Background: The human Naa50p N-terminal acetyltransferase acetylates proteins on the α-amino group of methionine residues to regulate genome integrity and has elevated activity in cancer.
Results: We report the structure of the Naa50p·CoA·peptide complex and related biochemistry.
Conclusion: We reveal the molecular basis for substrate-specific acetylation by Naa50p.
Significance: We provide a molecular scaffold for the design of Naa50p-specific inhibitors with possible therapeutic applications.
Keywords: Enzyme Catalysis, Enzyme Structure, Post-translational Modification, Protein Complexes, Translation Control, Acetyltransferase
Abstract
The co-translational modification of N-terminal acetylation is ubiquitous among eukaryotes and has been reported to have a wide range of biological effects. The human N-terminal acetyltransferase (NAT) Naa50p (NAT5/SAN) acetylates the α-amino group of proteins containing an N-terminal methionine residue and is essential for proper sister chromatid cohesion and chromosome condensation. The elevated activity of NATs has also been correlated with cancer, making these enzymes attractive therapeutic targets. We report the x-ray crystal structure of Naa50p bound to a native substrate peptide fragment and CoA. We found that the peptide backbone of the substrate is anchored to the protein through a series of backbone hydrogen bonds with the first methionine residue specified through multiple van der Waals contacts, together creating an α-amino methionine-specific pocket. We also employed structure-based mutagenesis; the results support the importance of the α-amino methionine-specific pocket of Naa50p and are consistent with the proposal that conserved histidine and tyrosine residues play important catalytic roles. Superposition of the ternary Naa50p complex with the peptide-bound Gcn5 histone acetyltransferase revealed that the two enzymes share a Gcn5-related N-acetyltransferase fold but differ in their respective substrate-binding grooves such that Naa50p can accommodate only an α-amino substrate and not a side chain lysine substrate that is acetylated by lysine acetyltransferase enzymes such as Gcn5. The structure of the ternary Naa50p complex also provides the first molecular scaffold for the design of NAT-specific small molecule inhibitors with possible therapeutic applications.
Introduction
Protein acetylation is one of the most common covalent modifications found in the proteome (1). In this process, an acetyl group is transferred from acetyl-CoA to an α-amino group at the N terminus of a protein or an ϵ-amino group on a lysine side chain. Lysine acetyltransferase enzymes have been more extensively characterized than N-terminal acetyltransferase (NAT)2 enzymes at the biochemical, enzymological, and structural levels (2). To date, only one structure of a prokaryotic NAT has been reported (3), and the mechanisms of substrate-specific recognition and catalysis used by these enzymes remain poorly understood.
In eukaryotes, N-terminal acetylation displays a wide range of functional effects varying on a case-by-case basis (4). Recently, this modification was shown to serve as a degradation signal for individual proteins (5) and also to prevent proper post-translational translocation through the endoplasmic reticulum (6). In eukaryotes, multiple protein complexes are capable of performing N-terminal acetylation, the most promiscuous and well characterized of which is the NatA complex (7–9). This protein complex contains two catalytic subunits, Naa10p (ARD1) and Naa50p, and two auxiliary subunits, Naa15p (NATH (N-acetyltransferase human)) and HYPK, which associate with the ribosome (9–11). The human and fruit fly Naa50p enzymes are essential for normal sister chromatid cohesion and chromosome condensation, but the NatA complex was not implicated in this phenotype, suggesting that this is attributed solely by Naa50p-mediated NAT activity independent of the complex (12–14). However, the responsible substrate potentially requiring N-terminal acetylation for proper functioning remains to be identified. Interestingly, NatA subunits have been shown to be overexpressed in a number of cancer cell lines, and NatA knockdown results in a decrease in cellular proliferation and induction of apoptosis (15–18). NATs have therefore become an attractive chemotherapeutic target, further driving the need for a molecular understanding of how these enzymes carry out their functions.
NAT enzymes are differentiated based on their ability to acetylate substrate peptides that are of a specific sequence, with a particular dependence placed on the identity of the first two residues. Naa10p, which is normally active in the presence of Naa15p, is relatively promiscuous and able to acetylate α-amino groups of peptides containing Ser, Ala, Gly, Val, Cys, Thr, Asp, or Glu at the N terminus (19, 20). The identities of downstream residues are not crucial for activity but can influence catalysis in some cases. Naa50p is only able to process substrate peptides that contain a methionine residue at the N terminus and is able to do so in vitro in the absence of other regulatory protein subunits (20, 21). In vivo knockdown of Naa10p and Naa15p indirectly destabilizes human Naa50p, resulting in down-regulation of N-terminal acetylation of heterogeneous nuclear ribonucleoprotein F, which may represent a true in vivo substrate (19). Kinetic analysis of recombinant Naa50p has been carried out using a 24-residue peptide that shares sequence identity with the first seven residues of heterogeneous nuclear ribonucleoprotein F. This study revealed that point mutations of any of the first four residues of the heterogeneous nuclear ribonucleoprotein peptide can have a detrimental effect on Naa50p activity, but only the first methionine residue is strictly required for catalysis. Many proteins known to contain acetylation-susceptible internal lysines have also been screened with Naa50p, and no significant acetylation has been detected (21). Notably, both Naa50p and Naa10p have been reported to have the ability to acetylate themselves in vitro; however, it is widely accepted that NAT enzymes are only minimally active as side chain lysine acetyltransferases and select for N-terminal acetylation through an unknown mechanism (21, 22).
