Background: A growing number of proteins are degraded by the proteasome in a ubiquitin-independent manner.
Results: Ubiquitin-independent degradation requires an intrinsically disordered region in cooperation with an α-helix.
Conclusion: Overall conformation, rather than a specific primary sequence, underlies the function of these elements.
Significance: This provides mechanistic insight into an important aspect of intracellular protein degradation.
Keywords: Proteasome, Protein Degradation, Protein Stability, Protein Structure, Ubiquitination, Intrinsically Disordered Regions, Human Thymidylate Synthase (hTS), Ubiquitin-independent, Degron
Abstract
The 26 S proteasomal complex, which is responsible for the bulk of protein degradation within the cell, recognizes its target substrates via covalently linked polyubiquitin moieties. However, a small but growing number of proteasomal substrates are degraded without a requirement for ubiquitinylation. One such substrate is the pyrimidine biosynthetic enzyme thymidylate synthase (EC 2.1.1.45), which catalyzes the synthesis of TMP and is the sole de novo source of TTP for DNA replication and repair. Previous work showed that intracellular proteolysis of human thymidylate synthase is directed by a degron at the polypeptide's N-terminal end, composed of an intrinsically disordered region (IDR) followed by a highly conserved amphipathic α-helix (hA). In the present report, we show that the hA helix does not function simply as an extension or scaffold for the IDR; rather, it provides a specific structural component that is necessary for degradation. Furthermore, its helical conformation is required for this function. We demonstrate that small domains from heterologous proteins can substitute for the IDR and the hA helix of human thymidylate synthase, indicating that the degradation-promoting function of these regions is not sequence-specific. The results, in general, indicate that cooperation between intrinsically disordered domains and α-helical segments is required for ubiquitin-independent degradation by the proteasome. There appears to be little sequence constraint on the ability of these regions to function as degron constituents. Rather, it is the overall conformation (or lack thereof) that is critical.
Introduction
The proteasome is a large multisubunit complex of several dozen proteins that is responsible for the bulk of protein degradation within the cell (1–3). Breakdown of proteasomal substrates occurs within a chamber formed by 28 subunits organized into four, stacked heptameric rings (4–6). The entrance to this chamber, denoted the 20 S complex, is protected by a small pore capped by regulatory complexes, the nature of which varies among proteasomal isoforms. For the 26 S proteasome, which has been examined in most detail, the 20 S complex is capped by the 19 S regulatory particle (or PA700) that includes six ATPases organized into a hexameric ring interacting directly with the outer heptameric ring of the 20 S core (3–5). The 19 S particle exhibits a chaperone-like activity that recognizes and unfolds substrates as they “thread” their way through the pore and into the proteolytic chamber (4, 7, 8).
Typically, polyubiquitin chains covalently attached to the target substrate mediate the latter's recognition of and docking to the proteasome (1–3). However, in recent years, a number of proteins have been shown to undergo proteasomal degradation in the absence of ubiquitin modification (9, 10). Included among the known ubiquitin-independent substrates are the polyamine biosynthetic enzyme ornithine decarboxylase (11–13), proto-oncoprotein c-FOS (14, 15), the cyclin-dependent kinase inhibitor p21Waf1/Cip1 (16, 17), the F protein of hepatitis C virus (18), and the NFκB inhibitor IκBα (19, 20). Recent proteomic studies have indicated that as many as 20% of cellular proteins may be degraded in the absence of a requirement for ubiquitin modification (21), suggesting that many more ubiquitin-independent substrates remain to be discovered.
