Abstract
When grown in green light, Fremyella diplosiphon strain UTEX 481 produces the red-colored protein phycoerythrin (PE) to maximize photosynthetic light harvesting. PE is composed of two subunits, CpeA and CpeB, which carry two and three phycoerythrobilin (PEB) chromophores, respectively, that are attached to specific Cys residues via thioether linkages. Specific bilin lyases are hypothesized to catalyze each PEB ligation. Using a heterologous, coexpression system in Escherichia coli, the PEB ligation activities of putative lyase subunits CpeY, CpeZ, and CpeS were tested on the CpeA and CpeB subunits from F. diplosiphon. Purified His6-tagged CpeA, obtained by coexpressing cpeA, cpeYZ, and the genes for PEB synthesis, had absorbance and fluorescence emission maxima at 566 and 574 nm, respectively. CpeY alone, but not CpeZ, could ligate PEB to CpeA, but the yield of CpeA-PEB was lower than achieved with CpeY and CpeZ together. Studies with site-specific variants of CpeA(C82S and C139S), together with mass spectrometric analysis of trypsin-digested CpeA-PEB, revealed that CpeY/CpeZ attached PEB at Cys82 of CpeA. The CpeS bilin lyase ligated PEB at both Cys82 and Cys139 of CpeA but very inefficiently; the yield of PEB ligated at Cys82 was much lower than observed with CpeY or CpeY/CpeZ. However, CpeS efficiently attached PEB to Cys80 of CpeB but neither CpeY, CpeZ, nor CpeY/CpeZ could ligate PEB to CpeB.
Keywords: Cyanobacteria, Fluorescence, Photosynthesis, Photosynthetic Pigments, Post-translational Modification, Bilin Lyase, Phycobiliproteins, Phycoerythrin, Phycoerythrobilin
Introduction
The light-harvesting antennae in cyanobacteria and red algae are supramolecular complexes, phycobilisomes (PBS),2 composed of water-soluble and brilliantly colored phycobiliproteins (PBPs) and linker polypeptides (1–3). The covalent attachment of phycobilin chromophores to specific Cys residues, usually by enzymes called bilin lyases, results in highly fluorescent holo-PBPs (4–8). The attached phycobilin chromophores transfer excitation energy with high quantum efficiency to photosynthetic reaction centers (2, 9–17).
The major PBPs, each consisting of α- and β-subunits, in cyanobacteria are the aqua-colored allophycocyanin (AP) (λmax ∼650 nm), the blue-colored phycocyanin (PC) (λmax ∼620 nm), and the red-colored phycoerythrin (PE) (λmax ∼560 nm) (18). The spectroscopic properties of these proteins are determined primarily by their bilin chromophore(s), which are attached to specific Cys residues. The bilin lyases responsible for the phycocyanobilin (PCB) ligation at the binding sites for all PBPs in Synechococcus sp. strain PCC 7002 have been characterized (19). Thus far, four types of bilin lyases are known, and each has different characteristics and amino acid sequences. The first bilin lyases to be discovered belonged to the E/F family and are typified by the CpcE and CpcF proteins; these proteins form heterodimeric enzymes that can attach PCB to Cys82 of CpcA (α-PC subunit) and can also remove bilins from holosubunits or transfer bilins from a holosubunit to an aposubunit (5, 20, 21). Recent studies in both cyanobacteria (22) and Escherichia coli (23) have demonstrated that the CpcE/CpcF lyase can attach noncognate bilins (e.g. phycoerythrobilin (PEB) or phytochromobilin) to CpcA. Paralogs of the cpcE and cpcF genes, usually encoded in operons with phycobiliprotein subunit genes, are known (24, 25). Some of these paralogs, such as pecE/pecF and rpcG, have been shown to be involved in the attachment and isomerization of bilin chromophores to Cys residues, which usually occur on the α-subunit of some PBPs (e.g. PecA or RpcA) (6, 26–28). Similar to CpcE/CpcF, these lyases appear to be capable of attaching noncognate bilins to noncognate PBP subunits (23).
The second family of bilin lyases belong to the S/U family and are typified by CpeS- and CpcS-like proteins, which can be active as monomers (29, 30), homodimers (31), or heterodimers (CpcS/CpcU) (32, 33). Members of the S/U lyase family do not appear to catalyze the transfer or removal of bilins from holo-PBP subunits, but their protein substrate specificity seems to be broader than that of other lyase types (7, 19). They can typically recognize many different PBP substrates and attach bilins at their Cys82 equivalent positions (30). The third family of lyases is called the T-type and is typified by CpcT, an enzyme that attaches PCB at Cys153 of CpcB (8, 34). Last, there is a family of PBPs that autocatalytically ligate bilin chromophores. Currently, the sole representative of this family is ApcE (19, 35). It has been reported that an allophycocyanin subunit (ApcA) is able to self-ligate its bilin chromophore (36), but phenotypic analyzes of bilin lyase mutants in cyanobacteria strongly suggest that bilin lyases such as CpcSU are required for this activity in vivo (32). It should also be noted that phytochrome (37) and other related photoreceptors (38, 39) also add their chromophores autocatalytically.
PEs are a diverse family of PBPs with extensive variation in subunit composition and spectral properties. Insertions have occurred in the primary amino acid sequences of PEs that have created more chromophore-binding sites than occur in other PBP subunits. In the PE of the freshwater cyanobacterium F. diplosiphon, five PEB chromophores are ligated to Cys residues at α82, α139, β80, β165, and β48/β59 (doubly linked at rings A and D) (40, 41). The PEs of marine cyanobacteria are even more complex and diverse; they have additional bilin attachment sites that can carry either PEB or phycourobilin (25, 42).
Previous studies have suggested that bilin lyases are also required for bilin attachment to PEs (41, 43). Kahn et al. (44) found that a transposon insertion into the cpeY gene resulted in diminished levels of PE in Fremyella diplosiphon cells grown in green light. The cpeY and cpeZ genes, encoded in the PE operon cpeBAYZ, are paralogs of the cpcE and cpcF lyase genes (24, 44, 45). Kahn et al. (44) suggested that CpeY and CpeZ function in PE biosynthesis, possibly as a lyase in the attachment of PEB to the α- or β-subunits. Zhao et al. (30) later showed that the CpcS-type lyase from Nostoc sp. PCC 7120 (an organism that does not contain PE) was capable of attaching PEB to Cys82 on CpeA and CpeB from F. diplosiphon; this result suggested that this type of lyase had broad substrate specificity and might recognize Cys82 on any type of PBP subunit (including CpcB, ApcA, ApcB, ApcD, ApcF, CpeA, and CpeB) with the exception of CpcA, RpcA, and PecA.
In this study, we have used a biochemical approach with recombinant enzymes to characterize the roles of CpeY, CpeZ, and CpeS in PEB addition to PE subunits in F. diplosiphon strain UTEX 481. CpeY alone could ligate PEB to apo-CpeA, but the yield was lower (∼60%) than when both CpeY and CpeZ were present. Site-directed mutagenesis of cysteine residues on CpeA and mass spectrometry showed that CpeY alone and CpeY/CpeZ ligate PEB to Cys82 of CpeA. A very small amount of PEB was found attached to Cys139 in this sample as well, but CpeY/CpeZ was unable to attach PEB to the CpeA(C82S) mutant. CpeS was capable of PEB attachment to Cys80 of CpeB as well as to both Cys82 and Cys139 of CpeA; however, lower amounts of PEB were ligated to the apo-CpeA than obtained with CpeY/CpeZ. This result strongly suggested that the main function of CpeS is to attach PEB to Cys80 of CpeB.
EXPERIMENTAL PROCEDURES
Construction of Expression Vectors
Plasmids used in this study are listed in supplemental Table S1. Some of the expression vectors used in this study were previously described (19, 46–49). All expression constructs newly produced for this study were sequenced at the W. M. Keck Conservation and Molecular Genetics Laboratory (University of New Orleans) to confirm that no mutations had been introduced during PCR amplification and cloning.
Each gene was amplified by PCR from F. diplosiphon chromosomal DNA using the primers listed in supplemental Table S2, and each resulting amplicon was cloned into a Duet vector as listed under supplemental Table S1 (Novagen, Madison, WI) after digestion with restriction enzymes (engineered into the primers; underlined in the sequences in supplemental Table S2). As indicated in supplemental Table S1, a His6 tag was engineered into the constructs for producing CpeA, CpeB, and CpeZ. The plasmid pPebS was a generous gift from Dr. Nicole Frankenberg-Dinkel; it contains the ho1 (heme oxygenase) and pebS (PEB synthase) genes from a myovirus that infects Prochlorococcus spp. 48. E. coli strains harboring this plasmid produce PEB from heme.
Site-directed Mutagenesis of cpeA and cpeB
Plasmid pCpeA (supplemental Table S1) was used as a template for generating mutations in cpeA. The TransformerTM Site-directed Mutagenesis Kit from Clontech Laboratories, Inc. was used to create mutated genes for the production of three CpeA variants: CpeA(C82S), CpeA(C139S), and CpeA(C82S/C139S). The primers used were CpeA(C82S), CpeA(C139S), and pETDuet (XhoI del) (supplemental Table S1). Plasmid pCpeB (supplemental Table S1) was used as a template for generating mutations in cpeB by the same method. Three variants of CpeB were produced: CpeB(C80S), CpeB(C165S), and CpeB(C48S/C59S). The primers used were CpeB(C80S), CpeB(C165S), CpeB(C48S/C59S), and pETDuet-1 (XhoI del; supplemental Table S1).