To understand the molecular basis for how NAT proteins such as Naa50p mediate catalysis and selectively target the α-amino group of N-terminal residues over side chain lysine residues, we determined the x-ray crystal structure of a ternary Naa50p complex with coenzyme A and a cognate N-terminal substrate peptide and carried out structure-based mutagenesis as well as accompanying biochemical and enzymatic characterization.
EXPERIMENTAL PROCEDURES
Naa50p Expression and Purification
A pETM-GST vector (G. Stier, European Molecular Biology Laboratory, Heidelberg, Germany) containing DNA encoding full-length human Naa50p (amino acids 1–169) fused to GST through a tobacco etch virus protease-cleavable site was transformed into BL21(DE3) cells. The transformed cells were cultured at 37 °C until the cell A600 reached ∼0.7, induced for protein expression with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside, and grown overnight at 18 °C. Cells were isolated by centrifugation and lysed by sonication in lysis buffer containing 25 mm HEPES (pH 7.5), 100 mm NaCl, and 10 mm 2-mercaptoethanol. The supernatant was isolated and passed over GST-binding resin (Clontech). Unbound proteins were washed off the resin with lysis buffer, and tobacco etch virus protease was added directly to the resin and allowed to incubate overnight at room temperature. Untagged Naa50p was washed off the resin with lysis buffer; collected; dialyzed into buffer containing 25 mm HEPES (pH 7.5), 50 mm NaCl, and 10 mm 2-mercaptoethanol; and bound to a 5-ml HiTrap SP ion exchange column (GE Healthcare). The protein was eluted in the same buffer using a salt gradient of 50–750 mm NaCl and further purified to homogeneity using a Superdex 75 gel filtration column (GE Healthcare) in storage buffer containing 25 mm HEPES (pH 7.5), 100 mm NaCl, and 10 mm dithiothreitol. Peak fractions containing protein that was >95% pure as judged by SDS-PAGE were concentrated to ∼12 mg/ml using a 10-kDa protein concentrator (Millipore) and stored at −70 °C until used. All protein mutants were generated using the QuikChange protocol from Stratagene (23). Protein harboring point mutations was obtained following the expression and purification protocols described above.
Crystallization and Data Collection
The peptides and CoA used in crystallization were purchased from GenScript and Sigma-Aldrich, respectively. The ternary Naa50p·CoA·peptide complex was prepared for crystallization by mixing in a 1:3:3 molar ratio, respectively. The length of the substrate peptide was varied between 7 and 20 amino acids in crystallization trials. The protein concentration used in crystallization was 12 mg/ml, and the peptide that produced the best diffracting crystals was a 10-mer with the sequence MLGPEGGRWG. Crystals of the ternary Naa50p·CoA·peptide complex were grown by hanging drop vapor diffusion at 20 °C over 1–3 days against reservoir solution containing 16% PEG 8000, 20% glycerol, and 40 mm potassium phosphate (monobasic; pH 5.0). Crystals were transferred to harvest solution containing well solution supplemented with 25% glycerol prior to flash-freezing in liquid nitrogen. Data were collected at beamline X6A at the National Synchrotron Light Source (Brookhaven National Laboratory) and processed using HKL2000 (24).
Structure Determination and Refinement
The structure of the ternary Naa50p·CoA·peptide complex was determined by molecular replacement with Phaser in the CCP4 suite of programs using the binary Naa50p·CoA complex (Protein Data Bank code 2PSW) as a search model (25). Solvent molecules and CoA were excluded from the search model. Structure refinement was performed by initially applying 3-fold non-crystallographic symmetry and running phenix.refine with group atomic displacement parameters for the initial cycles (26). Subsequently, CoA and substrate peptide were built into the resulting Fo − Fc electron density map, and additional refinement was performed manually using the molecular graphics program Coot (27). In advanced stages of refinement, non-crystallographic symmetry restraints were released, individual atomic displacement parameters were refined, and solvent was added. The final model was checked for errors against a composite omit map generated in CNS (28). Refinement statistics can be found in Table 1. The final model and structure factors were submitted to the Protein Data Bank with code 3TFY (Research Collaboratory for Structural Bioinformatics code RCSB067433).
TABLE 1.
Data collection and refinement statistics for the Naa50p·CoA·peptide complex
Values in parentheses are for the highest resolution shell. Rfree was calculated using 10% of the reflection data chosen randomly and omitted at the start of refinement. r.m.s.d., root mean square deviation.
