Abstract
A carbapenem-resistant Serratia marcescens strain, 10mdr148, was identified in a Japanese hospital in 2010. The carbapenem resistance of this strain was attributed to the production of a novel metallo-β-lactamase (MBL), named SMB-1 (Serratia metallo-β-lactamase). SMB-1 possessed a zinc binding motif, H(Q)XHXDH (residues 116 to 121), H196, and H263 and was categorized as a member of subclass B3 MBL. SMB-1 has 75% amino acid identity with the most closely related MBL, AMO1, of uncultured bacterium, recently identified through the metagenomic analysis of apple orchard soil. The introduction of blaSMB-1 into Escherichia coli conferred resistance to a variety of β-lactam antibiotics, penicillins, cephalosporins, and carbapenems, but not aztreonam, a resistance pattern consistent with those of other MBLs. SMB-1 demonstrated high kcat values of >500 s−1 for carbapenems, resulting in the highest hydrolyzing efficiency (kcat/Km) among the agents tested. The hydrolyzing activity of SMB-1 was well inhibited by chelating agents. The blaSMB-1 gene was located on the chromosome of S. marcescens strain 10mdr148 and at the 3′ end of the ISCR1 element in complex with a typical class 1 integron carrying aac(6′)-Ib and catB3 gene cassettes. Downstream of blaSMB-1, the second copy of the 3′conserved segment and ISCR1 were found. To our knowledge, this is the first subclass B3 MBL gene associated with an ISCR1 element identified in an Enterobacteriaceae clinical isolate. A variety of antibiotic resistance genes embedded with ISCR1 have been widely spread among Enterobacteriaceae clinical isolates, thus the further dissemination of blaSMB-1 mediated by ISCR1 transposition activity may become a future concern.
INTRODUCTION
The emergence of carbapenem resistance in Enterobacteriaceae clinical isolates is becoming a substantial clinical concern, because carbapenem antibiotics remain important agents for the treatment of infectious diseases caused by pathogenic Enterobacteriaceae in clinical settings (2, 7). The carbapenem resistance of these bacterial strains is due mostly to the production of horizontally acquired β-lactamases that are capable of hydrolyzing carbapenems, like IMP-1 metallo-β-lactamase (MBL), which was first characterized in a Serratia marcescens clinical isolate in Japan (18), and a KPC β-lactamase was first identified from a Klebsiella pneumoniae clinical isolate in the United States (31). As for MBLs, the identification of IMP- and VIM-type MBL genes, mediated by specific genetic elements like integrons, have increasingly been reported worldwide. In addition, SPM-1, SIM-1, GIM-1, KHM-1, and DIM-1 MBL genes have been found sporadically in members of the family Enterobacteriaceae, Pseudomonas spp., and Acinetobacter baumannii (4, 16, 20, 22, 28).
Recently, a novel MBL, NDM-1, was identified from a K. pneumoniae strain recovered from a Swedish patient who had come back from India (32). After that, several reports indicate the further worldwide dissemination of NDM-1 producers (5, 15, 19, 26), and it is becoming a great threat to human health together with the fact that NDM-1 producers also often possess a multidrug-resistant nature (14, 19). In consideration of the rapid worldwide spread of NDM-1 producers, a nationwide survey in Japan was performed to determine whether or not the NDM-1-producing bacterial strain resided in imipenem-nonsusceptible or ceftazidime-resistant Enterobacteriaceae strains collected from clinical facilities in Japan between September and December in 2010. In this survey, the presence of blaNDM-1, blaKPC, blaIMP-1, blaIMP-2, and blaVIM-2 was detected by PCR in the collected strains. As a result, a small number of blaNDM-1-positive strains were identified, although most of the MBL genes found in the collected strains were IMP-1-type MBL genes (unpublished data).
Several strains, including S. marcescens strain 10mdr148, were found to be negative for the five carbapenemase genes described above, despite showing resistance to carbapenems. These results indicate the possibility that the carbapenem resistance of these isolates, including S. marcescens strain 10mdr148, depends on an unknown molecular mechanism, such as the production of a novel MBL. This study aimed to characterize the molecular mechanism underlying the carbapenem resistance found in the S. marcescens 10mdr148 clinical isolate.
MATERIALS AND METHODS
Bacterial strain.
The clinical isolate S. marcescens 10mdr148 was identified using the API-20E system (bioMérieux) and VITEK2 system (bioMérieux). The chromosomally encoded ampC β-lactamase gene and 16S rRNA gene of S. marcescens strain 10mdr148 was amplified with the primers ampC-F and ampC-R as well as 16S rRNA-10F and 16S rRNA-800R, respectively (Table 1). The sequence of the amplified products was determined.
Table 1.