Thymidylate synthase (TS)3 (EC 2.1.1.45) catalyzes the reductive methylation of dUMP to form dTMP and is essential for the de novo biosynthesis of dTTP during DNA replication and repair (22, 23). Previous studies in our laboratory have identified the human enzyme (denoted hTS) as a ubiquitin-independent proteasomal substrate (24, 25). Degradation of hTS is governed by a 45-residue region at its N-terminal end, which is composed of a flexible, intrinsically disordered region (IDR) spanning the first 27 amino acids, followed by an amphipathic α-helix (helix A (hA)) at residues 31–42 (26). The region functions as an independent degradation signal, or degron, having the ability to destabilize a heterologous polypeptide to which it is attached (26). Studies focusing on the role of the IDR have shown that a free, unmodified amino group at the N terminus and an Arg-Arg dipeptide motif at residues 10–11 are critical to the function of the hTS degron (26, 27). The mechanisms by which these residues mediate ubiquitin-independent degradation of hTS are not well defined.
In the present report, we have examined the roles of both the IDR and the hA helix in more detail. We find that the helix contributes a structural role, rather than simply an extension or scaffold for the IDR; indeed, an α-helical conformation is required for the segment's activity. Both the IDR and the hA helix can be replaced with similarly structured elements from other proteins, indicating that it is conformation (or lack thereof) rather than primary sequence that is required for degron function. The implications of these findings for the mechanism of ubiquitin-independent degradation by the proteasome are discussed.
EXPERIMENTAL PROCEDURES
Plasmids
All constructs for mutant analysis were generated using standard molecular biology techniques and were verified directly by DNA sequencing. The parental expression plasmid for hTS, which was used to construct various mutant enzymes, was pJZ205, which contains a full-length hTS cDNA under the control of an SV40 promoter. For expression of enhanced green fluorescent protein (eGFP), plasmid pMP610, containing a full-length eGFP cDNA under control of a cytomegalovirus promoter, was utilized; the plasmid contains an Escherichia coli TS cDNA (optimized for expression in mammalian cells) under control of a murine stem cell virus LTR promoter. Mutagenesis was carried out by standard PCR-based protocols. Details of all plasmid constructions are available upon request.
Cell Culture
Cell lines were maintained at 37 °C in a humidified 5% CO2 atmosphere. Cell line RJK88.13, which is a TS-deficient derivative of V79 Chinese hamster lung cells (28), was maintained in Dulbecco's modified Eagle's medium (Cellgro) containing 4.5 g/liter glucose and supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals) and 10 μm thymidine.
Stable Transfection and Determination of Protein Half-life
RJK88.13 cells were transfected with the indicated expression plasmids using Lipofectamine 2000 or LTX (Invitrogen) according to the manufacturer's instructions. Stable transfectants were selected in thymidine-free medium containing the nucleoside transport inhibitor dipyridamole (5 μm; Sigma-Aldrich). Transfectants were pooled and maintained in mass culture.
Protein half-lives were analyzed following the addition of 50 μg/ml cycloheximide (CHX; Acros Organic) to the growth medium for the indicated times. Cells were harvested by scraping and lysed either by sonication (3 × 10 s) in NET2 buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.05% Nonidet P-40, 10 mm DTT, 2 mm β-mercaptoethanol, 5 mm PMSF, 200 μg/ml aprotinin, 100 μg/ml pepstatin, and 50 μg/ml leupeptin) or resuspension in M-Per® buffer (Pierce). Crude lysates were centrifuged at 15,000 × g for 1 h at 4 °C, and protein concentrations in the resulting extracts were quantified using the Bio-Rad assay reagent with bovine serum albumin as a standard. Immunoblotting was performed by standard techniques. Probes were monoclonal antibodies to hTS (provided by Dr. Sondra Berger, University of South Carolina), eGFP (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) catalog no. sc-9996), or ubiquitin (Santa Cruz Biotechnology, Inc., catalog no. sc-8017). To control for equal loading, blots were reprobed with an anti-actin monoclonal antibody (Sigma-Aldrich, Clone AC-40). The antigen-antibody complexes were visualized using appropriate secondary antibodies with the ECL chemiluminescence kit (Amersham Biosciences). Densitometry was carried out using ImageJ software maintained by the National Institutes of Health. A 2-fold dilution series of each extract was included on the blots for calibration and to correct for film exposure times. All values were normalized to actin concentrations on the same blots. Data were plotted according to first order decay kinetics, and half-lives were determined from the slopes of the resulting lines. Experiments were carried out at least twice and many more times in several cases.
