0.45 ± 0.10 | 0.52 ± 0.10 | 0.74 ± 0.15 |
| STAT1 | 0.54 ± 0.07 | 0.37 ± 0.10 | 0.23 ± 0.06 | 0.50 ± 0.13 | 0.35 ± 0.10 | 0.45 ± 0.07 |
| TLR3 | 0.18 ± 0.03 | 0.27 ± 0.06 | 0.30 ± 0.07 | 0.47 ± 0.10 | 0.28 ± 0.07 | 0.51 ± 0.12 |
| RIG-I | 0.32 ± 0.04 | 0.46 ± 0.07 | 0.24 ± 0.04 | 0.50 ± 0.11 | 0.18 ± 0.03 | 0.67 ± 0.11 |
| MDA5 | 0.38 ± 0.11 | 0.38 ± 0.05 | 0.29 ± 0.05 | 0.64 ± 0.12 | 0.18 ± 0.03 | 0.69 ± 0.09 |
| PKR | 0.55 ± 0.05 | 0.65 ± 0.17 | 0.32 ± 0.06 | 1.19 ± 0.13 | 0.39 ± 0.11 | 0.46 ± 0.13 |
| TRAIL | 0.44 ± 0.15 | 0.35 ± 0.20 | 0.18 ± 0.06 | 0.16 ± 0.08 | 0.13 ± 0.02 | 0.33 ± 0.14 |
| XAF1 | 0.14 ± 0.09 | 0.36 ± 0.13 | 0.08 ± 0.03 | 0.29 ± 0.25 | 0.12 ± 0.07 | 0.51 ± 0.26 |
RNA expression levels of the different genes were determined by qPCR using PGK1 as a reference gene and are presented relative to the levels in NHK cells, which were set to 1.
Reduced pathogen receptor levels correlate with a lower induction of IFNs in HR-HPV-positive cells.
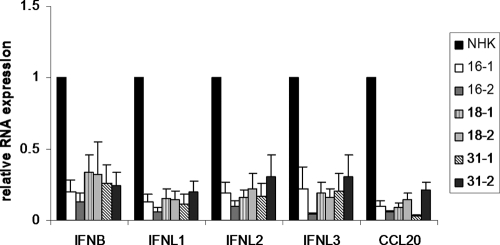
To test whether the repression of pathogen receptor expression has functional consequences for IFN expression, the different HPV-positive cell lines or normal keratinocytes were incubated with 10 μg/ml poly(I·C), which activates the TLR3, RIG-I, and MDA5 pathways in human keratinocytes (23). After 6 h, RNA was isolated and analyzed by qRT-PCR for the expression of IFN-β, IFN-λ1, IFN-λ2, IFN-λ3, and CCL20, an inflammatory cytokine. In normal keratinocytes poly(I·C) induced the expression of IFN-β 1,500-fold, that of IFN-λ1 3,000-fold, that of IFN-λ2 40-fold, that of IFN-λ3 22-fold, and that of CCL20 110-fold. In contrast, the induction of all analyzed cytokines was greatly impaired in HR-HPV-positive cell lines to similar extents (Fig. 1). IFN-β induction was reduced 5- and 7-fold by HPV16, 3-fold by HPV18, and 4-fold by HPV31. IFN-λ1 induction was reduced 8- and 16-fold by HPV16, 7-fold by HPV18, and 5- and 9-fold by HPV31. IFN-λ2 induction was inhibited 5- and 10-fold by HPV16, 5-fold by HPV18, and 3- and 6-fold by HPV31. The inhibition of IFN-λ3 was similar to that of IFN-λ2. CCL20 induction was impaired 10- and 16-fold by HPV16, 7- and 11-fold by HPV18, and 5- and 30-fold by HPV31 (Fig. 1). This suggested that the decrease in pathogen receptor expression results in a strongly reduced expression of different inducible IFNs and inflammatory cytokines.
Fig. 1.
Induction of IFN-β, IFN-λ1, IFN-λ2, IFN-λ3, and CCL20 by poly(I·C) is inhibited in HR-HPV-positive cells. Keratinocytes containing HPV16, -18, or -31 genomes or normal human keratinocytes (NHKs) were treated with poly(I·C) (10 μg/ml) for 6 h. RNA was analyzed by qRT-PCR for IFN-β, IFN-λ1, IFN-λ2, IFN-λ3, and CCL20 expression. Relative expression values were calculated using expression values for NHKs as a reference, which were set to 1, and PGK1 as a reference gene. The data represent the averages from at least four independent experiments, and error bars indicate the standard errors of the means.
IFN-κ expression is inhibited by HR-HPVs.
