SUMMARY
Mechanisms that control mRNA metabolism are critical for cell function, development and stress response. The S. cerevisiae mRNA-binding protein Ssd1 has been implicated in mRNA processing, aging, stress response and maintenance of cell integrity. Ssd1 is a substrate of the LATS/NDR tumor suppressor orthologue Cbk1 kinase. Previous data indicate that Ssd1 localizes to the cytoplasm, however biochemical interactions suggest that Ssd1 at least transiently localizes to the nucleus. We therefore explored whether nuclear localization is important for Ssd1 cytoplasmic functions. We identified a functional NLS in the N-terminal domain of Ssd1. An Ssd1 derived NLS-GFP fusion protein and several C-terminally truncated Ssd1 proteins, which presumably lack nuclear export sequences, accumulate in the nucleus. Alanine substitution of the Ssd1 NLS prevents Ssd1 nuclear entry, mRNA binding and disrupts Srl1 mRNA localization. Moreover, Ssd1-NLS mutations abolish Ssd1 toxicity in the absence of Cbk1 phosphorylation and cause Ssd1 to localize prominently to cytoplasmic puncta. These data indicate that nuclear shuttling is critical for Ssd1 mRNA binding and Ssd1-mRNA localization in the cytoplasm. Collectively these data support the model that Ssd1 functions analogously to hnRNPs, which bind mRNA co-transcriptionally, are exported to the cytoplasm and target mRNAs to sites of localized translation and P-bodies.
Keywords: Cbk1 kinase, RAM signaling network, morphogenesis, cell integrity, P-bodies
INTRODUCTION
mRNA binding proteins (mRNPs) are a functionally and structurally diverse group of proteins that play critical roles in gene transcription, mRNA processing, mRNA nuclear export, mRNA translation and mRNA decay (Goler-Baron et al., 2008, Chartrand et al., 2001, Oeffinger et al., 2007). Many mRNA binding proteins first associate with their mRNA targets co-transcriptionally (Aguilera, 2005, Kelly & Corbett, 2009). These include mRNA processing proteins, mRNA transport proteins, reversible translational repressors and mRNA decay mediators (Giorgi & Moore, 2007, Besse & Ephrussi, 2008, Shen et al., 2009, Shen et al.). The functions of many mRNPs are dependent on nucleo-cytoplasmic shuttling (Chartrand et al., 2001, Rodriguez et al., 2004). Defects in mRNP nuclear import or export can disrupt gene expression and alter the fate of nascent mRNAs. mRNA-protein complexes are exported to the cytoplasm through the nuclear pores by means of adaptor proteins and mRNA export proteins, such as NXF1/Mex67 (Terry & Wente, 2009, Carmody & Wente, 2009, Erkmann & Kutay, 2004). In the cytoplasm, mRNA-mRNP complexes are passively or actively delivered to destinations where their mRNAs are translated, sequestered or degraded, depending on mRNP complex composition and the physiological state of the cell (Rodriguez et al., 2008).
Most mRNAs are delivered to ribosomes for translation, however some mRNA-mRNP complexes are actively transported to discrete cortical localizations where their associated mRNAs are stored or translated (Martin & Ephrussi, 2009, Rodriguez et al., 2008). Mechanisms that govern asymmetric mRNA localization and translation contribute to axis development, cell fate determination, asymmetric cell division, cell motility and neuronal development (Martin & Ephrussi, 2009, Smith, 2004, Johnstone & Lasko, 2001). For example, during C. elegans and Drosophila development, polar mRNP granules dictate axis formation and cell fate determination (Johnstone & Lasko, 2001, Anderson & Kedersha, 2009). Similarly, many mRNAs are transported to developing dendritic spines and synapses, where they contribute to neuronal development and signaling (Rodriguez et al., 2008). Some mRNAs are delivered or recruited to cytoplasmic mRNA granules, such as mRNA processing bodies (P-bodies) and stress granules, where they are transiently sequestered or degraded (Anderson & Kedersha, 2009, Balagopal & Parker, 2009, Rodriguez et al., 2008). The mechanisms of mRNA recruitment to P-bodies and stress granules are not fully understood, but are critical for adapting to cellular stress (Sheth & Parker, 2006, Anderson & Kedersha, 2008, Buchan et al., 2008).
The S. cerevisiae mRNA-binding protein Ssd1 provides insight to the mechanisms governing the cytoplasmic fate of mRNAs. SSD1 is genetically linked to a variety of functions, including mRNA processing, translation, aging, cell integrity and stress response (Sutton et al., 1991, Chiannilkulchai et al., 1992, Turcq et al., 1992, Stettler et al., 1993, Uesono et al., 1994, Tsuchiya et al., 1996, Luukkonen & Seraphin, 1999, Moriya & Isono, 1999, Kaeberlein & Guarente, 2002, Wheeler et al., 2003, Kaeberlein et al., 2004, Tanaka et al., 2007, Mir et al., 2009, Ohyama et al., 2010). Ssd1 function is directly linked to the RAM (regulation of Ace2 and polarized morphogenesis) signaling network, which regulates polarized growth, differential gene expression and maintenance of cell integrity (Kurischko et al., 2011, Racki et al., 2000, Bidlingmaier et al., 2001, Weiss et al., 2002, Nelson et al., 2003, Bourens et al., 2009). Notably, Ssd1 binds to and is phosphorylated by the RAM kinase and LATS/NDR tumor suppressor orthologue Cbk1 (Kurischko et al., 2011, Jansen et al., 2009, Racki et al., 2000). Moreover, Ssd1 expression is toxic in cells harboring RAM mutations (Jorgensen et al., 2002, Du & Novick, 2002, Kurischko et al., 2005).
Recent data implicate Ssd1 and Cbk1 in the spatial regulation of mRNAs in the cytoplasm (Kurischko et al., 2011). Ssd1 specifically associates with a subset of mRNAs, including several mRNAs that encode cbk1 dosage suppressors (Uesono et al., 1997, Hogan et al., 2008, Jansen et al., 2009, Kurischko et al., 2011). At least two Ssd1-associated mRNAs, the cbk1 dosage suppressors SRL1 and UTH1, localize asymmetrically to the bud tip during polarized growth (Hasegawa et al., 2008, Shepard et al., 2003). Significantly, Srl1 mRNA polarity is diminished by ~10 fold in ssd1Δ mutants, indicating a role for Ssd1 in asymmetric mRNA localization (Kurischko et al., 2011). Moreover, this Ssd1 function appears dependent on Cbk1 phosphorylation, since phosphomimetic Ssd1 enhances Srl1 mRNA polarity and phospho-deficient Ssd1 abolishes Srl1 mRNA polarity (Kurischko et al., 2011).
Ssd1 localization changes dramatically in response to cellular stress and Cbk1 inhibition (Kurischko et al., 2011). In most cells Ssd1 localizes diffusely to the cytoplasm, however some Ssd1 can be observed at the bud and bud neck. Strikingly, glucose depletion, heat shock and salt stress cause Ssd1 to localize to P-bodies and stress granules (Kurischko et al., 2011, Jansen et al., 2009). Similarly, in the absence of Cbk1 phosphorylation, Ssd1 and Srl1 mRNA constitutively localize to P-bodies, suggesting that the toxicity of Ssd1 in cbk1 mutants is due to the translational repression of Ssd1-associated mRNAs (Kurischko et al., 2011). In agreement, Cbk1 inhibition correlates with a decreased fraction of Ssd1-associated mRNAs on polysomes (Jansen et al., 2009). These data suggest that Ssd1 mediates P-body and stress granule-dependent translational repression of a subset of mRNAs.
Although Ssd1 influences mRNA localization in the cytoplasm (Kurischko et al., 2011), Ssd1 was not previously observed in the nucleus by microscopy. Nevertheless, several lines of evidence indicate that Ssd1 must have nuclear functions. Notably, Ssd1 binds the phosphorylated C-terminal domain of RNA polymerase II and associates with pre-spliced mRNAs, suggesting that Ssd1 binds its target mRNAs co-transcriptionally (Hogan et al., 2008, Phatnani et al., 2004, Rodriguez-Gil et al., Ohyama et al., 2010). Based on these data, we hypothesize that nucleo-cytoplasmic shuttling is important for Ssd1 function.
Here, we provide evidence that Ssd1 contains a functional nuclear localization sequence (NLS). Our data support the model that Ssd1 nucleo-cytoplasmic shuttling is critical for mRNA binding and the subsequent transport of Ssd1-mRNA complexes to sites of polarized growth or to sites of translational repression (P-bodies, stress granules), depending on the physiological state of the cell.
RESULTS
Ssd1 contains a functional nuclear localization sequence (NLS)
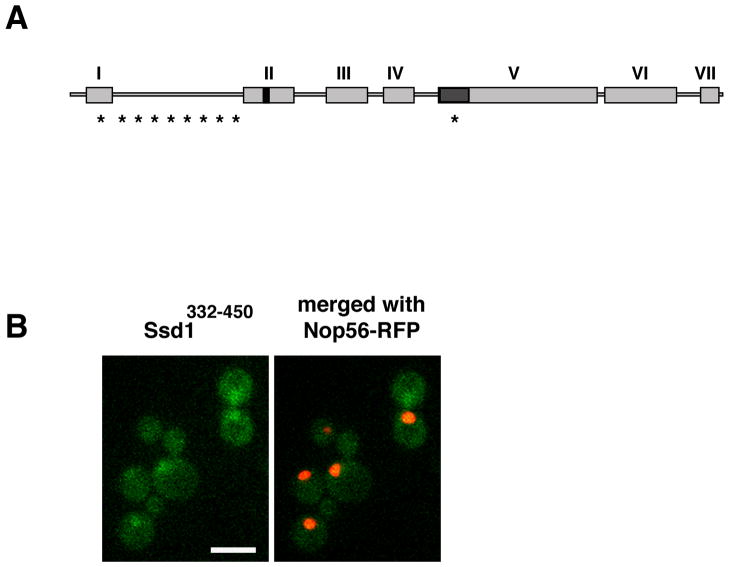
To test the hypothesis that nucleo-cytoplasmic shuttling is important for Ssd1 function, we surveyed Ssd1 for nuclear shuttling sequences. We identified a putative lysine-rich nuclear localization sequence (NLS; amino acids 417–427) in Ssd1 domain II (Fig. 1A). To test if the candidate Ssd1 NLS was capable of targeting GFP to the nucleus, we constructed a fusion protein consisting of GFP and the conserved domain II (designated Ssd1332–450-GFP) and analyzed its localization in yeast. We observed that Ssd1332–450-GFP accumulated in the nuclei of all cells, confirming the functionality of the Ssd1 NLS (Fig. 1B).
Figure 1. Ssd1 contains a functional NLS.
A) Ssd1 consists of several domains that are highly conserved among yeast and fungi. Domain I (aa 31–78) contains a Q-rich stretch and is part of the RNA-pol II phospho-CTD binding domain (aa 1–160); domain II (aa 332–450) contains an NLS at aa 417–427; domain III (aa 490–570) and domain VII (aa 1170–1250) contain no obvious protein motifs; domain IV (aa 579–670) and domain VI (aa 1015–1150) have homology to bacterial VacB, an RNase II family member and domain V (aa 713–1014) has homology to E. coli RNB, an RNase II family member. The RNA binding domain (aa 686–788) overlaps with domain V. Asterisks denote putative Cbk1 phosphorylation sites.
B) Ssd1332–450-GFP accumulates in the nucleus. A plasmid expressing the Ssd1 NLS containing domain (Ssd1332–450) fused to GFP (FLE1067) was transformed into ssd1Δ cells containing the nucleolar marker Nop56-RFP (FLY3074). Images represent single optical sections captured via wide field fluorescence microscopy. Scale bar = 8 um.