Primers used in this study
| Primer | Sequencea | Nucleotide positionb |
|---|---|---|
| ampC-F | 5′-ATG ACG AAA GTG AAC CGC-3′ | 1-18 |
| ampC-R | 5′-CCT GGA CGA TGT GGT AAG-3′ | 1103-1120 |
| CTX-M-3F | 5′-ACC GTC ACG CTG TTG TTA G-3′ | 47-64 |
| CTX-M-3R | 5′-CTT TCT GCC TTA GGT TGA G-3′ | 806-824 |
| IMP-1F | 5′-ACC GCA GCA GAG TCT TTG CC-3′ | 49-68 |
| IMP-1R | 5′-ACA ACC AGT TTT GCC TTA CC-3′ | 616-635 |
| IMP-2F | 5′-GTT TTA TGT GTA TGC TTC C-3′ | 16-34 |
| IMP-2R | 5′-AGC CTG TTC CCA TGT AC-3′ | 677-693 |
| VIM-2F | 5′-ATG TTC AAA CTT TTG AGT AAG-3′ | 1-21 |
| VIM-2R | 5′-CTA CTC AAC GAC TGA GCG-3′ | 784-801 |
| NDM-1F | 5′-TTG CCC AAT ATT ATG CAC CC-3′ | 7-26 |
| NDM-1R | 5′-ATT GGC ATA AGT CGC AAT CC-3′ | 407-426 |
| 16S rRNA-10F | 5′-GTT TGA TCC TGG CTC A-3′ | 11-26 |
| 16S rRNA-800R | 5′-TAC CAG GGT ATC TAA TCC-3′ | 785-802 |
| SMB-F | 5′-CAG CAG CCA TTC ACC ATC TA-3′ | 79-98 |
| SMB-R | 5′-GAA GAC CAC GTC CTT GCA CT-3′ | 551-570 |
| SMB-CloF | 5′-CCC AAGCTT TCC GCC GAC TTG GCG CAG-3′ | |
| SMB-CloR | 5′-GGGGTACCA AGA CCG ATT TAG CCG GC-3′ | |
| PET-1 | 5′-GGA ATT CCATATGAA AAT CAT CGC TTC CC-3′ | |
| PET-2 | 5′-CCC AAGCTT TCA GCG TTT CTC GCT GGC C-3′ |
Underlines indicate the sites for restriction endonuclease.
Position numbers correspond to the nucleotides of the coding sequences. Position numbers are assigned to the primers that amplify the internal region of the coding sequences.
PCR.
The primers used for the detection of β-lactamase genes are listed in Table 1.
Susceptibility testing.
The production of MBL was detected using a disk containing sodium mercaptoacetic acid (SMA) (Eiken) (23) and an Etest MBL IP/IPI strip (bioMérieux). The MICs of various β-lactam antibiotics were determined with the agar dilution method according to the CLSI guideline (6). The MICs of amikacin, gentamicin, ciprofloxacin, moxifloxacin, and tigecycline were determined by Etest.
Conjugation.
Escherichia coli strain DH10B was used as the recipient. Conjugation was performed as described elsewhere (30). The conjugants were selected on LB agar plates containing streptomycin (100 μg/ml) and ceftazidime (2 μg/ml).
Cloning of blaSMB-1.
The total DNA of S. marcescens strain 10mdr148 was extracted using the Wizard genomic DNA purification kit (Promega) and partially digested with Sau3AI. The digested fragments were ligated to pCL1920 cloning vector previously digested with BamHI, dephosphorylated, and transformed into E. coli strain KAM32. The transformants were selected on LB agar plates supplemented with streptomycin (25 μg/ml) and ceftazidime (2 μg/ml). The fragments on the two obtained recombinant plasmids (pCL1 and pCL2) were sequenced. The blaSMB-1 gene and its putative promoter region were amplified with the primers SMB-CloF and SMB-CloR (Table 1) with total DNA of S. marcescens strain 10mdr148 as the template and then cloned into a pCL1920 vector. The constructed plasmid (pCL-SMB) was transformed into E. coli KAM32.
Overexpression and purification of SMB-1.
The blaSMB-1 gene was amplified with the primers PET-1, which introduced an NdeI restriction site at the 5′ end, and PET-2, which introduced a HindIII site at the 3′ end (Table 1). The amplified fragments were digested with the endonucleases and ligated into pET30a vector (Novagen). The recombinant plasmid pET-SMB was introduced into E. coli BL21(DE3)pLysS by electroporation. The cells were cultured in 2 liters of LB broth supplemented with chloramphenicol (30 μg/ml) and kanamycin (30 μg/ml) at 37°C. Isopropyl-β-d-thiogalactopyranoside (IPTG) (final concentration, 0.5 mM) was added when the culture reached an optical density at 600 nm of 0.5, and the culture was incubated for an additional 3 h at 37°C. Cells were washed with 50 mM morpholineethanesulfonic acid (MES) buffer (pH 6.0), disrupted with a French press, and centrifuged at 100,000 × g for 30 min. The supernatant containing recombinant protein was loaded onto a HiTrap SP HP column (GE Healthcare) preequilibrated with 50 mM MES buffer (pH 6.0) and eluted with a linear gradient of 0 to 0.5 M NaCl. The fraction containing the protein was concentrated to a volume of 2 ml using an Amicon Ultra-15 Centricon (Millipore), loaded onto a HiLoad 16/60 Superdex 200 pg column (GE Healthcare), and eluted with 20 mM Tris-HCl (pH 7.5) buffer containing 0.2 M NaCl. The fraction containing the protein was concentrated and further loaded onto a HiPrep 16/60 Sephacryl S-200 HR column (GE Healthcare) and eluted with 20 mM Tris-HCl (pH 7.5) buffer containing 0.2 M NaCl. The eluted protein was stored at −80°C until use. The purity of the protein was estimated by SDS-PAGE and Coomassie brilliant blue (CBB) staining. The N-terminal sequence of the purified SMB-1 was obtained by Edman degradation with a model Shimadzu PPSQ-33A automated protein sequencer. The SMB-1 was subjected to isoelectric focusing (IEF) analysis with a CleanGel IEF (GE Healthcare).