Heterologous Expression and Purification of Recombinant Proteins
Expression plasmids were co-transformed into E. coli BL21(DE3) cells as required, and colonies were selected on Luria-Bertani (LB) plates in the presence of the appropriate combination of antibiotics (see supplemental Table S1) at the following concentrations: ampicillin (Ap: 100 μg ml−1), chloramphenicol (Cm: 34 μg ml−1), kanamycin (Km: 50 μg ml−1), and spectinomycin (Sp: 100 μg ml−1). To produce PEB using the pPebS expression plasmid, a 50-ml overnight starter culture was added to 1 liter of LB medium with the appropriate combination of antibiotics. This culture was shaken at 37 °C for 4 h until the optical density reached A600 nm = 0.6. Production of T7 RNA polymerase was induced by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were incubated with shaking at 190 rpm at 18 °C for another 16 h before they were harvested by centrifugation at 10,000 × g for 10 min. Cell pellets were stored at −20 °C until required.
E. coli cells containing recombinant proteins were thawed and resuspended in buffer O (20 mm Tris-HCl, 100 mm NaCl, pH 8.0) at 2.5 ml g−1 (wet weight) along with protease inhibitor mixture tablets (“Complete Mini” from Roche Applied Science). The cells were lysed, and the His6-tagged recombinant proteins were purified as previously described (8). The recombinant protein(s) were exhaustively dialyzed with buffer O containing 10 mm 2-mercaptoethanol overnight at 4 °C to remove the imidazole introduced during elution.
Fluorescence Emission and Absorbance Spectra
Fluorescence emission spectra were recorded with a PerkinElmer LS55 fluorescence spectrophotometer with slit widths set at 10 nm (excitation and emission). For recombinant PBPs, the excitation wavelength was set at 490 nm and samples were diluted to achieve a standard absorbance level ∼0.05 OD (at λmax) prior to recording the fluorescence emission spectra. Negative control samples (e.g. no lyase addition), which had little or no attached chromophore, were not diluted, as their OD values were generally less than 0.05. Absorbance spectra were acquired using a λ35, dual-beam UV-visible spectrophotometer (PerkinElmer Life Sciences). To compare the amount of relative fluorescent CpeA produced in the presence of the CpeY/CpeZ, CpeY, or CpeS lyases, these proteins were purified from the same volume of E. coli cultures (the pellets obtained were within 5% of each other in wet weight). The CpeA protein concentration was also estimated for each of these on SDS-polyacrylamide gels. The relative fluorescence intensity was multiplied by the dilution factor used, and then this was divided by the CpeA concentration to estimate the proportion of CpeA produced that was fluorescent. The value obtained for CpeY/CpeZ was set to 100%, and the other values were scaled accordingly (Table 1).
TABLE 1.
Comparison of spectral properties for various PE subunits produced with bilin lyases
| Plasmids used for Apoproteins | Plasmids used for bilin lyases | λmax (Q Vis/UV)a | Fluorescence emission, λmax | Фf | Fluorescence emission |
|---|---|---|---|---|---|
| nm | % | ||||
| pCpeA | pCpeYZb | 566/410 (5.4) | 574 | 0.72 | 100 |
| pCpeA(C82S) | pCpeYZb | NAc | NA | ND | NDd |
| pCpeA(C139S) | pCpeYZb | 566/410 (5.6) | 574 | ND | ND |
| pCpeA(C82S/C139S) | pCpeYZb | NA | NA | ND | ND |
| pCpeA | pCpeZb | NA | NA | ND | 0 |
| pCpeA | pCpeYb | 566/410 (15.6) | 574 | ND | 60 |
| pCpeA(C82S) | pCpeYb | NA | NA | ND | ND |
| pCpeA(C139S) | pCpeYb | 566/410 (14.8) | 574 | ND | ND |
| pCpeA(C82S/C139S) | pCpeYb | NA | NA | ND | ND |
| pCpeA | pCpeSb | 561/410 (0.315) | 574 | 0.89 | 0.8 |
| pCpeA(C82S) | pCpeSb | 550/398 (0.6) | 562 | ND | ND |
| pCpeA(C139S) | pCpeSb | 562/410 (0.6) | 574 | ND | ND |
| pCpeA(C82S/C139S) | pCpeSb | NA | NA | ND | ND |
| pCpeB | pCpeSb | 560/412 (5.2) | 571 | ND | ND |
| pCpeB(C80S) | pCpeSb | NA | NA | ND | ND |
| pCpeB(C165S) | pCpeSb | 560/412 (5.4) | 571 | ND | ND |
| pCpeB(C48S/C59S) | pCpeSb | 560/398 (5.3) | 571 | ND | ND |
| Native PE (αβ)6e | 563/374 (9.5) | 573 | ND | ND | |
a Q Vis/UV denotes the absorbance ratio of the visible and near-UV bands.
b Coexpressed with pPebS.
c Not applicable because there was no fluorescent product produced.
d ND, not determined.
e Holo-PE purified from F. diplosiphon.
Protein and Bilin Analysis
Polypeptides were resolved by PAGE (15%, w/v) in the presence of SDS, and visualized by staining with Coomassie Blue as described (8). To detect PEB linked to proteins, gels were soaked in 100 mm ZnSO4 for ∼5 min (50, 51) and the zinc-enhanced fluorescence, indicative of bilin attachment, was visualized using an FX imaging system (Bio-Rad) with excitation at 532 nm.
Calculating Fluorescence Quantum Yield
The fluorescence quantum yield relative to cresyl violet (in ethanol Φf = 0.59; Sigma) of CpeA-PEB or CpeB-PEB was calculated as described (52, 53) using a PerkinElmer LS55 fluorescence spectrophotometer with slit widths set at 10 nm (excitation and emission). The fluorescence emission spectrum was acquired from 570 to 800 nm. The fluorescence quantum yield of the sample was calculated in comparison with the standard using Equation 1,
where A is the absorbance value at the maximum, ϵ is the area of the fluorescence emission spectrum from 570 nm to 800 nm, and Φf is the fluorescence quantum yield.
Protein-Protein Interaction Assays
Pulldown assays between HT-CpeZ and CpeY were performed using whole cell extracts from a CpeY expression culture (pCpeY) and purified HT-CpeZ (from a pCpeZ culture) as described (33).
Immunoblotting Analysis
Antibodies against recombinant F. diplosiphon CpeA-PEB (produced with CpeY/CpeZ) and CpeB-PEB (produced with CpeS) were generated in rabbits (YenZym Antibodies, South San Francisco, CA). Immunoblotting analysis was performed as described (32) using antisera at a 1:5000 dilution.
Tryptic Digestion of Proteins
Purified CpeA-PEB or CpeB-PEB was dialyzed against 2 mm sodium phosphate buffer, pH 7.0, 1 mm 2-mercaptoethanol, concentrated by ultrafiltration through an Amicon YM10 (Millipore, Billerica, MA), and subjected to digestion with trypsin following the protocol described in Ref. 4. The reaction was quenched by adding 30% (v/v) glacial acetic acid, followed by passage through a C-18 Sep-Pak (Waters Corp., Milford, MA) cartridge as described (4). The eluted sample was vacuum dried and stored at −20 °C for HPLC analysis as described (8).
High Performance Liquid Chromatography
Tryptic peptides were separated on a C18 reverse-phase HPLC column (5 μm × 10 mm × 250 mm; Waters Corp., Milford, MA) using a Waters HPLC equipped with a 600E pump and a photodiode array detector. The peptide separation was carried out as described by Arciero et al. (4), using 0.1 m sodium phosphate, pH 2.1, as Solvent A and acetonitrile as Solvent B. The bilin peptides were eluted with an increasing concentration of acetonitrile (35 to 100%) and were monitored at 560 nm. The eluted samples were vacuum-dried and kept at −20 °C for mass spectrometric analysis.
Mass Spectrometry
MALDI MS and tandem MALDI MS/MS experiments were performed on an Applied Biosystems (Foster City, CA)/MDS Sciex (Concord, Ontario) 4800 MALDI TOF/TOF spectrometer. Mass spectral acquisitions were obtained in the reflectron mode using a Nd:YAG laser operated at 355 nm. The matrix used was α-cyanohydroxycinnamic acid (Sigma) at 15 mg ml−1 in 50% (v/v) acetonitrile (Sigma), 0.1% (v/v) trifluoroacetic acid (Sigma). An aliquot (2 μl) from each fraction was mixed with matrix (2 μl); the mixture was homogenized, and an aliquot (0.75 μl) was spotted on a MALDI 384-well plate and air dried prior to analysis.
RESULTS
Characterization of Bilin Lyase Activity of CpeY and CpeZ with CpeA
The cpeY and cpeZ genes occur downstream of the cpeBA genes, which encode the α- and β-subunits of PE, respectively. Based upon their sequence similarity, CpeY and CpeZ belong to the CpcE/CpcF family of bilin lyases. CpeY and CpcE (from Synechocystis strain PCC 6803) are 22% identical and 32% similar. CpeZ and CpcE are 25% identical and 38% similar, whereas CpeZ and CpcF (from Synechocystis strain PCC 6803) are 23% identical and 37% similar. Transposon mutants and complementation studies in F. diplosiphon suggested that these two proteins are involved in PE biogenesis, but their specific roles were not elucidated (44). An in vivo E. coli heterologous coexpression system was used to test whether either of these genes encodes a bilin lyase. Constructs used in this study are listed in supplemental Table S1.