| Crystal parameters | |
| Space group | P21 |
| Unit cell dimensions | a = 48.299 Å (α = 90.0°), b = 104.103 Å (β = 106.3°), c = 68.247 Å (γ = 90.0°) |
| Data collection | |
| Resolution range (Å) | 33.70–2.75 |
| Unique reflections | 16,010 |
| Multiplicity | 3.6 (3.2) |
| Completeness (%) | 94.7 (84.0) |
| Rmerge (%) | 12.9 (40.9) |
| I/σ | 10.2 (2.1) |
| Refinement statistics | |
| Rwork/Rfree (%) | 19.0/25.4 |
| r.m.s.d. | |
| Bonds (Å) | 0.008 |
| Angles | 1.596° |
| Average B-factor (Å2) | |
| Protein | 54.6 |
| CoA | 50.4 |
| Peptide | 73.9 |
| Water | 56.3 |
| Ramachandran statistics (%) | |
| Favored | 96.9 |
| Allowed | 3.1 |
| Outliers | 0 |
Acetyltransferase Activity Assays
Naa50p acetyltransferase assays were carried out with 100 nm untagged full-length enzyme at 37 °C for 40 min in 100 mm Tris (pH 8.0) and 50 mm NaCl unless stated otherwise. A saturating amount of acetyl-CoA (300 μm) was used in all enzymatic reactions, and the substrate peptide (+NH3-MLGPEGGRWGRPVGRRRRP-COO−; GenScript) concentration was varied to determine steady-state catalytic parameters. In the assay, radiolabeled [14C]acetyl-CoA (4 mCi/mmol; PerkinElmer Life Sciences) was mixed with the substrate peptide and allowed to incubate with enzyme in a 25-μl reaction volume. To quench the reaction, 20 μl of the reaction mixture was added to P81 paper discs (Whatman), and the paper discs were immediately placed in wash buffer. Washes were then carried out three times in 10 mm HEPES (pH 7.5), with each wash lasting 5 min, to remove unreacted acetyl-CoA. The papers were then dried with acetone and added to 4 ml of scintillation fluid, and the signal was measured using a Packard Tri-Carb 1500 liquid scintillation analyzer. Background control reactions were performed in the absence of enzyme. Reactions were also performed in the absence of the substrate peptide to ensure that any possible signal due to autoacetylation was negligible. Substrate peptide Km and Vmax parameters were derived by titrating the substrate peptide at eight different concentrations ranging from 100 to 1500 μm in the presence of 300 μm acetyl-CoA. Complications caused by TFA from the addition of high concentrations of the substrate peptide prevented point mutant catalytic assays from reaching a saturating amount of peptide, so we instead analyzed the catalytic efficiency (kcat/Km) of these mutants. By keeping the substrate concentration far below Km, we were able to plot velocity versus [peptide] and use the slope of the resulting line to obtain kcat/Km values for several of these mutants based on the following equation: v = (kcat/Km)[E][S]. These results allowed us to compare quantitatively the catalytic efficiencies of each mutant with the wild-type enzyme. The acetyl-CoA Km value was determined by titrating a range of acetyl-CoA concentrations (25–500 μm) into a reaction containing 2.5 mm peptide in buffer containing 50 mm NaCl and 200 mm Tris (pH 8.0). The high concentration of Tris is required to counter the pH effect caused by TFA carried into the reaction with high concentrations of peptide. A pH-rate profile was determined using a three-component buffer system able to maintain constant ionic strength at all pH values used in this study (29). All reactions were performed in triplicate. All radioactive count values were converted to molar units using a standard curve created with known concentrations of radioactive acetyl-CoA added to scintillation fluid.
RESULTS
Naa50p Shows a Gcn5-related N-Acetyltransferase (GNAT) Fold with a More Constricted Protein Substrate-binding Site
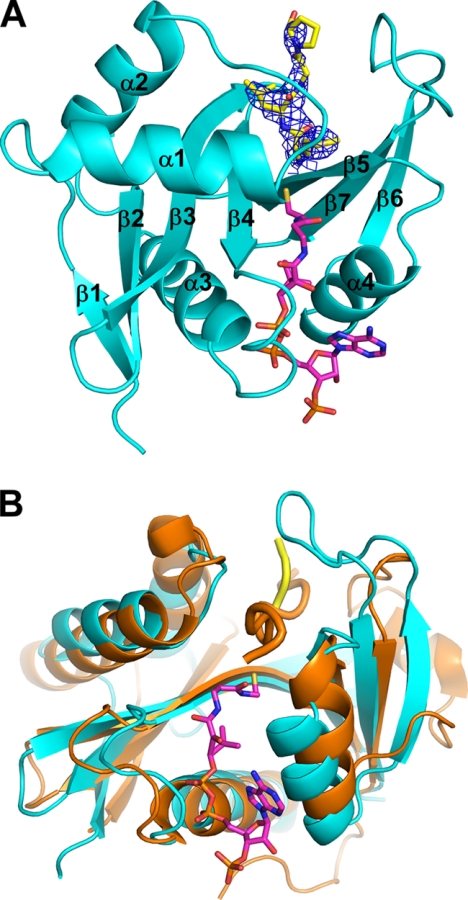
Crystals of the ternary Naa50p·CoA·substrate peptide complex formed in space group P21 with three molecules in the asymmetric unit and diffracted to a 2.75-Å resolution limit. The structure was refined to an Rwork/Rfree of 19.0/25.4% with good stereochemistry and with 96.9% of the residues in the favored region of the Ramachandran plot (Table 1). The structure reveals a mixed α/β-fold with a conserved acetyl-CoA-binding core region composed of one α-helix (α3) and three β-strands (β2–β4) as shown in Fig. 1. This motif is characteristic of GNATs and allows us to confidently place Naa50p into this family of enzymes (30). Indeed, Naa50p superimposes well with the histone acetyltransferase (HAT) domain of Gcn5, with a root mean square deviation of Cα atoms of 4.1 Å (Fig. 1B). In addition to a high degree of superposition within the core domain, the entire β-sheet that cuts through the center of the structure (β1–β5 and β7) and the α1-turn-α2 that flanks one side of the protein substrate-binding site superimpose well (Fig. 1B). The greatest regions of divergence between the two proteins map to C-terminal segments of the two proteins that flank the opposing side of the respective protein substrate-binding site, which also maps to the largest region of sequence and structural divergence between the GNAT proteins (30, 31). In the case of Gcn5, this region forms a loop-helix-loop-strand motif that facilitates the formation of a relatively wide protein substrate-binding site, whereas in the case of Naa50p, this regions forms a β-hairpin (β6-loop-β7) containing a relatively long 14-residue loop that blocks off part of the protein substrate-binding site that is otherwise accessible in Gcn5 (Fig, 1B).
FIGURE 1.