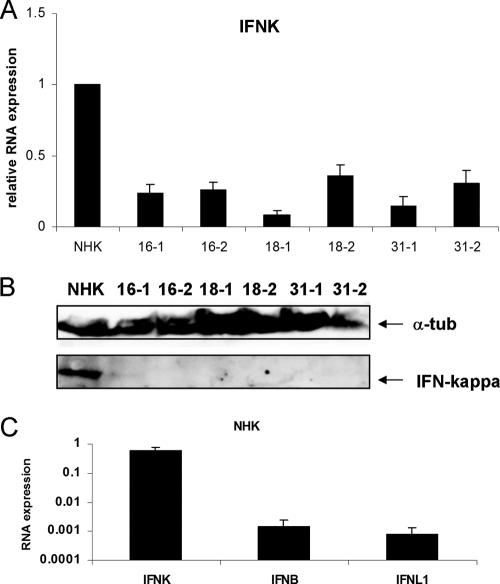
The expression levels of IFN-κ were also analyzed, as it is constitutively expressed in human keratinocytes and was suggested to be regulated by HPV16 in cancer cells (27, 37). In contrast to IFN-β or IFN-λ, IFN-κ was only very weakly induced by poly(I·C) treatment (∼1.5-fold) in normal keratinocytes. When comparing the basal mRNA levels of IFN-κ in normal keratinocytes with HR-HPV-positive cells, a 3- to 5-fold reduction in all HR-HPV-positive cell lines was observed (Fig. 2A). Consistent with this, IFN-κ protein levels determined by immunoblotting were also lower in HPV-positive cells than in NHKs (Fig. 2B). In order to compare the basal expression levels of the different interferons, qPCR analyses were carried out. A single preparation of total cellular DNA from 16-1 cells was used to generate standard curves for the IFN-κ, IFN-β, IFN-λ1, and PGK1 primers to allow for absolute quantification and thus a direct comparison of the different IFN expression levels. Using these standard curves, IFN-κ, IFN-β, and IFN-λ1 mRNA levels were determined from unstimulated NHK cells derived from five different donors, using PGK1 as a reference gene. This analysis revealed that basal IFN-β and IFN-λ1 levels are very similar in NHK cells (Fig. 2C). In contrast, IFN-κ RNA levels were 408-fold and 725-fold higher than IFN-β and IFN-λ1 levels, respectively (Fig. 2C). This strongly suggested that IFN-κ is the predominant interferon present in unstimulated keratinocytes.
Fig. 2.
Reduced IFN-κ levels in HPV-positive cells. (A) RNA from keratinocytes containing HPV16, -18, or -31 genomes or normal human keratinocytes (NHKs) was analyzed by qRT-PCR for IFN-κ expression. Relative expression values were calculated using expression values of NHK as a reference, which were set to 1, and PGK1 as a reference gene. The data represent the averages from at least four independent experiments, and error bars indicate the standard errors of the means. (B) Western blot analysis of IFN-κ expression. Whole-cell lysates were prepared and analyzed by immunoblotting for the presence of IFN-κ and α-tubulin as a loading control. (C) Comparison of IFN-κ, IFN-β, and IFN-λ1 RNA levels in NHK cells. Standard curves were generated for IFN-β, IFN-κ, IFN-λ1, and PGK1 using total DNA isolated from 16-1 cells and then used to determine copy numbers for IFN-β, IFN-κ, IFN-λ1, and PGK1 present in 25 ng of cDNA derived from NHKs. RNA copy numbers were then corrected for the corresponding PGK1 value. The data represent the averages of measurements of five independently isolated RNAs from NHK cells, and error bars indicate the standard errors of the means.
IFN-κ regulates ISG and pathogen receptor expression in HPV-positive cells.