Bioinformatics
Secondary structure analyses were carried out using two prediction tools: Jpred (29) maintained by the University of Dundee and PSIpred (30) from the University College London. Generation of the image depicting the three-dimensional structure of the hTS dimer was done using Protein Workshop (31), which is maintained on the RCSB Protein Data Bank server.
RESULTS
The hA Helix at Residues 31–42 of hTS Is Required for Degron Function
In earlier studies (26), we identified the N-terminal region of hTS as a degron by ligating amino acids 1–45, spanning the IDR (residues 1–28) and an hA (residues 31–42), to the N terminus of eGFP. CHX chase analysis indicated that this protein, termed N30-hA-eGFP, has a half-life of ∼9–10 h in transfected mammalian cells, as compared with >24 h for the parental eGFP reporter polypeptide (26). Deletion of the hA helix resulted in a molecule, N30-eGFP, that was ∼5-fold more stable than its parent, indicating that the helical segment is required for full activity (26). As shown in supplemental Fig. 1, treatment with the proteasome inhibitor MG132 causes stabilization of N30-hA-eGFP; in addition, no ubiquitinylated forms can be detected even under conditions where proteasome activity is impeded. Thus, degradation of the reporter substrate is, as expected, proteasome-mediated and ubiquitin-independent.
Our earlier studies did not determine whether or not the helix simply provides a scaffold-like extension function to the IDR, as opposed to playing a more specific structural role. To distinguish between these possibilities, we prepared a construct in which the hA segment of N30-hA-eGFP was replaced with a second copy of the IDR, producing two tandem copies of the IDR fused to the N terminus of eGFP. The resulting plasmid, termed N30-N30-eGFP was transfected into the TS-deficient Chinese hamster lung cell line RJK88.13 (26, 28); an E. coli TS gene contained within the construct was used to select stable transfectants, based upon their ability to grow in the absence of exogenous thymidine (see “Experimental Procedures” for details). Analysis of the intracellular degradation rate of the N30-N30-eGFP polypeptide showed it to have a half-life of >24 h, which is similar to the unfused eGFP reporter and significantly longer than that for N30-hA-eGFP (Fig. 1, A and C). Thus, a second copy of the IDR, which creates a long, 60-residue disordered domain, does not replace the hA helix in promoting degradation, indicating that the helical segment must provide more than a simple extension function in promoting full degron activity. Some aspect of its sequence and/or structure is likely to be critical.
FIGURE 1.
Role of helix hA in degron function. Plasmid constructs expressing native eGFP or various eGFP fusion proteins were stably transfected into cell line RJK88.13 and treated with CHX. Decay of protein expression over the indicated times was monitored by Western blotting (see “Experimental Procedures” for details). Schematic diagrams of the constructs are shown, along with representative blots. The half-lives (mean ± S.D.) are shown to the far right. A, constructs encoding eGFP, fusion protein N30-hA-eGFP, or fusion protein N30-N30-eGFP; B, constructs expressing fusion protein N30-hAP40/P41-eGFP or N30-hAP34/P35-eGFP; C and D, first order decay plots of the data in A and B, respectively, from which half-lives were determined. Error bars, S.D.
Perturbation of Helical Conformation Alters Degron Function
An important question relates to whether or not the secondary structure of the hA segment (i.e. its α-helical conformation) is required for degron function. To address this, we disrupted the helix via targeted introduction of proline residues. Proline is a well known “destabilizer” of helical structure and has been utilized to test the role of α-helices in protein function (32–34). Indeed, analysis of 1,131 α-helices in 205 globular proteins has revealed that proline residues are particularly disfavored at the C-terminal end of helical segments (33). We predicted, therefore, that Pro substitutions at the C-terminal end of the hA helix should result in profound destabilization of helical conformation.
