Abstract
The hepatitis C virus (HCV) NS3/4A protein has several essential roles in the virus life cycle, most probably through dynamic interactions with host factors. To discover cellular cofactors that are co-opted by HCV for its replication, we elucidated the NS3/4A interactome using mass spectrometry and identified Y-box-binding protein 1 (YB-1) as an interacting partner of NS3/4A protein and HCV genomic RNA. Importantly, silencing YB-1 expression decreased viral RNA replication and severely impaired the propagation of the infectious HCV molecular clone JFH-1. Immunofluorescence studies further revealed a drastic HCV-dependent redistribution of YB-1 to the surface of the lipid droplets, an important organelle for HCV assembly. Core and NS3 protein-dependent polyprotein maturation were shown to be required for YB-1 relocalization. Unexpectedly, YB-1 knockdown cells showed the increased production of viral infectious particles while HCV RNA replication was impaired. Our data support that HCV hijacks YB-1-containing ribonucleoparticles and that YB-1–NS3/4A–HCV RNA complexes regulate the equilibrium between HCV RNA replication and viral particle production.
INTRODUCTION
Hepatitis C virus (HCV) infection is a growing public health problem, since it affects 170 to 200 million people worldwide and is the major cause of chronic liver diseases, including cirrhosis and hepatocellular carcinoma (45). The standard treatment, involving pegylated interferon and ribavirin administration, is not well tolerated and provides limited efficacy (38, 44). Hence, there is an urgent need for the development of novel anti-HCV therapies.
HCV is a single-stranded positive RNA enveloped virus which belongs to the Hepacivirus genus in the Flaviviridae family. The viral RNA (vRNA) is 9.6 kb long and encodes a 3,000-amino-acid polyprotein whose translation is controlled by the internal ribosome entry site (IRES)-containing 5′-untranslated region (UTR). HCV polyprotein is posttranslationally processed by cellular and viral proteases into structural (core, E1, and E2) and nonstructural (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) viral proteins, respectively. Core protein binds HCV RNA, and through its multimerization it mediates the encapsidation process by packaging the plus-strand genomic RNA into the viral capsid. Additional assembly processes involving the glycoproteins E1 and E2 are required in the final assembly of enveloped virions and infectious particles for entry and spread into new host cells. NS2 harbors a protease activity responsible for processing at the NS2-NS3 junction. NS3 serine protease and its cofactor, NS4A, mediate the maturation of the polyprotein at all processing sites C terminal to NS3. NS5B is an RNA-dependent RNA polymerase that replicates viral RNA within detergent-resistant, endoplasmic reticulum (ER)-derived replication complexes (RC). This process is regulated by the phosphoprotein NS5A and requires the helicase and NTPase activities of NS3. NS4B induces the formation of a membranous web that is believed to be important for RC formation and activity (1, 5, 40).
The discovery of the JFH-1 and intergenotypic chimeric clones that produce infectious virions in cell culture allowed the molecular dissection of HCV assembly, the mechanistic details of which still remain poorly understood (31, 52). Several studies have reported that lipid droplets constitute a critical cell compartment for HCV particle production (7, 39, 46) despite being a likely transit platform for the capsid assembly process. A role of p7 protein has been proposed for the further continuation of the assembly process (46, 48) within a cell compartment and most likely is linked with very-low-density lipoprotein biogenesis (8, 21). Nonstructural proteins, with the exception of NS5B, also are key players in the assembly process. Notably, NS5A, possibly via its phosphorylation by casein kinase II, participates in viral capsid assembly through a core-dependent corecruitment with vRNA on lipid droplets (2, 37, 50). Moreover, a function of NS2 also is proposed during later stages of the assembly process. Indeed, deleterious NS2 mutations can abrogate particle production without affecting core sedimenting properties and the loading of NS5A/core complexes on the lipid droplets (56). Finally, HCV must regulate in time and space the selection of the HCV genome (positive strand) for encapsidation into nascent capsids in order to not deplete the pool of genomes that serve in replication/translation processes. The intracellular site, determinants, and control of this viral step still remain unclear. In addition, very little is known about how HCV orchestrates host machineries for the transition of RNA synthesis in RCs at endoplasmic reticulum (ER)-like membranes to the site of the encapsidation of genomic positive-strand RNAs at lipid droplets leading to virus particles. Indeed, it is proposed that the intricate organization and close proximity of the ER-membrane network and the lipid droplets favor the transfer of vRNA from the RCs to the assembling virus capsid (49).
In addition to polyprotein processing and RNA replication functions, a role of NS3 was recently proposed in HCV particle assembly (36). Indeed, Ma et al. showed that the Q221L mutation within the helicase domain can rescue the infectivity of the chimeric clone HJ3 (36). The NS3-dependent process is important for the generation of high-density rapidly sedimenting capsids at a step subsequent to the loading of core/NS5A complex on lipid droplets. It is proposed that NS3 influences HCV assembly via an association with host factors, most likely before the contribution of NS2. In addition, NS3 interferes with TLR3 and RIG-I signaling pathways by cleaving the adaptor proteins TRIF and MAVS, respectively, leading to the inhibition of type I interferon (IFN) and IFN-stimulated genes (9, 17, 29, 30, 33). Such virus-host interactions inhibit cellular antiviral immunity and confer an advantage to HCV infection. These observations demonstrate that NS3 protein takes on multiple roles during the HCV life cycle by interacting with cellular machineries and networks and contributes to a favorable environment for virus infection. Hence, identifying NS3-host protein interactions that are critical for HCV infection could lead to the development of new therapeutic strategies.
In this report, we identified Y-box-binding protein-1 (YB-1) as a novel interacting partner of the HCV NS3/4A protein using a proteomic approach. The YB-1–NS3/4A complex specifically associates with HCV RNA in an HCV genotype-independent infectious context. We demonstrated a crucial dependency of YB-1 expression for the maintenance and propagation of HCV in culture. We showed that gene silencing of YB-1 affects intracellular HCV RNA levels and the production of virus particles. Finally, we reported the redistribution of YB-1 to the lipid droplets upon HCV infection and the expression of core protein, which is accordance with a regulatory role of YB-1 in the viral RNA replication and assembly processes for the timely production of infectious particles.
MATERIALS AND METHODS
Cell culture.
All cells lines were cultured in Dulbecco's modified essential medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 1% penicillin-streptomycin antibiotics (Invitrogen), and 1% nonessential amino acids (Invitrogen). For 293EcR cells, the media contained 30 μg/ml bleocin (Calbiochem) to maintain ecdysone receptor expression. Subgenomic replicon-containing cells (32) were cultured with 500 μg/ml G418 to maintain HCV replication. Cells were transfected with Lipofectamine 2000 (Invitrogen) by following the manufacturer's instructions.
Expression plasmids.
The construction of pEF/JFH1-Rz/Neo and pEF/cJFH1 GND-Rz/Neo were previously described (23). For the construction of pEF/JFH1-del153-167-Rz/Neo, we performed a two-step recombinant PCR with Platinum Pfx DNA polymerase (Invitrogen) involving four oligonucleotides. Internal primers were designed to generate a 15-amino-acid deletion in the core coding sequence. The final PCR product was cloned into the FspAI/BsiWI cassette of pEF/JFH1-Rz/Neo. The same approach was used to generate p7-, NS2-, and NS5A-mutated PCR products that were cloned in the ClaI (p7 and NS2) or SanDI/BsrGI (NS5A) cassette of pEF/JFH1-Rz/Neo. NS3/4- and NS5A-encoding DNAs (genotype 1b) were PCR amplified using the replicon-encoding HCVAB12 Luc plasmid (53) and core-, p7-, and NS2-encoding DNAs (genotype 2a) using pFK-i389Luc-E1-core-3′-JFH-1wt (52). Resulting PCR products were cloned into the EcoRV/HindIII cassette of pcDNA3-Hygro-Flag-MCS and pcDNA3-Hygro-eYFP-MCS (4). YB-1 and hnRNP A1 cDNA were obtained from Origene. Coding sequences were amplified using PCR, and resulting products were cloned in the BamHI restriction site of pcDNA3-Hygro-Flag-MCS (4).
TAP and mass spectrometry.
To generate TAP-NS3/4A-expressing plasmid, NS3/4A sequence was PCR amplified using the replicon-encoding HCVAB12Luc plasmid (53). The resulting PCR fragment was inserted into the NotI/XbaI cassette of the tandem affinity purification (TAP) tag-expressing plasmid AB0411 (a kind gift from Benoît Coulombe).