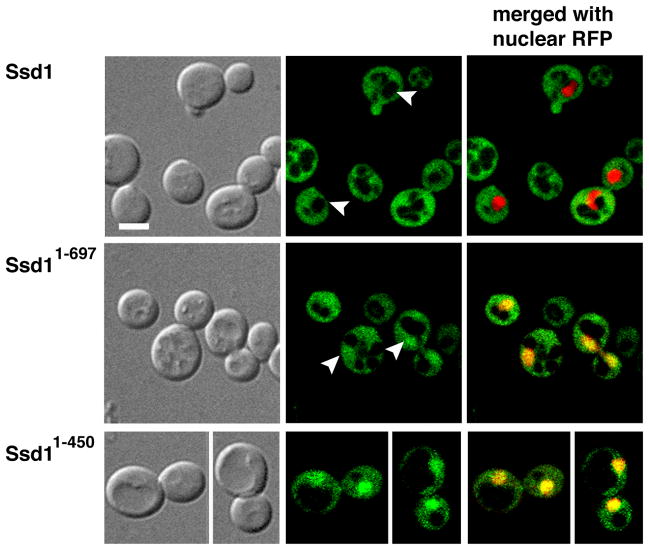
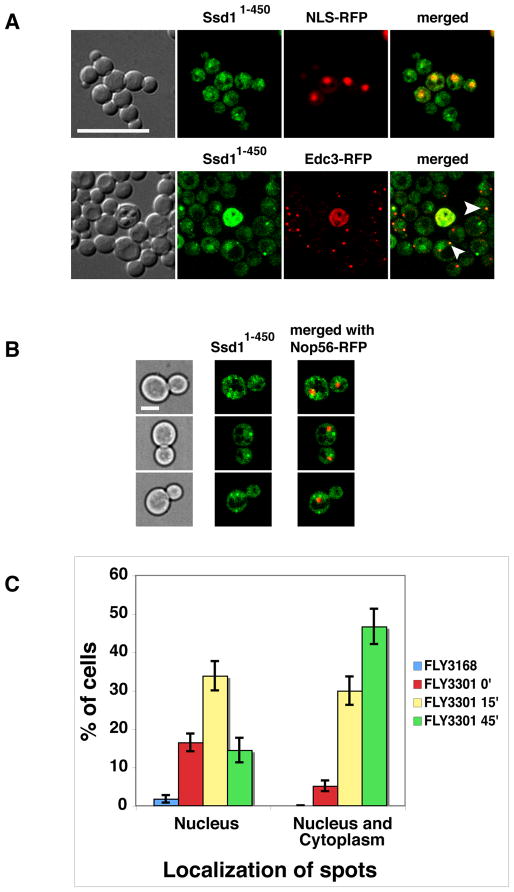
Several common laboratory strains contain the ssd1-d loss-of-function allele, which harbors a nonsense mutation that causes Ssd1 to be truncated at amino acid 697 (Ssd11–697) (Sutton et al., 1991, McDonald et al., 2002). Since Ssd11–697 contains the NLS, it seemed possible that it and other NLS-containing Ssd1 mutants can localize to the nucleus. We therefore tested if Ssd11–697 and Ssd11–450, which are truncated after the NLS-containing domain, accumulate in the nucleus. We constructed Ssd11–697-GFP and Ssd1–450-GFP strains by integrating GFP cassettes into the SSD1 ORF. Although Ssd11–697-GFP and Ssd1–450-GFP localized diffusely to the cytoplasm, each protein was readily detectable in the nuclei at all stages of the cell cycle, in contrast to wild type (full length) Ssd1, which was only apparent in the cytoplasm (Fig. 2). We confirmed the nuclear localization of truncated Ssd1 in cells expressing a plasmid based RFP-tagged nuclear marker, NLS-RFP (pRS425-NLS-mRFP). We observed similar results when truncated Ssd1 was modestly overexpressed via GPD promoter driven expression (Fig. 3A). These data indicate that the N-terminal NLS is functional and establish Ssd1 as a nucleo-cytoplasmic protein that preferentially localizes to the cytoplasm. Furthermore, because wild type Ssd1 is not detectable in the nucleus by these methods, these data suggest that C-terminal Ssd1 domains are important for nuclear export.
Figure 2. C-terminal truncated Ssd1 is detectable in the nucleus.
The C-terminal truncated Ssd11–697 and Ssd11–450 accumulate in both the mother and daughter nuclei, compared to wild type Ssd1. Logarithmically growing cells expressing Ssd1-GFP (FLY1593), Ssd11–697-GFP (FLY3149) and Ssd11–450-GFP (FLY3168) co-expressing an RFP-tagged nuclear marker (NLS-RFP, pKW1219) were observed by spinning disk fluorescence microscopy. Arrowheads indicate nuclei. Nuclear/cytoplasmic fluorescence ratios are provided in Fig. 8B. Images represent single optical sections. Scale bar = 4 um.
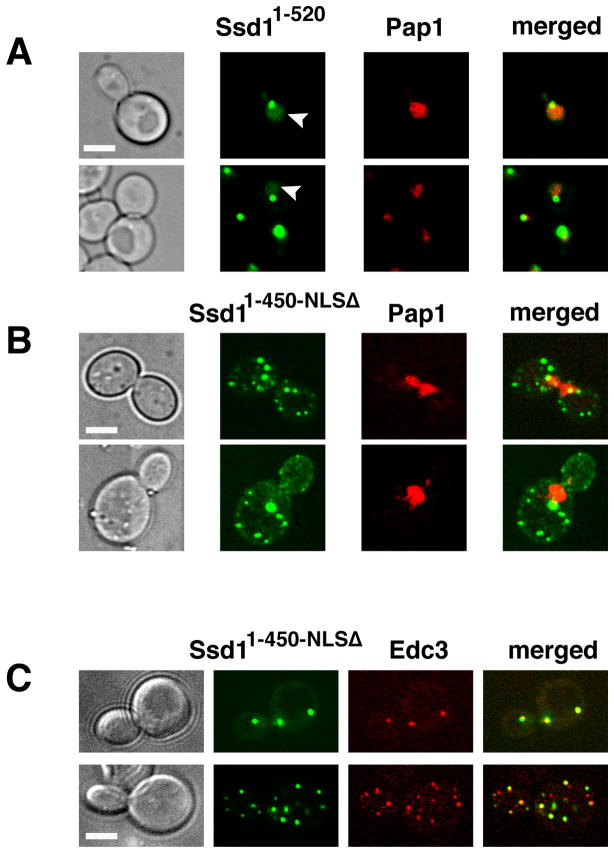
Figure 3. Ssd11–450-NLSΔ does not localize to nuclei.
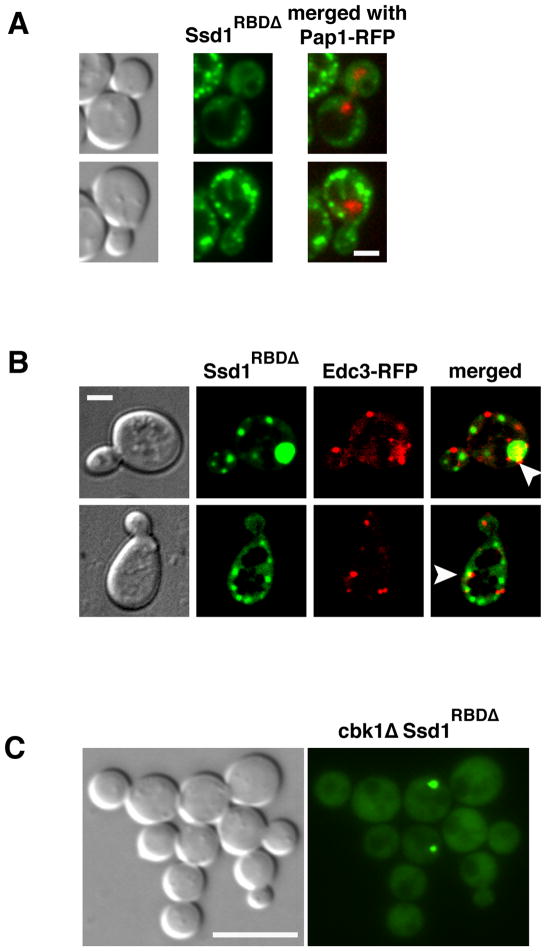
A) Truncated Ssd11–520-GFP, which contains the NLS domain, accumulates in the nucleus in all cells (n=34). Ssd11–520-GFP was expressed in ssd1Δ cells (FLY3365) via the GPD promoter on low copy plasmid (FLE1020). Ssd11–520-GFP nuclear localization was determined by co-localization with the nuclear marker Pap1-RFP (arrowhead). Ssd11–520-GFP localizes diffusely to the nucleus in 82.4% of the cells (n=34). Among these, 85.7% also contain 1–2 prominent nuclear/perinuclear puncta. The remaining cells (17.6%) contain single puncta localized near the nuclear periphery. Ssd11–520-GFP localizes to 1–3 cytoplasmic puncta in ~15% of all cells. B) A plasmid encoding Ssd11–450-NLSΔ-GFP (FLE1211) was introduced into ssd1Δ cells expressing the nuclear marker Pap1-RFP (FLY3365). Ssd11–450-NLSΔ fails to localize to nuclei and principally localizes to prominent cytoplasmic puncta, some of which are juxtanuclear. Ssd11–450-NLSΔ localizes to >5 cytoplasmic puncta in all (100%) cells. A 3-D model of the cell in the top panel is presented in Suppl. Fig. S1 and Suppl. Movie 1. C) The cytoplasmic Ssd11–450-NLSΔ-GFP puncta colocalize with the P-body marker Edc3-RFP (78.9%, n=38). All images were captures via by spinning disk fluorescence microscopy and represent a single optical section. Scale bars = 4 um.
NLS mutations impair Ssd1 localization
If the putative Ssd1 NLS is responsible for nuclear import in vivo, then NLS mutations should prevent the nuclear accumulation of truncated Ssd11–450. Thus, we converted the NLS in Ssd11–450-GFP to alanines and monitored the localization of the mutated fusion protein (Ssd11–450-NLSΔ-GFP) in vivo. We expressed Ssd11–450-NLSΔ-GFPin yeast via low copy plasmids under the control of the constitutive GPD promoter. As expected, Ssd11–450-NLSΔ failed to accumulate in the nucleus, indicating that the NLS is essential for nuclear import of truncated Ssd1 (Fig. 3B). Notably, Ssd11–450-NLSΔ localized prominently to bright cytoplasmic puncta in all cells (n=37 cells). 3-D projections of the image data indicate that some of the cytoplasmic Ssd11–450-NLSΔ puncta are perinuclear (Suppl. Fig. S1, Supp Movie 1). The Ssd11–450-NLSΔ cytoplasmic puncta were similar to the P-body-associated Ssd1 puncta that were observed in stressed cells and in the absence of Cbk1 phosphorylation (Kurischko et al., 2011). Thus, we tested whether Ssd11–450-NLSΔ co-localized with the P-body protein Edc3. As previously observed for full length Ssd1 (Kurischko et al., 2011), most (~78%) of the Ssd11–450-NLSΔ puncta co-localized with Edc3 (n=38 puncta, 8 cells), even in the absence of stress (Fig. 3C). These data indicate that Ssd1 can be targeted to P-bodies in the absence of nuclear import. Furthermore, since Ssd11–450-NLSΔ lacks the mRNA-binding domain as defined by (Uesono et al., 1997)(Fig. 1A), these data also indicate that Ssd1 recruitment to P-bodies is independent of RNA binding.
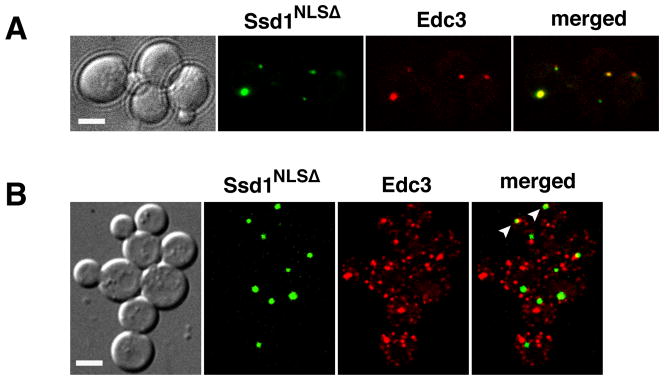
To test if NLS mutations affect the cytoplasmic localizations of full length Ssd1, we introduced plasmids encoding GPD-driven Ssd1-NLSΔ-GFP into ssd1Δ cells. Ssd1-NLSΔ-GFP localized predominantly to 1–3 cytoplasmic puncta in all cells (n=71) (Fig. 4A). Moreover, the majority of the Ssd1-NLSΔ puncta (80%, n=15) co-localized with the P-body marker Edc3 (Fig. 4A). In contrast, similarly expressed wild type Ssd1 localizes to puncta (P-bodies) in less than 60% of cells (Kurischko et al., 2011). These data suggest that if Ssd1 cannot enter the nucleus, it predominantly localizes to P-bodies.