Kinetic parameters.
The kinetic parameters of the purified SMB-1 against various β-lactams were measured at 30°C in 50 mM HEPES-NaOH buffer (pH 7.5) with an Ultrospec 3000 spectrophotometer (Pharmacia Biotech). The concentration of enzyme used was 1.6 nM to 1 μM in this assay. The values of the kinetic parameters Km and kcat were calculated from a Michaelis-Menten plot of the initial steady-state velocities (29). The kinetic values in this study are the means from at least three independent measurements. At least six different concentrations were used to determine the kinetic parameters for each substrate.
Inactivation of SMB-1 by chelating agents.
The inactivation of SMB-1 by removing Zn2+ was carried out in the presence of EDTA (Wako), dipicolinic acid (Sigma), and 1,10-o-phenanthroline (Sigma) at different concentrations. Meropenem (100 μM) was used as the reporter substrate in the present study. The 50% inhibitory concentration (IC50) was determined by preincubating 1.6 nM enzyme with various concentrations of chelating agents in HEPES-NaOH buffer (pH 7.5) for 5 min at 30°C before adding meropenem.
PFGE and Southern hybridization.
Plugs containing total DNA of the S. marcescens 10mdr148 clinical isolate were digested with I-CeuI overnight. The digested DNAs were subjected to electrophoresis with a contour-clamped homogeneous electric field (CHEF) DRIII drive module (Bio-Rad), with pulses ranging from 2.9 to 33.7 s at 6 V/cm for 27 h at 14°C. The DNA was transferred to Zeta-Probe blotting membranes (Bio-Rad) and hybridized with two probes: a 492-bp probe specific for blaSMB-1 (primers SMB-F and SMB-R) and a 792-bp probe specific for the 16S rRNA gene (primers 16S rRNA-10F and 16S rRNA-800R) (Table 1). The generation of probes and signal detection were carried out using the DIG High Prime DNA labeling and detection starter kit II (Roche).
Determination of genetic environment of blaSMB-1.
Plugs containing total DNA of the S. marcescens 10mdr148 clinical isolate were digested with SpeI. The plugs were completely melted with incubation at 70°C, and β-agarase (Takara) was added. After digestion, DNA was subjected to electrophoresis with a 1% agarose gel, and fragments of 9 to 23 kb were purified with a Wizard SV gel and PCR clean-up system (Promega). The purified fragments were ligated to the pMCL210 vector and transformed into E. coli DH10B by electroporation. The transformants were selected on LB agar plates supplemented with chloramphenicol (15 μg/ml) and ceftazidime (2 μg/ml). The cloned fragments were sequenced.
Nucleotide sequence accession number.
The nucleotide sequence of blaSMB-1 presented in this study has been deposited in GenBank under accession no. AB636283.
RESULTS AND DISCUSSION
Clinical isolate.
S. marcescens strain 10mdr148 was isolated in 2010 from the urine of an inpatient in a Japanese hospital. This strain showed a high level of resistance to various β-lactams, including penicillins, cephalosporins, carbapenems, and aztreonam, but it was susceptible to aminoglycosides, fluoroquinolones, and tigecycline (FDA breakpoints), as shown in Table 2. An apparent expansion of the growth-inhibitory zone around the imipenem disk was observed when the SMA disk was closely placed, and the >64-fold reduction in the MIC of imipenem was observed when using an Etest IP/IPI strip (data not shown). These results indicated the possibility that carbapenem resistance of S. marcescens strain 10mdr148 was attributed to the production of some kind of MBL. However, the preliminary PCR detection of the MBL genes blaIMP-1, blaIMP-2, blaVIM-2, and blaNDM-1 that have been found so far in clinical isolates in Japan gave no positive result.
Table 2.