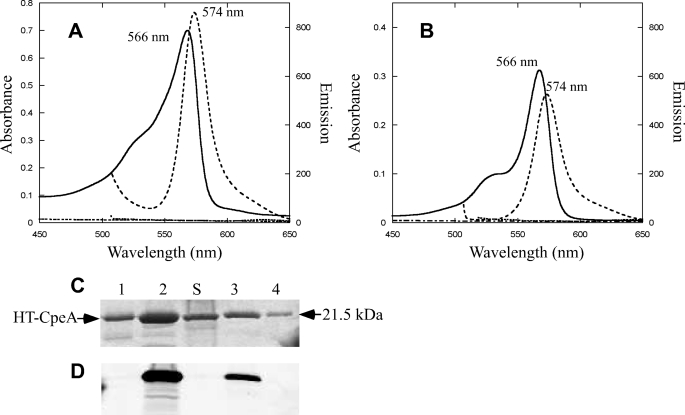
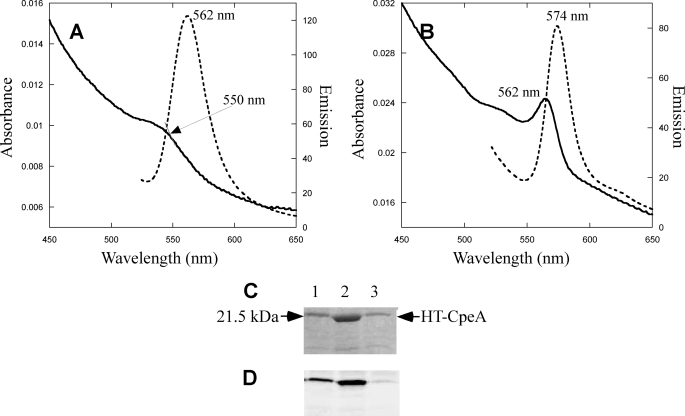
E. coli cells containing plasmids pCpeA and pPebS (i.e. no lyase present) had no significant color after induction with isopropyl 1-thio-β-d-galactopyranoside (data not shown), but cells containing these two plasmids in addition to pCpeYZ were bright pinkish-red (supplemental Fig. S1). CpeA-PEB purified from these cells had an absorbance maximum at 566 nm (Fig. 1A) and was intensely fluorescent with an emission maximum at 574 nm (Fig. 1A), whereas the purified protein obtained from the cells containing only pCpeA and pPebS did not have any significant absorbance or fluorescence emission (Fig. 1A). CpeZ and CpeY were also tested individually to determine whether either protein alone could attach PEB to CpeA. The fluorescence emission amplitude for CpeA-PEB purified from cells containing pCpeA, pPebS, and pCpeY showed that CpeY had significant activity by itself, but the amount of fluorescent product was lower than when both CpeY and CpeZ were present (Fig. 1B). The relative yields of the CpeA-PEB fluorescent product purified after coexpression with PebS along with either CpeY or CpeZ are given in Table 1. When CpeA was co-expressed with CpeY, the CpeA-PEB product was highly fluorescent (Fig. 1B, dashed lines); however, the CpeA product from coexpression with the other subunit, CpeZ, was not fluorescent (Fig. 1B, dashed dotted line, and Table 1). The three CpeA samples purified from E. coli cells were analyzed by SDS-PAGE (Fig. 1C). Bilin addition to CpeA was detected by zinc staining of the gel to enhance bilin fluorescence (Fig. 1D); subsequent staining of the same gel with Coomassie Blue revealed the protein content (Fig. 1C). The CpeA purified from cells expressing both CpeY and CpeZ was highly fluorescent after zinc staining (Fig. 1D, lane 2), but CpeA purified from cells containing no lyase subunit or with CpeZ alone was not fluorescent after zinc staining. Thus, little or no ligation of PEB occurred in the absence of the CpeY subunit. (Fig. 1D, lanes 1 and 4, respectively). However, CpeA purified from cells coexpressing CpeY produced a fluorescent product with a yield that was ∼60% of that achieved in the presence of both CpeY and CpeZ (Table 1); this observation suggested that CpeZ enhances the PEB ligation activity of CpeY. When CpeA was coexpressed with CpeY or CpeY/CpeZ and the genes for PEB biosynthesis, it accumulated in a soluble form (Fig. 1C, lanes 2 and 3). However, immunoblotting analyses (see supplemental Fig. S2) with antibodies to CpeA showed that CpeA formed inclusion bodies when expressed in the absence of lyase subunits in E. coli.
FIGURE 1.
Analyses of CpeA produced with CpeY and CpeZ in E. coli. A, absorbance (solid line) and fluorescence emission (dashed line) spectra of CpeA purified from cells containing pCpeA, pPebS with pCpeYZ, and absorbance (dashed dotted line) and fluorescence (dotted line) without pCpeYZ are shown. B, absorbance (solid line) and fluorescence emission (dashed line) spectra of CpeA purified from cells containing pCpeA, pPebS with pCpeY, and absorbance (dashed dotted line) and fluorescence (dotted line) with pCpeZ are shown. To acquire the fluorescence emission spectra for the CpeA produced in the presence of pCpeYZ and pCpeY (dashed lines in panels A and B) the samples were diluted 15- and 8-fold, respectively, to A560 nm of 0.05; however, no dilution was performed on CpeA produced in the absence of a lyase or in the presence of pCpeZ (dotted lines in panels A and B). C, SDS-PAGE analysis of recombinant CpeA. Lane 1, CpeA purified from cells containing pCpeA, pPebS with no lyase; lane 2, CpeA purified from cells containing pCpeA, pPebS, and pCpeYZ; lane 3, CpeA purified from cells containing pCpeA, pPebS, and pCpeY; lane 4, CpeA purified from cells containing pCpeA, pPebS, and pCpeZ. Molecular mass standards are loaded in lane S, and the mass is indicated to the right. D, the zinc-enhanced fluorescence of the gel pictured in panel C.
Because the presence of CpeZ enhanced the bilin ligation activity of CpeY, and because other bilin lyases such as CpcE and CpcF have been shown to form heterodimers, we tested whether CpeY and HT-CpeZ interact. The CpeY and HT-CpeZ proteins coproduced in E. coli were soluble, but CpeY did not copurify with HT-CpeZ (supplemental Fig. S3). No copurification of CpeY was observed when it was incubated together with both CpeA-PEB and HT-CpeZ (data not shown).
Analysis of the Cysteine Residues on CpeA Chromophorylated by the CpeY/CpeZ Lyase
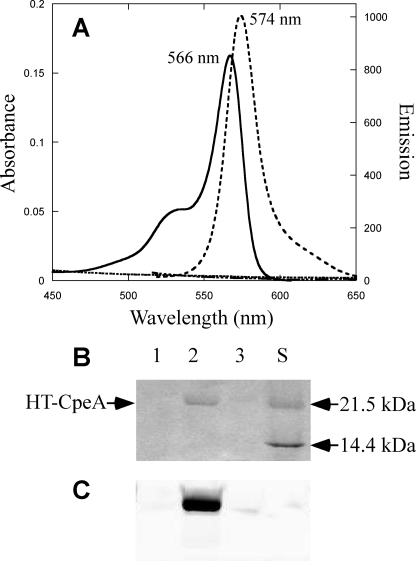
The holo-CpeA (α-PE subunit) isolated from F. diplosiphon carries PEB chromophores at Cys82 and Cys139 (41). To test the site specificity of the CpeY/CpeZ bilin lyase, site-specific variants of CpeA (C82S, C139S, and C82S/C139S) were produced in which cysteine residues were changed to serine. Each mutant gene was co-expressed with the CpeY/CpeZ lyase and the enzymes to synthesize PEB, and the CpeA produced was purified. The results of the absorbance and fluorescence emission measurements on these proteins are shown in Fig. 2 and Table 1. Only the C139S CpeA variant was a substrate for PEB ligation by CpeY/CpeZ, and the product had an absorption maximum at 566 nm and a fluorescence emission maximum at 574 nm (Fig. 2A). These values were identical to those for CpeA-PEB described above, and these results indicate that Cys82 is the residue that is chromophorylated with PEB by the CpeY/CpeZ lyase. The purified C82S and C82S/C139S variants of CpeA produced in the presence of the CpeY/CpeZ lyase and PEB synthesis enzymes had no significant fluorescence emission (Fig. 2A and Table 1). Similarly, no fluorescent products were observed when any of the variant proteins were produced in the absence of the lyase subunits (data not shown). The CpeA variants produced in these experiments were also analyzed by SDS-PAGE (Fig. 2, B and C). Bilin addition to each protein was examined by zinc-enhanced fluorescence of the gel (Fig. 2C). The purified C139S CpeA variant was fluorescent due to the presence of covalently attached PEB (Fig. 2C, lane 2). After staining the same gel shown in Fig. 2C with Coomassie Blue (Fig. 2B), it was apparent that CpeA only accumulated in the soluble fraction when PEB had been ligated to the protein (Fig. 2B, lane 2). As verified by immunoblot analyses using anti-CpeA antibodies, the nonchromophorylated CpeA variant proteins produced in these cells accumulated in an insoluble form in inclusion bodies, (data not shown, but results similar to those shown in supplemental Fig. S2A). From these experiments, we concluded that the CpeY/CpeZ bilin lyase specifically attaches PEB to Cys82 of CpeA. We will refer to this protein as CpeA-PEB to differentiate it from a true holo-CpeA carrying PEB at both Cys82 and Cys139.
FIGURE 2.