293EcR cells were transfected with TAP- and TAP-NS3/4A-expressing plasmids. Three hours later, the medium was replaced and cells were cultured in the presence of 4.5 μM ponasterone A (Invitrogene), an analogue of ecdysone, to induce TAP-NS3/4A expression. In one condition, the NS3/4A inhibitor BILN2061 (28) was added to the medium at a final concentration of 2 μM. Twenty-four hours posttransfection, cells were washed twice with cold phosphate-buffered saline (PBS), collected, pelleted, and resuspended in the lysis buffer (10 mM Tris-HCl, pH 8, 100 mM NaCl, 0.5% Triton X-100, 0.5 mM dithiothreitol [DTT], pH 8) containing 0.5 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, a protease inhibitor cocktail (Roche), and DMSO or 2 μM BILN2061. Lysates were cleared by centrifugation and quantitated. Twenty-seven mg of total protein was used for double purification. Lysates were incubated at 4°C during 1.5 h with 100 μl of IgG-Sepharose (GE Healthcare) prepared in TST buffer (50 mM Tris-HCl, 150 mM NaCl, 0.05% Tween 20, pH 7.6). Following this incubation, beads were extensively washed in IPP buffer (10 mM Tris-HCl, 100 mM NaCl, 0.1% Triton X-100, 10% glycerol, pH 8). Bound complexes were eluted from the resin twice by incubation at 4°C during 4.5 h and overnight with 100 and 30 U of AcTEV protease (Invitrogen), respectively, in TEV buffer (10 mM Tris-HCl, 100 mM NaCl, 0.1% Triton X-100, 0.5 mM EDTA, 10% glycerol, 1 mM DTT, pH 8, with 2 μM BILN2061 for one condition). Both elutions were combined and incubated during 2 h at 4°C with 50 μl of calmodulin-Sepharose (GE Healthcare) prepared in CBB buffer (10 mM Tris-HCl, 100 mM NaCl, 1 mM imidazole, 1 mM magnesium acetate, 2 mM CaCl2, 0.1% Triton X-100, 10% glycerol, 10 mM beta-mercaptoethanol, pH 8). CaCl2 was added to adjust the final concentration to approximately 2 mM. Following this incubation, the resin was extensively washed with CBB buffer. Triton X-100 was absent from the final wash. Three elutions were carried out by the sequential incubation of the resin with CEB buffer (10 mM Tris-HCl, 100 mM NaCl, 1 mM imidazole, 1 mM magnesium acetate, 4 mM EGTA). The eluates were combined and analyzed by silver staining and Western blotting using anti-NS3 antibodies (Vector Laboratories). The Proteomics Core Facility of the Institut de Recherche en Immunologie et Cancérologie (IRIC; Montréal, Canada) performed tryptic digestion on the TAP eluates and analyzed the resulting peptides by liquid chromatography combined with tandem mass spectrometry (LC-MS/MS) and database searches. Proteins were considered NS3/4A-associated proteins when the corresponding peptides were detected in the TAP-NS3/4A condition but were absent from the TAP negative control.
HCV replication, production, and infection.
Infectious JFH-1 viral particles were generated by transfecting Huh7.5 cells with pEF/JFH-1-Rz/Neo DNA using Lipofectamine 2000. Under the control of the elongation factor-1α promoter, this plasmid encodes the JFH-1 genomic RNA whose 5′ and 3′ termini are flanked by one ribozyme (23). Through their autocatalytic activity, the ribozymes generate the authentic ends of HCV RNA, which subsequently replicates. Three days posttransfection, the cell supernatants were collected, cleared with a 0.45-μm filter, and kept at −80°C for long-term storage. Cell extracts were prepared by lysis in RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, pH 8) containing proteases inhibitors (Roche) and then analyzed by Western blotting using mouse anti-core (Affinity BioReagents or Austral Biologicals) and anti-NS3 (Abcam or Biodesign). For viral infection, 500 μl of the JFH-1-containing supernatant was added to naïve Huh7.5 cells that were seeded in 12-well plates the day before. The medium was changed 5 to 6 h later. As a negative control for JFH-1 infection, BILN2061 was added to the culture medium at a final concentration of 2 μM. Three to 5 days later, cells were collected and analyzed by Western blotting and qRT-PCR for their viral content.
9-13 Huh7 cells were previously described (32) and stably express the genotype 1b Con1 subgenomic replicon. This bicistronic replicon expresses the neomycin phosphotransferase through the HCV IRES, while NS3-NS5B polyprotein production is under the control of the encephalomyocarditis virus (EMCV) IRES. The reporter Huh7 cells stably express the Con1 subgenomic replicon as well as both neomycin resistance and Firefly luciferase (32). Luciferase assays were performed as previously described (4).
Coimmunoprecipitation assays.
For immunoprecipitation directed against the Flag-tagged proteins, transfected cells were washed twice, collected, and lysed in 10 mM Tris-HCl, 100 mM NaCl, 0.5% Triton X-100, pH 7.6, with protease inhibitors (Roche). One to 2 mg of resulting cell extract was subjected to immunoprecipitation by adding 40 μl of anti-Flag affinity gel (Sigma-Aldrich) prepared in TBS buffer (50 mM Tris-HCl, 150 mM NaCl, pH 7.4) as described by the manufacturer. Following 2 h of incubation at 4°C, immunoprecipitates were washed three times with TBS. Immune complexes were eluted from the resin during 45 min at 4°C using the Flag peptide (Sigma-Aldrich) at a concentration of 250 ng/μl. For coimmunoprecipitation under HCV infection, cell extracts were prepared as described above and precleared for 1 to 2 h at 4°C using a 50:50 slurry of protein G-Sepharose (Sigma-Aldrich) prepared in the lysis buffer. Precleared lysates were subjected to an overnight immunoprecipitation using 1 μg of rabbit anti-YB-1 (Abcam), anti-RHA (Abcam), anti-hnRNP U (Abcam), anti-RNA polymerase II (Santa Cruz Biotechnology), or anti-hemagglutinin (HA) (Santa Cruz Biotechnology) antibody. The day after, samples were incubated with 35 μl of the 50:50 slurry of protein G-Sepharose for 1 to 2 h at 4°C. Immunoprecipitates were washed three times in lysis buffer and eluted in SDS-containing loading buffer. Eluates were analyzed by Western blotting using mouse anti-Flag (Sigma-Aldrich), anti-NS3 (Abcam, Vector Laboratories or Biodesign), anti-green fluorescent protein (GFP) (Roche), anti-hnRNP A1 (Abcam), anti-hnRNP U (Abcam), rabbit anti-YB-1 (Abcam), anti-RHA (Abcam), anti-ribosomal protein S6 (Cell Signaling), and anti-pol II (Santa Cruz Biotechnology) antibodies.
Gene silencing by lentivirus-mediated RNA interference.
For lentivirus production, 293T cells were transfected with pRSV-REV, pMDLg/pRRE, pMD2-VSVG, and various short hairpin RNA (shRNA)-expressing pLKO.1-puro constructs (Sigma-Aldrich) using Lipofectamine 2000. Two days posttransfection, cell media were collected and cleared with a 0.45-μm filter. Huh7.5 cells were infected overnight by adding the lentivirus preparation to the medium containing 8 μg/ml Polybrene. The day after, puromycin was added to the medium at a concentration of 3 μg/ml to select lentivirus-infected cells. Five shRNAs (Sigma-Aldrich) per gene were tested, and their respective knockdown efficiencies were assessed by Western blotting on extracts from infected cells using anti-YB-1 (Abcam) and anti-RHA antibodies. The shRNAs TRCN0000007952 (shYB-1) and TRCN0000001212 (shRHA) were chosen for YB-1 and RHA depletion, respectively. Twenty-four hours postinfection, cells were transfected with JFH-1-encoding plasmids.
Immunofluorescence analysis.
Huh7.5 cells were transfected with expression plasmids in 6-well plates. Two to 3 days posttransfection, cells were trypsinized and reseeded in coverslip-containing 24-well plates. Twenty-four hours later, cells were washed twice with PBS, fixed with 4% paraformaldehyde-containing PBS for 20 min at room temperature, and then permeabilized in 0.2% Triton X-100–PBS for 15 min. Blocking was performed in PBS with 10% normal goat serum, 5% bovine serum albumin (BSA), and 0.02% sodium azide for 45 min at room temperature. Following three rapid washes, cells were labeled with mouse anti-core (Affinity BioReagents), anti-NS3 (Abcam), and rabbit anti-YB-1 (Abcam) primary antibodies diluted in 5% BSA–0.02% sodium azide–PBS for 2 h. Slides were washed three times in PBS and then labeled with Alexa Fluor 488 and Alexa Fluor 594 secondary antibodies (Invitrogen) diluted in 5% BSA–0.02% sodium azide–PBS for 1 h. Cells were extensively washed and incubated with Hoechst dye (Invitrogen) at a final concentration of 1 μg/ml in PBS. Following three rapid washes, 1 h of incubation was performed with HCS LipidTOX Deep Red (Invitrogen) diluted 1:1,000 in PBS to label the lipid droplets. Immediately afterwards the slides were mounted using DABCO (Sigma-Aldrich) as an anti-fading agent. Labeled cells then were examined by laser-scanning microscopy using an LSM510 confocal microscope (Zeiss).