Overall structure of the ternary Naa50p·CoA·peptide complex. A, structure of the ternary complex showing Naa50p in teal; CoA as magenta sticks with carbon, nitrogen, and sulfur atoms in Corey-Pauling-Koltun coloring; and the substrate peptide as yellow sticks with carbon, nitrogen, and sulfur atoms in Corey-Pauling-Koltun coloring. Substrate peptide electron density obtained from a composite omit map (blue) is shown contoured to 1.5σ. B, superposition of the ternary Naa50p complex with the ternary Gcn5 HAT. The Gcn5 enzyme and accompanying substrate peptide (orange) are shown in schematic format. The substrate peptides for each enzyme are shown as thicker schematics for clarity.
In addition to well ordered electron density for the entire CoA molecule, we observed strong electron density in the Fo − Fc map corresponding to the first four residues of the 10-residue peptide that was used for the crystal structure (Fig. 2A). Following refinement, side chain density was visible for the first two residues of the peptide, whereas backbone density could be traced to the fourth residue (Fig. 1A). This correlates with the contacts formed between the peptide and the enzyme in the crystal structure (Fig. 2, B and C). Hydrogen bonding can be observed throughout the backbone for the first two residues, and the Met1 and Leu2 side chains both sit in hydrophobic pockets that appear to be tailored for these residues (Fig. 2, B and C).3 There are no substantial interactions beyond the leucine backbone carbonyl group seen in the structure. The overall thermal B-factor of the peptide is 73.9, which is high in relation to the overall thermal B-factor of the protein, 55. This discrepancy can be accounted for by the skewed nature of the B-factors of the individual residues of the peptide. Met1 has a B-factor of 62, and this value increases for each residue of the peptide that extends out of the binding pocket, with the Pro4 B-factor reaching 93. For comparison, the atoms of CoA have an average B-factor of 50. Superposition of the ternary complex with the binary Naa50p·acetyl-CoA complex (Protein Data Bank code 2OB0) showed no significant structural rearrangement upon substrate peptide binding (supplemental Fig. 1), further revealing that Naa50p does not undergo significant structural change upon protein substrate binding.
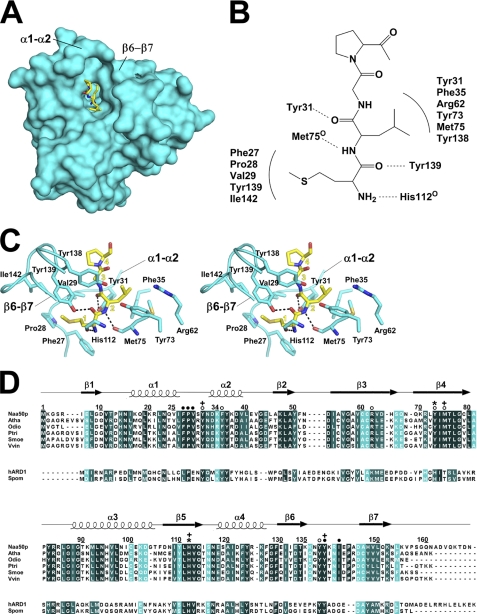
FIGURE 2.
Naa50p interactions with substrate peptide. A, surface of Naa50p highlighting the peptide substrate-binding site with the peptide shown in stick representation with the same color coding as described in the legend to Fig. 1A. B, ChemDraw representation summarizing all of the interactions between Naa50p and the peptide. Hydrogen bonding interactions are denoted by dotted lines, and van der Waals interactions are depicted with arches. C, stereo diagram of Naa50p interactions with the substrate peptide. Protein residues that make peptide interactions are highlighted. The His-112 side chain has been omitted for clarity. Substrate peptide resides are labeled with yellow numbers corresponding to their sequential order. D, sequence alignment of Naa50p orthologs from several species. The names of the orthologs are abbreviated as follows: Atha, Arabidopsis thaliana; Odio, Oikopleura dioica; Ptri, Populus trichocarpa; Smoe, Selaginella moellendorffii; and Vvin, Vitis vinifera. For comparison, two Naa10p orthologs are also shown. hARD1, human ARD1; Spom, Schizosaccharomyces pombe. Numbering and secondary structure elements for Naa50p are indicated above the Naa50p sequence. Residues of Naa50p that contact the peptide backbone (+), Met1 (●), Leu2 (○), and catalytic residues (*) are highlighted as indicated above the Naa50p sequence.
The Naa50p Protein Substrate-binding Site Is Ideally Suited to Accommodate an α-Amino Group of an N-terminal Methionine
The N-terminal peptide substrate points into the protein active site, with the backbone roughly perpendicular to the central β-sheet and parallel to the pantothenate arm of the CoA (Figs. 1B and 2A). The peptide is anchored to the protein through a series of hydrogen bonds between the side chains of Tyr-31 and Tyr-139 and the backbone carbonyl atoms of Met-75 and His-112 (Fig. 2C). The N-terminal methionine sits in a hydrophobic pocket composed of Phe-27, Pro-28, and Val-29 from the α1-α2 loop and Tyr-139 and Ile-142 from the β6-β7 hairpin loop, which each make van der Waals contacts with Met1 of the peptide. Leu2 also sits in a hydrophobic pocket formed by Tyr-31 and Phe-35 from the α1-α2 loop segment, Tyr-138 from the β6-β7 hairpin loop, and Tyr-73 and Met-75 from the β4 strand (Fig. 2C). There are also hydrophilic groups in the vicinity of Leu2 such as the hydroxyls of Tyr-73 and Tyr-138 and the guanidinium group of Arg-62 (Fig. 2C), which provides an explanation for the ability of Naa50p to acetylate a substrate with a hydrophilic residue at the second position.