Analyses of the specific cysteine residue on CpeA required for PEB addition by CpeY/CpeZ. A, absorbance (solid line) and fluorescence emission (dashed line) spectra of CpeA(C139S) purified from cells containing CpeA(C139S), pPebS with pCpeYZ, and the absorbance (dashed dotted line) and fluorescence (dotted line) spectra from cells containing CpeA(C82S) and pPebS with pCpeYZ are shown. To acquire the fluorescence emission spectra for the HTCpeA(C139S) produced in the presence of pCpeYZ (dashed lines in panel A), the sample was diluted 15-fold to A560 nm of 0.05; however, no dilution was performed on CpeA(C82S) (dotted line in panel A). B and C, SDS-PAGE analysis of CpeA variants; the gel was stained with Coomassie Blue (B) or visualized by zinc-enhanced fluorescence (C). CpeA variants were produced in cells that also contained pPebS and pCpeYZ. Lane 1, CpeA(C82S); lane 2, CpeA(C139S); lane 3, CpeA(C82S/C139S). Molecular mass standards are loaded in lane S, and masses are indicated to the right. C, zinc-enhanced fluorescence image of the gel pictured in panel B.
Mass Spectrometry of Tryptic Peptides
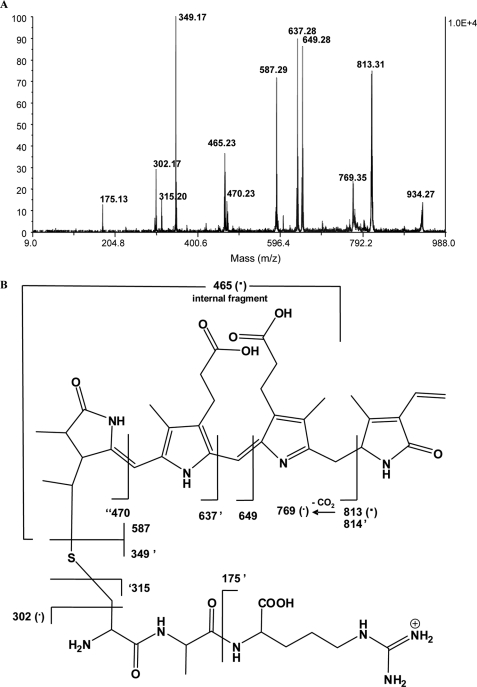
Because nonchromophorylated CpeA and its variants exhibited limited solubility when expressed in E. coli, it seemed plausible that a bound bilin at this central position within CpeA (at Cys82) increased the solubility and stability of CpeA in E. coli cells. Even though our site-specific variant experiments showed that CpeY/CpeZ ligates PEB at Cys82, this did not rule out the possibility that CpeY/CpeZ also ligates PEB at Cys139 but only after it attaches PEB at Cys82. Therefore, we analyzed the CpeA (nonvariant) produced in the presence of pCpeA, pCpeYZ, and pPebS to determine whether PEB was attached to more than one Cys on CpeA by subjecting the recombinant protein to tryptic digestion followed by mass spectrometry (54). The digested peptides were separated on a C18 reversed-phase HPLC (RP-HPLC) column. Two peaks were observed at 550 nm (specific for PEB); one major peak at 23.5 min and one minor peak at 23.0 min were collected (see supplemental Fig. S4). Each fraction obtained from HPLC separation was subjected to MALDI MS and tandem MS analysis to identify unambiguously the location of the covalently attached PEB on CpeA. Tandem mass spectrometry using MALDI MS of peptides resulting from the tryptic digestion of the covalent complex CpeA-PEB was performed to accomplish this. We sought to identify one or more peptides produced upon digestion that contained ligated PEB (Fig. 3). A peak at m/z 935 appeared in both fractions. Fig. 3A shows the MS/MS spectrum of the precursor at m/z 935. In particular, there are two main peaks of interest at m/z 587 and 349. The peak at m/z 587 is attributed to protonated PEB. The peak at m/z 349 matches a tripeptide, which has the sequence (K)CAR(D) and contains Cys82. The remaining major peaks were also assignable in a manner consistent with covalent attachment of PEB to Cys82. The scheme in Fig. 3B summarizes the structures of the assigned peaks.
FIGURE 3.
Mass spectrometric analyses of tryptic peptides of CpeA-PEB produced with CpeY and CpeZ. A, MALDI MS/MS spectrum of the m/z 935 precursor ion derived from peptides resulting from the tryptic digestion of the covalent complex CpeA-PEB. This m/z 935 precursor ion was deduced to be a peptide fragment with a covalently bound PEB chromophore. B, fragmentation pattern and corresponding mass assignments for data in panel A. A tick mark prior to number, e.g. ′470, indicates one hydrogen has been transferred to the departing neutral upon cleavage. A tick mark after number, e.g. 814′, indicates the transfer of one hydrogen to the formed ion. A (·) indicates a radical ion.
The above spectral interpretation suggests that, by applying sufficient collision energy in tandem mass spectrometry experiments, it was possible to break the thioether bond and separately detect the chromophore (m/z 587) and the peptide (m/z 349). The structure of the chromophore, which is highly conjugated, favored the formation of product ions that were stabilized by resonance. The large number of peaks enabled a thorough structural elucidation of the peptide-PEB covalent complex.
In a follow-up analysis, we specifically investigated whether PEB attachment occurred at Cys139. We were able to unambiguously identify a peptide at m/z = 503 that contained the unmodified cysteine at position 139 (sequence: (R)GCAPR(D)). If attachment of PEB occurred to Cys139, then it should have been possible to detect a chromopeptide with m/z = 1089 (503 + 586). An ion with this m/z ratio was indeed detected in one fraction. The product ion mass spectrum from this m/z 1089 precursor yielded a minor peak at m/z 587 and a peak at m/z 503 corresponding to the neutral loss of 586 (supplemental Fig. S5). This m/z 503 peak thus represents (R)GC139APR(D) that has lost the bilin moiety. Furthermore, a comparison of the peak heights observed in MALDI mass spectra that simultaneously contained m/z 1089 and 935 revealed that the former peak was quite minor compared with the latter. Although peak heights observed in MALDI mass spectra are not strictly reliable for quantification, these data strongly suggested CpeY/CpeZ predominantly attached PEB to Cys82.
Does CpeS Also Chromophorylate CpeA?
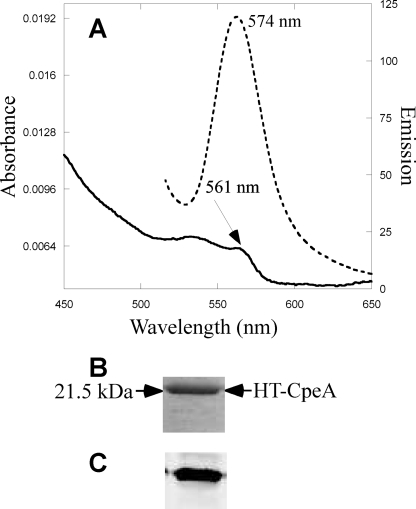
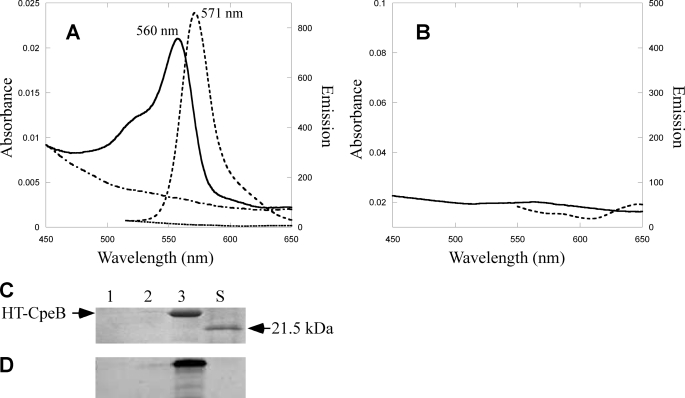
Using a recombinant E. coli in vivo assay system, Zhao et al. (30) reported that Nostoc sp. PCC 7120 CpcS (formerly denoted CpeS1 (29)) is a “near universal lyase” that adds bilins to Cys82 on most biliproteins, including the noncognate substrate, CpeA, from F. diplosiphon. (Note that Nostoc sp. PCC 7120 does not synthesize PE, so this bilin lyase should be designated as CpcS1 not as CpeS1.) Having demonstrated that the CpeY/CpeZ lyase attaches PEB primarily to Cys82 of CpeA, it was important to compare the activities of the CpeY/CpeZ and CpeS lyases on the substrate CpeA in E. coli. The cognate cpeS gene from F. diplosiphon was cloned to create plasmid pCpeS (see supplemental Table S1). Fig. 4A shows the absorbance and fluorescence emission spectra of CpeA purified from cells co-producing CpeA, CpeS, and enzymes for PEB synthesis. The yield of CpeA-PEB was much lower (supplemental Fig. S1 and Table 1) when the coproduced lyase was CpeS rather than CpeY/CpeZ. However, the absorbance and fluorescence properties of the resulting CpeA proteins were similar (Table 1). The CpeA-PEB produced in the presence of CpeS was analyzed by SDS-PAGE. The zinc-enhanced fluorescence in Fig. 4C shows that PEB is covalently attached to the CpeA protein, but the amount of fluorescent CpeA-PEB produced by CpeS in a 1-liter culture was estimated to be less than 1% of that obtained in the presence of CpeY/CpeZ (supplemental Fig. S1 and Table 1).
FIGURE 4.
Analyses of CpeA produced with CpeS in E. coli. A, absorbance (solid line) and fluorescence emission (dashed line) spectra of CpeA purified from cells containing pCpeA and pPebS with pCpeS. B, Coomassie-stained SDS-polyacrylamide gel containing CpeA purified from cells containing pCpeA, pPebS, and pCpeS. C, the zinc-enhanced fluorescence of the gel pictured in panel B.