RNA extraction and qRT-PCR.
Total cellular RNA was extracted by the lysis of the JFH-1-containing cells with RLT buffer (Qiagen) supplemented with 1% beta-mercaptoethanol. Total RNA was purified using the RNeasy kit (Qiagen). Coimmunoprecipitated RNAs were extracted using TRIzol LS (Invitrogen) according to the manufacturer's instructions. As a carrier, 7.5 μg of GlycoBlue (Ambion) was added during the isopropanol-mediated precipitation. RNAs then were treated with 0.25 U of amplification-grade RNase-free DNase I (Invitrogen) during 15 min at room temperature and subjected to RNA cleanup using the RNeasy minikit (Qiagen). HCV and actin RNAs were quantitated by quantitative real-time reverse transcription-PCR (qRT-PCR). One hundred to 200 ng of total RNA or 10 μl of coimmunoprecipitated RNA was reverse transcribed with antisense (5′-TGGTGCACGGTCTACGAGACCTC-3′ [for HCV RNA] and 5′-AGCACTGTGTTGGCGTACAG-3′ [for actin RNA]) oligonucleotides using the QuantiTect reverse transcription kit (Qiagen). As negative controls, reactions without RT were always included to monitor for DNA contamination. Resulting DNA was diluted 10 times in water, and 6 μl of this dilution was used for amplification by quantitative PCR using the QuantiTect SYBR green kit (Qiagen) and primer pairs 5′-CAAGCACCCTATCAGGCAGTA-3′/5′-TCTGCGGAACCGGTGAGTA-3′ (for HCV) and 5′-AGCACTGTGTTGGCGTACAG-3′/5′-GACTTCGAGCAAGAGATGG-3′ (for actin). Serial dilutions of JFH-1 and actin DNAs (6 × 102 to 6 × 106 copies) also were amplified to generate standard curves. Signal acquisitions were performed with the Rotor-Gene 3000 thermocycler (Corbett Research). HCV RNA content in the cells was always normalized with the actin RNA levels. To analyze the vRNA immunoprecipitation experiments together, we have arbitrarily set to 1 the absolute vRNA copy number for the anti-YB-1/JFH-1 condition and calculated the relative vRNA level per immunoprecipitation for each experiment. We used the same approach for the anti-Flag immunoprecipitation. Standard deviations were determined from three independent experiments.
MTT assays.
Cells were cultured in transparent 96-well plates (100 μl). Twenty μl of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] stock solution (5 mg/ml in PBS) was added to the cells that were incubated for 1 to 3 h at 37°C in the dark. After the removal of the medium, cells were incubated at room temperature with 150 μl of DMSO containing 2 mM glycine, pH 11. Absorbance was read at 570 nm.
RESULTS
YB-1 is an interacting partner of the HCV NS3/4A protein.
To elucidate the NS3/4A interactome and to identify host cofactors essential for HCV infection, we genetically engineered a plasmid expressing the recombinant NS3/4 protein fused by its N terminus to the tandem affinity purification (TAP) tag (Fig. 1 A). TAP-NS3/4A as well as the TAP tag alone were expressed at the expected molecular weight (Fig. 1B). Moreover, TAP-NS3/4A was properly processed at the NS3-NS4A junction and was able to cleave MAVS, since a faster-migrating form of MAVS was detectable when native or tagged NS3/4A was expressed (Fig. 1C). This suggests that the TAP tag does not significantly affect NS3/4A functions and interactions with cellular factors. The protease activity of TAP-NS3/4A was further confirmed following treatment with the specific NS3 protease inhibitor BILN2061 (28) that resulted in the appearance of the unprocessed TAP-NS3-4A precursor (Fig. 1D). Based on high affinity to IgG and calmodulin through the protein A and calmodulin-binding domain moieties, respectively, TAP-NS3/4A complexes were purified from 293-EcR cell extracts (Fig. 1D), and the resulting eluates were subjected to tryptic digestion and to LC-MS/MS analysis. The TAP-tag-only condition was used as a control for the identification of tag-associated proteins, which subsequently were excluded from the list of specific hits. We identified 23 proteins that were specific to the TAP-NS3/4A eluates, most of them strikingly harboring functions in RNA metabolism, including RNA translation, splicing, stability, and localization (Fig. 1E) (14, 35). Several of these associations were studied and confirmed using various fusion proteins, expression systems, purification procedures, and cell types. Flag-tagged NS3/4A-expressing 293T cell extracts were subjected to immunoprecipitation studies using antibodies directed against the Flag epitope (Fig. 2 A). Western blotting of the eluates revealed that YB-1, hnRNP A1, hnRNP U, and RHA proteins all were readily detectable in the immunoprecipitated Flag-tagged NS3/4A samples while being absent from the Flag control. Similar results were obtained with the expression of Flag-tagged NS3/4A in Huh7.5 liver cells (data not shown).
Fig. 1.
Expression of TAP-NS3/4A and identification of associated host factors. (A) Schematic representation of TAP-NS3/4A. 293EcR cells were transfected with NS3/4A-, TAP-, or TAP-NS3/4A-expressing plasmids. Twenty-four hours posttransfection, cell extracts were prepared and subjected to Western analysis using polyclonal rabbit antibodies that nonspecifically recognize the protein A moiety of the TAP tag and allow the visualization of TAP and TAP-NS3 proteins (B), as well as anti-NS3, anti-MAVS, and anti-GAPDH antibodies (C). The asterisks indicate the NS3 degradation products detected with the anti-NS3 antibodies with different exposures (expo.). (D) Cells were treated as described for panel B in the presence of DMSO or 2 μM the NS3 protease inhibitor BILN2061. Cell extracts were analyzed by Western blotting (WB) using anti-NS3 antibodies and subjected to TAP. Resulting eluates were analyzed by Western blotting using anti-NS3 antibodies and silver staining. (E) The host factors that were copurified with TAP-NS3/4A and absent from the TAP tag control eluate were identified using mass spectrometry.
Fig. 2.
YB-1 specifically interacts with HCV NS3 when expressed transiently or in the context of JFH-1 expression. (A) 293T cells were transfected with Flag-NS3/4A-expressing plasmid. Forty-eight hours posttransfection, cell extracts were prepared and subjected to immunoprecipitation (IP) using an anti-Flag-coupled resin. Resulting eluates as well as cell extracts were analyzed by Western blotting using anti-NS3, anti-YB-1, anti-hnRNP A1, anti-hnRNP U, anti-hnRNP A1, and anti-RHA antibodies. Anti-S6 was used as a loading control for cell extracts. (B) 293T cells were cotransfected with plasmids encoding Flag-YB-1 and several YFP-tagged HCV nonstructural proteins. Forty-eight hours posttransfection, cell extracts were prepared and subjected to immunoprecipitation using an anti-Flag-coupled resin. Resulting eluates as well as cell extracts were analyzed by Western blotting using anti-GFP antibodies. (C) Huh7.5 cells were transfected with control DNA (Mock) or JFH-1-expressing plasmid. Three days posttransfection, cell extracts were prepared and subjected to immunoprecipitation directed against the HA epitope and endogenous proteins YB-1 or RHA. Immune complexes were analyzed by Western blotting using anti-RHA, anti-YB-1, and anti-NS3 antibodies. IgG indicates nonspecific labeling due to the presence of antibodies in the loaded samples.
We then focused on YB-1, an RNA-binding protein that is found in several cell compartments, including P bodies, stress granules, and the nucleus (3, 24, 54). YB-1 is involved in CAP- and IRES-dependent translation as well as the stability and transcription of cellular RNAs (12, 14–16, 25, 47). To assess the specificity of NS3/4A–YB-1 interaction, Flag-tagged YB-1 was coexpressed with several yellow fluorescent protein (YFP)-fused HCV nonstructural proteins for coimmunoprecipitation studies in 293T cells (Fig. 2B). eYFP-NS3 was the only protein to coprecipitate with YB-1, in contrast to enhanced YFP (eYFP)-tagged p7, NS2, and NS5A, which were not detected in the eluates. To demonstrate the significance of YB-1–NS3/4A interaction in the HCV life cycle, we expressed the full-length infectious clone JFH-1 and performed an immunoprecipitation study using endogenous YB-1 as bait in Huh7.5 cells (Fig. 2C). NS3 was specifically detected upon JFH-1 expression, while it was absent from the anti-HA control condition (Fig. 2C). NS3 protein also was successfully pulled down with anti-RHA antibodies upon JFH-1 expression, as expected from the mass spectrometry analysis. Anti-YB-1 antibodies successfully pulled down RHA as expected from previous studies reporting the presence of RHA and YB-1 in the same protein complex for endogenous functions in RNA metabolism (26, 51). However, we were not able to detect YB-1 in anti-RHA immunoprecipitation, most probably because the detection was limited by the close proximity between YB-1 and the IgG heavy chains in SDS-containing gels. Overall, the data show that endogenous YB-1 specifically interacts with NS3 when ectopically expressed or upon HCV infection.
