Abstract
blaSIM-1 and blaOXA-23 were codetected in clinical carbapenem-resistant Acinetobacter baylyi strain NB09A30. Both of carbapenemase genes were located on a large plasmid (ca. 360 kb). blaSIM-1 was found as a gene cassette inserted into a class 1 integron identical to that determined in Acinetobacter sp. isolates from South Korea. The genetic structure of blaOXA-23 in NB09A30 was different from that in the prevalent Acinetobacter baumannii of clonal complex 92 (CC92) from the same hospital.
TEXT
Carbapenem-resistant Acinetobacter spp. are one of the most frightening threats in the current antibiotic era (14). The mechanisms of Acinetobacter sp. resistance to carbapenem are production of carbapenemases (the main mechanism), overexpression of efflux pumps, and reduced expression or loss of outer membrane proteins (8). The metallo-β-lactamase (MBL) gene, blaSIM-1, was first identified in Acinetobacter spp. from South Korea in 2005 (11) and then in Acinetobacter genomic species 10 from two hospitals of South Korea in 2010 (10) but, as far as we know, has not yet been detected in other countries. Here, we report the emergence of Acinetobacter baylyi coharboring blaSIM-1 and blaOXA-23 in China.
A 59-year-old male was admitted for cerebral hemorrhage at the Affiliated Hospital of Ningbo University in June 2009. After surgery to remove the intracerebral hematoma, he was transferred to the intensive care unit and received antibiotic therapy with cefminox, which was later replaced with cefodizime plus amikacin. Imipenem-resistant Acinetobacter sp. NB09A30 was isolated from the cloudy secretion of the surgical incision 10 days after the operation. MICs were determined by Etest and interpreted according to the CLSI 2011 recommendation (7). The strain was resistant to almost all β-lactams, including imipenem (MIC ≥ 32 μg/ml) and meropenem (MIC ≥ 32 μg/ml), but remained susceptible to minocycline, tigecycline, ciprofloxacin, levofloxacin, and colistin. The MICs of gentamicin and amikacin were ≥256 μg/ml and 16 μg/ml, respectively. Etest MBL strip testing against NB09A30 indicated the presence of metallo-β-lactamase (imipenem/imipenem plus EDTA MIC of >16 μg/ml). Surgical site infection was suspected, and the antibiotic regimen was changed to levofloxacin. The man was cured finally, and NB09A30 was no longer recovered.
PCR detection of the blaOXA-51-like gene in NB09A30 was negative, implying that NB09A30 is not an Acinetobacter baumannii strain (15). The 16S-23S rRNA intergenic spacer region (ISR) of NB09A30 was amplified and cloned into the pGEM-T Easy vector for sequencing (5). This showed that NB09A30 possessed ISRs with different lengths (long ISR, 650 bp; short ISR, 611 bp), a unique organization of the 16S-23S rRNA ISRs of A. baylyi (12). The ISR sequences were 94% identical to those of A. baylyi strain 93A2 (GenBank accession no. EU042163) and A. baylyi ADP1 (GenBank accession no. CR543861); they were less than 85% identical to those of the other genomic species, which indicated that NB09A30 is an A. baylyi strain (12).
PCR was performed to check for the presence of carbapenem-hydrolyzing class D β-lactamase (CHDL) genes (blaOXA-23, blaOXA-24, and blaOXA-58), MBL genes (blaIMP-type and blaVIM-type genes, blaSIM-1, blaSPM-1, blaGIM-1, and blaNDM-1), and the blaKPC-type gene, as previous reported (13, 16). blaSIM-1 and blaOXA-23 were detected and confirmed by sequencing.