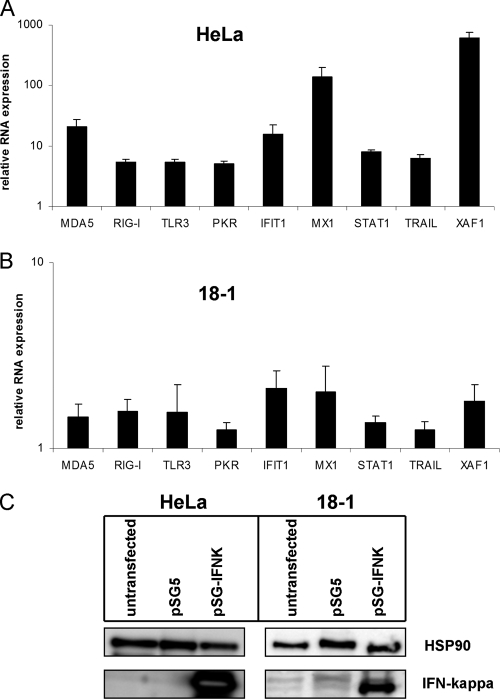
As the pathogen receptors TLR3, RIG-I, and MDA5 have been described to be IFN inducible in some cell systems (20, 24, 45), we hypothesized that the reduced IFIT1, MX1, PKR, STAT1, TRAIL, XAF1, and pathogen receptor levels in HR-HPV-positive cells are due to reduced IFN-κ levels. To test whether IFN-κ can induce the transcription of the respective genes, we cloned the human IFN-κ cDNA from normal keratinocytes into the eukaryotic expression vector pSG5 and performed transfection experiments with HPV18-positive HeLa cells, which have 10,000-fold less IFN-κ RNA than normal keratinocytes. Cells were harvested at 72 h posttransfection. Protein extracts were analyzed by immunoblotting for the expression of IFN-κ (Fig. 3C), and RNA was analyzed by qPCR for the expression of IFIT1, MDA5, MX1, RIG-I, STAT1, TLR3, TRAIL, and XAF1 transcripts (Fig. 3A and B). This revealed that IFIT1 was induced 15-fold, MX1 135-fold, STAT1 8-fold, TRAIL 6-fold, and XAF1 600-fold (Fig. 3A). Furthermore, MDA5 was induced 20-fold, RIG-I 5-fold, and TLR3 5-fold (Fig. 3A). To investigate whether the induction of ISGs and PRRs might be caused by an effect of IFN-κ expression on cell cycle progression, we analyzed the DNA content of transfected HeLa cells after IFN-κ expression by flow cytometry (see Fig. S1 in the supplemental material). No changes in the cell cycle profile or differences in the numbers of apoptotic cells were observed, suggesting that ISGs are directly activated by IFN-κ.
Fig. 3.
Overexpression of IFN-κ in HPV18-positive cells increases PRR and ISG transcription. HeLa (A and C) or 18-1 (B and C) cells were transfected with 1.5 μg of empty vector pSG5 or the expression vector for IFN-κ (pSG-IFNK). (A and B) PRR and ISG transcript levels were analyzed by qRT-PCR using PGK1 as a reference gene and are presented relative to those in pSG5-transfected cells. The data represent the averages from at least three independent experiments, and error bars indicate the standard errors of the means. (C) Western blot analysis of IFN-κ expression in HeLa and 18-1 cells. Whole-cell lysates were prepared and analyzed by immunoblotting for the presence of IFN-κ and HSP90 as a loading control.
To confirm that IFN-κ also regulated these genes in cells with complete HPV genomes, transfection experiments were carried out with the 18-1 cell line, which displayed low IFN-κ levels (Fig. 2). Consistent with the data obtained with HeLa cells, all investigated genes were induced by IFN-κ (Fig. 3B). However, the extent of induction (1.5- to 2-fold) was much lower than in HeLa cells. This is most likely due to the reduced transfection efficiency of 18-1 compared to HeLa cells, which is reflected by the lower IFN-κ protein amounts upon transfection (Fig. 3C), and possibly also due to the higher levels of endogenous IFN-κ in 18-1 cells. In summary, these data are consistent with IFN-κ being responsible for ISG and pathogen receptor expression in HPV-positive keratinocytes.
IFN-κ expression is inhibited by the E6 protein.
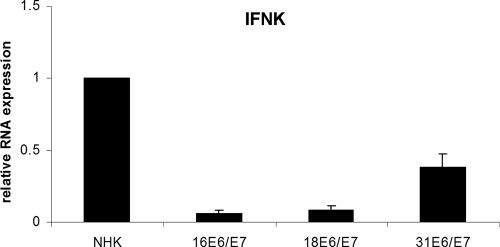
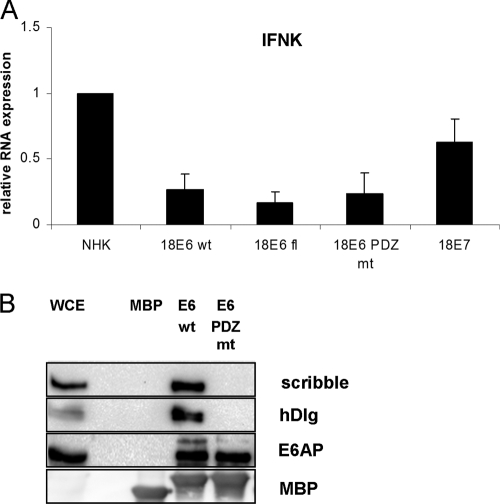
As ISG levels are reduced in HPV16 E6/E7 cells and the inhibition of IFN-κ in cervical cancer cells has been linked to the expression of HPV16 E6 (32, 37), we tested whether HR-HPV E6/E7 expression is sufficient to inhibit IFN-κ expression. Using cells from two donors, human keratinocyte lines that express HPV16 E6/E7, HPV18 E6/E7, and HPV31 E6/E7 from retroviral vectors were established and analyzed for IFN-κ expression. As can be seen in Fig. 4, IFN-κ expression is reduced ∼10-fold by HPV16 E6/E7, 7-fold by HPV18 E6/E7, and ∼2-fold by HPV31 E6/E7. This suggested that the expression of the HR-HPV oncogenes is sufficient to reduce IFN-κ expression in keratinocytes. To evaluate