YB-1 contributes to viral RNA replication of JFH-1 and genotype 1b subgenomic replicon.
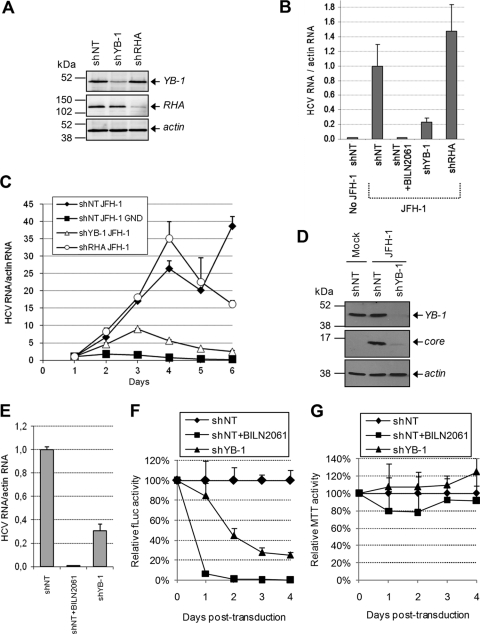
Since YB-1 associates with HCV NS3/4A protein, we determined if HCV replication is dependent on the host factor YB-1 and performed gene silencing studies using a lentivirus-based expression system allowing the transcription of short hairpin RNA (shRNA). We identified lentivirus-delivered shRNA that specifically reduced the expression of YB-1 (shYB-1) and of RHA (shRHA) compared to that of control nontarget shRNA (shNT) by Western blotting (Fig. 3 A). For all experiments, functional analyses were assessed in puromycin-selected cell populations, resulting in an efficient gene-silencing phenotype. To evaluate the effect of reduced YB-1 and RHA proteins on the genotype 2a HCV life cycle, transduced Huh7.5 cells were infected with JFH-1 virus preparations, and 5 days later HCV RNA levels were assessed by real-time qRT-PCR as a readout of viral replication (Fig. 3B). Compared to those of shNT-transduced cells, HCV RNA levels were decreased by 80% in YB-1-silenced Huh7.5 cells. Similar results were observed with two other shRNAs depleting YB-1, excluding an off-target effect of shYB-1 (data not shown). In contrast, the gene silencing of RHA did not significantly affect JFH-1 replication. The NS3/4A inhibitor BILN2061 was used as a control for HCV RNA replication (Fig. 3B). The data clearly indicate that YB-1 contributes significantly to viral RNA replication in JFH-1 infection.
Fig. 3.
YB-1 expression levels are important for HCV life cycle. (A) Huh7.5 cells were infected with lentiviruses that express shRNA silencing YB-1 (shYB-1) and RHA (shRHA). As a control, a nontarget shRNA (shNT) was used. Transduced cells were selected with puromycin, and knockdown efficiencies for YB-1 and RHA were confirmed by Western blotting. (B) shNT-, shRHA-, and shYB-1-expressing Huh7.5 cells were infected with JFH-1. Five days later, actin and JFH-1 RNAs were quantitated using qRT-PCR. HCV RNA levels were normalized with actin RNA content and arbitrarily set to 1 for the JFH-1-plus-shNT condition. Error bars represent standard deviations from biological triplicates. (C) Transduced Huh7.5 cells were transfected with JFH-1-expressing plasmids, collected each day, and analyzed for their content of actin and HCV RNA as described for panel B. The normalized levels of HCV RNA 1 day posttransfection were arbitrarily set to 1 for each condition to monitor viral RNA amplification. Error bars represent standard deviations from biological duplicates. (D) Huh7.5 cells were treated as described for panel C and analyzed 4 days posttransfection for their content in core protein, YB-1, and actin by Western blotting. (E) Genotype 1b subgenomic replicon-containing 9-13 cells were transduced with shRNA-expressing lentiviruses and were analyzed 5 days later for their content of HCV RNA using qRT-PCR. Error bars represent standard deviations from biological triplicates. (F and G) Huh7 cells that stably express a luciferase-producing Con1 subgenomic replicon were transduced with shRNA-expressing lentiviruses. Luciferase and MTT assays were performed each day following transduction as readouts of HCV replication (F) and cell viability (G), respectively. As a control, cells were transduced with shNT lentiviruses in the presence of BILN2061. Error bars represent standard deviations from four biological replicates.
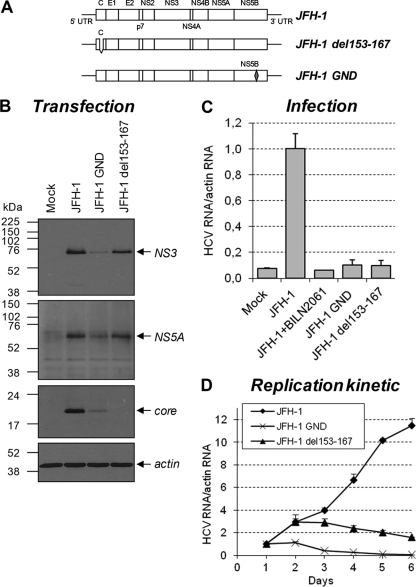
To gain further insight into the YB-1 dependency for HCV replication kinetics, we took advantage of the ribozyme-based JFH-1 infectious system developed by Kato et al. (23). In this experimental system, transcription from the transfected JFH-1 DNA plasmid generates low vRNA levels, which increase as a function of time as a consequence of viral replication and propagation by reinfection (Fig. 4 D). Control cells transfected with the replication-defective mutant JFH-1 GND (containing a point mutation in NS5B; Fig. 4A) showed the expected plasmid-dependent low HCV RNA levels from days 1 to 6 resulting from the lack of RNA polymerase activity (Fig. 4D). As expected, corresponding cell supernatants were not infectious (Fig. 4C), while wild-type JFH-1 viral preparations were able to infect naive Huh7.5 cells and to propagate in cell culture. In a second control, we transfected the JFH-1 del153-167 mutant, containing a previously reported deletion in the protein core, which consequently is unstable and undetectable in cell extracts (Fig. 4A and B) (41). This mutant showed viral RNA replication but no release of infectious particles, as determined by infection assays (Fig. 4C). HCV RNA levels in JFH-1 del153-167-transfected cells increased during the first 2 days posttransfection and then slowly decreased because of RNA replication in the absence of propagation by de novo infection (Fig. 4D). Thus, RNA-level amplification through viral spread by reinfection is detectable with the ribozyme-based JFH-1 system 3 days posttransfection.
Fig. 4.
Characterization of the ribozyme-based JFH-1 infectious system. (A) Schematic representation of the wild-type construct as well as assembly (del153-167)- and replication (GND)-deficient JFH-1 DNA constructs. (B) Huh7.5 cells were transfected with the different JFH-1 constructs represented. Seventy-two hours posttransfection, cell extracts were prepared and analyzed by Western blotting using anti-NS3, anti-NS5A, anti-core protein, and anti-actin antibodies. (C) Culture media from transfected cells described for panel B were used to infect naive Huh7.5 cells. After 4 days, cellular RNA was purified and actin and JFH-1 RNAs were determined using qRT-PCR. HCV RNA levels were normalized with actin RNA content and arbitrarily set to 1 for wild-type JFH-1. As a control for the complete inhibition of HCV replication, cells were cultured with 2 μM BILN2061. Error bars represent standard deviations from biological triplicates. (D) Huh7.5 cells were transfected as described for panel B. Transfected cells then were collected each day and analyzed for HCV RNA levels as described for panel C in a 6-day kinetic study. The normalized HCV RNA levels at day 1 posttransfection were arbitrarily set to 1 for each construct to monitor viral RNA amplification. Error bars represent standard deviations from biological duplicates.