The site-specific variants of CpeA were additionally used to investigate the activity of CpeS, and these results are shown in Fig. 5 and Table 1. Interestingly, small amounts of fluorescent product were obtained for both CpeA(C82S) and CpeA(C139S), but no significant fluorescence emission was observed when CpeA variants were coexpressed in the absence of the CpeS lyase (data not shown). These results suggested that CpeS could ligate PEB to both cysteines on CpeA but not very efficiently. The fluorescence emission maxima for the two variants were different; this suggested that PEB was bound in different protein environments within the two variants, thereby affecting its absorbance and emission properties (Table 1). In further support of this interpretation, two PEB-containing peptides were observed after tryptic digestion of CpeA-PEB chromophorylated in the presence CpeS (data not shown). Although CpeS can attach PEB to both Cys residues on CpeA, the low activity level suggested that CpeS is unlikely to be the cognate PEB lyase for either of these positions.
FIGURE 5.
Analyses of the cysteine residue on CpeA for PEB addition by CpeS. A, absorbance (solid line) and fluorescence emission spectra (dotted line) of CpeA(C82S) variants purified from cells containing pCpeS and pPebS. B, absorbance (solid line) and fluorescence emission spectra (dotted line) of the CpeA(C139S) variant purified from cells containing pCpeS and pPebS. C, Coomassie-stained, SDS-polyacrylamide gel analysis of CpeA variants purified from cells containing pPebS, pCpeS, and pCpeA(C82S) (lane 1), pCpeA(C139S) (lane 2), or pCpeA(C82S/C139S) (lane 3). D, the zinc-enhanced fluorescence of the gel in panel C.
Comparison of the PEB Ligation Activity of CpeY/CpeZ and CpeS Bilin Lyases with CpeB
Because CpeS is unlikely to be a cognate lyase for CpeA, we tested CpeS activity with CpeB as a substrate and investigated whether CpeY/CpeZ could also ligate PEB to CpeB. Holo-CpeB (β-PE) synthesized in F. diplosiphon has three PEB chromophores attached to four Cys residues: Cys80 and Cys165 carry single linked PEBs and Cys48 and Cys59 carry a double linked PEB (at C31 and C181 of the bilin) (41). Two different stereoisomers of PEB occur in CpeB; the R-isomer is present at C31 on Cys80 and the S-isomer is present at C31 on Cys165 and at C3′ and C18′ on Cys48/Cys59, respectively (55). Using the in vivo coexpression system, CpeB was coproduced with enzymes for PEB synthesis and either no lyase, CpeS, CpeY, CpeZ, or CpeY/CpeZ (supplemental Table S1). Fig. 6A shows the absorbance and fluorescence emission spectra of the resulting CpeB product after purification from cells producing no lyase or CpeS. The CpeB-PEB produced in the presence of CpeS had an absorbance maximum at 560 nm and a fluorescence emission maximum at 571 nm. No significant ligation of PEB to CpeB occurred in the absence of CpeS (Fig. 6A, dashed dotted line). When CpeB was purified from cells expressing CpeY and CpeZ in addition to the PEB synthesis enzymes, no significant absorbance or fluorescence emission of the product was observed as shown in Fig. 6B. The same result was observed when only CpeY or CpeZ was present as the bilin lyase subunit (data not shown). These data strongly suggested that CpeB is not a substrate for the CpeY/CpeZ lyase.
FIGURE 6.
Analyses of the CpeB produced in the presence of different lyases in E. coli. A, absorbance (solid line) and fluorescence emission (dashed line) spectra of CpeB purified from cells containing pCpeB, pPebS with pCpeS, and absorbance (dashed dotted line) and fluorescence (dotted line) without pCpeS (no lyase). B, absorbance (solid line) and fluorescence emission (dashed line) spectra of CpeB purified from cells containing pCpeB, pPebS, and pCpeYZ. To acquire the fluorescence emission spectra for the CpeB produced in the presence of pCpeS (dashed lines in panel A), the sample was diluted 15-fold to A560 nm of 0.05. No dilution was performed on CpeB produced in the absence of a lyase or with pCpeYZ (dotted lines in panels A and B). C, Coomassie-stained SDS-polyacrylamide gel containing CpeB purified from cells containing pCpeB, pPebS, and no lyase (lane 1) or from cells containing pCpeB, pPebS and pCpeYZ (lane 2), or from cells containing pCpeB, pPebS, and pCpeS (lane 3). Molecular mass standards are loaded in lane S, and mass is indicated to the right. D, zinc-enhanced fluorescence image of the gel pictured in panel C.
After separating the proteins on SDS-PAGE (Fig. 6, C and D), the bilin content of each protein was examined by zinc-enhanced fluorescence staining of the SDS-polyacrylamide gel (Fig. 6D), and proteins were detected after staining the same gel with Coomassie Blue (Fig. 6C). Very little apo-CpeB could be purified from the control reactions (no lyase present, lane 1 in Fig. 6, C and D) or under conditions for which no bilin was attached by CpeY/CpeZ (lane 2 in Fig. 6, C and D). CpeB only accumulated in soluble form in E. coli after PEB was covalently attached in the presence of CpeS (Fig. 6, C and D, lane 3). Immunoblotting analyses showed that CpeB was mainly in inclusion bodies unless CpeS was also coexpressed (supplemental Fig. S2B).
Analysis of Specific Cys Residue(s) of CpeB Chromophorylated by CpeS
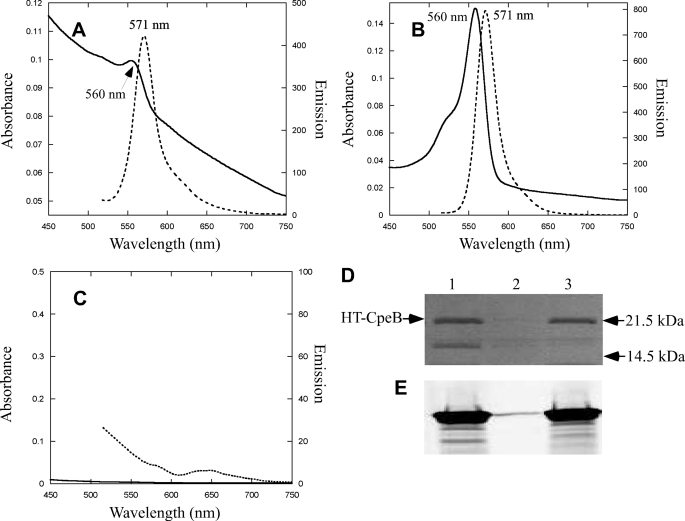
To determine the site specificity of the CpeS bilin lyase, site-specific variants of CpeB (C80S, C165S, and C48S/C59S) were produced as substrates for CpeS. After coproduction of each site-specific variant with CpeS and the enzymes for PEB synthesis, the CpeB product was purified, and the results of absorbance and fluorescence emission measurements are shown in Fig. 7. The C165S and C48S/C59S CpeB variants had absorbance maxima at 560 nm and fluorescence emission maxima at 571 nm (Fig. 7, A and B, respectively), but the CpeB(C80S) variant had no significant absorbance or fluorescence emission (Fig. 7C). Control experiments without the CpeS lyase with all CpeB variants were also performed, and in all cases no fluorescent product was observed (data not shown). The CpeB produced in these experiments was analyzed by SDS-PAGE. Bilin addition to each protein was examined by zinc-enhanced fluorescence (Fig. 7E). The CpeB variants (C165S and C48S/C59S) were fluorescent due to the presence of covalently attached PEB (Fig. 7E, lanes 1 and 3), whereas the CpeB(C80S) variant had no attached PEB (Fig. 7E, lane 2). After staining the same gel shown with Coomassie Blue (see Fig. 7D), it was apparent that CpeB only accumulated in a soluble form when PEB had been ligated to it (Fig. 7D, lanes 1 and 3). The CpeB (C80S) variant produced in the presence of CpeS and PEB synthesis enzymes accumulated as the apoprotein in inclusion bodies as judged by immunoblotting (data not shown, but results similar to those in supplemental Fig. S2B). From these experiments, we conclude that the CpeS bilin lyase attaches PEB to Cys80 of CpeB, but it does not play a significant role in PEB attachment to CpeA.
FIGURE 7.
Analyses of the specific cysteine residue on CpeB required for PEB addition by CpeS. A, absorbance (solid line) and fluorescence emission spectra (dotted line) of CpeB obtained by coexpressing pCpeB(C165S), pCpeS, and pPebS. The samples have been diluted 15-fold to an absorbance of 0.05. B, absorbance (solid line) and fluorescence emission spectra (dotted line) of CpeB obtained by coexpressing pCpeB(C48S/C59S), pCpeS, and pPebS. C, absorbance (solid line) and fluorescence emission spectra (dotted line) of CpeB obtained by coexpressing pCpeB(C80S), pCpeS, pPebS. D, Coomassie Blue-stained SDS-polyacrylamide gel loaded with CpeB purified from cells containing pCpeB(C48S/C59S), pPebS and pCpeS (lane 1), pCpeB(C80S), pCpeS and pPebS (lane 2), or pCpeB(C165S), pPebS, and pCpeS (lane 3). The electrophoretic mobility positions of the molecular mass standards are indicated to the right. E, zinc-enhanced fluorescence image of the gel pictured in panel D.
Because CpeB does not accumulate in a soluble form in E. coli when no PEB is attached at Cys80, we wondered whether CpeS was able to ligate a chromophore to any other site after it attached PEB at Cys80. Recombinant CpeB-PEB (nonvariant) produced with CpeS was subjected to trypsin digestion. The resulting tryptic peptides were separated by HPLC chromatography on a reversed-phase C18 column. In the chromatogram monitored at 550 nm to detect peptides with bound PEB, two peaks, eluting at 23 and 24 min, were observed and collected (see supplemental Fig. S6). MALDI MS and tandem MS was used to identify the peptides from these two peaks (56). Supplemental Fig. S7A shows the MS/MS spectrum of the precursor at m/z 1250. The peaks at m/z 587 and 664 were most informative. The m/z 587 peak corresponds to protonated PEB as previously discussed. The peak at m/z 664 matched a peptide containing a cysteine at position 80, (R)MAACLR(D). The scheme in supplemental Fig. S7B summarizes the structures of the assigned peaks. A review of the tandem mass spectra did not show an attachment to any other peptide. These results confirm that the CpeS bilin lyase specifically attaches PEB to Cys80 of CpeB and to no other Cys residues.