the individual contributions of the E6 and E7 oncoproteins, we generated keratinocytes that express only HPV18 E6 or E7, as the HPV16 E6/E7 proteins have been shown to have additional activities compared to HPV18 E6/E7 that interfere with innate immunity pathways (18, 38). HPV18 E6 and E7 were introduced by retroviral delivery into normal human keratinocytes from different donors, and upon drug selection RNA was isolated to analyze IFN-κ expression. Both E6 and E7 reduced IFN-κ levels compared to those in normal keratinocytes (Fig. 5A). However, E6 reduced IFN-κ levels to 27%, whereas E7 inhibited levels to only 62%. This suggested that IFN-κ levels are regulated mainly by E6. To gain further insight into the molecular mechanism, we constructed an E6 mutant (E6fl) which can produce only full-length E6 and not the short E6* due to the mutation of the internal splice donor site and an E6 mutant (E6 PDZ mt) in which the C-terminal binding domain for cellular PDZ proteins is mutated. Pulldown experiments using recombinant E6 proteins from Escherichia coli confirmed that the PDZ binding domain mutation specifically abolishes the interaction of E6 with PDZ proteins hDlg and scribble but not that with E6AP (Fig. 5B). Inhibition of IFN-κ expression was further reduced to 16% when only the full-length form of E6 was expressed but was unchanged by the inactivation of the PDZ domain (Fig. 5A). This strongly suggests that the full-length form of E6 is responsible for IFN-κ inhibition and that PDZ binding partners of HPV18 E6 do not contribute to inhibition.
Fig. 4.
Reduced IFN-κ levels in keratinocytes expressing HPV16, -18, or -31 E6/E7 proteins. RNA from keratinocytes containing HPV16, -18, or -31 E6/E7 genes or normal human keratinocytes (NHKs) was analyzed by qRT-PCR for IFN-κ expression. Relative expression values were calculated using expression values of NHK as a reference, which were set to 1, and PGK1 as a reference gene. The data represent the averages from four independent experiments, and error bars indicate the standard errors of the means.
Fig. 5.
HPV18E6 prevents IFN-κ expression. (A) RNA from keratinocytes expressing HPV18 E7, E6, or E6 mutant genes E6fl and E6 PDZmt or from normal human keratinocytes (NHKs) was analyzed by qRT-PCR for IFN-κ expression. Relative expression values were calculated using expression values of NHK as a reference, which were set to 1, and PGK1 as a reference gene. The data represent the averages from three independent experiments, and error bars indicate the standard errors of the means. (B) Mutation of the 18E6 PDZ domain inhibits binding to scribble and hDlg proteins. MBP alone or MBP-HPV18E6 or -HPV18E6 PDZ mt fusion proteins were purified from E. coli and incubated with whole-cell extracts (WCE) from C33a cells as described previously (31). Bound proteins were eluted and analyzed by immunoblotting for scribble, hDlg, E6AP, or MBP.
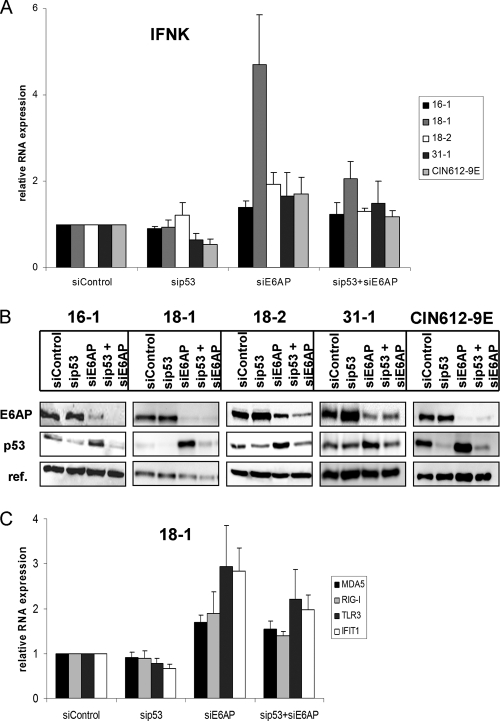
It has been suggested that all transcriptional effects of E6 in cervical cancer cells are due to the recruitment of the E3 ubiquitin ligase E6AP (26). We therefore tested the effect of an E6AP knockdown by siRNA on IFN-κ transcription in several HPV-positive cell lines. In all cell lines, E6AP protein levels were reduced by siE6AP and there was an increase in p53 protein levels, indicating that the reduction of E6AP levels was sufficient to impede E6 activity (Fig. 6B). In the three cell lines 16-1, 18-2, and 31-1, very moderate increases (1.4- to 1.9-fold) of IFN-κ transcripts were observed (Fig. 6A). A comparable 1.7-fold induction of IFN-κ by siE6AP was obtained with the HPV31-positive CIN612-9E cell line, which is derived from a CIN1 lesion and displays even lower levels of IFN-κ transcripts than the HPV-positive keratinocytes (Fig. 6). In contrast, a highly