We used this ribozyme/HCV system to further characterize the YB-1 dependency of viral replication in 6-day kinetic studies. Huh7.5 cells expressing shNT, shYB-1, or shRHA were transfected with the JFH-1-expressing plasmid, collected each day, and analyzed for their HCV RNA content by qRT-PCR. In shYB-1-expressing cells, we observed a drastic decline in HCV RNA levels from day 3, the time from which HCV RNA levels mostly increase from viral replication and reinfection (Fig. 3C). HCV proteins also were reduced in shYB-1-expressing cells, as illustrated by the core protein levels at day 4 (Fig. 3D). In contrast, shRHA expression had no major effect on HCV RNA levels (Fig. 3C). Viral RNA levels were comparable to those of the shNT condition at day 1, suggesting that the gene silencing of YB-1 did not influence plasmid-derived JFH-1 transcription. Additionally, the transfection efficiency of JFH-1 was controlled by the cotransfection of a reporter plasmid, and similar GFP levels were detected in both shNT and shYB-1 conditions (data not shown).
The effect of YB-1 gene silencing also was evaluated in 9-13 cells stably expressing a genotype 1b Con1 subgenomic HCV replicon that encodes the NS3-NS5B replicative unit and confers neomycin resistance (32). In this model, YB-1 silencing also significantly reduced viral RNA replication, as measured in a qRT-PCR assay (Fig. 3E). Similar results were obtained with the silencing of YB-1 in a Huh7 stable cell line containing a luciferase-encoding Con1b subgenomic replicon (Fig. 3F) where Firefly luciferase activity is dependent on HCV RNA replication (32). Cell proliferation assessed by MTT assays was carried out in parallel and revealed no effect from shYB-1-expressing cells during a 4-day period (Fig. 3G). Overall, these results show that host factor YB-1 contributes to viral RNA replication from both an HCV genotype 2a infectious clone and an HCV genotype 1b subgenomic replicon.
YB-1 associates with vRNA during HCV life cycle.
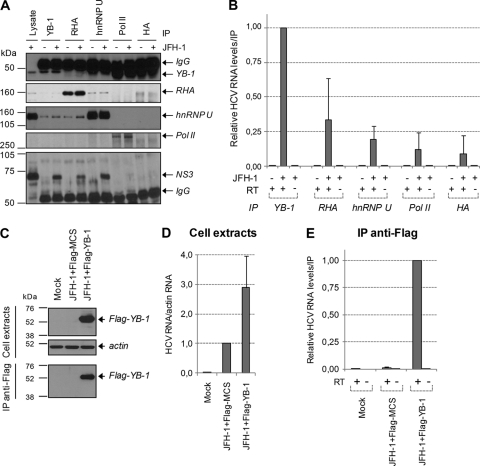
NS3/4A-associated host proteins identified by mass spectrometry are related to RNA metabolism and harbor RNA-binding activities. Moreover, YB-1, RHA, hnRNP A1, and hnRNP U were previously reported to interact with untranslated regions of HCV in vitro (20, 22, 34, 42). To determine if interacting partners of NS3/4A were associated with HCV RNA, JFH-1-expressing cell extracts were subjected to immunoprecipitation directed against endogenous YB-1, RHA, and hnRNP U. The presence of NS3 protein was confirmed in these eluates by Western blot analysis (Fig. 5 A), while they were undetectable in negative-control experiments with anti-polymerase II and anti-HA antibodies. Again, we detected YB-1 as well as hnRNP U in RHA immunoprecipitates independently of HCV expression, confirming that these proteins interact in normal cells. The eluates then were analyzed for their content for vRNA by qRT-PCR (Fig. 5B). vRNA was readily detected in YB-1 immunoprecipitates, while undetectable vRNA levels were obtained with control Pol II and HA, attesting to the specificity of the procedure. We also detected vRNA with the coimmunoprecipitation of RHA protein, but it was evident to a lesser extent than that of YB-1. Interestingly, very little, if any, HCV RNA was detected with the immunoprecipitation of hnRNP U that associated efficiently with NS3/4A protein. This result suggests that determinants other than the RNA-binding property of NS3 are involved in the association of HCV RNA with YB-1–NS3/4A complex.
Fig. 5.
YB-1 associates with HCV RNA. (A) Huh7.5 cells were transfected with control and JFH-1-expressing plasmid. Seventy-two hours posttransfection, cell extracts were prepared and subjected to immunoprecipitation directed against YB-1, RHA, hnRNP U, and RNA polymerase II (Pol II) endogenous proteins. Antibodies against the HA epitope were used for negative-control immunoprecipitations. Immune complexes were analyzed by Western blotting using anti-NS3, anti-YB-1, anti-RHA, anti-hnRNP U, and anti-Pol II antibodies. IgG indicates nonspecific labeling due to the antibodies loaded on the gel. (B) Immune complexes from panel A were subjected to RNA extraction, and HCV RNA from each immunoprecipitate was quantitated using qRT-PCR. Controls without reverse transcriptase (RT) were included to monitor for plasmid DNA contamination. (C) Huh7.5 cells were cotransfected with plasmids encoding JFH-1 and Flag-tagged YB-1. Seventy-two hours posttransfection, cell extracts were prepared and subjected to immunoprecipitation using an anti-Flag-coupled resin. Resulting eluates as well as cell extracts were analyzed by Western blotting using anti-Flag and anti-actin antibodies. RNA was purified from cell extracts (D) and immunoprecipitates (E), and HCV RNA was quantitated using qRT-PCR. All error bars represent standard deviations from three independent experiments.
To further confirm the association of HCV RNA within a YB-1-containing complex, we exogenously expressed Flag-tagged YB-1 and performed coimmunoprecipitation studies directed against the Flag epitope. Figure 5C shows that Flag–YB-1 protein was expressed and efficiently immunoprecipitated. Consistently with a positive role of YB-1 in HCV replication, HCV RNA was more abundant in cells that overexpressed Flag-YB-1 than in control cells (Fig. 5D). Again, analyses of the eluates confirmed that HCV RNA coimmunoprecipitated with Flag–YB-1 (Fig. 5E). The interaction of HCV RNA with YB-1 seemed independent of viral genotype and of the assembly process, since both are coimmunoprecipitated in subgenomic replicon-containing cells and assembly-deficient JFH-1 del153-167-expressing cells (data not shown). Overall, the data demonstrate that NS3/4A–YB-1 complex specifically associates with HCV RNA, possibly within RNP during the virus life cycle, and suggest a role of YB-1 in viral processes involving both NS3/4A and vRNA.
JFH-1 infection induces YB-1 redistribution to core-containing lipid droplets.
To better confirm viral replication dependency on YB-1 and interaction with HCV (Fig. 2, 3, and 5), JFH-1-expressing Huh7.5 cells were analyzed by laser-scanning confocal microscopy following the fluorescent labeling of endogenous YB-1 and HCV proteins. We also stained the lipid droplets (LD) with LipidTOX, as NS3 is recruited by core protein to this organelle during HCV assembly (39). In noninfected cells, YB-1 clearly harbored a nucleocytoplasmic distribution, including small cytosolic puncta corresponding to P bodies (54). In JFH-1-expressing cells, core protein perfectly colocalized to the surface of LD, and this cell compartment likely is a crucial platform for HCV assembly (2, 37, 39, 46, 50, 56). Strikingly, YB-1 redistributed to the perinuclear region within ring-shaped structures that colocalized with core protein and NS3 at the surface of LD (Fig. 6 A, yellow arrows and inset, and 7 C). Such YB-1 distribution was observed with both plasmid-derived JFH-1 transfection and authentic JFH-1 infection (data not shown). Hence, these data strongly suggest that YB-1 is recruited upon HCV infection within assembly complexes. Additionally, a subset of YB-1 also relocalized within structures morphologically distinct from the LD (Fig. 6A, white arrows). Core and NS3 proteins were not detected in these structures, whose nature and composition remain to be determined. Surprisingly, JFH-1 infection did not alter the cellular distribution of other NS3/4A-interacting partners, such as RHA, hnRNP A1, and hnRNP U (Fig. 6B and data not shown), demonstrating that the observed JFH-1-induced LD redistribution phenotype is specific to YB-1.
Fig. 6.
HCV induces YB-1 relocalization to core-containing lipid droplets. Control and JFH-1-expressing Huh7.5 cells were fixed and probed with rabbit anti-YB-1 (A), anti-RHA (B) (green), and mouse anti-core (red) antibodies. Nuclei and lipid droplets were stained with Hoechst dye (white) and LipidTOX (blue), respectively. Yellow arrows, core-containing ring-like structures; white arrows, core-free large structures. Unmerged and merged higher-magnification images of the red selected area are shown. The white asterisks indicate untransfected cells.
Core protein is required for JFH-1-induced redistribution of YB-1.