Result of susceptibility testing
| Antimicrobial agent | MIC (μg/ml) against strain: |
||
|---|---|---|---|
| S. marcescens 10mdr148 | E. coli KAM32 (pCL-SMB) | E. coli KAM32 (pCL1920) | |
| Ampicillin | >256 | >256 | 2 |
| Piperacillin | >256 | >256 | 0.25 |
| Cephalothin | >256 | 256 | 8 |
| Cephaloridine | >256 | 64 | 2 |
| Cefazolin | >256 | 256 | 1 |
| Cefuroxime | >256 | >256 | 0.25 |
| Cefotaxime | >256 | 16 | ≤0.06 |
| Ceftazidime | 256 | 256 | 0.25 |
| Cefepime | >256 | 0.5 | ≤0.06 |
| Cefoxitin | >256 | >256 | 2 |
| Cefmetazole | >256 | 256 | 0.5 |
| Flomoxef | >256 | 64 | ≤0.06 |
| Aztreonam | >256 | 0.13 | 0.13 |
| Imipenem | >32 | 8 | 0.25 |
| Meropenem | >32 | 16 | 0.03 |
| Panipenem | >32 | 32 | 0.5 |
| Biapenem | >32 | 16 | 0.25 |
| Amikacin | 12 | NDa | ND |
| Gentamicin | 1.5 | ND | ND |
| Ciprofloxacin | 1 | ND | ND |
| Moxifloxacin | 0.5 | ND | ND |
| Tigecycline | 1 | ND | ND |
ND, not determined.
The CTX-M-3-type β-lactamase gene and ampC β-lactamase gene were detected by PCR. The sequence of the amplified fragments (nucleotide positions 65 to 805) for the CTX-M-3-type β-lactamase gene was 100% identical to the corresponding region of blaCTX-M-3 of Enterobacter aerogenes (GenBank accession no. AB432919). The sequence of the amplified fragments (nucleotide positions 19 to 1102) for the ampC β-lactamase gene was 99% identical to the corresponding region of the chromosomally encoded ampC β-lactamase gene of S. marcescens (GenBank accession no. AY524276).
Cloning and characterization of an MBL gene.
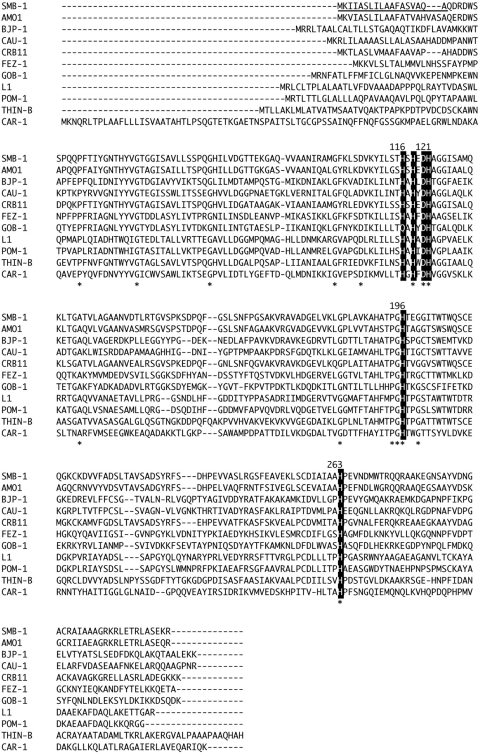
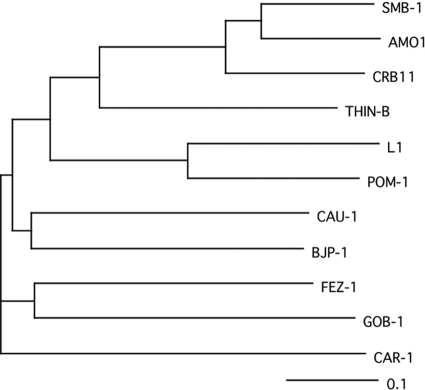
Although the conjugation experiment was performed to transfer the β-lactam resistance of S. marcescens strain 10mdr148 to E. coli DH10B, a transconjugant could not be obtained under the experimental conditions in this study. Thus, we attempted to isolate the genes responsible for β-lactam resistance by a shotgun cloning experiment using total DNA of S. marcescens strain 10mdr148. As a result, two recombinant plasmids of different sizes were obtained: pCL1, carrying a ca. 2-kb fragment, and pCL2, carrying a ca. 6-kb fragment. Both plasmids carried the same 843-bp open reading frame (ORF) encoding a protein consisting of 280 amino acids. This protein was assigned to be a member of Ambler class B β-lactamases through database homology searching, and it was named SMB-1 (Serratia metallo-β-lactamase). SMB-1 possessed a zinc binding motif, H(Q)XHXDH (residues 116 to 121), H196, and H263, which were well conserved in MBLs belonging to the subclass B3 MBL group (Fig. 1) (13). Thus, SMB-1 could be categorized as a member of subclass B3 MBL. SMB-1 exhibited 75 and 71% amino acid identity to AMO1 and CRB11 MBLs, respectively, of uncultured bacterium recently identified through the metagenomic analysis of apple orchard soil (10), and it exhibited 42% identity to THIN-B MBL of Janthinobacterium lividum (Fig. 2) (21). The G+C content of blaSMB-1 was 63%, slightly higher than that (60%) of the S. marcescens strain Db11 genome (http://www.sanger.ac.uk/resources/downloads/bacteria/serratia-marcescens.html).