reproducible 4.7-fold increase of IFN-κ transcripts was observed upon E6AP knockdown in the 18-1 cell line (Fig. 6). As recruitment of E6AP by 18E6 results in the degradation of p53 (22), the influence of siRNA-mediated knockdown of p53 on IFN-κ transcription was investigated. As can be seen in Fig. 6A, the knockdown of p53 in the presence of E6AP had a weak effect (<2-fold) on IFN-κ transcription in HPV31-positive cell lines but not in other cell lines. The combination of siE6AP and sip53 reduced IFN-κ transcript levels from 4.7-fold to 2-fold in 18-1 cells, whereas in the other cell lines only very moderate effects were observed (Fig. 6A). This suggests that in the 18-1 cell line, IFN-κ levels are controlled by E6AP and p53. To further investigate whether the reactivation of IFN-κ transcription by E6AP knockdown in 18-1 cells results in the induction of ISG and PRR transcription, qPCR analyses were carried out (Fig. 6C). This revealed that TLR3 and IFIT1 transcription is induced ∼3-fold and Mda5 and RIG-I transcription ∼2-fold by E6AP knockdown (Fig. 6C). This suggests that E6AP-mediated inhibition of IFN-κ transcription contributes to the inhibition of ISG and PRR transcription in 18-1 cells.
Fig. 6.
Influence of E6AP and p53 knockdown of IFN-κ expression in HPV16-, HPV18-, and HPV31-positive cells. Cell lines with HPV16 (16-1), HPV18 (18-1 and 18-2), or HPV31 (31-1 and CIN612-9E) genomes were transfected with control siRNA (siControl), sip53, siE6AP, or a combination of sip53 and siE6AP and were harvested at 72 h posttransfection. (A) RNA was analyzed by qRT-PCR for IFN-κ expression using PGK1 as a reference gene. Results are presented relative to control siRNA transfected cells. The data represent the averages from four independent experiments, and error bars indicate the standard errors of the means. (B) Whole-cell lysates were analyzed by immunoblotting for E6AP, p53, and a reference protein (18-1 and CIN612-9E, HSP90; 16-1, 18-2, and 31-1, α-tubulin) as a loading control. (C) RNA obtained from 18-1 cells after siRNA treatment was analyzed by qRT-PCR for MDA5, RIG-I, TLR3, and IFIT1 expression as described above. The data represent the averages from four independent experiments, and error bars indicate the standard errors of the means.
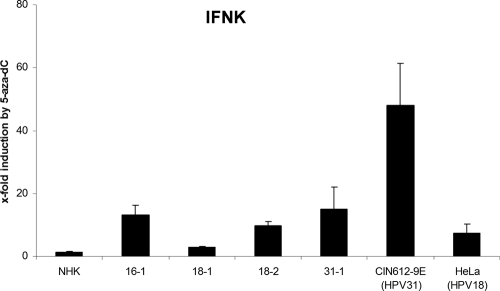
To address possible differences between 18-1 cells and the other HPV-positive cell lines, we investigated the effects of a DNA methyltransferase inhibitor, as it had been suggested that IFN-κ transcription is controlled in HPV-positive cervical cancer cell lines by methylation of the IFN-κ promoter (37). In addition to 16-1, 18-1, 18-2, 31-1, and CIN612-9E cells, normal human keratinocytes and HeLa cells as controls were incubated with a solvent control or the DNA methyltransferase inhibitor 5-aza-dC, and IFN-κ transcript levels were determined (Fig. 7). No change was observed in normal keratinocytes. In contrast, in all HPV-positive cell lines IFN-κ transcription could be induced, but to different extents (Fig. 7). In 16-1, 18-2, 31-1, and HeLa cells, IFN-κ could be induced 9- to 13-fold (Fig. 7). In CIN612-9E cells, IFN-κ transcription was activated 47-fold. Interestingly, in 18-1 cells, IFN-κ could be increased only 2.5-fold by 5-aza-dC. This suggested that IFN-κ transcription is regulated in HPV-positive cells by DNA methylation.
Fig. 7.
5-Aza-dC activates IFN-κ expression in HPV-positive cells. HPV-positive cell lines 16-1, 18-1, 18-2, 31-1, CIN612-9E, HeLa, and NHK were treated with 10 μM 5-aza-dC or vehicle, and 96 h later RNA was isolated and analyzed by qRT-PCR for IFN-κ expression and PGK1 as a reference gene. Data are presented relative to vehicle-treated cells. Values represent the averages from three independent experiments, and error bars indicate the standard errors of the means.