Analysis of the Ability of CpeY/CpeZ and CpeS to Attach Alternative Bilins to PE Subunits
Because both PCB and PEB are synthesized in F. diplosiphon when cells are grown in green light, we tested whether the CpeY/CpeZ lyase could attach PCB to CpeA to determine the specificity of this lyase for bilin substrates. CpeA, which was produced in cells containing pPcyA and pCpeA with and without pCpeYZ, was analyzed by absorbance and fluorescence spectroscopy (supplemental Fig. S8A). The CpeA produced in the absence of a lyase had no absorbance or fluorescence as expected. However, there was a small amount of absorbance and fluorescence of the CpeA product as a result of ligation of PCB to CpeA by CpeY/CpeZ. When these proteins were analyzed by SDS-PAGE (supplemental Fig. 8B) and zinc-enhanced bilin fluorescence (supplemental Fig. S8C), it was clear that although the amount of CpeA produced in these cells was very low, PCB was only attached to CpeA in the presence of the CpeY/CpeZ enzyme. Therefore, although this enzyme is capable of attaching PCB to CpeA, the amount of ligation was very low in comparison to its PEB ligation activity. When the ability of CpeS to ligate PCB to CpeB was tested in the same way, no absorbance or fluorescence was detected (data not shown).
DISCUSSION
This study compared the activities of three putative bilin lyase subunits on CpeA and CpeB substrates derived from F. diplosiphon, a filamentous cyanobacterium capable of Type III complementary chromatic acclimation (57–59). This is the first examination of the bilin lyase specificity for both α- and β-subunits of PE in which the enzymes and substrates were derived from the same organism. PEs are present in the distal ends of the peripheral rods of PBS, and they exhibit the most complex patterns of bilin-binding sites that occur in PBPs. PE conjugates are also widely used as fluorescence markers for cell sorting (60), so an understanding of their biosynthesis could have biotechnological implications. The fluorescence quantum yield for the CpeY/CpeZ-generated CpeA was 0.72 and for the CpeS-generated CpeB-PEB was 0.89 (Table 1); most PE subunits (which may have two or three bilins attached) have quantum yields ranging between 0.84 and 0.98 (61), which makes them excellent fluorescent markers.
Mass spectrometric data and attachment assays conducted with site-specific variants allowed confirmation of the site specificity of CpeY/CpeZ for PEB ligation to Cys82 of the α-subunit, and the site specificity of CpeS for linking PEB to Cys80 of the β-subunit. Zhao and co-workers (26, 30) reported that CpcS from Nostoc sp. strain PCC 7120 had broad PBP substrate recognition and might attach all chromophores at position Cys82 except for those of CpcA, PecA, and RpcA. However, because Nostoc sp. PCC 7120 does not synthesize PE or PEB, a more thorough examination of the substrate specificity of bilin lyases for PE subunits within one organism seemed important. Other studies have shown that some bilin lyases are promiscuous with respect to both the bilin and PBP substrates (22, 23). The studies reported here showed that CpeY/CpeZ, and not CpeS, is the principal bilin lyase responsible for attachment of PEB at Cys82 on CpeA. We did detect a small amount of PEB ligation at Cys139 on CpeA using mass spectrometry (supplemental Fig. S5), but the amount of attachment at Cys139 in the presence of CpeY/CpeZ was very small compared with that at Cys82; these data suggested that CpeY/CpeZ is not the lyase for this position. This conclusion is also supported by the fact that CpeY/CpeZ did not attach PEB to the CpeA(C82S) variant, whereas a small amount of PEB ligation by CpeS on the CpeA(C82S) variant was observed. Whereas some PEB ligation to Cys139 on CpeA by both CpeY/CpeZ and by CpeS was detected, the amounts were extremely low, and neither of these lyases seems likely to be responsible for PEB attachment at this Cys residue. Although CpeS could ligate PEB to Cys82 on CpeA, a comparison of the yields obtained with CpeS and CpeY/CpeZ proteins in E. coli strongly suggested that CpeY/CpeZ is more important in ligating PEB to CpeA to Cys82. Consistent with the data we show here, preliminary analyses of a F. diplosiphon cpeY deletion mutant, which avoids polarity effects that were likely present in the original cpeY transposon mutant isolated by Kahn et al. (44), have shown that it produces very little PE in green light and that the PE that is synthesized has a defect in CpeA.3
Wiethaus et al. (31) showed that CpeS from Prochlorococcus marinus MED4 can ligate PEB to Cys82 of CpeB. This organism is unusual in the sense that cells are devoid of phycobilisomes, and furthermore, they lack CpeA; the function of this degenerated form of CpeB is unknown (62, 63). The F. diplosiphon CpeS bilin lyase is a polypeptide of 222 amino acids and is 42% similar to CpeS from P. marinus MED4, and it appears that both CpeS lyases are capable of ligating PEB to Cys80 (equivalent) of CpeB. The studies reported here are the first to characterize a CpeS bilin lyase from a cyanobacterium containing PE in its phycobilisome rods. Unexpectedly, it was found that CpeS could also ligate PEB to Cys139 of CpeA in addition to Cys82 on CpeA and Cys80 on CpeB. However, based upon the very low levels of chromophorylation by CpeS at these positions on CpeA, it seems unlikely that CpeS is the lyase that catalyzes these reactions in cyanobacteria. Several other bilin lyase candidates are currently being tested for PEB ligation at Cys139 on CpeA.
At 429 amino acids, CpeY is much larger than typical members of the E/F lyase family, and it appears that it might have resulted from a fusion of ORFs encoded by a cpcE and cpcF-like gene. CpeY aligned well with the concatenated sequences of CpcE and CpcF of Synechocystis sp. strain PCC 6803 and with RpcG from Synechococcus WH8102 (supplemental Fig. S9). This could also explain why CpeY has significant activity in the absence of CpeZ. Individual CpcE and CpcF subunits usually exhibit low levels of ligation activity when assayed separately (5, 21). For example, compared with PecE/PecF together, PecE from Mastigocladus laminosus had 10% PCB ligation activity on PecA (64). RpcG is also a larger bilin lyase that appears to have resulted from a fusion of genes encoding RpcE and RpcF (25); RpcG is involved in PEB chromophore ligation and isomerization to phycourobilin on RpcA (26).
CpeZ is 205 amino acids in length and is most similar to CpcE-like, HEAT repeat proteins that are found in cyanobacteria and in other bacteria that do not contain PBPs. All CpcE/CpcF-type bilin lyases contain 5–6 HEAT repeat motifs; these motifs, which occur in many proteins in diverse eukaryotic organisms, are thought to facilitate protein-protein interactions (65, 66). CpeZ increased the PEB ligation activity of CpeY, but no evidence for a stable interaction between CpeY and CpeZ was detected using pulldown assays (supplemental Fig. S3). Likewise, no demonstrable interaction between CpeA and either CpeY or CpeZ was observed (data not shown). Nevertheless, CpeZ may play a chaperone-type role by assisting in the interactions of CpeA with other bilin lyases or with NblA/proteases (67–70).
Fairchild and Glazer (41) produced recombinant apo-CpeA and apo-CpeB from F. diplosiphon in E. coli and found that both proteins were insoluble. They were able to partially renature CpeA but not CpeB from inclusion bodies, and they demonstrated some autocatalytic ligation of PEB to CpeA in vitro, but the spectrum of the product did not match that of native, holo-α-PE. When R-PE subunits were expressed in E. coli, they were also found to be insoluble unless fused to the maltose-binding protein (43). In the experiments reported here, only a small amount of apo-CpeA was soluble, but PEB ligation increased its solubility. Coexpression of subunits of PC and AP increases their solubility in the apo-form (19), but coexpression of cpeB with cpeA without bilin attachment did not increase solubility (data not shown). However, chromophorylation at the position equivalent to Cys82 is obviously an important factor determining the solubility and accumulation of folded PBPs in E. coli. Although CpeB was slightly soluble at 18 °C (see lane 3 in supplemental Fig. S2B), we were unable to purify it in its apo-form. It became much more soluble when coexpressed with the CpeS lyase. Bilin deletion mutants in PC (where Cys were mutated to Ala) in cyanobacteria showed lower stability in vivo (71). The absence of bilins at various positions reduces the strength of α/β interactions in the heterodimers, and the authors suggested that these mutants were diverted to degradation pathways in cyanobacteria (71, 72).
Because the CpcE/CpcF bilin lyases have been shown to have bilin removal activity (21), it seemed logical to test whether CpeY/CpeZ possessed such an activity. This assay is normally performed as a transfer assay using holo-PBP subunits and apo-PBP subunits that are either His6-tagged or bound to beads. Unfortunately, because apo-CpeA is not very soluble in E. coli, it was not possible to perform the PEB transfer assay. Both CpcE/CpcF and PecE/PecF subunits copurify with their respective holo-PBP substrates (21, 49, 73), but CpeY (nontagged) did not. This suggests that CpeY may not have a transferase activity.