Figure 4. Ssd1-NLSΔ-GFP localizes to P-bodies and is not toxic to cbk1Δ cells.
A) Ssd1-NLSΔ-GFP was expressed in ssd1Δ cells (FLY2184) via low copy plasmids (FLE1213). Ssd1-NLSΔ-GFP localizes to 1–3 cytoplasmic puncta per cell (n=71). ~80% of the cytoplasmic Ssd1-NLSΔ puncta colocalize with the P-body protein Edc3 (n=15). B) Ssd1-NLSΔ-GFP localizes to cytoplasmic puncta in cbk1Δ cells (FLY2293). Ssd1-NLSΔ is not toxic to cbk1Δ cells and localizes similarly in cbk1Δ as in CBK1 cells. Arrowheads point to some Ssd1 puncta that colocalize with Edc3. All cells were monitored by spinning disk fluorescence microscopy and each image represents a single optical section. Scale bars = 4 um.
Ssd1 nuclear import is essential for Ssd1 function
It is well established that Ssd1 expression is lethal to cbk1Δ mutants, thus providing a simple assay for Ssd1 function. Previous data suggest that wild type Ssd1 is lethal in cbk1Δ cells because in the absence of Cbk1 phosphorylation, Ssd1 constitutively localizes to P-bodies and stress granules, leading to the translational repression of its associated mRNAs (Kurischko et al., 2011, Jansen et al., 2009). In support, Ssd1-9A, which lacks Cbk1 phosphorylation sites, constitutively localizes to P-bodies and is dominantly toxic (Kurischko et al., 2011). If nuclear entry is essential for Ssd1 function, then Ssd1-NLSΔ expression should not be toxic to cbk1Δ cells. Alternatively, if Ssd1 toxicity in cbk1Δ cells is exclusively due to cytoplasmic functions, then Ssd1-NLSΔ expression will be lethal to cbk1Δ cells. To distinguish between these possible outcomes, we introduced Ssd1-NLSΔ plasmids into cbk1Δ ssd1Δ mutant cells and assayed viability. Significantly, Ssd1-NLSΔ expression did not impair viability or growth of cbk1Δ ssd1Δ cells (data not shown and Fig. 4B). Moreover, cbk1 deletion did not significantly affect Ssd1-NLSΔ localization (Fig. 4B). These data indicate that even though Ssd1-NLSΔ can localize to P-bodies, it likely does not repress mRNA translation in cbk1Δ cells. Taken together, these data indicate that Ssd1-NLSΔ is not fully functional and suggest that Ssd1-NLSΔ does not bind mRNA. Although unlikely, it is formally possible that modest alanine substitutions in the Ssd1 NLS disrupt Ssd1 protein conformation.
We recently established that Ssd1 influences the cytoplasmic localization of Srl1 mRNA, which encodes a cell wall mannoprotein (Kurischko et al., 2011). The polarized localization of Srl1 mRNA is significantly diminished (by ~10 fold) in ssd1Δ cells in comparison to corresponding wild type cells and is restored (and modestly enhanced) by phosphomimetic Ssd1 (Kurischko et al., 2011). To determine if Ssd1 nuclear import is essential for Srl1 mRNA localization, we analyzed Srl1 mRNA in Ssd1-NLSΔ cells, using previously published methods (Kurischko et al., 2011, Haim-Vilmovsky & Gerst, 2009). We observed that Srl1 mRNA polarity was significantly diminished in Ssd1-NLSΔ cells (Suppl. Figure S2). Notably, Srl1 mRNA only localized exclusively to the bud in 13.9% of budded Ssd1-NLSΔ cells (n=226 cells), compared to >40% of wild type cells (Kurischko et al., 2011). These data indicate that Ssd1 nuclear import is critical for influencing the cytoplasmic localizations of at least one Ssd1-associated mRNA.
Ssd1 nuclear import is crucial for mRNA binding
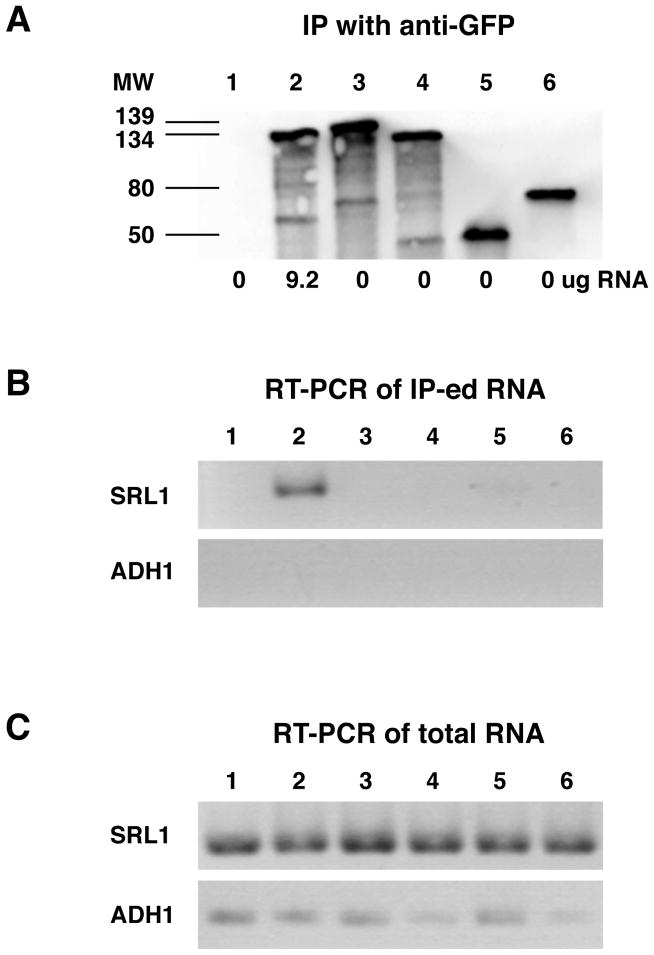
Our data suggest that Ssd1 must enter the nucleus in order to bind mRNA, as shown for other mRNA-binding proteins (Kelly & Corbett, 2009, Aguilera, 2005). If this hypothesis is correct, then mutations that impair Ssd1 nuclear localization will prevent mRNA binding. To test if Ssd1-NLSΔ binds mRNA in vivo, we immunoprecipitated GFP-tagged Ssd1-NLSΔ from cells and assayed for associated mRNAs by spectroscopy and RT-PCR. We did not observe any detectable RNA associated with immunoprecipitated Ssd1-NLSΔ by UV spectroscopy, however wild type Ssd1 consistently precipitated RNA. Specifically, ~9.2 ug RNA associated with immunoprecipitated Ssd1-GFP from 525 ug of yeast extract. Since RT-PCR is a more sensitive method for detecting specific mRNAs than UV spectroscopy, we probed immunoprecipitated Ssd1 complexes for associated mRNAs (Srl1, Ccw12, Sim1 mRNAs) via RT-PCR, as described (Kurischko et al., 2011). We discovered that none of the probed mRNAs were detectable in immunoprecipitated Ssd1-NLSΔ complexes (Fig. 5, and data not shown). In contrast, wild type Ssd1 specifically co-precipitated with Srl1 mRNA and several other Ssd1-associated mRNAs (Fig. 5; and (Kurischko et al., 2011). As a control for specificity we also probed immunoprecipitated Ssd1 for Adh1 mRNA, which does not specifically interact with Ssd1. As expected, neither wild type Ssd1 nor Ssd1-NLSΔ precipitated Adh1 mRNA. Thus, Ssd1 nuclear import is critical for mRNA binding.
Figure 5. Ssd1-NLSΔ does not bind mRNA in vivo.
GFP-tagged Ssd1 proteins were immunoprecipitated from 525 ug yeast lysate and assayed for associated mRNAs by UV spectroscopy and RT-PCR. A) Immunoblot of immunoprecipitated Ssd1-GFP probed with anti-GFP antibody. Lane 1: wild type cells (no GFP); lane 2: Ssd1-GFP; lane 3: Ssd1-NLSΔ; lane 4: Ssd1-RBDΔ; lane 5: Ssd11–450-NLSΔ; lane 6: Sec63-GFP. Sec63 does not bind RNA and thus serves as a negative control. The numbers below each lane represent the relative amount (ug) of total RNA associated with the immunoprecipitated proteins. Three independent experiments yielded the same results. B) Immunoprecipitated Ssd1 complexes (from A) were probed for SRL1 and ADH1 mRNA by RT-PCR. ADH1 mRNA serves as a negative control since it does not bind Ssd1. C) Total cell extracts (starting material) were probed for SRL1 and ADH1 mRNA by RT-PCR. For these experiments, the strain FLY2184 was transformed with plasmids FLE1019, FLE1210, FLE1211, FLE1213 and pJK59.
Ssd1 mRNA binding is essential for Ssd1 function but does not influence its nuclear export
The nuclear localizations of truncated Ssd1–450-GFP and Ssd11–697 (Fig. 2) clearly establish that the Ssd1 mRNA binding domain and other C-terminal domains (III–VII) are not critical for nuclear import. However because full length Ssd1 does not accumulate in the nucleus, it seemed possible that mRNA binding facilitates efficient Ssd1 nuclear export. To test this hypothesis we deleted the Ssd1 RNA binding domain (amino acids 686–788) and assayed the activities and localization of the mutant Ssd1-RBDΔ in vivo. To confirm that Ssd1-RBDΔ does not bind mRNA, we immunoprecipitated Ssd1-RBDΔ from yeast and assayed for associated RNA, using the methods outlined above for Ssd1-NLSΔ. As expected, Ssd1-RBDΔ did not precipitate any RNA (Fig. 5, lane 4). In addition, Ssd1-RBDΔ expression could not suppress the diminished polarity of Srl1 mRNA in ssd1Δ cells, consistent with its inability to bind mRNA (Kurischko et al. 2011 and Suppl. Figure S2).
To test if Ssd1-RBDΔ is toxic to cbk1Δ cells, which presumably reflects the translational repression of Ssd1-associated mRNAs, we introduced plasmids into cbk1Δ ssd1Δ cells and assayed viability. We found that Ssd1-RBDΔ expression was not toxic to cbk1Δ cells, indicating that the RBD (amino acids 686–788) is critical for Ssd1 function (Kurischko et al., 2011) and see Fig. 6C). Thus as predicted, the Ssd1 RNA binding domain is essential for RNA binding and for Ssd1 function, in agreement with (Uesono et al., 1997).
Figure 6. Ssd1-RBDΔ localizes to the cytoplasm.
A) A plasmid encoding GFP-tagged Ssd1-RBDΔ (which lacks Ssd1 amino acids 686–788; FLE1210) was introduced into A) ssd1Δ Pap1-RFP, B) ssd1Δ Edc3-RFP and C) cbk1Δ ssd1Δ cells (FLY3365, FLY2184 + pRP1574, FLY2293). In CBK1 cells, Ssd1-RBDΔ-GFP does not accumulate in the nucleus, but localizes diffusely to the cytoplasm (in all cells) and to multiple cytoplasmic puncta in ~50% of cells, as previously reported for GPD-driven Ssd1 (Kurischko et al., 2011). Some (~23%), but not all, Ssd1-RBDΔ-GFP puncta co-localize with Edc3 (see arrowheads). C) Ssd1-RBDΔ expression is not toxic to cbk1Δ cells, consistent with the observation that Ssd1-RBDΔ does not bind mRNA in vivo (see Fig. 5). All cell images represent single optical sections captured via spinning disk (A and B) and wide field (C) fluorescence microscopy. Scale bars = 2 um (A, B) and 4 um (C).
How does the RBD domain influence Ssd1 localization? If mRNA binding facilitates Ssd1 nuclear export, then Ssd1-RBDΔ should accumulate in the nucleus. Thus, we introduced Ssd1-RBDΔ-GFP plasmids into ssd1Δ cells that co-express Pap1-RFP or Edc3-RFP and monitored Ssd1-RBDΔ localization by fluorescence microscopy. When expressed via the constitutive GPD promoter, Ssd1-RBDΔ-GFP localized diffusely to the cytoplasm in all cells (n=83) and to bright cytoplasmic puncta in ~50% of cells (Fig. 6A, B), as described for GPD-driven wild type Ssd1 (Kurischko et al., 2011). Approximately 25% of the Ssd1-RBDΔ puncta co-localized with the P-body protein Edc3 (Fig. 6B, see arrowheads). Significantly, Ssd1-RBDΔ did not accumulate in the nucleus, suggesting that the mRNA binding domain, and hence mRNA binding, is not essential for Ssd1 nuclear export (Fig. 6A). These data indicate that mRNA binding is critical for Ssd1 function, but is not essential for efficient nuclear export.