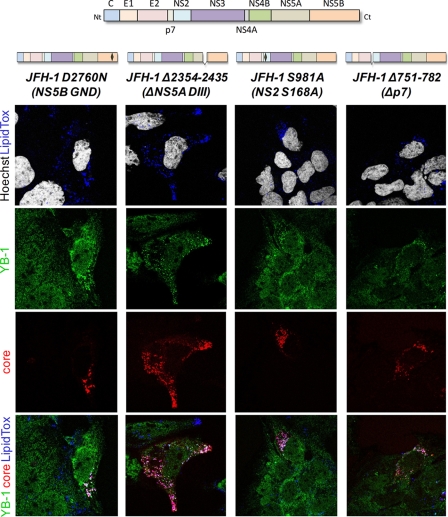
To test if YB-1 relocalization to LD was dependent on the assembly process, we studied YB-1 cellular distribution in subgenomic replicon-containing Huh7 9-13 cells (Fig. 7B). As a control, cells were treated with the NS3 inhibitor BILN2061, leading to the complete disappearance of viral proteins (Fig. 7A and B) and the inhibition of the NS3-mediated MAVS cleavage (Fig. 7A). YB-1 cellular distribution was clearly similar in replicon-containing cells, parental Huh7.5 cells (compare Fig. 7B to mock-treated cells in Fig. 6), and BILN2061-treated replicon-cured cells. This suggests that the assembly process is a prerequisite for YB-1 recruitment to LD. To rule out that the lack of YB-1 relocalization was due to the HCV genotype (1b versus 2a), we studied HCV-mediated YB-1 distribution upon the expression of the assembly-incompetent JFH-1 del153-167 mutant (Fig. 7C). The cells containing the JFH-1 mutant were identified using anti-NS3 antibodies, since the del153-167 core protein is not detectable as previously described (41). Although YB-1 redistributed to LDs and partially colocalized with NS3 (Fig. 7C, inset) in wild-type JFH-1-expressing cells, the JFH-1 del153-167 mutant failed to recruit YB-1 to LD, and the cellular distribution of YB-1 was comparable to that in mock-transfected cells (compare Fig. 7C with mock-treated cells in Fig. 6). Notably, YB-1 recruitment to LDs required, in addition to core protein, the formation of mature viral NS proteins. Indeed, the treatment of JFH-1-expressing cells with the NS3/4A inhibitor BILN2061 did not induce the relocalization of YB-1 (Fig. 7D). However, the replication-defective GND mutant retained its capacity to relocalize YB-1 to LDs, showing that this process does not require NS5B-mediated vRNA replication (Fig. 8). Finally, HCV-induced YB-1 hijacking appeared to be specific to core-dependent steps of assembly, since p7-, NS2-, and NS5A-mutated JFH-1 clones, which were previously reported to be defective for other steps of viral particle assembly (2, 48, 56), did not significantly affect YB-1 distribution to LD (Fig. 8). Taken together, these results suggest a functional link between the JFH-1-induced recruitment of YB-1 to LD and the core-dependent early steps of HCV assembly.
Fig. 7.
Core protein is necessary for HCV-induced YB-1 relocalization to lipid droplets. (A) Huh7.5 and subgenomic replicon-containing 9-13 cells were cultured in the presence of DMSO or 2 μM BILN2061 for 5 days. Cell extracts were prepared and analyzed by Western blotting using anti-NS3, anti-YB-1, anti-MAVS, and anti-GAPDH antibodies. GAPDH was used as a loading control. (B) 9-13 cells were grown on coverslips as described for panel A. After 5 days of culture, cells were fixed and probed with rabbit anti-YB-1 (green) and mouse anti-NS3 (red) antibodies. (C) Huh7.5 cells were transfected with plasmids encoding either wild-type JFH-1 or the del153-167 JFH-1 mutant. Following 4 days of culture, cells were fixed and probed with rabbit anti-YB-1 (green) and mouse anti-NS3 (red) antibodies. Nuclei and lipid droplets were stained with Hoechst dye (white) and LipidTOX (blue), respectively. Unmerged and merged higher-magnification images of the red selected area are shown. Yellow arrows indicate YB-1/NS3 colocalization foci. (D) Huh7.5 cells were treated as described for panel C in the presence of 1 μM BILN2061 in the culture media.
Fig. 8.
YB-1 localization phenotype of various replication- and assembly-defective JFH-1 mutants. A schematic representation of each JFH-1 mutant is shown above the corresponding panel. JFH-1 mutant-expressing Huh7.5 cells were fixed and probed with rabbit anti-YB-1 (green) and mouse anti-core (red) antibodies. Nuclei and lipid droplets were stained with Hoechst (white) and LipidTox (blue) dyes, respectively. Merged images are shown.
YB-1 restrains assembly of infectious HCV particles.
Since YB-1 is recruited by HCV assembly complexes to LDs (Fig. 6 and 7), we evaluated the potential role of YB-1 in HCV assembly by assessing shYB-1-transduced cells for their capacity to produce particles from residual viral replication. Cell homogenates and virus-containing supernatants were analyzed for their HCV RNA content. Under YB-1 knockdown, we observed a 50% reduction of intracellular vRNA levels (Fig. 9 A), as previously demonstrated (Fig. 3C). Strikingly, the corresponding cell supernatants contained significantly higher levels of vRNA than those of shNT-transduced cells (Fig. 9B). As a control, the supernatants of defective del153-167 and GND JFH-1 mutants contain low levels of HCV RNA that we attributed to DNase-resistant plasmid amplified during the PCR procedure. As the silencing of YB-1 significantly improved extracellular RNA levels, these results strongly suggested that YB-1 restrains virus assembly and regulates the release of virus particles. To test if the increased extracellular HCV RNA levels correlate with an enhancement of the supernatant infectivity, we measured the de novo infection of naïve Huh7.5 cells at 72 h using intracellular HCV RNA and NS3 levels as readouts. As seen in Fig. 9C, supernatants originating from YB-1-depleted cells were significantly more infectious than those in the shNT condition. This shows that YB-1 knockdown led to an increased production of infectious particles. As expected, control supernatants of del153-167- and GND-expressing cells did not produce infectious particles.
Fig. 9.
YB-1 expression knockdown significantly increases the production of infectious viral particles. shNT- and shYB-1-expressing Huh7.5 cells were transfected with wild-type and mutant JFH-1-expressing plasmids. (A) Two days posttransfection, cells were collected and analyzed for their content in YB-1 and HCV RNA using Western blotting and qRT-PCR, respectively. The corresponding cell supernatants were collected, analyzed for their content in HCV RNA (B), and used to infect naive Huh7.5 cells (C). Three days postinfection, naive cells were collected and analyzed for their HCV NS3 and RNA content using Western blotting and qRT-PCR, respectively. HCV RNA levels were normalized with actin RNA content and arbitrarily set to 1 for the JFH-1-plus-shNT condition. All error bars represent standard deviations from biological triplicates.
Overall, the data show that YB-1 is involved in optimal HCV RNA replication and also negatively regulates infectious particle production. This dual role argues in favor of an important function of YB-1 in the control of the equilibrium between HCV replication and assembly.
DISCUSSION
Using a powerful TAP approach and mass spectrometry, we identified novel interacting partners of the HCV NS3/4A protein. No overlap was observed between these NS3/4A-associated host factors (with the exception of tubulin) and those previously identified (27), although most were RNA-binding proteins involved in RNA metabolism, splicing, localization, stability, and translation. Because of the technical limitations associated with an inducible TAP tag expression system, we used the nonhepatic 293EcR cell line that is not permissive for HCV replication. Hence, it is possible that we did not identify liver cell-specific NS3 protein partners. Unfortunately, attempts to purify NS3/4A interactome using Huh7 cell extracts resulted in yields and purity that were not satisfying for mass spectrometry analysis. However, most of the NS3/4A-interacting partners identified in this study have been validated in the Huh7.5 liver cell line. New purification strategies from liver cells will have to be elaborated and optimized for a complete analysis of the NS3/4A interactome. We focused this study on the potential role of YB-1 in the life cycle of HCV following the confirmation of its association with NS3/4A protein in an infectious model (Fig. 2). The functional relevance of the NS3/4A–YB-1 interaction was first established in the context of subgenomic replicon and infectious JFH-1 replication. Indeed, the efficient RNA interference (RNAi)-mediated reduction of YB-1 levels severely impaired HCV RNA replication regardless of the viral genotype. YB-1 dependency of HCV contrasts with the YB-1 restrictive role observed during Dengue virus infection (43). Differences between YB-1 roles in the virus replication of the Flaviviridae family members will have to be investigated further. Notably, YB-1 knockdown did not influence NS3-4A autoprocessing and MAVS cleavage by NS3 (L. Chatel-Chaix and D. Lamarre, unpublished results). Additionally, no NS3/4A protease-dependent cleavage of YB-1 was observed in any experiment, in contrast to results for MAVS (Fig. 7A). Thus, the role of YB-1 in the HCV life cycle does not seem to be related to NS3/4A proteolytic activity per se.