Our mass spectrometry results build upon previous studies investigating the location of the attachment site of phycobilins (74, 75). The PEB-peptide resulting from tryptic digestion of α-PE (detected at m/z 935) and from tryptic digestion of β-PE (detected at m/z 1250) were reported (30, 76). Fragments resulting from tandem mass spectrometric experiments such as protonated free PEB (m/z 587) (75, 77), and the tri-pyrrole fragment that results from the loss of pyrrole ring D (m/z 464) were also reported (77). Using an off-line MALDI TOF/TOF method, very recently Wiethaus and co-workers (31) were able to locate the site of ligation of PEB on the β-PE subunit from P. marinus based upon a major peak corresponding to PEB loss from a sequenced tryptic peptide containing Cys82.
The molecular structure of the tetrapyrrole, PEB, has an extended “π-conjugated” system, and may exist as three different tautomers. These isomeric structures, which exist in a dynamic equilibrium, differ only in that the pyrrole rings carrying the imino and amino nitrogens have changed. Fig. 3 and supplemental Figs. 5 and S7 show structures of PEB attached to tryptic peptides derived from CpeA and CpeB. The tautomeric forms shown are based upon previously published work (74, 75, 77) that relied upon NMR to assign the predominant tautomer.
Why are different lyases needed for ligation of bilins to Cys82 of the α- and β-subunits of PBPs that occur in the rods of PBS (e.g. CpcA/CpcB, PecA/PecB, and CpeA/CpeB)? Because lyases of the CpcS/CpcU family are capable of chromophorylating both β- and α-subunits such as ApcA, ApcB, CpcB, and minor AP subunits (19, 30, 32–34, 47), it seems plausible that CpcA was originally chromophorylated by a lyase of this type. Therefore, it seems likely that the CpcE/CpcF lyase family evolved later, perhaps to perform a specialized function. For example, some members of the E/F family of bilin lyases can remove and transfer bilins (5, 21), and the α-subunits of PBP have been shown to interact with NblA near the binding site for the chromophore at Cys82 of the PC α-subunit (70). Thus, it has been speculated that these lyases might have a unique role in biliprotein degradation/turnover during nutrient starvation conditions (30). Another possibility is that the evolution of organisms producing PEs and PEBs required greater specificity in the existing lyases, especially those involved in the core energy transfer reactions (i.e. ApcA, ApcB, and CpcB), to prevent the misattachment of PEB at sites that would greatly reduce energy transfer efficiency. The postulated greater specificity may have come at the expense of slow chromophore ligation on some apoproteins, e.g. CpcA. Together with the necessity to add PEB chromophores to the apo-PE subunits, cyanobacteria evolved new lyases to accommodate the greater complexity of chromophore-substrate possibilities. Whatever the true origin of this lyase specialization, once it occurred, nature took advantage of this to diversify PCs by gene duplication and divergence. In extant cyanobacteria, all four known chromophores (PCB, PEB, phycourobilin, and phycoviolobilin) can occur at Cys82 in the α-subunits of peripheral rod proteins. The production of two of these chromophores, phycoviolobilin and phycourobilin, requires a bilin lyase/isomerase, like PecE/PecF or RpcG (6, 26, 27), a capability that evolved by duplication and divergence from the CpcE/CpcF lyases. Finally, chromophores at the Cys82 position of α-subunits transfer energy to the terminal acceptor bilin present at Cys82 on β-subunits within trimers (αβ)3, so there appears to be more flexibility for differences in chromophore type on the α-subunits. Although this is not a reason why a specialized lyase evolved, it may have facilitated such a development.
The fluorescence quantum yields of CpeA-PEB and CpeB-PEB were 0.72 and 0.89, respectively; these values are much higher than the quantum yield of 0.60 obtained from the best mutant of green fluorescent protein (GFP) (78). They are also larger than the values reported for CpeA-PEB (0.51) and CpeB-PEB (0.63) produced in E. coli with CpcS1 (30). Therefore, these recombinant proteins may be useful for biotechnological applications as fluorescent probes or for therapeutic purposes, because R-PE has been used as a photosensitizer in cancer cells (79). Finally, with this report and that of Wiethaus et al. (31), details of how these complex yet important PEs are biosynthesized are finally emerging. However, many unresolved questions remain. What is the exact role of CpeZ in PE biosynthesis? Which bilin lyases ligate PEB to the other Cys residues, including α-Cys139, β-Cys48,59, and β-Cys165? We are approaching these questions by using a combination of a reverse-genetics approach and biochemical analyses of recombinant proteins, and answers should soon be forthcoming.
Supplementary Material
Acknowledgments
We thank the W. M. Keck Foundation for equipment utilized for this study, located in the Keck Conservation and Molecular Genetics lab at the University of New Orleans. We thank Yasmin Vasquez for technical assistance and Corry Paul for input in editing the manuscript. We thank Dr. Nicole Frankenberg-Dinkel for providing plasmid pPebS.
This work was supported by National Science Foundation Grants MCB-0843664 (to W. M. S.), DBI-0619272 and CHE-1058764 (to R. B. C.), and MCB-0519743 and MCB-1021725 (to D. A. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S9.
A. Biswas, A. Gutu, D. Kehoe, and W. M. Schluchter, unpublished data.
- PBS
- phycobilisome(s)
- PBP
- phycobiliprotein(s)
- PC
- phycocyanin
- PCB
- phycocyanobilin
- PE
- phycoerythrin
- PEB
- phycoerythrobilin
- AP
- allophycocyanin.
REFERENCES
- 1. Sidler W. A. (1994) in The Molecular Biology of Cyanobacteria (Bryant D. A. ed) pp. 139–216, Kluwer Academic, Dordrecht, The Netherlands [Google Scholar]
- 2. Bryant D. A. (1991) in Cell Culture and Somatic Cell Genetics of Plants (Bogarad L., Vasil I. L. eds) pp. 257–300, Academic Press, San Diego, CA [Google Scholar]
- 3. Glazer A. N. (1989) J. Biol. Chem. 264, 1–4 [PubMed] [Google Scholar]
- 4. Arciero D. M., Bryant D. A., Glazer A. N. (1988) J. Biol. Chem. 263, 18343–18349 [PubMed] [Google Scholar]
- 5. Fairchild C. D., Glazer A. N. (1994) J. Biol. Chem. 269, 8686–8694 [PubMed] [Google Scholar]
- 6. Zhao K. H., Deng M. G., Zheng M., Zhou M., Parbel A., Storf M., Meyer M., Strohmann B., Scheer H. (2000) FEBS Lett. 469, 9–13 [DOI] [PubMed] [Google Scholar]
- 7. Scheer H., Zhao K. H. (2008) Mol. Microbiol. 68, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shen G., Saunée N. A., Williams S. R., Gallo E. F., Schluchter W. M., Bryant D. A. (2006) J. Biol. Chem. 281, 17768–17778 [DOI] [PubMed] [Google Scholar]
- 9. Gómez-Lojero C., Pérez-Gómez B., Shen G., Schluchter W. M., Bryant D. A. (2003) Biochemistry 42, 13800–13811 [DOI] [PubMed] [Google Scholar]
- 10. Gindt Y. M., Zhou J., Bryant D. A., Sauer K. (1994) Biochim. Biophys. Acta 1186, 153–162 [DOI] [PubMed] [Google Scholar]
- 11. Gindt Y. M., Zhou J., Bryant D. A., Sauer K. (1992) J. Photochem. Photobiol. B 15, 75–89 [DOI] [PubMed] [Google Scholar]
- 12. Maxson P., Sauer K., Zhou J. H., Bryant D. A., Glazer A. N. (1989) Biochim. Biophys. Acta 977, 40–51 [DOI] [PubMed] [Google Scholar]
- 13. de Lorimier R., Bryant D. A., Stevens S. E., Jr. (1990) Biochim. Biophys. Acta 1019, 29–41 [DOI] [PubMed] [Google Scholar]
- 14. de Lorimier R., Bryant D. A., Porter R. D., Liu W. Y., Jay E., Stevens S. E., Jr. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 7946–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bryant D. A., de Lorimier R., Guglielmi G., Stevens S. E., Jr. (1990) Arch. Microbiol. 153, 550–560 [DOI] [PubMed] [Google Scholar]
- 16. Bryant D. A. (1988) in Light-Energy Transduction in Photosynthesis: Higher Plant and Bacterial Models (Stevens S. E. J., Bryant D. A. eds) pp. 62–90, American Society of Plant Physiologists, Rockville, MD [Google Scholar]
- 17. Bryant D. A., Stirewalt V. L., Glauser M., Frank G., Sidler W., Zuber H. (1991) Gene 107, 91–99 [DOI] [PubMed] [Google Scholar]
- 18. Glazer A. N. (1988) Methods Enzymol. 167, 291–303 [DOI] [PubMed] [Google Scholar]
- 19. Biswas A., Vasquez Y. M., Dragomani T. M., Kronfel M. L., Williams S. R., Alvey R. M., Bryant D. A., Schluchter W. M. (2010) Appl. Environ. Microbiol. 76, 2729–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou J., Gasparich G. E., Stirewalt V. L., de Lorimier R., Bryant D. A. (1992) J. Biol. Chem. 267, 16138–16145 [PubMed] [Google Scholar]