Cbk1 influences the nuclear and cytoplasmic localizations of Ssd11–450
Cbk1 kinase clearly influences Ssd1 localization in the cytoplasm (Kurischko et al., 2011). Notably, Cbk1-mediated phosphorylations prevent Ssd1 from localizing to P-bodies (Kurischko et al., 2011). To test if Cbk1 regulates Ssd1 nuclear localization, we monitored Ssd11–450-GFP in cbk1 mutant cells. Ssd11–450-GFP was integrated and expressed under control of the physiological SSD1 promoter. As noted above, Ssd11–450-GFP localizes to nuclei in wild type cells (Fig. 2) and is not lethal to cbk1Δ cells (see Fig. 7A). Significantly, Ssd11–450 localized to nuclei in cbk1Δ cells, as in wild type cells. In addition, some Ssd11–450 localized to nuclear and cytoplasmic puncta (~1–4 puncta/cell) in cbk1Δ cells (Fig. 7A). About half of the cytoplasmic puncta colocalize with Edc3 (Fig. 7A, arrowheads). These data suggest that Cbk1 influences Ssd1 cytosolic and nuclear protein interactions.
Figure 7. Nuclear and cytoplasmic Ssd11–450 is aberrant in cbk1 mutants.
A) The localization of physiologically expressed Ssd11–450-GFP was monitored in cbk1Δ cells (FLY3202) co-expressing an RFP-tagged nuclear marker (NLS-RFP, pKW1219) or P-body protein (Edc3-RFP, pRP1574). Ssd11–450-GFP accumulates in nuclei in cbk1Δ cells to the same extent as it does in CBK1 cells (compare to Fig. 2). In addition, Ssd11–450 localizes to a few nuclear and cytoplasmic puncta in cbk1Δ cells. Approximately half of the Ssd11–450 cytoplasmic puncta colocalize with Edc3 (see arrowheads). B) Physiologically expressed Ssd11–450-GFP was monitored in analog-sensitive cbk1-as mutants that express the nucleolar marker Nop56-RFP (FLY3301). Micrographs were taken 25 min after addition of the Cbk1-as inhibitor 1NA-PP1. C) Graphical representation of the cbk1-as data. Upon Cbk1 inhibition, the number of cells with nuclear and cytoplasmic Ssd11–450-GFP puncta increases over time. In the absence of 1NA-PP1, cbk1-as cells exhibit enhanced level of nuclear puncta compared to corresponding CBK1+ cells (FLY3168). The cells were monitored by spinning disk fluorescence microscopy and all images represent single optical sections. Scale bars = 8 um (A) and 4 um (B).
As a second method to investigate the role of Cbk1 for nuclear Ssd1 localization, we monitored Ssd11–450-GFP localization upon Cbk1 inhibition in analog-sensitive cbk1-as cells (Weiss et al., 2002, Kurischko et al., 2008). After 15 min of Cbk1 kinase inhibition (via 1NA-PP1 addition), some Ssd11–450-GFP redistributed to a prominent nuclear spot and to cytoplasmic puncta (Fig. 7B, C). Prior to Cbk1 kinase inhibition, ~15% of cbk1-as cells contained nuclear and/or cytoplasmic Ssd11–450-GFP puncta, compared to <2% in wild type cells. Upon 45 min of Cbk1 kinase inhibition, the percentage of cells with nuclear or cytoplasmic puncta increased to 60% (Fig. 7C). These data indicate that Cbk1 kinase is critical for maintaining diffuse Ssd1 nuclear localization, perhaps by stabilizing Ssd1 nuclear interactions or by preventing Ssd1 protein aggregation.
Ssd1 nuclear export is Crm1- and Mex67-independent
Our data indicate that Ssd1 must enter the nucleus in order to bind mRNA. Thus, the cytoplasmic functions of Ssd1-mRNA complexes require nuclear export. Published data suggest the export factors Crm1/Xpo1 and Mex67 as prime candidates for mediating Ssd1-mRNA nuclear export. Crm1/Xpo1 mediates the export of at least one Cbk1 substrate, Ace2, and several mRNP proteins, including Dbp5, Npl3 (Stade et al., 1997, Watanabe et al., 1999, Hutten & Kehlenbach, 2007, Weiss et al., 2002, Bourens et al., 2008, Mazanka et al., 2008, Sbia et al., 2008). The Mex67-Mtr2/NXF1-p15 heterodimer complex is required for the nuclear export of most mRNAs (Stewart, 2007, Erkmann & Kutay, 2004, Cook et al., 2007). We therefore monitored Ssd1-GFP localization in conditional crm1 and mex67 mutants to determine if Crm1 or Mex67 mediates Ssd1 nuclear export. Full length Ssd1 did not accumulate in the nuclei of temperature sensitive or leptomycin B-sensitive crm1 mutants (xpo1-1, xpo1-lpmS) or mex67-5 mutants at permissive or restrictive conditions, suggesting that Ssd1 export is Crm1 and Mex67-independent (data not shown).
Ssd1 C-terminal domains influence Ssd1 nuclear export
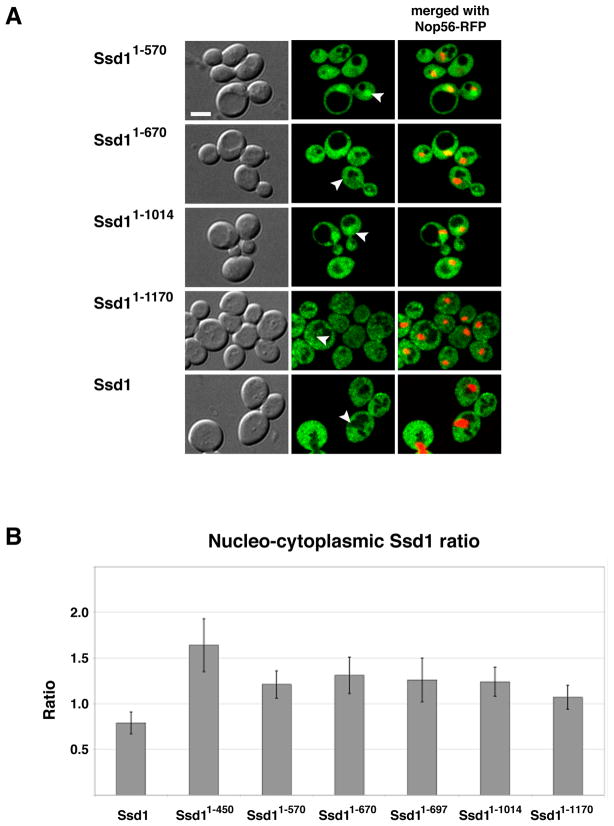
In principle, mutations that delete or disrupt nuclear export domains will cause more Ssd1 to accumulate in the nucleus than corresponding wild type Ssd1. Thus, we hypothesize that there are sequences within the C-terminal domains (III –VII) that function to keep most Ssd1 out of the nucleus. In support, wild type Ssd1 preferentially localizes to the cytoplasm, yet truncated Ssd11–450 and Ssd11–697 are readily detectable in the nucleus (Fig. 2). Note the absence of full length (wild type) Ssd1-GFP fluorescence in the nucleus compared to the surrounding diffuse cytoplasmic signal (Fig. 2). We calculated the nuclear-cytoplasmic fluorescence ratios to compare relative nuclear Ssd1 for each strain. The mean nuclear-cytoplasmic fluorescence ratios of GFP-tagged Ssd1, Ssd11–450 and Ssd11–697 are ~0.75, 1.6 and 1.25.
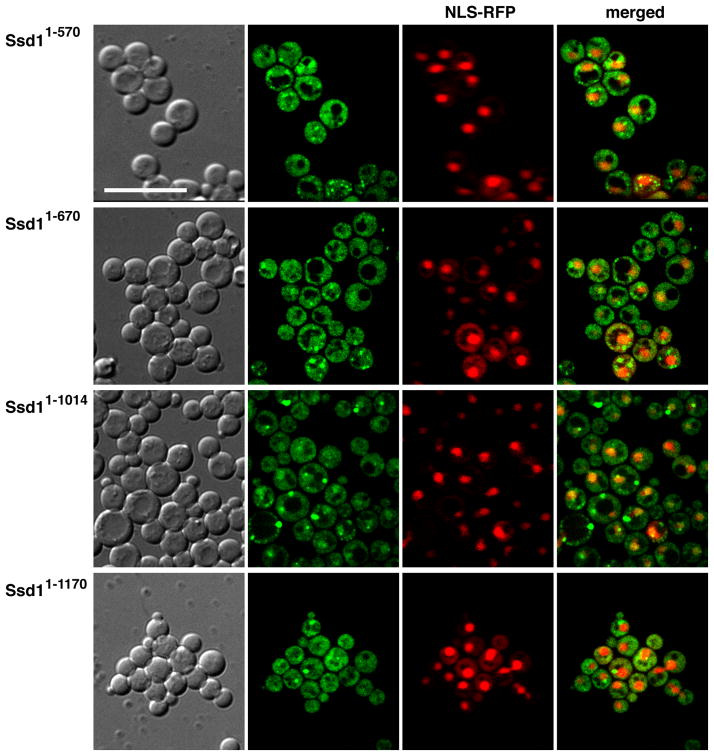
To identify Ssd1 nuclear export domains, we analyzed the localizations of several C-terminally truncated Ssd1 proteins. Ssd1 truncations were constructed by the targeted integration of a GFP cassette into the SSD1 ORF after distinct conserved domains (Fig. 1A). Thus, all fusion proteins for these assays were expressed under the control of the physiological SSD1 promoter. In contrast to full length Ssd1, truncated Ssd11–570, Ssd11–670 and Ssd11–1014 (which lack IV–VII, V–VII and VI–VII domains respectively) moderately accumulate in the nucleus (Fig. 8), suggesting that they are exported from the nucleus less efficiently than wild type Ssd1. Notably, Ssd11–1170, which lacks the C-terminal 80 amino acids of Ssd1 (domain VII), is less abundant in the nuclei than other C-terminal truncated Ssd1 (Ssd11–450, Ssd11–570, Ssd11–670 and Ssd11–1014) (Fig. 8A). Nevertheless, close inspection reveals that Ssd11–1170-GFP accumulates in the nucleus to a greater degree than wild type Ssd1-GFP (Fig. 8B). The nuclear to cytoplasmic ratios for all truncated Ssd1 were consistently greater than that of full length Ssd1 (Fig. 8B).
Figure 8. Nuclear localization of Ssd1 C-terminal truncations in wild type cells.
A) Physiologically expressed Ssd11–570, Ssd11–670, Ssd11–1014, Ssd11–1170 and full length Ssd1-GFP were monitored in wild type cells co-expressing the nucleolar marker Nop56-RFP. Each image represents a single optical section captured via by spinning disk fluorescence microscopy. Arrowheads indicate nuclei. Scale bar = 4 um. B) Mean values and standard deviations of nuclear-cytoplasmic fluorescence ratios were plotted for full length and truncated Ssd1-GFP (n=16–26 cells per strain). Measurements and calculations were performed as described in Material and Methods. Nuclei were defined by NLS-RFP (pKW1219) fluorescence. All truncated Ssd1-GFP proteins accumulate in the nuclei to a greater degree than full length Ssd1 (p<10−7). The raw data and p values for statistical significance are provided in Supplement Table 1. See Table I for the strains used in these experiments.
Collectively, these data suggest that domain VI (amino acids 1015–1170) and to a lesser degree domain VII are important for Ssd1 nuclear export. Our data support the model that Ssd1 domain VI contains sequences that facilitate binding to nuclear export factors. Ssd1 domain VII (amino acids 1171–1250) may influence nucleo-cytoplasmic shuttling by contributing to efficient nuclear export in conjunction with domain VI or by influencing the efficiency of nuclear import.