We also investigated the role of YB-1 in the regulation of HCV translation and/or stability. Indeed, this host factor, which associated with vRNA in HCV-infected cells (Fig. 5), previously was shown to bind exogenous HCV 5′- and 3′-untranslated regions in vitro (20, 34) and to positively regulate the IRES-controlled translation of the Myc family genes (12). In addition, a YB-1 restrictive role was observed in Dengue virus infection by its binding to the viral 3′-untranslated region (43). However, our data indicate that YB-1 knockdown or its overexpression in Huh7.5 cells did not significantly influence HCV IRES-mediated gene expression from the bicistronic construct and in vitro-transcribed RNA (data not shown). Additionally, HCV RNA, NS3, and core protein levels of the replication-defective GND JFH-1 mutant were not decreased following YB-1 knockdown (data not shown), strongly suggesting that YB-1 does not negatively influence HCV RNA stability and translation. We rather favor the idea that YB-1 is involved in the function of the HCV replication complex per se.
YB-1 associates with vRNA and NS3 protein in subgenomic 1b replicon-containing cells and in JFH-1-expressing or JFH-1 del153-167 mutant-expressing cells (Fig. 5 and data not shown), illustrating that viral assembly is not a prerequisite for the formation of YB-1–NS3/4A–vRNA complexes. It is conceivable that these interactions occur before viral assembly, and that the decreased HCV replication observed with YB-1 depletion is due to the disruption of these complexes. Finally, as other interacting partners of NS3/4A do not bind HCV RNA as efficiently as YB-1, we favor the direct interaction of YB-1 with HCV RNA either alone or in complex with NS3/4A. At this point we cannot exclude the RNA-binding property of NS3 as the unique determinant for the association of HCV RNA with YB-1–NS3/4A complexes.
YB-1 is specifically recruited upon JFH-1 infection to the surface of LD within complexes that contain core and NS3 proteins and presumably are viral particle assembly sites (Fig. 6 and 7). Core protein is essential but not sufficient for the YB-1 relocalization to LD, which also requires mature nonstructural proteins, as evidenced by the loss of phenotype with the BILN2061 NS3 protease inhibitor. In addition, our data suggest that NS2, NS5A, and p7 do not contribute to the recruitment of YB-1, as evidenced by the presence of the LD-associated phenotype in JFH-1-expressing mutants that are functionally defective in downstream steps of assembly (Fig. 8). In contrast, RHA, hnRNP U, and hnRNP A1 distribution remained unaltered upon JFH-1 infection (Fig. 6B and data not shown), although associations were observed with NS3 (Fig. 2 and 5). This supports that the observed JFH-1-induced redistribution phenotype is specific to YB-1. Our data favor the idea that YB-1–NS3/4A–vRNA complexes are co-opted by HCV capsids upon a rate-limiting transition step from RC to LD sites, since (i) this requires mature NS protein and RC formation but not the RNA replicative activity of NS5B polymerase (see data for BILN2061-treated cells and the JFH-1 GND mutant in Fig. 7D and 8) and (ii) the transient expression of either core or NS3/4A protein alone is unable to efficiently relocalize YB-1 to LDs (data not shown). Although YB-1 recruitment strictly required the presence of core protein, we never detected an interaction between core and YB-1 proteins using coimmunoprecipitation and bioluminescence resonance energy transfer assays (data not shown). This could be explained by (i) a very transient core–YB-1 interaction, (ii) an association which requires a three-dimensional structure of oligomeric HCV core subunits, or (iii) an indirect interaction for YB-1 recruitment to LD. Indeed, core protein has been shown to induce LD redistribution to the perinuclear region by a microtubule-dependent mechanism involving the motor protein dynein (6). Strikingly, both NS3/4A and YB-1 were shown to associate with microtubules, the latter harboring the potency to increase tubulin nucleation in vitro (10, 11, 27). Hence, core protein could favor the recruitment of tubulin-bound YB-1–NS3/4A RNA complexes by modulating the dynamics of cytoskeleton-associated LDs.
Finally, we showed that YB-1 could influence the release of infectious particles from plasmid-derived JFH-1-expressing cells under defined conditions. Unexpectedly, an experimental setup in which YB-1 expression knockdown decreased viral replication by 50% (Fig. 3C, day 2 posttransfection, and 9A) allowed us to visualize a significant increase of extracellular HCV RNA levels and numbers of infectious particles (Fig. 9). Thus, YB-1 can influence the equilibrium between RNA replication and virus assembly, which argues in favor of an important spatiotemporal function of YB-1 in the fine-tuned control of the HCV life cycle. Clearly, a YB-1-mediated role in particle production is transient and cannot compensate for its anti-replicative effect, since YB-1 depletion is almost completely lethal for HCV after 6 days in culture (Fig. 3C). These separate viral functions most probably are achieved through the HCV-mediated remodelling of YB-1 RNP. Indeed, our preliminary experiments involving quantitative mass spectrometry show that HCV induces drastic changes in protein composition and the stoichiometry of YB-1-containing RNP (L. Chatel-Chaix and D. Lamarre, unpublished). To understand how YB-1 is able to achieve separate viral functions and to identify new host factors exploited by HCV, we are performing a comparative proteomic study analyzing the YB-1 interactome under conditions in which wild-type, nonreplicative, and assembly-deficient JFH-1 are expressed.
During the HCV life cycle, vRNA is translated, replicated, and encapsidated into newly formed viral particles. These processes obviously cannot occur simultaneously and must be precisely fine-tuned in time and space with respect to the stoichiometry of the viral components. Very little is known about how HCV controls the dynamics between these processes. This study strongly supports a role of YB-1 at the equilibrium between RNA replication and capsid formation by limiting HCV assembly until the RNA genome is fully replicated and available for encapsidation into neosynthesized capsids. Unfortunately, it remains unknown what step of the assembly is controlled by YB-1. YB-1 depletion does not influence core loading to the LD surface and the further recruitment of NS5A to this compartment (L. Chatel-Chaix and D. Lamarre, unpublished). Thus, YB-1 might participate in the transfer of the vRNA from the ER-associated RC to the LD-associated assembly complex as proposed for the NS5A domain III (37). Efficient anti-core antibody in immunoprecipitation assays may determine if YB-1 depletion affects the levels of capsid-associated vRNA in JFH-1-infected cells. Alternatively, we favor the idea that the core-dependent recruitment of YB-1 in association with a subcellular NS3 fraction with LD regulates NS3 function during assembly. Indeed, Ma et al. proposed that NS3 helicase achieves its functions during HCV assembly through virus-host interactions (36).
Finally, YB-1 is involved in the expression of genes implicated in growth and survival and is associated with tumor progression (13, 16, 18, 19, 25, 55). Consequently, YB-1 has been proposed to represent a marker for cancer aggressiveness. Of particular importance, YB-1 has been shown to be overexpressed in hepatocellular carcinoma (55) that is induced in patients chronically infected with HCV. It is conceivable that the selection of hepatocytes with increased levels of YB-1 favors basal HCV RNA replication, hence they participate in chronicity in HCV-infected patients.
In conclusion, we have identified the host factor YB-1 as a novel NS3 partner which is recruited to the surface of LD by the assembling capsid in HCV infection. Our data highlight a dual role during the HCV life cycle for YB-1 that represents, to our knowledge, the first host factor reported to influence the equilibrium between RNA replication and particle production. Understanding how HCV hijacks cellular machineries such as YB-1 RNP to control its life cycle will improve our knowledge of HCV biology and may allow the design of novel antiviral strategies.
ACKNOWLEDGMENTS
We thank Jake Liang for pEF/JFH1-Rz/Neo, pEF/cJFH1 GND-Rz/Neo, pEF/CG-Rz/SEAP, and pEF/CG GND-Rz/SEAP plasmids; Takaji Wakita for the use of JFH-1; Benoît Coulombe for AB0411 plasmid and the 293EcR cell line; Judith Caron for the construction of TAP-NS3/4A; Ralf Bartenschlager (University of Heidelberg) for anti-NS5A antibodies, luciferase-encoding Con1b subgenomic replicon, 9-13 cells, and JFH-1-Luc plasmid; Darius Moradpour for anti-NS5B antibodies; and Christopher D. Richardson for HCVAB12Luc plasmid. We are grateful to the IRIC's Bio-Imagery Core Facility and Christian Charbonneau for confocal microscopy; IRIC's Genomics Core Facility, Raphaëlle Lambert, and Caroline Paradis for sequencing and real-time PCR; and IRIC's Proteomics Core Facility, Pierre Thibault, and Éric Bonneil for mass spectrometry and results analysis.