- 21. Fairchild C. D., Zhao J., Zhou J., Colson S. E., Bryant D. A., Glazer A. N. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 7017–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alvey R. M., Biswas A., Schluchter W. M., Bryant D. A. (2011) J. Bacteriol. 193, 1663–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alvey R. M., Biswas A., Schluchter W. M., Bryant D. A. (2011) Biochemistry 50, 4890–4902 [DOI] [PubMed] [Google Scholar]
- 24. Wilbanks S. M., Glazer A. N. (1993) J. Biol. Chem. 268, 1226–1235 [PubMed] [Google Scholar]
- 25. Six C., Thomas J. C., Garczarek L., Ostrowski M., Dufresne A., Blot N., Scanlan D. J., Partensky F. (2007) Genome Biol. 8, R259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blot N., Wu X. J., Thomas J. C., Zhang J., Garczarek L., Böhm S., Tu J. M., Zhou M., Plöscher M., Eichacker L., Partensky F., Scheer H., Zhao K. H. (2009) J. Biol. Chem. 284, 9290–9298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Storf M., Parbel A., Meyer M., Strohmann B., Scheer H., Deng M. G., Zheng M., Zhou M., Zhao K. H. (2001) Biochemistry 40, 12444–12456 [DOI] [PubMed] [Google Scholar]
- 28. Jung L. J., Chan C. F., Glazer A. N. (1995) J. Biol. Chem. 270, 12877–12884 [DOI] [PubMed] [Google Scholar]
- 29. Zhao K. H., Su P., Li J., Tu J. M., Zhou M., Bubenzer C., Scheer H. (2006) J. Biol. Chem. 281, 8573–8581 [DOI] [PubMed] [Google Scholar]
- 30. Zhao K. H., Su P., Tu J. M., Wang X., Liu H., Plöscher M., Eichacker L., Yang B., Zhou M., Scheer H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14300–14305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wiethaus J., Busch A. W., Kock K., Leichert L. I., Herrmann C., Frankenberg-Dinkel N. (2010) J. Biol. Chem. 285, 37561–37569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shen G., Schluchter W. M., Bryant D. A. (2008) J. Biol. Chem. 283, 7503–7512 [DOI] [PubMed] [Google Scholar]
- 33. Saunée N. A., Williams S. R., Bryant D. A., Schluchter W. M. (2008) J. Biol. Chem. 283, 7513–7522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao K. H., Zhang J., Tu J. M., Böhm S., Plöscher M., Eichacker L., Bubenzer C., Scheer H., Wang X., Zhou M. (2007) J. Biol. Chem. 282, 34093–34103 [DOI] [PubMed] [Google Scholar]
- 35. Zhao K. H., Su P., Böhm S., Song B., Zhou M., Bubenzer C., Scheer H. (2005) Biochim. Biophys. Acta 1706, 81–87 [DOI] [PubMed] [Google Scholar]
- 36. Hu I. C., Lee T. R., Lin H. F., Chiueh C. C., Lyu P. C. (2006) Biochemistry 45, 7092–7099 [DOI] [PubMed] [Google Scholar]
- 37. Wu S. H., Lagarias J. C. (2000) Biochemistry 39, 13487–13495 [DOI] [PubMed] [Google Scholar]
- 38. Ulijasz A. T., Cornilescu G., von Stetten D., Cornilescu C., Velazquez Escobar F., Zhang J., Stankey R. J., Rivera M., Hildebrandt P., Vierstra R. D. (2009) J. Biol. Chem. 284, 29757–29772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hirose Y., Narikawa R., Katayama M., Ikeuchi M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 8854–8859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sidler W., Kumpf B., Rüdiger W., Zuber H. (1986) Biol. Chem. Hoppe-Seyler 367, 627–642 [DOI] [PubMed] [Google Scholar]
- 41. Fairchild C. D., Glazer A. N. (1994) J. Biol. Chem. 269, 28988–28996 [PubMed] [Google Scholar]
- 42. Ong L. J., Glazer A. N. (1991) J. Biol. Chem. 266, 9515–9527 [PubMed] [Google Scholar]
- 43. Isailovic D., Sultana I., Phillips G. J., Yeung E. S. (2006) Anal. Biochem. 358, 38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kahn K., Mazel D., Houmard J., Tandeau de Marsac N., Schaefer M. R. (1997) J. Bacteriol. 179, 998–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mazel D., Guglielmi G., Houmard J., Sidler W., Bryant D. A., Tandeau de Marsac N. (1986) Nucleic Acids Res. 14, 8279–8290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miller C. A., Leonard H. S., Pinsky I. G., Turner B. M., Williams S. R., Harrison L., Jr., Fletcher A. F., Shen G., Bryant D. A., Schluchter W. M. (2008) J. Biol. Chem. 283, 19293–19300 [DOI] [PubMed] [Google Scholar]
- 47. Shen G. Z., Saunee N. A., Gallo E., Begovic Z., Schluchter W. M., Bryant D. A. (2004) in Photosynthesis 2004 Light-Harvesting Systems Workshop (Niederman R. A., Blankenship R. E., Frank H., Robert B., van Grondelle R. eds) pp. 14–15, Saint Adele, Quebec, Canada [Google Scholar]
- 48. Dammeyer T., Bagby S. C., Sullivan M. B., Chisholm S. W., Frankenberg-Dinkel N. (2008) Curr. Biol. 18, 442–448 [DOI] [PubMed] [Google Scholar]
- 49. Tooley A. J., Cai Y. A., Glazer A. N. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10560–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Berkelman T. R., Lagarias J. C. (1986) Anal. Biochem. 156, 194–201 [DOI] [PubMed] [Google Scholar]
- 51. Raps S. (1990) Plant Physiol. 92, 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Parker C. A., Rees W. T. (1960) Analyst 85, 587–600 [Google Scholar]
- 53. Lakowicz J. R. (1983) Principles of Fluorescence Spectroscopy, Plenum Press, New York [Google Scholar]
- 54. Boutaghou M. N., Biswas A., Cole R. B., Schluchter W. M. (2010) in Proceedings of the 58th ASMS Conference on Mass Spectrometry and Allied Topics, Salt Lake City, UT, May 26, 2010, American Society for Mass Spectrometry, Santa Fe, NM [Google Scholar]
- 55. Ficner R., Lobeck K., Schmidt G., Huber R. (1992) J. Mol. Biol. 228, 935–950 [DOI] [PubMed] [Google Scholar]
- 56. Boutaghou M. N., Biswas A., Schluchter W. M., Cole R. B. (2011) in Proceedings of the 59th ASMS Conference on Mass Spectrometry and Allied Topics, Denver, CO, June 9, 2011, American Society for Mass Spectrometry, Santa Fe, NM [Google Scholar]
- 57. Kehoe D. M., Gutu A. (2006) Annu. Rev. American Society for Mass Spectrometry santa Fe, NM Plant Biol. 57, 127–150 [DOI] [PubMed] [Google Scholar]
- 58. Tandeau de Marsac N., Houmard J. (1988) Methods Enzymol. 167, 318–328 [DOI] [PubMed] [Google Scholar]
- 59. Federspiel N. A., Grossman A. R. (1990) J. Bacteriol. 172, 4072–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Glazer A. N. (1994) J. Appl. Phycol. 6, 105–112 [Google Scholar]
- 61. Nguyen D. C., Keller R. A., Jett J. H., Martin J. C. (1987) Anal. Chem. 59, 2158–2161 [DOI] [PubMed] [Google Scholar]
- 62. Hess W. R., Partensky F., van der Staay G. W., Garcia-Fernandez J. M., Börner T., Vaulot D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11126–11130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Steglich C., Frankenberg-Dinkel N., Penno S., Hess W. R. (2005) Environ. Microbiol. 7, 1611–1618 [DOI] [PubMed] [Google Scholar]
- 64. Böhm S., Endres S., Scheer H., Zhao K. H. (2007) J. Biol. Chem. 282, 25357–25366 [DOI] [PubMed] [Google Scholar]
- 65. Takano H., Gusella J. (2002) BMC Neurosci. 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Andrade M. A., Petosa C., O'Donoghue S. I., Müller C. W., Bork P. (2001) J. Mol. Biol. 309, 1–18 [DOI] [PubMed] [Google Scholar]
- 67. Collier J. L., Grossman A. R. (1994) EMBO J. 13, 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Baier K., Lehmann H., Stephan D. P., Lockau W. (2004) Microbiology 150, 2739–2749 [DOI] [PubMed] [Google Scholar]
- 69. Dines M., Sendersky E., David L., Schwarz R., Adir N. (2008) J. Biol. Chem. 283, 30330–30340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bienert R., Baier K., Volkmer R., Lockau W., Heinemann U. (2006) J. Biol. Chem. 281, 5216–5223 [DOI] [PubMed] [Google Scholar]
- 71. Toole C. M., Plank T. L., Grossman A. R., Anderson L. K. (1998) Mol. Microbiol. 30, 475–486 [DOI] [PubMed] [Google Scholar]
- 72. Anderson L. K., Toole C. M. (1998) Mol. Microbiol. 30, 467–474 [DOI] [PubMed] [Google Scholar]
- 73. Tooley A. J., Glazer A. N. (2002) J. Bacteriol. 184, 4666–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Klotz A. V., Glazer A. N., Bishop J. E., Nagy J. O., Rapoport H. (1986) J. Biol. Chem. 261, 6797–6805 [PubMed] [Google Scholar]
- 75. Bishop J. E., Lagarias J. C., Nagy J. O., Schoenleber R. W., Rapoport H., Klotz A. V., Glazer A. N. (1986) J. Biol. Chem. 261, 6790–6796 [PubMed] [Google Scholar]
- 76. Isailovic D., Li H. W., Yeung E. S. (2004) J. Chromatogr. A 1051, 119–130 [PubMed] [Google Scholar]
- 77. Bishop J. E., Rapoport H., Klotz A. V., Chan C. F., Glazer A. N., Füglistaller P., Zuber H. (1987) J. Am. Chem. Soc. 109, 875–881 [Google Scholar]
- 78. Tsien R. Y. (1998) Annu. Rev. Biochem. 67, 509–544 [DOI] [PubMed] [Google Scholar]
- 79. Huang B., Wang G. C., Zeng C. K., Li Z. G. (2002) Cancer Biother. Radiopharm. 17, 35–42 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.