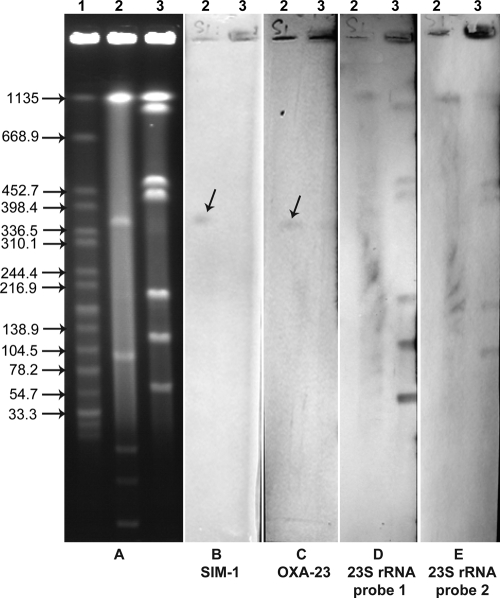
Repeated mating-out assays and electrotransformation to transfer blaSIM-1 and blaOXA-23 failed. Therefore, we used an S1 nuclease assay and an I-CeuI assay to identify the locations of the two carbapenemase genes as previously described (2). Two 23S rRNA gene probes were designed to target the sequences upstream and downstream of the I-CeuI cut site, respectively, according to the genomic sequence of A. baylyi strain ADP1 (GenBank accession no. CR543861). All seven I-CeuI-generated DNA fragments can be hybridized with 23S rRNA gene probe 1 plus 23S rRNA gene probe 2 but did not cohybridize with blaSIM-1 or blaOXA-23 probes, excluding chromosomal locations for the two carbapenemase genes (Fig. 1). In contrast, hybridization signals of blaSIM-1 and blaOXA-23 probes were codetected in a ca. 360-kb large plasmid after S1 nuclease treatment, suggesting plasmid locations for the two carbapenemase genes (Fig. 1). blaSIM-1 was confirmed to have a chromosomal location in Acinetobacter spp. of South Korea (10, 11), but it has a large-plasmid location in A. baylyi strain NB09A30. Using the A. baumannii plasmid replicon typing scheme constructed by Bertini et al. (4), we detected four replicase (rep) gene groups (GRs), GR2, GR3, GR7, and GR13, in A. baylyi strain NB09A30. Further hybridization with corresponding probes is needed to define the large plasmid's rep group.
Fig. 1.
Analysis of the locations of blaSIM-1 and blaOXA-23 in A. baylyi strain NB09A30 using the S1 nuclease assay and I-CeuI assay. Shown are pulsed-field gel electrophoresis (PFGE) profiles of total DNA after digestion (A) and Southern blotting and hybridization results with blaSIM-1 (B), blaOXA-23 (C), 23S rRNA gene probe 1 (D), and 23S rRNA gene probe 2 (E). The largest chromosomal fragment of the I-CeuI profile was not hybridized using 23S rRNA gene probe 1 (D). The smallest chromosomal fragment was not hybridized using 23S rRNA gene probe 2 (E). Arrows indicate the hybridization signals of blaSIM-1 and blaOXA-23. Lane 1, Salmonella enterica serotype Braenderup strain H9812 DNA digested by XbaI, used as a molecular marker (kb); lanes 2, A. baylyi strain NB09A30 digested by S1 nuclease; lanes 3, A. baylyi strain NB09A30 DNA digested by I-CeuI.
Primers targeting 5′ and 3′ conserved segments of the class I integron were used for PCR amplification (11). blaSIM-1 of A. baylyi strain NB09A30 was found to be class I integron borne. It is interesting that the sequence of the entire integron was 100% identical to that in Acinetobacter sp. isolates from South Korea (GenBank accession no. EF125010) (11). The genetic structure of blaOXA-23 was determined by a cloning experiment as previously reported (1). blaOXA-23 in A. baylyi strain NB09A30 was part of transposon Tn2008 (1), which was inserted into a noncoding region and flanked by two copies of an 11-bp direct repeat (ATATTCTGTTT).
In order to trace the source of blaSIM-1 and blaOXA-23, we further screened for the presence of the two carbapenemases in clinical A. baumannii isolates. From October 2008 to June 2009, we collected 29 carbapenem-resistant A. baumannii isolates from the same hospital. Multilocus sequence typing (MLST) identified an identical sequence type, ST138 (http://pubmlst.org/abaumannii/), in all isolates (3); this sequence type was clustered into clonal complex 92 (CC92), the dominant clonal complex of carbapenem-resistant A. baumannii in China (9). blaOXA-23 was identified in all isolates, but blaSIM-1 was absent. The patient in this case had no history of visiting South Korea. Moreover, we failed to detect blaSIM-1 in A. baumannii isolates prevalent in the hospital at the time. Therefore, the source of blaSIM-1 in A. baylyi NB09A30 is still unknown. The genetic structure of blaOXA-23 in the prevalent A. baumannii isolates with ST138 included Tn2009, a novel blaOXA-23-containing transposon also identified in CC92 A. baumannii strain MDR-ZJ06 from China (17). Distinct from Tn2008, which was flanked on one side by ISAba1, Tn2009 was a composite transposon flanked by two ISAba1 sequences with identical orientations (17), implying different origins of blaOXA-23 in A. baylyi NB09A30 and A. baumannii ST138.