DISCUSSION
Genome-wide transcriptome studies of HR-HPV-positive cells indicated that the inhibition of interferon-stimulated genes is common to HPV16, -18, and -31 (8, 25, 32). Our data demonstrate that the different HPV types inhibit the same set of ISGs via the inhibition of the constitutive transcription of IFN-κ.
Among these ISGs are not only genes with demonstrated antiviral activity (IFIT1 and MX1) and genes involved in IFN-signaling (STAT1) but also proapoptotic genes (TRAIL and XAF1) and pathogen recognition receptors (TLR3, RIG-I, and MDA5). The reduction of pathogen recognition receptors levels correlated with decreased IFN-β, IFN-λ, and CCL20 transcription upon stimulation, suggesting that the reduction of pathogen recognition receptors further ensures that expression of inducible IFNs is minimized in order to prevent expression of antiviral ISGs such as IFIT1. A reduction of IFN-β induction upon virus infection in human keratinocytes by HPV16 E6 has been recently linked to the interaction of E6 with IRF3 (38). However, IRF3 interacted strongly only with HPV16 E6 and not with HPV18 E6, indicating that the impaired IFN induction in HPV18-positive cells is a consequence of reduced PRR levels (38). Conflicting results on whether retroviral expression of HPV16 E6 and E7 affects CCL20 induction by poly(I·C) in human keratinocytes have been reported (16, 18). Using cell lines established by immortalization with complete HPV genomes, we observe a strongly reduced CCL20 expression. This may indicate that CCL20 expression is prevented both by reducing PRR levels and by interference at the transcriptional level by HPV.
It is currently unknown whether PRR molecules are activated by HPV in infected keratinocytes. It has been demonstrated that viral DNA in the cytoplasm can be recognized by RNA polymerase III and transcribed into immunostimulatory RNA which then activates RIG-I to trigger IFN transcription (1, 10). Thus, if not all HPV genomes are bound to mitotic chromosomes during mitosis, these genomes might be recognized via RIG-I after cell division. TLR3 has been implicated in the recognition of herpesviruses (36) and therefore might also be a sensor of HPV. Interestingly, TLR3 also detects cellular RNA from injured cells in order to induce normal inflammation after injury (28). Thus, downregulation of TLR3 by HPV might prevent antiviral-cytokine production induced by wounding.
Our analyses revealed that the constitutive transcription of IFN-κ is inhibited in HPV-positive keratinocytes, which is consistent with recent in situ analyses of HPV-positive biopsy specimen material and findings in HPV-positive cervical cancer cells (12, 37). Since a good correlation between ISGs reduced in HPV-positive cells (Table 1) and ISGs induced by IFN-κ in HPV18-positive cells can be observed, we postulate that the major cause for the reduction of ISGs in HPV-positive cells is the inhibition of IFN-κ transcription. The antiviral activity of IFN-β against HPV has been linked to the induction of IFIT1/ISG56, which interferes with the replication activity of HPV E1 (41). IFIT1 expression is strongly reduced in all HPV-positive cells and can be ∼10-fold induced by IFN-κ, suggesting that IFN-κ serves to maintain high IFIT1 levels in keratinocytes.
Using retroviral expression, we have demonstrated that the E6/E7 genes of HPV16, -18, or -31 are sufficient to inhibit IFN-κ transcription. The use of single expression constructs for HPV18 E6 and E7 identified HPV18 E6 as the major inhibitor of IFN-κ transcription, and this is most likely conserved among HR-HPVs, as HPV16 E6 shows similar effects (37). To understand the molecular mechanism of E6-mediated IFN-κ inhibition, mutations in the E6 gene were introduced. This suggested that IFN-κ inhibition requires neither the short form of E6, E6*, nor binding of HPV18 E6 to PDZ proteins. It had been suggested that transcriptional changes by E6 are due to the recruitment of E6AP by E6 (14, 26). RNA interference (RNAi)-mediated knockdown of E6AP failed in the majority of tested cell lines (4 out of 5) to upregulate IFN-κ transcription strongly (<1.9-fold). Only in one cell line (18-1) was an increase (4.7-fold) of IFN-κ transcription observed (Fig. 6). Interestingly, this increase was dependent on p53, suggesting that IFN-κ is regulated by p53 in the 18-1 cell line. However, this is not a specific feature of HPV18, as 18-2 cells behaved similarly to the HPV16- and HPV31-positive cell lines. These differences appear not to be due to differences in the efficacy of the siRNAs, as in all cell lines similar reductions of E6AP and increasing p53 protein levels were observed. It has been recently shown that IFN-κ transcription is regulated in some HPV-positive cervical cancer cells by DNA methylation of the IFN-κ promoter (37). The comparison of HeLa cervical cancer cells with CIN612-9E cells, derived from a cervical intraepithelial neoplasia 1 lesion, revealed 1,000-fold lower IFN-κ transcript levels in HeLa cells, which could be activated only 9-fold by DNA methyltransferase inhibitors. This suggests that DNA methylation contributes to the reduced IFN-κ transcription in HeLa cervical cancer cells but that additional factors are affected that are required for a high level of IFN-κ transcription in the absence of DNA methylation. The inhibition of DNA methyltransferases by 5-aza-dC resulted in an induction of IFN-κ transcription in all cell lines with complete HPV genomes. This was specific for the presence of HPV, as normal keratinocytes did not respond to 5-aza-dC treatment. Interestingly, an inverse correlation was found between the response to E6AP knockdown and 5-aza-dC treatment. IFN-κ was strongly induced by DNA methylation inhibitors only in cell lines in which the knockdown of E6AP had very weak effects, whereas in 18-1 cells 5-aza-dC led to only a 2.5-fold induction of IFN-κ transcripts. Future studies are required to determine how HR-HPVs induce DNA methylation through their E6 proteins.