Fig. 1.
Amino acid alignments of SMB-1 sequence with those of other subclass B3 MBLs. The residues involved in zinc binding are highlighted with a dark background. The signal peptide of SMB-1 is shown with an underline. An asterisk indicates amino acid residues conserved among all subclass B3 MBLs. Proteins (GenBank accession no.) are the following: SMB-1 (AB636283), AMO1 (ACS83721), BJP-1 (NP772870), CAU-1 (CAC87665), CRB11 (ACS83724), FEZ-1 (CAB96921), GOB-1 (ABO21417), L1 (ABO60992), POM-1 (ADC79555), THIN-B (CAC33832), and CAR-1 (Q6D395).
Fig. 2.
Tree view exhibiting the similarity of SMB-1 with the other subclass B3 MBLs. The tree was constructed using ClustalW, version 1.83 (http://clustalw.ddbj.nig.ac.jp/top-j.html), and was provided by the DNA Data Bank of Japan (DDBJ). Sequences incorporated to draw the tree were the same as those used for Fig. 1. The 0.1 scale represents a genetic unit reflecting 10% of the amino acid substitutions, and it was calculated with the ClustalW program.
Susceptibility testing of the transformant producing SMB-1.
In the present study, the E. coli transformant that produces SMB-1, encoded on the recombinant plasmid (pCL-SMB) carrying blaSMB-1 and its putative promoter region, was subjected to susceptibility testing. The transformant producing SMB-1 showed resistance to a variety of β-lactams, except aztreonam (Table 2). The SMB-1 production yielded a >8-fold increase in the MIC of cefepime, but cefepime remained fully active against the SMB-1-producing transformant. This trend was observed in the transformant that produces the THIN-B subclass B3 MBL (21). On the one hand, most of the transformants producing the subclass B3 MBLs, such as CAR-1 and BJP-1, could not confer resistance to cefepime (24, 25). The high MICs of cefepime and aztreonam observed for the parent S. marcescens strain 10mdr148 probably were attributable to the production of CTX-M-3-type β-lactamase and/or AmpC β-lactamase, not to SMB-1 MBL.
Biophysical characterization of SMB-1.
E. coli BL21(DE3)pLysS and the pET30a expression vector were used for the overexpression and purification of SMB-1. E. coli BL21(DE3)pLysS carrying pET30a was susceptible to ceftazidime (MIC, ≤0.06 μg/ml), while E. coli BL21(DE3)pLysS carrying pET-SMB showed a reduction in susceptibility to ceftazidime (MIC, 8 μg/ml). This result indicated that the recombinant SMB-1 produced was functional and responsible for β-lactam resistance in E. coli BL21(DE3)pLysS. An optimized culture condition yielded 22 mg of purified protein per 2 liters of bacterial culture, and the purified enzyme gave a single band on SDS-PAGE with CBB staining (data not shown). The N-terminal sequence of mature SMB-1 was determined to be QDRDW by Edman degradation, and this corresponds to the sequence after the cleavage of signal peptide predicted by GENETYX-MAC version 14.0.1. The native SMB-1 was determined to be a monomeric form by gel filtration. The pI of SMB-1 was estimated to be 7.4 by isoelectric focusing.
Kinetic parameters of SMB-1.
The results of the kinetic parameters of SMB-1 against representative β-lactams are shown in Table 3. SMB-1 was capable of hydrolyzing penicillins, most cephalosporins (except cefepime), and carbapenems. The hydrolyzing efficiency (kcat/Km) of SMB-1 against cefepime was low due to its higher Km value (747 μM) and the lower kcat value (2.7 s−1), corroborating the MIC of cefepime conferred by SMB-1 production (Table 2). The poor hydrolytic efficiency (kcat/Km, <104 M−1·s−1) of SMB-1 against cefepime was very similar to those of other subclass B3 MBLs, such as BJP-1, FEZ-1, CAR-1, and THIN-B (8, 17, 24, 25). SMB-1 demonstrated higher kcat values (>500 s−1) against carbapenems, imipenem, and meropenem, resulting in high hydrolytic efficiency. The properties of high hydrolytic efficiency (kcat/Km, >105 M−1·s−1) against carbapenems of SMB-1 are similar to those of BJP-1, L1, GOB-1, FEZ-1, CAU-1, and THIN-B (1, 8, 9, 11, 17, 25). Against carbapenems, a significant substrate preference, like meropenem over imipenem, as observed in BJP-1, L1, and GOB-1, was not identified in SMB-1 (1, 11, 12, 25). SMB-1 exhibited no measurable hydrolyzing activity against aztreonam, in accordance with previous biochemical studies of other MBLs (3). The inhibition profile determined, the IC50 with meropenem as the substrate, revealed that the activity of SMB-1 was well inhibited by the chelating agents dipicolinic acid, 1,10-o-phenanthroline, and EDTA, as shown in Table 3.
Table 3.