Consistent with the importance of residues that contact the peptide, each of these residues is highly conserved within the Naa50p orthologs from other species (Fig. 2D). In particular, the residues that contact the peptide backbone (Tyr-31, Met-75, His-112, and Tyr-139) and Met1 (Phe-27, Pro-28, Val-29, Tyr-139, and Ile-142) are strictly conserved, and residues that contact Leu2 (Tyr-31, Phe-35, Tyr-73, Met-75, and Tyr-138) are strictly or highly conserved (Fig. 2D).
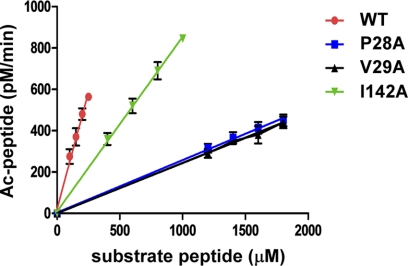
To establish directly the functional importance of Naa50p residues that appear to make important peptide substrate contacts in the crystal structure, we targeted several of these residues for mutagenesis, followed by enzymatic characterization of the mutant proteins. Each mutated residue was changed to alanine, and the catalytic efficiency of each mutant was determined and compared with that of the wild-type protein. Each mutant was purified to homogeneity and exhibited gel filtration chromatography elution profiles identical to that of the wild-type construct. To confirm further that the mutant proteins were folded properly, they were subjected to circular dichroism experiments, each revealing a similar spectrum to the wild-type protein (data not shown). Notably, each of the mutant proteins showed defects in catalytic efficiency, with most of the mutant proteins showing no detectable activity (Fig. 3 and Table 2). Of the residues that contact the peptide backbone, Tyr-31, His-112, and Tyr-139 were mutated, and each of these mutants showed no detectable activity, arguing for the importance of these residues for backbone interaction, although Tyr-31 also participates in Leu2 peptide contact, and His-112 likely participates directly in catalysis, as will be described below (Fig. 3 and Table 2). Of the residues that contact Met1 of the peptide, the Phe-27 mutant showed no detectable activity, whereas the Pro-28 and Val-29 mutants showed at least an 85% reduction in activity. The Tyr-139 mutant showed no detectable activity, and the Ile-142 mutant retained ∼40% activity (Fig. 3 and Table 2). Mutants of Leu2 peptide contact residues Tyr-31, Phe-35, and Tyr-73 each showed no detectable activity (Fig. 3 and Table 2). Taken together, the results from the structure-based mutagenesis are consistent with the functional importance of the protein-peptide substrate interactions observed in the crystal structure.
FIGURE 3.
Catalytic efficiency of Naa50p and mutant proteins. Four data points were collected for each enzyme in triplicate. The straight line formed by these four data points crosses through the origin, indicative of the fact that their slope can be used to calculate catalytic efficiency. Ac, acetyl.
TABLE 2.
Catalytic efficiency of the wild-type Naa50p enzyme and mutants
| Enzyme | kcat/Km | Wild-type efficiency |
|---|---|---|
| μm−1min−1 | % | |
| Wild-type | 1.94 ×10−2 | 100 |
| F27A | NDa | <10 |
| P28A | 2.59 ×10−3 | 13.4 |
| V29A | 2.38 ×10−3 | 12.3 |
| Y31A | ND | <10 |
| F35A | ND | <10 |
| Y73A | ND | <10 |
| Y73F | ND | <10 |
| H112A | ND | <10 |
| H112F | ND | <10 |
| Y139A | ND | <10 |
| I142A | 8.18 ×10−3 | 42.2 |
a ND, mutants that did not yield detectable activity at 2.0 mm substrate peptide and a saturating amount of acetyl-CoA.
Tyr-73 and His-112 Likely Play Catalytic Roles through an Ordered Water Molecule
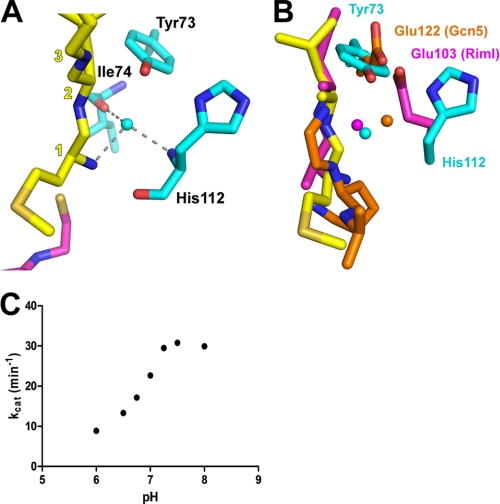
Analysis of the Naa50p active site revealed that only two residues are in position to function in catalysis, Tyr-73 and His-112 (Fig. 4A). Although the functional groups of these residues could participate in proton extraction of the amino nitrogen substrate, the hydroxyl of Tyr-73 and the imidazole nitrogen of His-112 are too far for direct proton extraction. To potentially circumvent this issue, a well ordered water molecule, with a B-factor of 48, is in a position to mediate proton abstraction. This molecule is held in place via hydrogen bonds with the peptide N-terminal α-amino group and the backbone carbonyl and nitrogen of Ile-74 and His-112, respectively (Fig. 4A). Supporting the role of such a water molecule, the GNAT histone acetyltransferase Gcn5 has been shown to employ a glutamate residue to mediate proton abstraction from the substrate peptide through a well ordered water molecule (32). An active site alignment of these two proteins showed that Tyr-73 of Naa50p aligns very well with the catalytic Glu-122 of Gcn5 (Fig. 4B), which supports a role of Tyr-73 of Naa50p as participating in catalysis. Superposition of the Naa50p active site with the active site of the bacterial NAT protein RimI (3) further supports the catalytic roles of Tyr-73 and His-112 of Naa50p (Fig. 4B). This superposition illustrates that the catalytic glutamate general base residue of RimI sits roughly midway between Tyr-73 and His-112 of Naa50p and that RimI also contains a well ordered water molecule in a similar position to the corresponding water molecule of Naa50p.