In conclusion, our study identified blaSIM-1 for the first time in China. Since the potential source and details of the epidemiological situation are unclear, more study should be carried out to survey the presence of blaSIM-1 in Acinetobacter spp. A. baylyi used to be considered an environment species, existing in soil and activated sludge, and nonpathogenic (8). Though nosocomial infections caused by A. baylyi have been reported (6), carbapenem resistance is still rare. More attention should be paid to the emergence of carbapenem resistance in this potential pathogen.
Nucleotide sequence accession numbers.
The nucleotide sequences of the 16S-23S rRNA intergenic spacer region, the blaSIM-1-containing class I integron, Tn2008 of A. baylyi strain NB09A30, and Tn2009 of the prevalent A. baumannii isolates of ST138 are deposited in the GenBank database under accession no. JF731031 to JF731040, JF731030, JF731029, and JF731028, respectively.
Acknowledgments
This study was supported by research grants from National Natural Science Foundation of China (no. NSFC30970113), the Ministry of Health of the People's Republic of China (no. 200802107), and the Natural Science Foundation of Zhejiang Province (no. Y207477).
Footnotes
Published ahead of print on 29 August 2011.
REFERENCES
- 1. Adams-Haduch J. M., et al. 2008. Genetic basis of multidrug resistance in Acinetobacter baumannii clinical isolates at a tertiary medical center in Pennsylvania. Antimicrob. Agents Chemother. 52:3837–3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arias C. A., Panesso D., Singh K. V., Rice L. B., Murray B. E. 2009. Cotransfer of antibiotic resistance genes and a hylEfm-containing virulence plasmid in Enterococcus faecium. Antimicrob. Agents Chemother. 53:4240–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartual S. G., et al. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bertini A., et al. 2010. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54:4168–4177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang H. C., et al. 2005. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J. Clin. Microbiol. 43:1632–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen T. L., et al. 2008. Acinetobacter baylyi as a pathogen for opportunistic infection. J. Clin. Microbiol. 46:2938–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Dijkshoorn L., Nemec A., Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939–951 [DOI] [PubMed] [Google Scholar]
- 9. Fu Y., et al. 2010. Wide dissemination of OXA-23-producing carbapenem-resistant Acinetobacter baumannii clonal complex 22 in multiple cities of China. J. Antimicrob. Chemother. 65:644–650 [DOI] [PubMed] [Google Scholar]
- 10. Lee K., et al. 2010. Characteristics of clinical isolates of Acinetobacter genomospecies 10 carrying two different metallo-beta-lactamases. Int. J. Antimicrob. Agents 36:259–263 [DOI] [PubMed] [Google Scholar]
- 11. Lee K., et al. 2005. Novel acquired metallo-beta-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother. 49:4485–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maslunka C., Gurtler V., Carr E. L., Seviour R. J. 2008. Unique organization of the 16S-23S intergenic spacer regions of strains of Acinetobacter baylyi provides a means for its identification from other Acinetobacter species. J. Microbiol. Methods 73:227–236 [DOI] [PubMed] [Google Scholar]
- 13. Moubareck C., Bremont S., Conroy M. C., Courvalin P., Lambert T. 2009. GES-11, a novel integron-associated GES variant in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:3579–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Towner K. J. 2009. Acinetobacter: an old friend, but a new enemy. J. Hosp. Infect. 73:355–363 [DOI] [PubMed] [Google Scholar]
- 15. Turton J. F., et al. 2006. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44:2974–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yong D., et al. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou H., et al. 25 July 2011. Genomic analysis of the multidrug-resistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob. Agents Chemother. doi: 10.1128/AAC.01134-10 [DOI] [PMC free article] [PubMed] [Google Scholar]