Kinetic parameters and inhibition profile of SMB-1a
| Substrate or chelating agent | Km (μM) | kcat (s−1) | kcat/Km (M−1·s−1) | Relative kcat/Kmd | IC50 (μM) |
|---|---|---|---|---|---|
| Ampicillin | 102 | 247 | 2.4 × 106 | 100 | |
| Piperacillin | 380 | 68 | 1.8 × 105 | 7.5 | |
| Cephalothin | 15 | 28 | 1.9 × 106 | 79 | |
| Cefuroxime | 22 | 30 | 1.4 × 106 | 58 | |
| Cefotaxime | 35 | 31 | 8.9 × 105 | 37 | |
| Ceftazidime | 57 | 4.4 | 7.7 × 104 | 3.2 | |
| Cefepime | 747 | 2.7 | 3.6 × 103 | 0.15 | |
| Cefoxitin | 26 | 39 | 1.5 × 106 | 63 | |
| Aztreonam | NHb | NDc | ND | ND | |
| Imipenem | 133 | 518 | 3.9 × 106 | 163 | |
| Meropenem | 144 | 604 | 4.2 × 106 | 175 | |
| Dipicolinic acid | 2.2 | ||||
| 1,10-o-Phenanthroline | 156 | ||||
| EDTA | 14 |
Standard deviations for each parameter were below 10%.
NH, no measurable hydrolysis detected with 1 μM enzyme.
ND, not determined.
Relative kcat/Km value was expressed compared to that of ampicillin, which was assigned 100.
Localization of blaSMB-1 in S. marcescens strain 10mdr148.
The I-CeuI digestion of total DNA of S. marcescens strain 10mdr148 yielded six fragments of different sizes under the experimental conditions employed in this study (Fig. 3). Southern hybridization revealed that five of the six fragments hybridized with the probes specific for the 16S rRNA gene (Fig. 3). The signal by the blaSMB-1 probe was detected with the ca. 730-kb fragment (Fig. 3), which also was hybridized with the probes specific for the 16S rRNA gene. Therefore, it was found that the blaSMB-1 gene was located on the chromosome of S. marcescens strain 10mdr148.
Fig. 3.
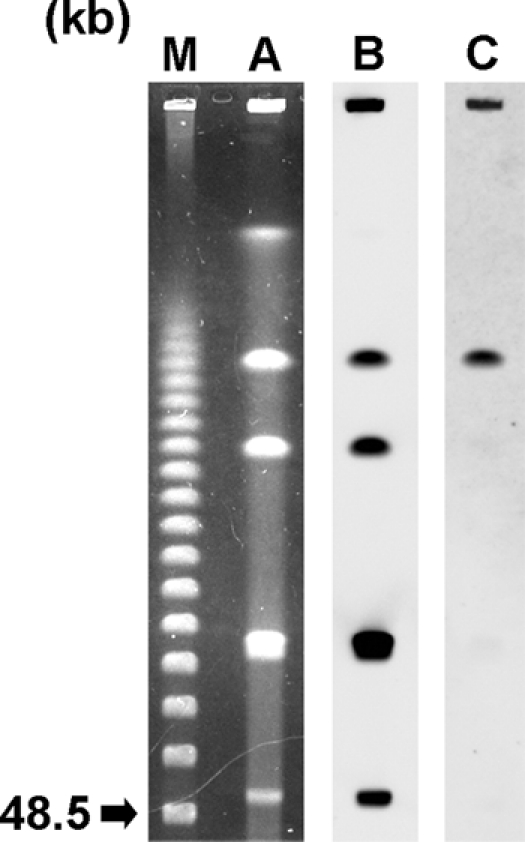
Localization of the blaSMB-1 gene on I-CeuI-digested total DNA of S. marcescens strain 10mdr148 separated by PFGE. Lane: M, CHEF DNA size standard marker (Bio-Rad); A, I-CeuI-digested total DNA of S. marcescens strain 10mdr148 stained with ethidium bromide; B, hybridization of I-CeuI-digested total DNA of S. marcescens strain 10mdr148 with probe specific for 16S rRNA gene; and C, hybridization of I-CeuI-digested total DNA of S. marcescens strain 10mdr148 with probe specific for the blaSMB-1 gene.
Genetic environment of blaSMB-1.
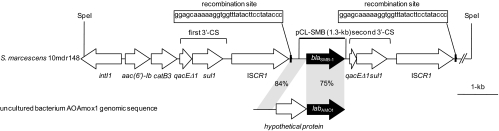
To characterize the genetic context of blaSMB-1, the sequence of the SpeI fragment cloned from genomic DNA of S. marcescens strain 10mdr148 was partially determined. The genetic structure of flanking regions of blaSMB-1 is shown in Fig. 4. The blaSMB-1 gene was located downstream of the ISCR1 element (previously called orf513) that was frequently linked to antibiotic resistance genes such as class A β-lactamase genes (blaCTX-M-2, blaCTX-M-9, and blaPER-1), class C β-lactamase genes (blaCMY-9 and blaDHA-1), a plasmid-mediated quinolone resistance gene (qnrA), and an aminoglycoside-resistant 16S rRNA methyltransferase gene (armA) (27). The ISCR1 element upstream of blaSMB-1 typically was associated with a class 1 integron carrying the gene cassettes aac(6′)-Ib and catB3, which were involved in kanamycin and chloramphenicol resistance, respectively.