This work was supported by grants from the Canadian Institute of Health Research (MOP-86485) and by the Novartis/Canadian Liver Foundation Hepatology Research Chair to D.L. L.C.-C. was a recipient of the Michel Saucier Postdoctoral Fellowship (Michel Saucier and the Canadian Louis Pasteur Foundation).
Footnotes
Published ahead of print on 17 August 2011.
REFERENCES
- 1. Appel N., Schaller T., Penin F., Bartenschlager R. 2006. From structure to function: new insights into hepatitis C virus RNA replication. J. Biol. Chem. 281:9833–9836 [DOI] [PubMed] [Google Scholar]
- 2. Appel N., et al. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bargou R. C., et al. 1997. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat. Med. 3:447–450 [DOI] [PubMed] [Google Scholar]
- 4. Baril M., Racine M. E., Penin F., Lamarre D. 2009. MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J. Virol. 83:1299–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartenschlager R., Frese M., Pietschmann T. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71–180 [DOI] [PubMed] [Google Scholar]
- 6. Boulant S., et al. 2008. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic 9:1268–1282 [DOI] [PubMed] [Google Scholar]
- 7. Boulant S., Targett-Adams P., McLauchlan J. 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 88:2204–2213 [DOI] [PubMed] [Google Scholar]
- 8. Chang K. S., Jiang J., Cai Z., Luo G. 2007. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 81:13783–13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng G., Zhong J., Chisari F. V. 2006. Inhibition of dsRNA-induced signaling in hepatitis C virus-infected cells by NS3 protease-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. U. S. A. 103:8499–8504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chernov K. G., et al. 2008. Atomic force microscopy reveals binding of mRNA to microtubules mediated by two major mRNP proteins YB-1 and PABP. FEBS Lett. 582:2875–2881 [DOI] [PubMed] [Google Scholar]
- 11. Chernov K. G., et al. 2008. YB-1 promotes microtubule assembly in vitro through interaction with tubulin and microtubules. BMC Biochem. 9:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cobbold L. C., et al. 2008. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol. Cell. Biol. 28:40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dahl E., et al. 2009. Nuclear detection of Y-box protein-1 (YB-1) closely associates with progesterone receptor negativity and is a strong adverse survival factor in human breast cancer. BMC Cancer 9:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Evdokimova V., Ovchinnikov L. P., Sorensen P. H. 2006. Y-box binding protein 1: providing a new angle on translational regulation. Cell Cycle 5:1143–1147 [DOI] [PubMed] [Google Scholar]
- 15. Evdokimova V., et al. 2001. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J. 20:5491–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Evdokimova V., et al. 2009. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell 15:402–415 [DOI] [PubMed] [Google Scholar]
- 17. Ferreon J. C., Ferreon A. C., Li K., Lemon S. M. 2005. Molecular determinants of TRIF proteolysis mediated by the hepatitis C virus NS3/4A protease. J. Biol. Chem. 280:20483–20492 [DOI] [PubMed] [Google Scholar]
- 18. Finkbeiner M. R., et al. 2009. Profiling YB-1 target genes uncovers a new mechanism for MET receptor regulation in normal and malignant human mammary cells. Oncogene 28:1421–1431 [DOI] [PubMed] [Google Scholar]
- 19. Gluz O., et al. 2009. Y-box-binding protein YB-1 identifies high-risk patients with primary breast cancer benefiting from rapidly cycled tandem high-dose adjuvant chemotherapy. J. Clin. Oncol. 27:6144–6151 [DOI] [PubMed] [Google Scholar]
- 20. Harris D., Zhang Z., Chaubey B., Pandey V. N. 2006. Identification of cellular factors associated with the 3′-nontranslated region of the hepatitis C virus genome. Mol. Cell. Proteomics 5:1006–1018 [DOI] [PubMed] [Google Scholar]
- 21. Huang H., et al. 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 104:5848–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Isken O., et al. 2007. Nuclear factors are involved in hepatitis C virus RNA replication. RNA 13:1675–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kato T., et al. 2007. Production of infectious hepatitis C virus of various genotypes in cell cultures. J. Virol. 81:4405–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kedersha N., Anderson P. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 431:61–81 [DOI] [PubMed] [Google Scholar]
- 25. Kohno K., Izumi H., Uchiumi T., Ashizuka M., Kuwano M. 2003. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays 25:691–698 [DOI] [PubMed] [Google Scholar]
- 26. Kozak S. L., Marin M., Rose K. M., Bystrom C., Kabat D. 2006. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 281:29105–29119 [DOI] [PubMed] [Google Scholar]
- 27. Lai C. K., Jeng K. S., Machida K., Lai M. M. 2008. Association of hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5A. J. Virol. 82:8838–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lamarre D., et al. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186–189 [DOI] [PubMed] [Google Scholar]
- 29. Li K., et al. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. U. S. A. 102:2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li X. D., Sun L., Seth R. B., Pineda G., Chen Z. J. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. U. S. A. 102:17717–17722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lindenbach B. D., et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 32. Lohmann V., et al. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 33. Loo Y. M., et al. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 103:6001–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu H., Li W., Noble W. S., Payan D., Anderson D. C. 2004. Riboproteomics of the hepatitis C virus internal ribosomal entry site. J. Proteome Res. 3:949–957 [DOI] [PubMed] [Google Scholar]
- 35. Lunde B. M., Moore C., Varani G. 2007. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8:479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ma Y., Yates J., Liang Y., Lemon S. M., Yi M. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 82:7624–7639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Masaki T., et al. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McHutchison J. G., Fried M. W. 2003. Current therapy for hepatitis C: pegylated interferon and ribavirin. Clin. Liver Dis. 7:149–161 [DOI] [PubMed] [Google Scholar]
- 39. Miyanari Y., et al. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097 [DOI] [PubMed] [Google Scholar]
- 40. Moradpour D., Penin F., Rice C. M. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453–463 [DOI] [PubMed] [Google Scholar]
- 41. Murray C. L., Jones C. T., Tassello J., Rice C. M. 2007. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J. Virol. 81:10220–10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pacheco A., Reigadas S., Martinez-Salas E. 2008. Riboproteomic analysis of polypeptides interacting with the internal ribosome-entry site element of foot-and-mouth disease viral RNA. Proteomics 8:4782–4790 [DOI] [PubMed] [Google Scholar]
- 43. Paranjape S. M., Harris E. 2007. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J. Biol. Chem. 282:30497–30508 [DOI] [PubMed] [Google Scholar]
- 44. Pereira A. A., Jacobson I. M. 2009. New and experimental therapies for HCV. Nat. Rev. Gastroenterol. Hepatol. 6:403–411 [DOI] [PubMed] [Google Scholar]
- 45. Poynard T., Yuen M. F., Ratziu V., Lai C. L. 2003. Viral hepatitis C. Lancet 362:2095–2100 [DOI] [PubMed] [Google Scholar]
- 46. Shavinskaya A., Boulant S., Penin F., McLauchlan J., Bartenschlager R. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282:37158–37169 [DOI] [PubMed] [Google Scholar]
- 47. Skabkina O. V., et al. 2003. Poly(A)-binding protein positively affects YB-1 mRNA translation through specific interaction with YB-1 mRNA. J. Biol. Chem. 278:18191–18198 [DOI] [PubMed] [Google Scholar]
- 48. Steinmann E., et al. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 3:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Targett-Adams P., Boulant S., McLauchlan J. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J. Virol. 82:2182–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tellinghuisen T. L., Foss K. L., Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Villacé P., Marion R. M., Ortin J. 2004. The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res. 32:2411–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wakita T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wilson J. A., et al. 2003. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl. Acad. Sci. U. S. A. 100:2783–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang W. H., Bloch D. B. 2007. Probing the mRNA processing body using protein macroarrays and “autoantigenomics.” RNA 13:704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yasen M., et al. 2005. The up-regulation of Y-box binding proteins (DNA binding protein A and Y-box binding protein-1) as prognostic markers of hepatocellular carcinoma. Clin. Cancer Res. 11:7354–7361 [DOI] [PubMed] [Google Scholar]
- 56. Yi M., Ma Y., Yates J., Lemon S. M. 2009. Trans-complementation of an NS2 defect in a late step in hepatitis C virus (HCV) particle assembly and maturation. PLoS Pathog. 5:e1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]