To determine if the C-terminally truncated Ssd1 proteins were functional, we introduced GFP-tagged Ssd11–570, Ssd11–670, Ssd11–1014 and Ssd11–1170 into cbk1Δ cells. None of the C-terminally truncated Ssd1 proteins were toxic to cbk1Δ cells, indicating that they are not fully functional in vivo (Fig. 9). These data likely indicate that the truncated proteins do not constitutively repress translation of Ssd1-associated mRNAs (see discussion). All C-terminal truncated proteins (from Ssd11–450 to Ssd11–1170) localize to nuclei in cbk1Δ cells to the same degree as in corresponding CBK1 cells, indicating that Cbk1 is not essential for Ssd1 nuclear import (Fig. 9). However, in 20–30% of cbk1Δ cells, the truncated Ssd1 proteins localize to nuclear and cytoplasmic puncta. The percentage of cbk1Δ cells with nuclear and cytoplasmic puncta varies among the C-terminal truncations (see legend for Suppl. Fig. S3). Some, but not all, Ssd1 cytoplasmic puncta co-localize with the P-body protein Edc3 (Suppl. Fig. S3). These data support the model that Cbk1 influences nuclear and cytoplasmic Ssd1 interactions and support the conclusion that the Ssd1 N-terminal region (domains I – II) is sufficient for P-body recruitment, as described in (Kurischko et al., 2011).
Figure 9. Expression of C-terminal truncations of Ssd1-GFP is not lethal in cbk1Δ cells.
Physiologically expressed Ssd11–570, Ssd11–670, Ssd11–1014 and Ssd11–1170 were monitored in cbk1Δ cells containing the nuclear marker NLS-RFP (pKW1219). None of the truncated Ssd1 proteins is toxic, in contrast to full length Ssd1 (not shown). Truncated Ssd1 proteins localize diffusely to the nucleus and to a few prominent foci, many of which localize in or near the nucleus. Up to 50% of the cytoplasmic puncta co-localize with P-body protein Edc3 (see Supplemental Fig. S3). See Table I for the strains used in these experiments. The cells were monitored by spinning disk fluorescence microscopy and each image represents a single optical section. Scale bar = 8 um.
Discussion
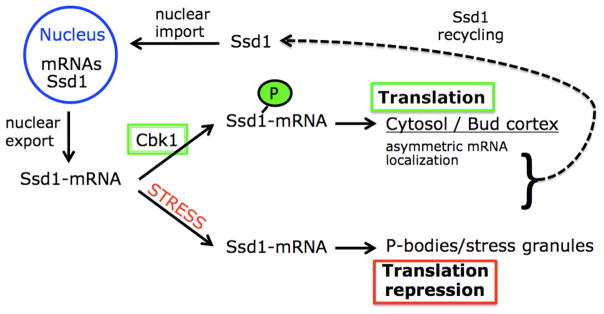
Our data provide strong evidence that Ssd1 is subject to nucleo-cytoplasmic shuttling that is critical for mRNA binding and all Ssd1-mRNA functions (see working model Fig. 10). Significantly, we established that Ssd1 nuclear import is essential for mRNA binding and for dictating the cytoplasmic localizations of an Ssd1-associated mRNA. Our data support the model that Ssd1-mRNA interactions are established co-transcriptionally and that Ssd1 nuclear interactions are critical for defining the cytoplasmic fate of Ssd1-mRNA complexes.
Figure 10. Working model of Ssd1 function.
See Discussion for details.
Role of Cbk1 on Ssd1 function
We previously established that Cbk1 kinase regulates Ssd1 localization in the cytoplasm (Kurischko et al., 2011). During logarithmic growth, most Ssd1 localizes diffusely to the cytoplasm and some accumulates at sites of polarized growth. Upon Cbk1 inhibition, Ssd1 redistributes to cytoplasmic puncta that partly co-localize with P-bodies and stress granules, which are known to repress translation (Kurischko et al., 2011). These data support the model that Cbk1 negatively regulates Ssd1 function at P-bodies and therefore indirectly promotes the translation of Ssd1 associated mRNAs (Fig. 10). Thus, we surmise that in the absence of Cbk1, cells die because Ssd1 targets its associated mRNAs to P-bodies, where they are translationally repressed. In support, moderate overexpression of several genes encoding Ssd1-associated mRNAs rescues the lethality of conditional cbk1 and dominant Ssd1-9A mutants (Kurischko et al., 2008, Kurischko et al., 2005, Kurischko et al., 2011).
In this study, we demonstrate that Cbk1 also influences the intra-nuclear localization of Ssd1. When Cbk1 is inhibited, most Ssd11–450 rapidly redistributes from a more diffuse nuclear localization to a punctate nuclear and cytoplasmic distribution. Likewise, CBK1 deletion causes Ssd11–450 and other C-terminally truncated Ssd1 proteins to localize to nuclear and cytoplasmic puncta. Notably, only a fraction of the truncated Ssd1 cytoplasmic puncta co-localize with P-bodies (Suppl. Fig. S3). Thus, Cbk1 inhibition or deletion stimulates the appearance of at least two novel classes of Ssd1 puncta: nuclear/perinuclear puncta and P-body independent cytoplasmic puncta. These data suggest that Cbk1 promotes or stabilizes Ssd1 nuclear interactions and localization, perhaps indirectly by preventing Ssd1 from localizing to P-bodies or other puncta.
It is possible that some of the observed Ssd1 puncta are protein aggregates and that Cbk1 is important to prevent Ssd1 aggregation. In support, a recent study suggests that Ssd1 has the capacity to form prions (Alberti et al., 2009). Moreover, Ssd1 has potential prion-promoting sequences in the N-terminal Q-rich domain I and the Cbk1 phosphorylation domain (Alberti et al., 2009). Many newly identified prion-forming proteins are involved in gene expression and RNA regulation, similar to Ssd1 (Alberti et al., 2009). Ssd1 is also required for Hsp104-mediated disaggregation of protein aggregates (Mir et al., 2009). Collectively, these data may anticipate a role for Cbk1 and Ssd1 in regulating gene expression via modulation of protein aggregate/prion development.
Ssd1 nuclear interactions influence mRNA localization
Cbk1 mediated Ssd1 phosphorylation appears to stimulate the polarized localization and translation of an Ssd1-associated mRNA (Kurischko et al., 2011). Thus, Ssd1 belongs to a class of mRNA binding proteins that influence asymmetric mRNA localization (Czaplinski & Singer, 2006). At least two Ssd1-associated mRNAs, Srl1 and Uth1 mRNA, are delivered to the bud via the locasome complex, which is comprised of the mRNP She2, the adapter protein She3 and type V myosin (Sil & Herskowitz, 1996, Bohl et al., 2000, Beach & Bloom, 2001, Oeffinger et al., 2007, Shepard et al., 2003, Paquin & Chartrand, 2008). In the absence of Ssd1, Srl1 mRNA polarity is diminished by ~10 fold and restored by phosphomimetic Ssd1-9D (Kurischko et al., 2011). Likewise, in the absence of the Ssd1 NLS or RBD, Srl1 mRNA polarity is significantly diminished (Suppl. Fig. S2). Thus, Cbk1 phosphorylation of Ssd1 prevents Ssd1-P-body association and promotes asymmetric mRNA localization and translation (Fig. 10). Furthermore, our data suggest that Ssd1 must shuttle into and out of the nucleus in order to bind mRNAs (presumably during transcription) and deliver them to cytoplasmic locales where they will be translated.
It is tempting to speculate that Ssd1 must enter the nucleus to associate with proteins that are necessary for asymmetric delivery of mRNAs. Indeed She2, the best-studied mRNA-binding protein of its class, localizes to the nucleus, binds mRNAs co-transcriptionally and recruits other mRNP proteins, including Loc1p and the translation repressor Puf6p (Olivier et al., 2005, Shen et al., 2009, Shen et al., Hasegawa et al., 2008, Paquin & Chartrand, 2008). Once in the cytoplasm, She2-mRNA complexes bind the adapter protein She3, which binds myosin V Myo4, and are transported along actin cables to the bud tip (Bohl et al., 2000, Beach & Bloom, 2001, Darzacq et al., 2003, Olivier et al., 2005). Ssd1 may promote asymmetric Srl1 mRNA localization by enhancing the functions of She2 or other locasome-associated proteins.
How is Ssd1 nuclear export regulated?
Since Ssd1 nuclear import is essential for mRNA binding, Ssd1-mRNA complexes must be exported from the nucleus. Some mRNP proteins, such as S. cerevisiae She2 and Drosophila Staufen 2, both of which influence cytoplasmic mRNA localization, must bind mRNA in order to be exported from the nucleus (Du et al., 2008, Macchi et al., 2004). In contrast, Ssd1-RBDΔ, which contains a functional NLS but cannot bind mRNA, does not accumulate in the nucleus, suggesting that RNA binding is not a prerequisite for Ssd1 nuclear export.
Our data regarding the localization of truncated derivatives of Ssd1, suggest that Ssd1 domains VI and possibly VII play a prominent role in the efficient export of Ssd1 from the nucleus. Deletion of Ssd1 domains VI and VII leads to prominent nuclear enrichment, whereas deletion of VII alone (amino acids 1171–1250) leads to a much more modest nuclear accumulation (Fig. 8). Thus we hypothesize that Ssd1-mRNA nuclear export involves protein interactions at domains VI and VII. Intriguingly, Ssd1 domains VI and VII are highly conserved among fungi, suggesting that they are functionally important. Nevertheless, domains VI and VII do not contain known nuclear export sequences or other functional motifs. The identification of proteins associated with Ssd1 domains VI and VII is likely to provide further insight into the mechanism of Ssd1 nuclear export.
Ssd1-like mRNA functions in other organisms
Ssd1 is the first example of a yeast mRNP that shuttles into and out of the nucleus and has a role both in polarized mRNA localization and in translational repression. This complexity of Ssd1 function coupled with the diversity of its associated mRNAs likely account for the large variety of SSD1 genetic interactions (Sutton et al., 1991, Chiannilkulchai et al., 1992, Turcq et al., 1992, Stettler et al., 1993, Uesono et al., 1994, Tsuchiya et al., 1996, Luukkonen & Seraphin, 1999, Moriya & Isono, 1999, Kaeberlein & Guarente, 2002, Wheeler et al., 2003, Kaeberlein et al., 2004, Tanaka et al., 2007, Mir et al., 2009).
Ssd1 may be functionally related to hnRNPs (heterogeneous nuclear ribonucleoproteins) in other organisms, which bind mRNA in the nucleus and influence cytoplasmic mRNA localization and translation (Krecic & Swanson, 1999, Percipalle et al., 2009). These proteins are referred to as zip code binding proteins as they recognize specific 3′ or 5′ UTR sequences in their target mRNAs (Mili & Macara, 2009). Ssd1 also recognizes consensus sequences in the 5′ UTR of associated mRNAs (Hogan et al., 2008, Ohyama et al., 2010). It is possible that these sequences function as zip codes for Ssd1-dependent mRNA localizations.
Ssd1 may function analogously to both Drosophila hnRNP-C and FMRP, which competitively bind to the same region of APP (amyloid precursor protein) mRNA (Lee et al., Zalfa et al., 2006). hnRNP C promotes translation and FMRP represses translation and targets the mRNA complexes to P-bodies (Lee et al., 2010). In addition, Ssd1 may interact with cytoplasmic proteins that function analogously to neuronal NXF7 (nuclear export factor), which targets hnRNP A3 mRNA complexes to P-bodies or stress granules (Katahira et al., 2008).
Cbk1, Ssd1 and cancer
The functional relationship between Cbk1 and Ssd1 kinase may suggest a previously unanticipated mechanism for mammalian LATS/NDR tumor suppressor kinases in regulation of post-transcriptional gene expression. It is well established that the translational regulation of localized mRNA has a direct impact on cancer development and metastasis (Rodriguez et al., 2008). For example, the protein levels of the mRNA localization factors ZBP1 and IMP1 directly influence metastasis and tumor progression (Gu et al., 2008, Lapidus et al., 2007, Wang et al., 2004, Kobel et al., 2007, Dimitriadis et al., 2007). LATS/NDR kinases are implicated in cellular transformation and growth control (Hergovich et al., 2006, Hergovich & Hemmings, 2009). Despite their importance, the mechanisms by which they influence cancer progression are poorly understood (Hergovich & Hemmings, 2009). Our work suggests that a conserved function for LATS/NDR kinases is to regulate mRNA localization and metabolism via proteins such as Ssd1. Thus, further work on Cbk1 and Ssd1-dependent regulation of mRNA function may reveal insights into the LATS/NDR-mediated mechanisms of cancer development.