Fig. 4.
Schematic representation of the generic environment of the blaSMB-1 gene. The 5′ end of the qacEΔ1 gene in the second 3′-CS was deleted of 143 bp that are present in the first 3′-CS.
The blaSMB-1 gene was followed by the second 3′ conserved segment (CS) and another ISCR1 element. The 3′-CS was made of qacEΔ1 with a 143-bp deletion at the 5′ end and sul1 (Fig. 4). Both ISCR1 elements in the cloned SpeI fragment carried the same crossover recombination site containing oriIS at their 3′ ends. Toleman et al. proposed the model that ISCR1 mobilizes an adjacent DNA sequence bearing an antibiotic resistance gene using oriIS and an alternative termination site (terIS) (27). Therefore, it is suggested that the ISCR1 element plays a role in spreading antibiotic resistance genes among pathogenic Enterobacteriaceae clinical isolates. Although the detailed mechanism of transferring antibiotic resistance genes mediated by ISCR1 remains controversial, it is likely that the blaSMB-1 gene will be horizontally disseminated in the near future among pathogenic Enterobacteriaceae clinical isolates via the transposition activity of the second ISCR1 element downstream of blaSMB-1 (Fig. 4). Furthermore, the finding of a horizontally acquired subclass B3 MBL gene like blaSMB-1 would imply that the subclass B3 MBL gene will spread and become a great clinical concern, as did the subclass B1 MBL gene blaNDM-1.
The blaSMB-1 gene has 75% nucleotide identity with blaAMO1, which was isolated as a β-lactamase gene through the functional metagenomic analysis of apple orchard soil (10). In addition, the genetic region upstream of start codon ATG of blaSMB-1 and downstream of the first ISCR1 element shows 84% nucleotide identity with the 5′ end of the gene encoding a hypothetical protein upstream of blaAMO1 (Fig. 4). This genetic relatedness between blaSMB-1 and blaAMO1 presents the possibility that the blaSMB-1 gene found in S. marcescens strain 10mdr148 was derived from an environmental bacterial species that existed as a natural reservoir for this subclass B3 MBL gene. This is supported by the fact that subclass B3 MBL genes reported so far have been found mostly through the large-scale postgenomic analysis of environmental microbial genomes (9, 10, 21, 24, 25). For example, the origin of NDM-1 was speculated to be the genomic enzyme of some marine bacterium, such as Erythrobacter spp. or its family (33). Further metagenomic analyses of environmental microbial genomes will help identify the origin of the blaSMB-1 gene.
Conclusion.
To our knowledge, this is the first subclass B3 MBL gene in complex with an ISCR1 element to be identified in a human pathogenic Enterobacteriaceae clinical isolate. The majority of the horizontally acquired MBL genes identified so far in pathogenic Gram-negative microbes belong primarily to the subclass B1 MBL group, but here it is likely that pathogenic microbes producing subclass B3 MBLs, like SMB-1, which show high hydrolyzing activity against carbapenems, may emerge and become an actual concern in clinical settings. Therefore, special precautions must be taken continuously regarding the emergence of carbapenem-resistant pathogenic microbes in clinical settings and the molecular mechanisms underlying carbapenem resistance.
ACKNOWLEDGMENTS
This study was supported by the Ministry of Health, Labor, and Welfare of Japan (grant no. H21-Shinkou-Ippan-008).
We are grateful to Kai Kumiko, Yoshie Taki, and Yumiko Yoshimura for their technical assistance. We thank Tomofusa Tsuchiya, a professor of the faculty of Pharmaceutical Sciences, Okayama University, for donating E. coli KAM32. We thank the National BioResource Project (National Institute of Genetics, Japan) for providing the cloning vector pMCL210.
Footnotes
Published ahead of print on 29 August 2011.