The reduction of IFN-κ has been observed in malignant cervical cancer cells (37). Our results demonstrate that IFN-κ expression is already reduced in cell lines with persistently replicating HPV genomes and thus does not require malignant transformation of HPV-infected cells or overexpression of E6 from retroviral vectors (37). Consistent with a role early after infection, IFN-κ activates IFIT1, which has been demonstrated to inhibit HPV E1 replication proteins (41). Cell lines such as CIN612-9E or W12, which maintain replicating HPV genomes, are far more sensitive to IFN-β than cells with integrated HPV genomes or cervical cancer cells (9, 21). Thus, the constitutive IFN-κ expression in keratinocytes might pose a major hurdle for HPV genome replication which needs to be counteracted to achieve establishment of a productive infection. Interestingly, E6 has been shown to be involved in viral genome replication. HPV31 E6 knockout genomes fail to be maintained as extrachromosomal elements, and HPV18 E6 knockout genomes display a reduced amplification of genomes upon differentiation (42, 44). Future studies are required to determine whether a lack of interference of E6 with IFN-κ expression contributes to these phenotypes.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Galloway for providing the pLXSN16E6/E7 plasmid and L. A. Laimins for providing cell lines.
This work was supported by a grant from the Wilhelm-Sander-Stiftung (2011.007.1) to F.S.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 17 August 2011.
REFERENCES
- 1. Ablasser A., et al. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 10:1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Antonsson A., Payne E., Hengst K., McMillan N. A. 2006. The human papillomavirus type 16 E7 protein binds human interferon regulatory factor-9 via a novel PEST domain required for transformation. J. Interferon Cytokine Res. 26:455–461 [DOI] [PubMed] [Google Scholar]
- 3. Bedell M. A., et al. 1991. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J. Virol. 65:2254–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bodily J. M., Mehta K. P., Laimins L. A. 2011. Human papillomavirus E7 enhances hypoxia-inducible factor 1-mediated transcription by inhibiting binding of histone deacetylases. Cancer Res. 71:1187–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borden E. C., et al. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6:975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bowie A. G., Unterholzner L. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 8:911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buontempo P. J., et al. 2006. Antiviral activity of transiently expressed IFN-kappa is cell-associated. J. Interferon Cytokine Res. 26:40–52 [DOI] [PubMed] [Google Scholar]
- 8. Chang Y. E., Laimins L. A. 2000. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J. Virol. 74:4174–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang Y. E., Pena L., Sen G. C., Park J. K., Laimins L. A. 2002. Long-term effect of interferon on keratinocytes that maintain human papillomavirus type 31. J. Virol. 76:8864–8874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiu Y. H., Macmillan J. B., Chen Z. J. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cogliano V., et al. 2005. Carcinogenicity of human papillomaviruses. Lancet Oncol. 6:204. [DOI] [PubMed] [Google Scholar]
- 12. DeCarlo C. A., et al. 2010. IFN-kappa, a novel type I IFN, is undetectable in HPV-positive human cervical keratinocytes. Lab Invest. 90:1482–1491 [DOI] [PubMed] [Google Scholar]
- 13. Frattini M. G., Lim H. B., Laimins L. A. 1996. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. U. S. A. 93:3062–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gewin L., Myers H., Kiyono T., Galloway D. A. 2004. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 18:2269–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gilliet M., Cao W., Liu Y. J. 2008. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 8:594–606 [DOI] [PubMed] [Google Scholar]
- 16. Guess J. C., McCance D. J. 2005. Decreased migration of Langerhans precursor-like cells in response to human keratinocytes expressing human papillomavirus type 16 E6/E7 is related to reduced macrophage inflammatory protein-3alpha production. J. Virol. 79:14852–14862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Halbert C. L., Demers G. W., Galloway D. A. 1991. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 65:473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hasan U. A., et al. 2007. TLR9 expression and function is abolished by the cervical cancer-associated human papillomavirus type 16. J. Immunol. 178:3186–3197 [DOI] [PubMed] [Google Scholar]
- 19. Hebner C. M., Wilson R., Rader J., Bidder M., Laimins L. A. 2006. Human papillomaviruses target the double-stranded RNA protein kinase pathway. J. Gen. Virol. 87:3183–3193 [DOI] [PubMed] [Google Scholar]