EXPERIMENTAL PROCEDURES
Yeast growth conditions and strain construction
Standard yeast genetic and culture methods were used as described (Guthrie & Fink, 1991, Kurischko et al., 2005). The strains used in this study are listed in Table I. Strains expressing C-terminal truncations of Ssd1 (Ssd11–450, Ssd11–570, Ssd11–670, Ssd11–697, Ssd11–1014 and Ssd11–1170) were constructed by integration of PCR-based GFP cassettes, as described (Longtine et al., 1998). The parental Pap1-cherryFP::HIS3 strain was constructed by integration of Pap1-cherryFP::HIS3 PCR cassette, amplified from strain MS7839 (gift of Dr. P. Melloy, Farleigh Dickinson University). The parental Nop56/Sik1-RFP::KANMX strain for FLY3074 was obtained from Dr. James Falvo (UCSF). The oligonucleotides used in this study are listed in Table III.
Table I.
Yeast strains
| Strains | Relevant genotype |
|---|---|
| FLY1593 | MATa SSD1-GFP::KANMX |
| FLY2184 | MATα ssd1Δ::NATMX |
| FLY2293 | MATa ssd1Δ::NATMX cbk1Δ::KANMX |
| FLY3074 | MATα NOP56-RFP::KANMX ssd1Δ::NATMX |
| FLY3149 | MATa SSD11–697-GFP::KANMX |
| FLY3168 | MATa SSD11–450-GFP::KANMX |
| FLY3172 | MATa SSD11–570-GFP::KANMX |
| FLY3176 | MATa SSD11–670-GFP::KANMX |
| FLY3202 | MATa SSD11–450-GFP::KANMX cbk1Δ::KANMX |
| FLY3206 | MATa SSD11–570-GFP::KANMX cbk1Δ::KANMX |
| FLY3210 | MATa SSD11–670-GFP::KANMX cbk1Δ::KANMX |
| FLY3219 | MATα SSD11–450-GFP::KANMX NOP56-RFP::KANMX |
| FLY3222 | MATa SSD11–570-GFP::KANMX NOP56-RFP::KANMX |
| FLY3225 | MATa SSD11–670-GFP::KANMX NOP56-RFP::KANMX |
| FLY3290 | MATa SSD11–1014-GFP::KANMX |
| FLY3292 | MATa SSD11–1170-GFP::KANMX |
| FLY3301 | MATa NOP56-RFP::KANMX SSD11–450-GFP::KANMX cbk1-as-HIS3::cbk1Δ::KANMX |
| FLY3308 | MATα SSD11–1014-GFP::KANMX NOP56-RFP::KANMX |
| FLY3310 | MATa SSD11–1170-GFP::KANMX NOP56-RFP::KANMX |
| FLY3313 | MATα SSD11–1014-GFP::KANMX cbk1Δ::KANMX |
| FLY3316 | MATa SSD11–1170-GFP::KANMX cbk1Δ::KANMX |
| FLY3365 | MATa PAP1-RFP::HIS3 ssd1Δ::NATMX |
| FLY3433 | MATa/α SSD1-GFP::KANMX/SSD1 PAP1-RFP::HIS3/PAP1 |
| FLY3467 | MATa SSD11–697-GFP::KANMX PAP1-RFP::HIS3 |
| FLY3795 | MATa/α SSD1-GFP::KANMX/SSD1 NOP56-RFP::KANMX/NOP56 |
Table III.
Oligonucleotides
| Oligos | Sequence | Purpose | Gene |
|---|---|---|---|
| FLO499 | 5′-GAAAGTCTAGAATATAGGAGGAATTTTACGGACACTAATGAGGGTGGTCCCGGTGGTCGGATCCCCGGGTTAATTAA-3′ | tagging | SSD11–697 |
| FLO504 | 5′-ATTCCTTTGAACAGTAGTGACGATTACCACAACGATGCATCTGTTGGTGGTCCCGGTGGTCGGATCCCCGGGTTAATTAA-3′ | tagging | SSD11–450 |
| FLO505 | 5′-AGCGGTACATTAGGTTTGTTGAGACCATCCCAACAAGCTAATAGCGGTGGTCCCGGTGGTCGGATCCCCGGGTTAATTAA-3′ | tagging | SSD11–570 |
| FLO506 | 5′-AACAATTTTCTTTCGAATGAATATTTGGATCAAAAAAATCCGCAAGGTGGTCCCGGTGGTCGGATCCCCGGGTTAATTAA-3′ | tagging | SSD11–670 |
| FLO507 | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCAATAACGGAGGTGGACGTAAGTCCCTATT-3′ | Gateway cloning | SSD1332–450 |
| FLO508 | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCAACAGATGCATCGTTGTGGTAATCGTCACTACTGTTCAAAGGAAT-3′ | Gateway cloning | SSD11–450 |
| FLO520 | 5′-AGAAGATACGCTGATCATGTCGTTCATAGGCAATTAAAGGCCGTTGGTGGTCCCGGTGGTCGGATCCCCGGGTTAATTAA-3′ | tagging | SSD11–1014 |
| FLO521 | 5′-TATAGAAATTCCATTAAGAACAAATTCAGATCCACAGCCGCTGAGGGTGGTCCCGGTGGTCGGATCCCCGGGTTAATTAA-3′ | tagging | SSD11–1170 |
| FLO525 | 5′-GACCACAAAGTATTCAATCTGCCCTCCGCTTCGTAAAGGGTTACGATTTGCCAGATGAAGTTTTCGATGAAAATGAAAAGAGA CCATCAAAGAAGAGTA-3′ | tagging | PAP1-RFP |
| FLO526 | 5′-ATCCAGCTTGTAACACAAAATTCTCACCTAATGGTAAAATGTCGCTACACGCGAAAGAGCATATGTAGAGAGAATCTCGTAAAC GTTTTACTGCTTCGTA-3′ | tagging | PAP1-RFP |
| FLO527 | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAACAAAATGTCTAAAAATAGCAACGTTAACA-3′ | Gateway cloning | SSD11–450, SSD11–520 |
| FLO534 | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCACCTTCAACTTCAACATCGTCATT-3′ | Gateway cloning | SSD11–520 |
| FLO635 | 5′-TCCAAAGCCAAAGGCCAACG-3′ | RT-PCR | ADH1 |
| FLO636 | 5′-CCGATCTTCCAGCCCTTAACG-3′ | RT-PCR | ADH1 |
| FLO639 | 5′-ATGCTTCAATCCGTTGT-3′ | RT-PCR | SRL1 |
| FLO640 | 5′-CGTATTTGGGGTGACTG-3′ | RT-PCR | SRL1 |
Plasmid construction
The plasmids and oligonucleotides used in this study are listed in Table II and III. SSD1 was subcloned from YEp13-SSD1 (a gift from Dr. C. Boone, University of Toronto) into pUC19 for further manipulations. For SSD1-NLSΔ plasmids we obtained an SSD1 fragment with each NLS amino acid codon (amino acids 417–427: KKEKEEKKRRK) mutated to alanine from Mr. Gene GmbH (Regensburg, Germany). pUC19-SSD1-NLSΔ plasmid (FLE1180) was constructed by replacing the 266 bp NsiI-SpeI fragment of pUC19-SSD1 (FLE1069) with the corresponding SSD1-NLSΔ fragment. We subcloned the 1.6 kb NdeI-XbaI fragment of pUC19-SSD1-NLSΔ into the NdeI-XbaI sites of pENTRY-SSD1. We reinserted an internal 309 bp XbaI fragment that was lost during XbaI digest to yield pENTRY-SSD1-NLSΔ (FLE1212). pENTRY-SSD11–450-NLSΔ was constructed by PCR amplification of the N-terminal 1350 bp fragment of Ssd1 from pUC19-SSD1-NLSΔ (FLE1180) using oligos FLO508 and FLO527. SSD11–450-NLSΔ was subcloned into pDONR221 to create pENTRY-SSD11–450-NLSΔ (FLE1204). pENTRY-SSD1-RBDΔ vector (FLE1205) was constructed by deleting the 309 bp XbaI fragment from FLE1057, thereby creating an in-frame deletion of amino acids 686–788. All SSD1 derivatives in entry vectors were subcloned into Gateway-compatible GFP and TAP plasmids (C-terminally tagged), as described (Alberti et al., 2007). pDONR221 and all Gateway-compatible parent vectors were provided by Dr. Aaron Gitler (Univ. of Pennsylvania). All SSD1 constructs were confirmed by sequencing.
Table II.
Plasmids
| Plasmids | Alias/relevant markers | Source |
|---|---|---|
| FLE1019 | pAG415-GPDpr-SSD1-GFP | Kurischko et al., 2011 |
| FLE1020 | pAG415-GPDpr-SSD11–520-GFP | Kurischko et al., 2011 |
| FLE1057 | pENTRY-SSD1 | Kurischko et al., 2011 |
| FLE1066 | pENTRY-SSD1332–450 | This work |
| FLE1067 | pAG416-GPDpr-SSD1332–450-GFP | This work |
| FLE1069 | pUC19-SSD1 | Kurischko et al., 2011 |
| FLE1180 | pUC19-SSD1NLSΔ | This work |
| FLE1204 | pENTRY-SSD11–450-NLSΔ | This work |
| FLE1205 | pENTRY-SSD1RBDΔ | Kurischko et al., 2011 |
| FLE1210 | pAG415-GPDpr-SSD1RBDΔ-GFP | Kurischko et al., 2011 |
| FLE1211 | pAG415-GPDpr-SSD11–450-NLSΔ-GFP | This work |
| FLE1212 | pENTRY-SSD1NLSΔ | This work |
| FLE1213 | pAG415-GPDpr-SSD1NLSΔ-GFP | This work |
| FLE1276 | pAG415-GPDpr-SSD1RBDΔ-TAP | This work |
| FLE1277 | pAG415-GPDpr-SSD1NLSΔ-TAP | This work |
| pJK59 | pRS416-SEC63-GFP | Fehrenbacher et al., 2002 |
| pKW1219 | pRS425-NLS-mRFP | Madrid et al., 2006 |
| pRP1574 | pEDC3-chRFP-URA3 | Roy Parker, Univ. of Arizona |
Unless otherwise designated, all genes are expressed under the control of their physiological promoters.
The pENTRY vector (FLE1066) for the NLS-GFP reporter (FLE1067) was constructed by subcloning the PCR-amplified SSD1 fragment corresponding to amino acids 332–450 (bp 996–1350), using oligos FLO507 and FLO508, into pDONR221. The RFP-tagged Edc3 plasmid was obtained from Dr. Roy Parker, Howard Hughes Medical Institute, Univ. of Arizona. Plasmid pRS425-NLS-mRFP (pKW1219) was obtained from Dr. Karsten Weis, Univ. of California, Berkeley (Madrid et al., 2006).
Immunoprecipitation and immunoblot analysis
Immunoprecipitation and immunoblot analyses were conducted as described (Kurischko et al., 2011) using monoclonal anti-GFP and secondary anti-mouse AP-conjugated antibodies (Roche Applied Science and Promega). All yeast extracts were prepared in buffer containing the RNAse inhibitor RNAsin (Promega).
For protein-mRNA affinity precipitation experiments, GFP-tagged proteins were immunoprecipitated from 525 ug yeast extract that was pre-treated with 50 ug proteinase K at 70°C for 15 min. RNA was purified using the MasterPure kit (EPICENTRE Biotechnologies) and resuspended in 40 ul TE. 10 ul of each RNA sample was quantified by UV spectroscopy at 260 nm and 280 nm. The remaining RNA sample was used for RT-PCR procedures.
RT-PCR methods
RT-PCR with immunoprecipitated mRNAs was carried out as described (Peritz et al., 2006, Kurischko et al., 2011). Yeast RNA was purified using the MasterPure kit (EPICENTRE Biotechnologies) and cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science). The oligos used to amplify ADH1 and SRL1 are listed in Table III.
