Abstract
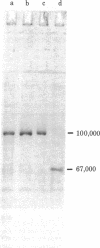
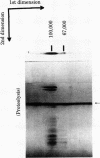
Two type I DNA topoisomerases have been purified to homogeneity from the nuclei of HeLa cells. One topoisomerase has a peptide molecular weight of 100,000 and the other, a molecular weight of 67,000. Several lines of evidence indicate that these two topoisomerases are closely related, (a) Both exhibit similar enzymatic activities on DNA. (b) The chromatographic properties of the two topoisomerases during purification are similar. (c) Mild proteolysis of the purified molecular weight 100,000 topoisomerase in vitro generates a group of protein bands of molecular weight approximately 67,000, and these bands retain topoisomerase activity. (d) The peptides formed by partial proteolysis of the 67,000 topoisomerase in the presence of NaDodSO4 form a subset of those produced from the 100,000 enzyme. The 100,000 topoisomerase is the major type I enzyme in the cell. The 67,000 topoisomerase, which may be identical to the previously identified "nicking-closing" enzyme [Champoux, J. J. & Dulbecco, R. (1972) Proc. Natl. Acad. Sci. USA 69, 143-146], is probably formed by proteolysis of the 100,000 enzyme.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldi M. I., Benedetti P., Mattoccia E., Tocchini-Valentini G. P. In vitro catenation and decatenation of DNA and a novel eucaryotic ATP-dependent topoisomerase. Cell. 1980 Jun;20(2):461–467. doi: 10.1016/0092-8674(80)90632-7. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brown P. O., Cozzarelli N. R. A sign inversion mechanism for enzymatic supercoiling of DNA. Science. 1979 Nov 30;206(4422):1081–1083. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- Brown P. O., Cozzarelli N. R. Catenation and knotting of duplex DNA by type 1 topoisomerases: a mechanistic parallel with type 2 topoisomerases. Proc Natl Acad Sci U S A. 1981 Feb;78(2):843–847. doi: 10.1073/pnas.78.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA--a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc Natl Acad Sci U S A. 1972 Jan;69(1):143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J., McConaughy B. L. Purification and characterization of the DNA untwisting enzyme from rat liver. Biochemistry. 1976 Oct 19;15(21):4638–4642. doi: 10.1021/bi00666a014. [DOI] [PubMed] [Google Scholar]
- Champoux J. J. Proteins that affect DNA conformation. Annu Rev Biochem. 1978;47:449–479. doi: 10.1146/annurev.bi.47.070178.002313. [DOI] [PubMed] [Google Scholar]
- Champoux J. J. Strand breakage by the DNA untwisting enzyme results in covalent attachment of the enzyme to DNA. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3800–3804. doi: 10.1073/pnas.74.9.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Cozzarelli N. R. DNA topoisomerases. Cell. 1980 Nov;22(2 Pt 2):327–328. doi: 10.1016/0092-8674(80)90341-4. [DOI] [PubMed] [Google Scholar]
- Depew R. E., Liu L. F., Wang J. C. Interaction between DNA and Escherichia coli protein omega. Formation of a complex between single-stranded DNA and omega protein. J Biol Chem. 1978 Jan 25;253(2):511–518. [PubMed] [Google Scholar]
- Fuller F. B. Decomposition of the linking number of a closed ribbon: A problem from molecular biology. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3557–3561. doi: 10.1073/pnas.75.8.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T., Brutlag D. ATP-dependent DNA topoisonmerase from D. melanogaster reversibly catenates duplex DNA rings. Cell. 1980 Aug;21(1):115–125. doi: 10.1016/0092-8674(80)90119-1. [DOI] [PubMed] [Google Scholar]
- Keller W. Characterization of purified DNA-relaxing enzyme from human tissue culture cells. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2550–2554. doi: 10.1073/pnas.72.7.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Nash H. A. Nicking-closing activity associated with bacteriophage lambda int gene product. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3760–3764. doi: 10.1073/pnas.76.8.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Wang J. C. Escherichia coli DNA topoisomerase I catalyzed linking of single-stranded rings of complementary base sequences. Nucleic Acids Res. 1978 Oct;5(10):3811–3820. doi: 10.1093/nar/5.10.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer K. N., Cozzarelli N. R. Formation and resolution of DNA catenanes by DNA gyrase. Cell. 1980 May;20(1):245–254. doi: 10.1016/0092-8674(80)90252-4. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Depew R. E., Wang J. C. Knotted single-stranded DNA rings: a novel topological isomer of circular single-stranded DNA formed by treatment with Escherichia coli omega protein. J Mol Biol. 1976 Sep 15;106(2):439–452. doi: 10.1016/0022-2836(76)90095-4. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Liu C. C., Alberts B. M. T4 DNA topoisomerase: a new ATP-dependent enzyme essential for initiation of T4 bacteriophage DNA replication. Nature. 1979 Oct 11;281(5731):456–461. doi: 10.1038/281456a0. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Liu C. C., Alberts B. M. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980 Mar;19(3):697–707. doi: 10.1016/s0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Interaction between DNA and Escherichia coli DNA topoisomerase I. Formation of complexes between the protein and superhelical and nonsuperhelical duplex DNAs. J Biol Chem. 1979 Nov 10;254(21):11082–11088. [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- Marini J. C., Miller K. G., Englund P. T. Decatenation of kinetoplast DNA by topoisomerases. J Biol Chem. 1980 Jun 10;255(11):4976–4979. [PubMed] [Google Scholar]
- Mizuuchi K., Fisher L. M., O'Dea M. H., Gellert M. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel L. M., Monty K. J. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966 Feb 7;112(2):346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Sternglanz R., DiNardo S., Voelkel K. A., Nishimura Y., Hirota Y., Becherer K., Zumstein L., Wang J. C. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc Natl Acad Sci U S A. 1981 May;78(5):2747–2751. doi: 10.1073/pnas.78.5.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetler G. L., King G. J., Huang W. M. T4 DNA-delay proteins, required for specific DNA replication, form a complex that has ATP-dependent DNA topoisomerase activity. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3737–3741. doi: 10.1073/pnas.76.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D. Purification of a DNA nicking-closing enzyme from mouse L cells. Nucleic Acids Res. 1978 Aug;5(8):2861–2875. doi: 10.1093/nar/5.8.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse Y., Wang J. C. E. coli and M. luteus DNA topoisomerase I can catalyze catenation of decatenation of double-stranded DNA rings. Cell. 1980 Nov;22(1 Pt 1):269–276. doi: 10.1016/0092-8674(80)90174-9. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971 Feb 14;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]