REFERENCES
- 1. Bellais S., Aubert D., Naas T., Nordmann P. 2000. Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing ß-lactamases in Chryseobacterium meningosepticum. Antimicrob. Agents Chemother. 44:1878–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bush K. 2010. Alarming ß-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 13:558–564 [DOI] [PubMed] [Google Scholar]
- 3. Bush K., Jacoby G. A. 2010. Updated functional classification of ß-lactamases. Antimicrob. Agents Chemother. 54:969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Castanheira M., Toleman M. A., Jones R. N., Schmidt F. J., Walsh T. R. 2004. Molecular characterization of a ß-lactamase gene, blaGIM-1, encoding a new subclass of metallo-ß-lactamase. Antimicrob. Agents Chemother. 48:4654–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chihara S., Okuzumi K., Yamamoto Y., Oikawa S., Hishinuma A. 2011. First case of New Delhi metallo-ß-lactamase 1-producing Escherichia coli infection in Japan. Clin. Infect. Dis. 52:153–154 [DOI] [PubMed] [Google Scholar]
- 6. Clinical Laboratory Standards Institute, 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 8th ed. Document M07-A8. CLSI, Wayne, PA [Google Scholar]
- 7. Cornaglia G., Rossolini G. M. 2010. The emerging threat of acquired carbapenemases in Gram-negative bacteria. Clin. Microbiol. Infect. 16:99–101 [DOI] [PubMed] [Google Scholar]
- 8. Docquier J. D., et al. 2004. Biochemical characterization of the THIN-B metallo-ß-lactamase of Janthinobacterium lividum. Antimicrob. Agents Chemother. 48:4778–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Docquier J. D., et al. 2002. CAU-1, a subclass B3 metallo-ß-lactamase of low substrate affinity encoded by an ortholog present in the Caulobacter crescentus chromosome. Antimicrob. Agents Chemother. 46:1823–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Donato J. J., et al. 2010. Metagenomic analysis of apple orchard soil reveals antibiotic resistance genes encoding predicted bifunctional proteins. Appl. Environ. Microbiol. 76:4396–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Felici A., Amicosante G. 1995. Kinetic analysis of extension of substrate specificity with Xanthomonas maltophilia, Aeromonas hydrophila, and Bacillus cereus metallo-ß-lactamases. Antimicrob. Agents Chemother. 39:192–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Felici A., et al. 1993. An overview of the kinetic parameters of class B ß-lactamases. Biochem. J. 291:151–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garau G., et al. 2004. Update of the standard numbering scheme for class B ß-lactamases. Antimicrob. Agents Chemother. 48:2347–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaase M., et al. 2011. NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J. Antimicrob. Chemother. 66:1260–1262 [DOI] [PubMed] [Google Scholar]
- 15. Kumarasamy K. K., et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee K., et al. 2005. Novel acquired metallo-ß-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother. 49:4485–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mercuri P. S., et al. 2001. Biochemical characterization of the FEZ-1 metallo-ß-lactamase of Legionella gormanii ATCC 33297T produced in Escherichia coli. Antimicrob. Agents Chemother. 45:1254–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Osano E., et al. 1994. Molecular characterization of an enterobacterial metallo-ß-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38:71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poirel L., Lagrutta E., Taylor P., Pham J., Nordmann P. 2010. Emergence of metallo-ß-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 54:4914–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poirel L., Rodriguez-Martinez J. M., Al Naiemi N., Debets-Ossenkopp Y. J., Nordmann P. 2010. Characterization of DIM-1, an integron-encoded metallo-ß-lactamase from a Pseudomonas stutzeri clinical isolate in the Netherlands. Antimicrob. Agents Chemother. 54:2420–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rossolini G. M., et al. 2001. Metallo-ß-lactamase producers in environmental microbiota: new molecular class B enzyme in Janthinobacterium lividum. Antimicrob. Agents Chemother. 45:837–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sekiguchi J., et al. 2008. KHM-1, a novel plasmid-mediated metallo-ß-lactamase from a Citrobacter freundii clinical isolate. Antimicrob. Agents Chemother. 52:4194–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shibata N., et al. 2003. PCR typing of genetic determinants for metallo-ß-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J. Clin. Microbiol. 41:5407–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stoczko M., Frere J. M., Rossolini G. M., Docquier J. D. 2008. Functional diversity among metallo-ß-lactamases: characterization of the CAR-1 enzyme of Erwinia carotovora. Antimicrob. Agents Chemother. 52:2473–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stoczko M., Frere J. M., Rossolini G. M., Docquier J. D. 2006. Postgenomic scan of metallo-ß-lactamase homologues in rhizobacteria: identification and characterization of BJP-1, a subclass B3 ortholog from Bradyrhizobium japonicum. Antimicrob. Agents Chemother. 50:1973–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tijet N., et al. 2011. New Delhi metallo-ß-lactamase, Ontario, Canada. Emerging Infect. Dis. 17:306–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Toleman M. A., Bennett P. M., Walsh T. R. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Toleman M. A., et al. 2002. Molecular characterization of SPM-1, a novel metallo-ß-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J. Antimicrob. Chemother. 50:673–679 [DOI] [PubMed] [Google Scholar]
- 29. Wachino J., et al. 2006. Horizontal transfer of blaCMY-bearing plasmids among clinical Escherichia coli and Klebsiella pneumoniae isolates and emergence of cefepime-hydrolyzing CMY-19. Antimicrob. Agents Chemother. 50:534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wachino J., et al. 2006. Novel plasmid-mediated 16S rRNA methylase, RmtC, found in a Proteus mirabilis isolate demonstrating extraordinary high-level resistance against various aminoglycosides. Antimicrob. Agents Chemother. 50:178–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yigit H., et al. 2003. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing ß-lactamase KPC-2. Antimicrob. Agents Chemother. 47:3881–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yong D., et al. 2009. Characterization of a new metallo-ß-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zheng B., et al. 2011. An unexpected similarity between antibiotic-resistant NDM-1 and ß-lactamase II from Erythrobacter litoralis. Protein Cell 2:250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]