FIGURE 4.
Naa50p active site. A, detailed view of the active site highlighting an ordered water molecule that plays a putative catalytic role along with Tyr-73 and His-112 in proton extraction from the α-amino group of the peptide substrate. Hydrogen bonds are depicted with gray dashed lines. Substrate peptide resides are labeled with yellow numbers corresponding to their sequential order. B, superposition of the Naa50p active site with the active sites of bacterial RimI and the Gcn5 HAT. Catalytic residues and water molecules of RimI and Gcn5 are shown in purple and orange, respectively. C, pH-rate profile for wild-type Naa50p. See “Experimental Procedures” for details.
To probe directly the functional importance of Naa50p Tyr-73 and His-112, we mutated these residues individually to both alanine and phenylalanine, purified these proteins to homogeneity, confirmed that these proteins were properly folded by circular dichroism, and analyzed their catalytic properties. As shown in Table 2, each of the mutants showed background levels of catalytic activity consistent with a role for Tyr-73 and His-112 in catalysis by Naa50p, although other non-catalytic effects of mutating Tyr-73 and His-112 cannot be ruled out. We also carried out a pH-rate profile of wild-type Naa50p from pH 8.0 to 6.0, the most acidic environment at which activity could still be measured (Fig. 4C). As shown in Fig. 4C, the wild-type enzyme showed optimal activity above pH 7.5, and the kcat decreased by ∼3-fold when the pH was decreased to 6.0. The inflection point at pH ∼7.0 could represent the pKa of the catalytic base or that of the substrate α-amino group in the active site. Although a pH of 7.0 is reasonable for the imidazole side chain, one would expect a more drastic catalytic effect once the side chain is fully protonated if the histidine was the only residue that participated in proton abstraction. Taken together, these data are consistent with the proposal that Tyr-73 and His-112 cooperate in functioning as general bases for catalysis that proceeds through the ordered water molecule observed in the crystal structure. Tyr-73 and His-112 do not appear to be in the proper position in the current structure to act as general acids for reprotonation of the CoA leaving group, although it is possible that one of these residues or a different protein residue might swing into place to play this role following the acetyl transfer.
Comparison with the Ternary Gcn5 Complex Suggests Why Naa50p Prefers α-Amino Versus Side Chain Lysine Substrates
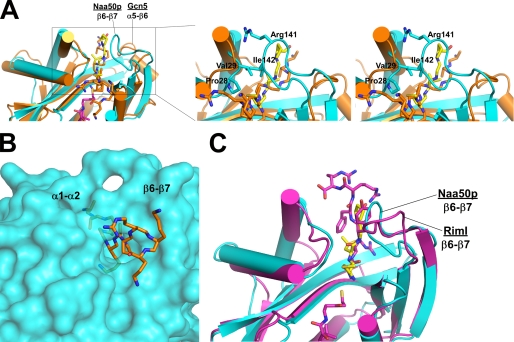
Although there have been several reports of NATs performing internal lysine acetylation, it is clear that the process is very slow in relation to N-terminal acetyltransferase activity (16, 21). To better understand the structural basis for Naa50p preference for N-terminal acetylation, we superimposed the ternary Naa50p complex with the Tetrahymena Gcn5·CoA·H3 peptide complex to identify key differences in substrate peptide-binding motifs. As noted above, the two structures share a similar overall fold, with the most pronounced differences mapping to the C-terminal segment of the proteins and particularly to the β6-β7 loop flanking one side of the protein peptide-binding site (Fig. 5A). As a result of this difference, one side of the Naa50p protein substrate-binding site is constricted such that a lysine side chain-containing peptide segment cannot sit across the substrate-binding groove (Fig. 5B). Indeed, Ile-142 of the β6-β7 loop makes van der Waals interactions with Pro-28 and Val-29 of the α1-α2 loop across the binding groove, and Arg-141 also sits across the binding groove (Fig. 5A). Because the more constricted Naa50p substrate-binding site cannot easily accommodate a peptide sitting across the substrate-binding site, it must accommodate a peptide substrate that enters the active site in a different orientation. This is consistent with the binding mode of the N-terminal peptide that enters the active site from the accessible end of the peptide-binding groove and at roughly a 90° angle relative to the histone H3 peptide bound to Gcn5 (Fig. 1B). Residues that participate in constricting one end of the Naa50p binding groove, Pro-28, Val-29, and Ile-142, also participate in N-terminal peptide binding (Fig. 2C). The unique features that differentiate Naa50p from Gcn5 are conserved in the RimI structure (Fig. 5C), and we extrapolate that they are conserved throughout the NATs to mediate N-terminal acetylation specificity.
FIGURE 5.
Structural comparison with other acetyltransferases. A, structural alignment of Naa50p with the Gcn5 HAT. The proteins are shown as a schematic with the peptide substrates shown in stick representation with Corey-Pauling-Koltun coloring. The CoA molecule from Naa50p is also shown in stick representation. The zoom view (rotated 30° clockwise on the y axis) shows a stereo image that includes the position of several side chains in the Naa50p substrate-binding loops. B, surface representation of Naa50p with a stick model of the Gcn5 substrate peptide docked into the Naa50p peptide-binding site. C, structural alignment of Naa50p with the bacterial NAT RimI. The Rim protein is shown in purple with the associated bisubstrate inhibitor shown as purple sticks with Corey-Pauling-Koltun coloring.