- 20. Heinz S., et al. 2003. Species-specific regulation of Toll-like receptor 3 genes in men and mice. J. Biol. Chem. 278:21502–21509 [DOI] [PubMed] [Google Scholar]
- 21. Herdman M. T., et al. 2006. Interferon-beta treatment of cervical keratinocytes naturally infected with human papillomavirus 16 episomes promotes rapid reduction in episome numbers and emergence of latent integrants. Carcinogenesis 27:2341–2353 [DOI] [PubMed] [Google Scholar]
- 22. Huibregtse J. M., Scheffner M., Howley P. M. 1991. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 10:4129–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kalali B. N., et al. 2008. Double-stranded RNA induces an antiviral defense status in epidermal keratinocytes through TLR3-, PKR-, and MDA5/RIG-I-mediated differential signaling. J. Immunol. 181:2694–2704 [DOI] [PubMed] [Google Scholar]
- 24. Kang D. C., et al. 2002. mda-5: an interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. U. S. A. 99:637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karstensen B., et al. 2006. Gene expression profiles reveal an upregulation of E2F and downregulation of interferon targets by HPV18 but no changes between keratinocytes with integrated or episomal viral genomes. Virology 353:200–209 [DOI] [PubMed] [Google Scholar]
- 26. Kelley M. L., Keiger K. E., Lee C. J., Huibregtse J. M. 2005. The global transcriptional effects of the human papillomavirus E6 protein in cervical carcinoma cell lines are mediated by the E6AP ubiquitin ligase. J. Virol. 79:3737–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. LaFleur D. W., et al. 2001. Interferon-kappa, a novel type I interferon expressed in human keratinocytes. J. Biol. Chem. 276:39765–39771 [DOI] [PubMed] [Google Scholar]
- 28. Lai Y., et al. 2009. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat. Med. 15:1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leaman D. W., et al. 2002. Identification of X-linked inhibitor of apoptosis-associated factor-1 as an interferon-stimulated gene that augments TRAIL Apo2L-induced apoptosis. J. Biol. Chem. 277:28504–28511 [DOI] [PubMed] [Google Scholar]
- 30. Li S., et al. 1999. The human papilloma virus (HPV)-18 E6 oncoprotein physically associates with Tyk2 and impairs Jak-STAT activation by interferon-alpha. Oncogene 18:5727–5737 [DOI] [PubMed] [Google Scholar]
- 31. Muench P., et al. 2009. Binding of PDZ proteins to HPV E6 proteins does neither correlate with epidemiological risk classification nor with the immortalization of foreskin keratinocytes. Virology 387:380–387 [DOI] [PubMed] [Google Scholar]
- 32. Nees M., et al. 2001. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-kappaB-responsive genes in cervical keratinocytes. J. Virol. 75:4283–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park J. S., et al. 2000. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. Implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J. Biol. Chem. 275:6764–6769 [DOI] [PubMed] [Google Scholar]
- 34. Perea S. E., Massimi P., Banks L. 2000. Human papillomavirus type 16 E7 impairs the activation of the interferon regulatory factor-1. Int. J. Mol. Med. 5:661–666 [DOI] [PubMed] [Google Scholar]
- 35. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rathinam V. A., Fitzgerald K. A. 2011. Innate immune sensing of DNA viruses. Virology 411:153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rincon-Orozco B., et al. 2009. Epigenetic silencing of interferon-kappa in human papillomavirus type 16-positive cells. Cancer Res. 69:8718–8725 [DOI] [PubMed] [Google Scholar]
- 38. Ronco L. V., Karpova A. Y., Vidal M., Howley P. M. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rozen S. S., Skaletsky H. J. (ed.). 2000. Primer3 on the WWW for general users and for biologist programmers. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 40. Stubenrauch F., Hummel M., Iftner T., Laimins L. A. 2000. The E8E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J. Virol. 74:1178–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Terenzi F., Saikia P., Sen G. C. 2008. Interferon-inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1. EMBO J. 27:3311–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thomas J. T., Hubert W. G., Ruesch M. N., Laimins L. A. 1999. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc. Natl. Acad. Sci. U. S. A. 96:8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Uze G., Monneron D. 2007. IL-28 and IL-29: newcomers to the interferon family. Biochimie 89:729–734 [DOI] [PubMed] [Google Scholar]
- 44. Wang H. K., Duffy A. A., Broker T. R., Chow L. T. 2009. Robust production and passaging of infectious HPV in squamous epithelium of primary human keratinocytes. Genes Dev. 23:181–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yoneyama M., et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.