Fluorescence microscopy
All microscopy was carried out as described in (Kurischko et al., 2011). Wide-field fluorescence microscopy was carried out with a Leica DMR5 fluorescence microscope equipped with a 100× PL APO 1.46 NA oil objective and an ImagEM 16-bit cooled Hamamatsu EMCCD camera, as described (Nelson et al., 2003, Kurischko et al., 2008). Image-capture was controlled by Volocity software (Perkin Elmer). Spinning disk confocal microscopy was conducted with Leica Inverted DMI4000 microscope equipped with a 100× HC× PL APO 1.46 NA oil objective, a Yokogawa CSU-10 spinning disk confocal system and an ImagEM 16-bit cooled Hamamatsu EMCCD camera. Laser excitation was provided by a 488 nm (Spectra Physics) and a 561 nm laser (Cobolt Jive) controlled through LMM5 (Spectral Applied Research). The emissions were collected at 503–552 nm for GFP and 583–650 nm for RFP. Z stacks were taken for a total thickness of 1.8–3.4 μm at a step size of 0.2 μm. Image-capture was controlled by Metamorph software (MDS Analytical Technologies). Data analysis and 3D modeling was conducted with Volocity software (Perkin Elmer).
SRL1 mRNA localization was monitored in Ssd1-NLSΔ-TAP and Ssd1-RBDΔ-TAP cells, as described (Kurischko et al., 2011). Nuclear-cytoplasmic ratios of Ssd1-GFP were obtained using Metamorph software. Nuclei were defined by colocalization with NLS-RFP. The average GFP fluorescence signals in a region of interest (nucleus and cytoplasm) were measured. For cytoplasmic GFP measurements, vacuoles were avoided to prevent underestimates. The background (external to cells) was subtracted from the nuclear and cytoplasmic fluorescence and the ratios were calculated for 16–26 cells per strain. The mean values with the corresponding standard deviations were calculated and plotted.
Supplementary Material
Supplemental Figure S1. Ssd11–450-NLSΔ fails to localize to the nucleus. Images of three cells expressing Ssd11–450-NLSΔ-GFP and Pap1-RFP (FLY3365 + FLE1211) captured by spinning disk confocal fluorescence microscopy. Left panels: brightfield images, middle panel: merged optical sections of GFP and RFP fluorescence (21 × 0.2um sections); right panels: 3-D models of image data generated by Volocity software. The 3-D models reveal that the Ssd11–450-NLSΔ puncta are extra-nuclear or perinuclear in localization. Scale bar = 4 um. The bottom cell is also depicted in Fig. 3B.
Supplemental Figure S2. SRL1 mRNA localization is aberrant in Ssd1-NLSΔ and Ssd1-RBDδ cells. SRL1 mRNA localization was analyzed in budded cells, as previously described (Kurischko et al., 2011). Cells (FLY3196) expressing wild type Ssd1 (SSD1; FLE1083), Ssd1-NLSΔ (SSD1-NLSΔ-TAP; FLE1277), Ssd1-RBDΔ (SSD1-RBDΔ-TAP; FLE1276) and empty vector (ssd1Δ; pRS415) exhibited 5 general patterns of SRL1 mRNA localization, as depicted in the graph. These include cells with no SRL1 mRNA spots (no spots), multiple faint spots, and 1–3 bright spots restricted to the mother (M), bud (D) or both mother and bud (M+D). The data for the wild type SSD1 and empty vector controls are from previously published experiments (Kurischko et al., 2011).
Supplemental Figure S3. C-terminal truncated Ssd1-GFP partially co-localizes with P-bodies in cbk1Δ cells. Physiologically expressed Ssd11–570, Ssd11–670, Ssd11–1014 and Ssd11–1170 were monitored in cbk1Δ cells that co-express Edc3-RFP. Some cytoplasmic Ssd1 puncta co-localize with P-body protein Edc3, see arrowheads (26.2% for Ssd11–570, 45.9% for Ssd11–670, 30.3% for Ssd11–1014 and 17.6% for Ssd11–1170 cells; n=100–300 cells for each). The strains used in these experiments were FLY3206, FLY3210, FLY3313 and FLY3316. All cells were monitored by spinning disk fluorescence microscopy and each image represents a single optical section. Scale bar = 8 um.
Supplemental Table 1. Nuclear-cytoplasmic ratios of C-terminal truncated Ssd1. Data were obtained by measuring the average Ssd1-GFP signal in the nucleus and cytoplasm, as described in the Material and Methods. Cells with large vacuoles were not taken in consideration. The data represent N (nucleus), C (cytoplasm), B (backgound), N-B, C-B, ratio N-B/C-B, number of nuclei (observations), median, mean, standard deviation, p values for pairwise comparisons of ratios between mutants and wild type.
The movie was generated from the upper 3-D model in Supplemental Fig. S1 and demonstrates that the Ssd11–450-NLSΔ puncta are extra-nuclear. 3-D projections and movie were generated by Volocity software.
Acknowledgments
The authors thank Dr. Lingli Zhang and Dr. Bruce Freedman (University of Pennsylvania School of Veterinary Medicine’s Core Imaging Facility), Andrea Stout (University of Pennsylvania School of Medicine Core Imaging Facility) and Phong Tran for help with confocal microscopy. We thank Dr. HongKyung Kim for constructing the SSD11–520 plasmid and for helpful discussions. We are grateful to Dr. Aaron Gitler (Univ. of Pennsylvania), Dr. Roy Parker (Univ. of Arizona), Dr. Karsten Weis (Univ. of California Berkeley) and Dr. Patricia Melloy (Farleigh Dickinson University) for providing various plasmids and reagents. We thank Drs. Erfei Bi, Aaron Gitler and Phong Tran (University of Pennsylvania) for many insightful discussions. This work was supported by grants from the National Institutes of Health (GM060575), the American Cancer Society (RSG0508401) and the University Research Foundation (UPenn) to F.C.L.
References
- Aguilera A. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr Opin Cell Biol. 2005;17:242–250. doi: 10.1016/j.ceb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Alberti S, Gitler AD, Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24:913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach DL, Bloom K. ASH1 mRNA localization in three acts. Mol Biol Cell. 2001;12:2567–2577. doi: 10.1091/mbc.12.9.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- Bidlingmaier S, Weiss EL, Seidel C, Drubin DG, Snyder M. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:2449–2462. doi: 10.1128/MCB.21.7.2449-2462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohl F, Kruse C, Frank A, Ferring D, Jansen RP. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. Embo J. 2000;19:5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourens M, Panozzo C, Nowacka A, Imbeaud S, Mucchielli MH, Herbert CJ. Mutations in the Saccharomyces cerevisiae kinase Cbk1p lead to a fertility defect that can be suppressed by the absence of Brr1p or Mpt5p (Puf5p), proteins involved in RNA metabolism. Genetics. 2009;183:161–173. doi: 10.1534/genetics.109.105130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourens M, Racki W, Becam AM, Panozzo C, Boulon S, Bertrand E, Herbert CJ. Mutations in a small region of the exportin Crm1p disrupt the daughter cell-specific nuclear localization of the transcription factor Ace2p in Saccharomyces cerevisiae. Biol Cell. 2008;100:343–354. doi: 10.1042/BC20070077. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody SR, Wente SR. mRNA nuclear export at a glance. J Cell Sci. 2009;122:1933–1937. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P, Singer RH, Long RM. RNP localization and transport in yeast. Annu Rev Cell Dev Biol. 2001;17:297–310. doi: 10.1146/annurev.cellbio.17.1.297. [DOI] [PubMed] [Google Scholar]
- Chiannilkulchai N, Moenne A, Sentenac A, Mann C. Biochemical and genetic dissection of the Saccharomyces cerevisiae RNA polymerase C53 subunit through the analysis of a mitochondrially mis-sorted mutant construct. J Biol Chem. 1992;267:23099–23107. [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Czaplinski K, Singer RH. Pathways for mRNA localization in the cytoplasm. Trends Biochem Sci. 2006;31:687–693. doi: 10.1016/j.tibs.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Darzacq X, Powrie E, Gu W, Singer RH, Zenklusen D. RNA asymmetric distribution and daughter/mother differentiation in yeast. Curr Opin Microbiol. 2003;6:614–620. doi: 10.1016/j.mib.2003.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadis E, Trangas T, Milatos S, Foukas PG, Gioulbasanis I, Courtis N, Nielsen FC, Pandis N, Dafni U, Bardi G, Ioannidis P. Expression of oncofetal RNA-binding protein CRD-BP/IMP1 predicts clinical outcome in colon cancer. Int J Cancer. 2007;121:486–494. doi: 10.1002/ijc.22716. [DOI] [PubMed] [Google Scholar]
- Du LL, Novick P. Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:503–514. doi: 10.1091/mbc.01-07-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du TG, Jellbauer S, Muller M, Schmid M, Niessing D, Jansen RP. Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep. 2008;9:781–787. doi: 10.1038/embor.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkmann JA, Kutay U. Nuclear export of mRNA: from the site of transcription to the cytoplasm. Exp Cell Res. 2004;296:12–20. doi: 10.1016/j.yexcr.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Moore MJ. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin Cell Dev Biol. 2007;18:186–193. doi: 10.1016/j.semcdb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Goler-Baron V, Selitrennik M, Barkai O, Haimovich G, Lotan R, Choder M. Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev. 2008;22:2022–2027. doi: 10.1101/gad.473608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Wells AL, Pan F, Singer RH. Feedback regulation between zipcode binding protein 1 and beta-catenin mRNAs in breast cancer cells. Mol Cell Biol. 2008;28:4963–4974. doi: 10.1128/MCB.00266-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. Methods in Enzymology. 1991;194:1–933. [PubMed] [Google Scholar]
- Haim-Vilmovsky L, Gerst JE. m-TAG: a PCR-based genomic integration method to visualize the localization of specific endogenous mRNAs in vivo in yeast. Nat Protoc. 2009;4:1274–1284. doi: 10.1038/nprot.2009.115. [DOI] [PubMed] [Google Scholar]
- Hasegawa Y, Irie K, Gerber AP. Distinct roles for Khd1p in the localization and expression of bud-localized mRNAs in yeast. Rna. 2008;14:2333–2347. doi: 10.1261/rna.1016508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A, Hemmings BA. Mammalian NDR/LATS protein kinases in hippo tumor suppressor signaling. Biofactors. 2009;35:338–345. doi: 10.1002/biof.47. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol. 2006;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S, Kehlenbach RH. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 2007;17:193–201. doi: 10.1016/j.tcb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Jansen JM, Wanless AG, Seidel CW, Weiss EL. Cbk1 regulation of the RNA-binding protein Ssd1 integrates cell fate with translational control. Curr Biol. 2009;19:2114–2120. doi: 10.1016/j.cub.2009.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Nelson B, Robinson MD, Chen Y, Andrews B, Tyers M, Boone C. High-Resolution Genetic Mapping With Ordered Arrays of Saccharomyces cerevisiae Deletion Mutants. Genetics. 2002;162:1091–1099. doi: 10.1093/genetics/162.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Andalis AA, Liszt GB, Fink GR, Guarente L. Saccharomyces cerevisiae SSD1-V confers longevity by a Sir2p-independent mechanism. Genetics. 2004;166:1661–1672. doi: 10.1534/genetics.166.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Guarente L. Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics. 2002;160:83–95. doi: 10.1093/genetics/160.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Miki T, Takano K, Maruhashi M, Uchikawa M, Tachibana T, Yoneda Y. Nuclear RNA export factor 7 is localized in processing bodies and neuronal RNA granules through interactions with shuttling hnRNPs. Nucleic Acids Res. 2008;36:616–628. doi: 10.1093/nar/gkm556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SM, Corbett AH. Messenger RNA export from the nucleus: a series of molecular wardrobe changes. Traffic. 2009;10:1199–1208. doi: 10.1111/j.1600-0854.2009.00944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobel M, Weidensdorfer D, Reinke C, Lederer M, Schmitt WD, Zeng K, Thomssen C, Hauptmann S, Huttelmaier S. Expression of the RNA-binding protein IMP1 correlates with poor prognosis in ovarian carcinoma. Oncogene. 2007;26:7584–7589. doi: 10.1038/sj.onc.1210563. [DOI] [PubMed] [Google Scholar]
- Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- Kurischko C, Kim HK, Kuravi VK, Pratzka J, Luca FC. The yeast Cbk1 kinase regulates mRNA localization via the mRNA-binding protein Ssd1. J Cell Biol. 2011;192:583–598. doi: 10.1083/jcb.201011061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurischko C, V, Kuravi K, Wannissorn N, Nazarov PA, Husain M, Zhang C, Shokat KM, McCaffery JM, Luca FC. The yeast LATS/Ndr kinase Cbk1 regulates growth via Golgi-dependent glycosylation and secretion. Mol Biol Cell. 2008;19:5559–5578. doi: 10.1091/mbc.E08-05-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurischko C, Weiss G, Ottey M, Luca FC. A role for the Saccharomyces cerevisiae regulation of Ace2 and polarized morphogenesis signaling network in cell integrity. Genetics. 2005;171:443–455. doi: 10.1534/genetics.105.042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus K, Wyckoff J, Mouneimne G, Lorenz M, Soon L, Condeelis JS, Singer RH. ZBP1 enhances cell polarity and reduces chemotaxis. J Cell Sci. 2007;120:3173–3178. doi: 10.1242/jcs.000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EK, Kim HH, Kuwano Y, Abdelmohsen K, Srikantan S, Subaran SS, Gleichmann M, Mughal MR, Martindale JL, Yang X, Worley PF, Mattson MP, Gorospe M. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat Struct Mol Biol. 2010;17:732–739. doi: 10.1038/nsmb.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Luukkonen BG, Seraphin B. A conditional U5 snRNA mutation affecting pre-mRNA splicing and nuclear pre-mRNA retention identifies SSD1/SRK1 as a general splicing mutant suppressor. Nucleic Acids Res. 1999;27:3455–3465. doi: 10.1093/nar/27.17.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchi P, Brownawell AM, Grunewald B, DesGroseillers L, Macara IG, Kiebler MA. The brain-specific double-stranded RNA-binding protein Staufen2: nucleolar accumulation and isoform-specific exportin-5-dependent export. J Biol Chem. 2004;279:31440–31444. doi: 10.1074/jbc.C400226200. [DOI] [PubMed] [Google Scholar]
- Madrid AS, Mancuso J, Cande WZ, Weis K. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J Cell Biol. 2006;173:361–371. doi: 10.1083/jcb.200506199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazanka E, Alexander J, Yeh BJ, Charoenpong P, Lowery DM, Yaffe M, Weiss EL. The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry. PLoS Biol. 2008;6:e203. doi: 10.1371/journal.pbio.0060203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald HB, Helfant AH, Mahony EM, Khosla SK, Goetsch L. Mutational analysis reveals a role for the C terminus of the proteasome subunit Rpt4p in spindle pole body duplication in Saccharomyces cerevisiae. Genetics. 2002;162:705–720. doi: 10.1093/genetics/162.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Macara IG. RNA localization and polarity: from A(PC) to Z(BP) Trends Cell Biol. 2009;19:156–164. doi: 10.1016/j.tcb.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir SS, Fiedler D, Cashikar AG. Ssd1 is required for thermotolerance and Hsp104-mediated protein disaggregation in Saccharomyces cerevisiae. Mol Cell Biol. 2009;29:187–200. doi: 10.1128/MCB.02271-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H, Isono K. Analysis of genetic interactions between DHH1, SSD1 and ELM1 indicates their involvement in cellular morphology determination in Saccharomyces cerevisiae. Yeast. 1999;15:481–496. doi: 10.1002/(SICI)1097-0061(199904)15:6<481::AID-YEA391>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Nelson B, Kurischko C, Horecka J, Mody M, Nair P, Pratt L, Zougman A, McBroom LD, Hughes TR, Boone C, Luca FC. RAM: A Conserved Signaling Network That Regulates Ace2p Transcriptional Activity and Polarized Morphogenesis. Mol Biol Cell. 2003;14:3782–3803. doi: 10.1091/mbc.E03-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods. 2007;4:951–956. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- Ohyama Y, Kasahara K, Kokubo T. Saccharomyces cerevisiae Ssd1p promotes CLN2 expression by binding to the 5′-untranslated region of CLN2 mRNA. Genes Cells. 2010;15:1169–1188. doi: 10.1111/j.1365-2443.2010.01452.x. [DOI] [PubMed] [Google Scholar]
- Olivier C, Poirier G, Gendron P, Boisgontier A, Major F, Chartrand P. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol Cell Biol. 2005;25:4752–4766. doi: 10.1128/MCB.25.11.4752-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin N, Chartrand P. Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol. 2008;18:105–111. doi: 10.1016/j.tcb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Percipalle P, Raju CS, Fukuda N. Actin-associated hnRNP proteins as transacting factors in the control of mRNA transport and localization. RNA Biol. 2009;6:171–174. doi: 10.4161/rna.6.2.8195. [DOI] [PubMed] [Google Scholar]
- Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiriksdottir E, Langel U, Eberwine J. Immunoprecipitation of mRNA-protein complexes. Nat Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Jones JC, Greenleaf AL. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry. 2004;43:15702–15719. doi: 10.1021/bi048364h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racki WJ, Becam AM, Nasr F, Herbert CJ. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. Embo J. 2000;19:4524–4532. doi: 10.1093/emboj/19.17.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AJ, Czaplinski K, Condeelis JS, Singer RH. Mechanisms and cellular roles of local protein synthesis in mammalian cells. Curr Opin Cell Biol. 2008;20:144–149. doi: 10.1016/j.ceb.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Stutz F. Nuclear export of RNA. Biol Cell. 2004;96:639–655. doi: 10.1016/j.biolcel.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gil A, Garcia-Martinez J, Pelechano V, Munoz-Centeno Mde L, Geli V, Perez-Ortin JE, Chavez S. The distribution of active RNA polymerase II along the transcribed region is gene-specific and controlled by elongation factors. Nucleic Acids Res. 2010;38:4651–4664. doi: 10.1093/nar/gkq215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbia M, Parnell EJ, Yu Y, Olsen AE, Kretschmann KL, Voth WP, Stillman DJ. Regulation of the yeast Ace2 transcription factor during the cell cycle. J Biol Chem. 2008;283:11135–11145. doi: 10.1074/jbc.M800196200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Paquin N, Forget A, Chartrand P. Nuclear shuttling of She2p couples ASH1 mRNA localization to its translational repression by recruiting Loc1p and Puf6p. Mol Biol Cell. 2009;20:2265–2275. doi: 10.1091/mbc.E08-11-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, St-Denis A, Chartrand P. Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Genes Dev. 2010;24:1914–1926. doi: 10.1101/gad.1937510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci U S A. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sil A, Herskowitz I. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84:711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- Smith R. Moving molecules: mRNA trafficking in Mammalian oligodendrocytes and neurons. Neuroscientist. 2004;10:495–500. doi: 10.1177/1073858404266759. [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Stettler S, Chiannilkulchai N, Hermann-Le Denmat S, Lalo D, Lacroute F, Sentenac A, Thuriaux P. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol Gen Genet. 1993;239:169–176. doi: 10.1007/BF00281615. [DOI] [PubMed] [Google Scholar]
- Stewart M. Ratcheting mRNA out of the nucleus. Mol Cell. 2007;25:327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Sutton A, Immanuel D, Arndt KT. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Yamada K, Yabumoto K, Fujii S, Huser A, Tsuji G, Koga H, Dohi K, Mori M, Shiraishi T, O’Connell R, Kubo Y. Saccharomyces cerevisiae SSD1 orthologues are essential for host infection by the ascomycete plant pathogens Colletotrichum lagenarium and Magnaporthe grisea. Mol Microbiol. 2007;64:1332–1349. doi: 10.1111/j.1365-2958.2007.05742.x. [DOI] [PubMed] [Google Scholar]
- Terry LJ, Wente SR. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell. 2009;8:1814–1827. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya E, Matsuzaki G, Kurano K, Fukuchi T, Tsukao A, Miyakawa T. The Saccharomyces cerevisiae SSD1 gene is involved in the tolerance to high concentration of Ca2+ with the participation of HST1/NRC1/BFR1. Gene. 1996;176:35–38. doi: 10.1016/0378-1119(96)00204-1. [DOI] [PubMed] [Google Scholar]
- Turcq B, Dobinson KF, Serizawa N, Lambowitz AM. A protein required for RNA processing and splicing in Neurospora mitochondria is related to gene products involved in cell cycle protein phosphatase functions. Proc Natl Acad Sci U S A. 1992;89:1676–1680. doi: 10.1073/pnas.89.5.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesono Y, Fujita A, Toh-e A, Kikuchi Y. The MCS1/SSD1/SRK1/SSL1 gene is involved in stable maintenance of the chromosome in yeast. Gene. 1994;143:135–138. doi: 10.1016/0378-1119(94)90618-1. [DOI] [PubMed] [Google Scholar]
- Uesono Y, Toh-e A, Kikuchi Y. Ssd1p of Saccharomyces cerevisiae associates with RNA. J Biol Chem. 1997;272:16103–16109. doi: 10.1074/jbc.272.26.16103. [DOI] [PubMed] [Google Scholar]
- Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, Singer RH, Segall JE, Condeelis JS. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Fukuda M, Yoshida M, Yanagida M, Nishida E. Involvement of CRM1, a nuclear export receptor, in mRNA export in mammalian cells and fission yeast. Genes Cells. 1999;4:291–297. doi: 10.1046/j.1365-2443.1999.00259.x. [DOI] [PubMed] [Google Scholar]
- Weiss EL, Kurischko C, Zhang C, Shokat K, Drubin DG, Luca FC. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J Cell Biol. 2002;158:885–900. doi: 10.1083/jcb.200203094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler RT, Kupiec M, Magnelli P, Abeijon C, Fink GR. A Saccharomyces cerevisiae mutant with increased virulence. Proc Natl Acad Sci U S A. 2003;100:2766–2770. doi: 10.1073/pnas.0437995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Achsel T, Bagni C. mRNPs, polysomes or granules: FMRP in neuronal protein synthesis. Curr Opin Neurobiol. 2006;16:265–269. doi: 10.1016/j.conb.2006.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Ssd11–450-NLSΔ fails to localize to the nucleus. Images of three cells expressing Ssd11–450-NLSΔ-GFP and Pap1-RFP (FLY3365 + FLE1211) captured by spinning disk confocal fluorescence microscopy. Left panels: brightfield images, middle panel: merged optical sections of GFP and RFP fluorescence (21 × 0.2um sections); right panels: 3-D models of image data generated by Volocity software. The 3-D models reveal that the Ssd11–450-NLSΔ puncta are extra-nuclear or perinuclear in localization. Scale bar = 4 um. The bottom cell is also depicted in Fig. 3B.
Supplemental Figure S2. SRL1 mRNA localization is aberrant in Ssd1-NLSΔ and Ssd1-RBDδ cells. SRL1 mRNA localization was analyzed in budded cells, as previously described (Kurischko et al., 2011). Cells (FLY3196) expressing wild type Ssd1 (SSD1; FLE1083), Ssd1-NLSΔ (SSD1-NLSΔ-TAP; FLE1277), Ssd1-RBDΔ (SSD1-RBDΔ-TAP; FLE1276) and empty vector (ssd1Δ; pRS415) exhibited 5 general patterns of SRL1 mRNA localization, as depicted in the graph. These include cells with no SRL1 mRNA spots (no spots), multiple faint spots, and 1–3 bright spots restricted to the mother (M), bud (D) or both mother and bud (M+D). The data for the wild type SSD1 and empty vector controls are from previously published experiments (Kurischko et al., 2011).
Supplemental Figure S3. C-terminal truncated Ssd1-GFP partially co-localizes with P-bodies in cbk1Δ cells. Physiologically expressed Ssd11–570, Ssd11–670, Ssd11–1014 and Ssd11–1170 were monitored in cbk1Δ cells that co-express Edc3-RFP. Some cytoplasmic Ssd1 puncta co-localize with P-body protein Edc3, see arrowheads (26.2% for Ssd11–570, 45.9% for Ssd11–670, 30.3% for Ssd11–1014 and 17.6% for Ssd11–1170 cells; n=100–300 cells for each). The strains used in these experiments were FLY3206, FLY3210, FLY3313 and FLY3316. All cells were monitored by spinning disk fluorescence microscopy and each image represents a single optical section. Scale bar = 8 um.
Supplemental Table 1. Nuclear-cytoplasmic ratios of C-terminal truncated Ssd1. Data were obtained by measuring the average Ssd1-GFP signal in the nucleus and cytoplasm, as described in the Material and Methods. Cells with large vacuoles were not taken in consideration. The data represent N (nucleus), C (cytoplasm), B (backgound), N-B, C-B, ratio N-B/C-B, number of nuclei (observations), median, mean, standard deviation, p values for pairwise comparisons of ratios between mutants and wild type.
The movie was generated from the upper 3-D model in Supplemental Fig. S1 and demonstrates that the Ssd11–450-NLSΔ puncta are extra-nuclear. 3-D projections and movie were generated by Volocity software.