DISCUSSION
Despite its biological importance, the process of N-terminal protein acetylation remains relatively poorly understood. In contrast, HATs that modify side chain lysine residues have been extensively characterized at both the biochemical and structural levels, and this understanding has led to the development of HAT-specific inhibitors (33–35). The ternary structure of Naa50p bound to CoA and a peptide fragment of its native substrate reported here now provides the first molecular insights into the mechanism of substrate binding and possibly also catalysis used by this class of enzymes and provides a molecular scaffold for the design of NAT-specific inhibitors.
The stringent requirement of Naa50p for a substrate containing an N-terminal methionine is well explained by the structure and accompanying mutagenesis and enzymology studies. The enzyme uses a hydrophobic pocket that forms van der Waals contacts with the methionine residue, which prevents the enzyme from interacting with all other potential N-terminal residues. BLAST alignments revealed that these residues are highly conserved among Naa50p but not Naa10p orthologs (Fig. 2D). It is likely that the corresponding residues in Naa10p are optimized to be more tolerant of amino acid variability in this position. Notably, most of the Naa50p residues that mediate peptide backbone interactions (Tyr-31, His-112, and Tyr-139) are conserved with the Naa10p proteins, suggesting that the mode of binding of the N-terminal protein segment to the Naa10p proteins might be similar to that of Naa50p. The interactions between Naa50p and peptide residues C-terminal to Met1 are less extensive, which allows for more flexibility in residue identity at these positions (Fig. 2C). Previous studies show that Naa50p activity is affected by the first four residues of the substrate peptide sequence (21). Here, we have been able to provide an explanation for the ability of the enzyme to accommodate both hydrophobic and hydrophilic residues at the second amino acid position of the substrate. The third and fourth substrate residues in the structure presented here are partially disordered, so it is difficult to draw conclusions regarding their role in substrate binding from the structure. A point mutation screen showed that conservative mutations in the third and fourth positions are tolerated quite well, with the most detrimental mutations at these positions exhibiting only a 5-fold reduction in Km and a modest effect on kcat (21). This is in contrast to the more drastic effects on mutations of Met1 or Leu2.
The GNAT family of enzymes has been shown to employ a variety of different chemical strategies to perform protein acetylation. For example, the Gcn5/PCAF (p300/CBP-associated factor) family of HATs uses a conserved glutamic acid residue to serve as a general base for a ternary complex mechanism (2); the p300/CBP (cAMP-responsive element-binding protein-binding protein) family does not employ a dedicated general base for catalysis but likely uses a tyrosine residue as a general acid to mediate a Theorell-Chance catalytic mechanism (36). In addition, the small molecule acetyltransferase serotonin acetyltransferase employs two histidine residues to act as general bases for catalysis (37). In light of the varied ways in which acetyltransferase enzymes carry out chemistry, it is not surprising that Naa50p appears to use yet another chemical strategy that likely involves histidine (His-112) and tyrosine (Tyr-73) general base residues, at least one of which might function as a general acid as well (Fig. 4A).
It is also of note that the in vitro catalytic parameters derived in this study are inefficient relative to those of other acetyltransferases (which are measured on a s−1 scale). The slower rate observed for recombinant Naa50p may be a consequence of the absence of the Naa15p or Naa10p subunits, which might function to elevate the catalytic rate. Consistent with this possibility, recombinant Naa10p has been shown to harbor a greater catalytic activity toward its classical substrates in the presence of the Naa15p subunit (20).
Finally, this study represents the first description of the molecular details that distinguish the substrate-binding modes between the N-terminal and side chain lysine protein acetyltransferases. There appear to be several features of the Naa50p protein substrate-binding site that center around an extended β6-β7 hairpin loop, which plays a particularly important role in biasing the specificity of the NAT proteins for N-terminal α-amino groups. Our results are therefore in agreement with previous studies that have shown that Naa50p is a very inefficient lysine side chain acetyltransferase (21). We also anticipate that the ternary Naa50p scaffold provided here, along with the distinguishing features between NATs and lysine acetyltransferases, may lead to the development of NAT-specific inhibitors that might have therapeutic applications.
Acknowledgments
We acknowledge the use of the Wistar Proteomics Core facility, which is supported in part by National Institutes of Health Grant CA010815. We also acknowledge Mike Brent, Marc Holbert, and Rolf Sternglanz for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM060293 (to R. M.) and Training Grant GM071339 (to G. L.). This work was also supported by the Research Council of Norway and the Norwegian Cancer Society (to T. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
The atomic coordinates and structure factors (code 3TFY) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The positions of residues in the peptide substrate are indicated by superscript numbers, and those in the protein enzyme are indicated by hyphenated full-size numbers.
- NAT
- N-terminal acetyltransferase
- GNAT
- Gcn5-related N-acetyltransferase
- HAT
- histone acetyltransferase.
REFERENCES
- 1. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 2. Hodawadekar S. C., Marmorstein R. (2007) Oncogene 26, 5528–5540 [DOI] [PubMed] [Google Scholar]
- 3. Vetting M. W., Bareich D. C., Yu M., Blanchard J. S. (2008) Protein Sci. 17, 1781–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arnesen T. (2011) PLoS Biol. 9, e1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hwang C. S., Shemorry A., Varshavsky A. (2010) Science 327, 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Forte G. M., Pool M. R., Stirling C. J. (2011) PLoS Biol. 9, e1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Starheim K. K., Gromyko D., Velde R., Varhaug J. E., Arnesen T. (2009) BMC Proc. 3, Suppl. 6, S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Polevoda B., Sherman F. (2003) Biochem. Biophys. Res. Commun. 308, 1–11 [DOI] [PubMed] [Google Scholar]
- 9. Arnesen T., Anderson D., Torsvik J., Halseth H. B., Varhaug J. E., Lillehaug J. R. (2006) Gene 371, 291–295 [DOI] [PubMed] [Google Scholar]
- 10. Arnesen T., Anderson D., Baldersheim C., Lanotte M., Varhaug J. E., Lillehaug J. R. (2005) Biochem. J. 386, 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gautschi M., Just S., Mun A., Ross S., Rücknagel P., Dubaquié Y., Ehrenhofer-Murray A., Rospert S. (2003) Mol. Cell. Biol. 23, 7403–7414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hou F., Chu C. W., Kong X., Yokomori K., Zou H. (2007) J. Cell Biol. 177, 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pimenta-Marques A., Tostões R., Marty T., Barbosa V., Lehmann R., Martinho R. G. (2008) Dev. Biol. 323, 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Williams B. C., Garrett-Engele C. M., Li Z., Williams E. V., Rosenman E. D., Goldberg M. L. (2003) Curr. Biol. 13, 2025–2036 [DOI] [PubMed] [Google Scholar]
- 15. Arnesen T., Gromyko D., Pendino F., Ryningen A., Varhaug J. E., Lillehaug J. R. (2006) Oncogene 25, 4350–4360 [DOI] [PubMed] [Google Scholar]
- 16. Lim J. H., Park J. W., Chun Y. S. (2006) Cancer Res. 66, 10677–10682 [DOI] [PubMed] [Google Scholar]
- 17. Ren T., Jiang B., Jin G., Li J., Dong B., Zhang J., Meng L., Wu J., Shou C. (2008) Cancer Lett. 264, 83–92 [DOI] [PubMed] [Google Scholar]
- 18. Yu M., Ma M., Huang C., Yang H., Lai J., Yan S., Li L., Xiang M., Tan D. (2009) Cancer Invest. 27, 978–983 [DOI] [PubMed] [Google Scholar]
- 19. Arnesen T., Van Damme P., Polevoda B., Helsens K., Evjenth R., Colaert N., Varhaug J. E., Vandekerckhove J., Lillehaug J. R., Sherman F., Gevaert K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8157–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van Damme P., Evjenth R., Foyn H., Demeyer K., De Bock P. J., Lillehaug J. R., Vandekerckhove J., Arnesen T., Gevaert K. (March 7, 2011) Mol. Cell. Proteomics 10, M110.004580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Evjenth R., Hole K., Karlsen O. A., Ziegler M., Arnesen T., Lillehaug J. R. (2009) J. Biol. Chem. 284, 31122–31129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seo J. H., Cha J. H., Park J. H., Jeong C. H., Park Z. Y., Lee H. S., Oh S. H., Kang J. H., Suh S. W., Kim K. H., Ha J. Y., Han S. H., Kim S. H., Lee J. W., Park J. A., Jeong J. W., Lee K. J., Oh G. T., Lee M. N., Kwon S. W., Lee S. K., Chun K. H., Lee S. J., Kim K. W. (2010) Cancer Res. 70, 4422–4432 [DOI] [PubMed] [Google Scholar]
- 23. Braman J., Papworth C., Greener A. (1996) Methods Mol. Biol. 57, 31–44 [DOI] [PubMed] [Google Scholar]
- 24. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 25. Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 26. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 29. Tanner K. G., Trievel R. C., Kuo M. H., Howard R. M., Berger S. L., Allis C. D., Marmorstein R., Denu J. M. (1999) J. Biol. Chem. 274, 18157–18160 [DOI] [PubMed] [Google Scholar]
- 30. Neuwald A. F., Landsman D. (1997) Trends Biochem. Sci. 22, 154–155 [DOI] [PubMed] [Google Scholar]
- 31. Trievel R. C., Rojas J. R., Sterner D. E., Venkataramani R. N., Wang L., Zhou J., Allis C. D., Berger S. L., Marmorstein R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8931–8936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rojas J. R., Trievel R. C., Zhou J., Mo Y., Li X., Berger S. L., Allis C. D., Marmorstein R. (1999) Nature 401, 93–98 [DOI] [PubMed] [Google Scholar]
- 33. Bowers E. M., Yan G., Mukherjee C., Orry A., Wang L., Holbert M. A., Crump N. T., Hazzalin C. A., Liszczak G., Yuan H., Larocca C., Saldanha S. A., Abagyan R., Sun Y., Meyers D. J., Marmorstein R., Mahadevan L. C., Alani R. M., Cole P. A. (2010) Chem. Biol. 17, 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu J., Wang J., Li M., Yang Y., Wang B., Zheng Y. G. (2011) Bioorg. Chem. 39, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Furdas S. D., Shekfeh S., Bissinger E. M., Wagner J. M., Schlimme S., Valkov V., Hendzel M., Jung M., Sippl W. (2011) Bioorg. Med. Chem. 19, 3678–3689 [DOI] [PubMed] [Google Scholar]
- 36. Liu X., Wang L., Zhao K., Thompson P. R., Hwang Y., Marmorstein R., Cole P. A. (2008) Nature 451, 846–850 [DOI] [PubMed] [Google Scholar]
- 37. Hickman A. B., Namboodiri M. A., Klein D. C., Dyda F. (1999) Cell 97, 361–369 [DOI] [PubMed] [Google Scholar]