We generated a derivative of N30-hA-eGFP, termed N30-hAP40/P41-eGFP, in which both Ile-40 and Leu-41 at the C-terminal end of the hA helix were converted to Pro. Two secondary structure prediction tools, Jpred (29) and PSIpred (30), were used to assess the potential impact of these substitutions. Both predicted an α-helical domain within the wild-type degron at residues 31–42, which is the precise location of the hA helix, as detected by x-ray crystallographic analysis (35, 36). In contrast, loss of helical conformation within the hA region was predicted for the I40P/L41P mutant. Thus, the structure of the hA helix is likely to be significantly disrupted in the N30-hAP40/P41-eGFP polypeptide. The half-life of this mutant was found to be ∼23 h, indicating it to be stable relative to its parent, N30-hA-eGFP (Fig. 1, B and D). This suggests that an α-helical conformation is necessary for full degron activity.
To verify this result, we generated a second mutant, N30-hAP34/P35-eGFP, containing Pro substitutions at positions 34 and 35 within the hA helix. Again, both Jpred and PSIpred predicted disruption of helical conformation within the mutant molecule's hA region. A half-life of ∼19 h was measured for this mutant, indicating it to be quite stable relative to N30-hA-eGFP (Fig. 1, B and D).
In sum, the observation that Pro substitutions within the hA segment result in stabilization of the reporter suggests that degron activity requires maintenance of helical propensity.
Impact of Amino Acid Substitutions on the Degradation Function of the hA Helix
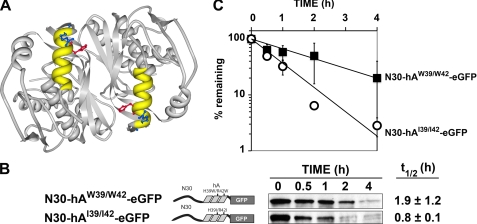
The amphipathic hA helix is highly conserved in mammals (23, 26). In the hTS dimer, the two helices lie along the edge of a cleft formed by the interface of the two subunits and are arranged in an antiparallel fashion relative to one another (Fig. 2A). Two charged amino acids near the C-terminal end of the helix (His-39 and Arg-42) face away from the body of the polypeptide into the space created by this cleft. To determine whether or not these residues contribute to degron function, we introduced tryptophan substitutions at both sites within N30-hA-eGFP. The resulting polypeptide, denoted N30-hAW39/W42-eGFP, exhibited a half-life of 1.9 h (Fig. 2, B and C), which is significantly reduced as compared with the 9.8-h half-life observed for N30-hA-eGFP (see Fig. 1). Similarly, substitution of isoleucine at these sites (N30-hAI39/I42-eGFP) also resulted in a very unstable molecule, having a half-life of <1 h (Fig. 2, B and C). Thus, hydrophobic residues at positions 39 and 42 result in marked destabilization of the reporter, indicating that the amino acids at these sites are important determinants of degradation-promoting activity. Apparently, the wild-type hTS degron, containing positively charged His and Arg at these positions, is less than maximally active.
FIGURE 2.
Impact of substitutions at residues 39 and 42 within the hA helix. A, three-dimensional structure of the hTS dimer, generated using Protein Workshop (31), is depicted. The two hA helices, shown in yellow, occur in an antiparallel configuration relative to one another; the side chains of His-39 and Arg-42, shown in red and blue, respectively, point into the cleft created by the interface of the two subunits. B, plasmids expressing fusion protein N30-hAW39/W42-eGFP or N30-hAI39/I42-eGFP were stably transfected into cell line RJK88.13 and treated with CHX. Decay of eGFP concentrations over the indicated times was monitored by Western blotting (see “Experimental Procedures” for details). Schematic diagrams of the constructs are shown, along with representative blots. The half-lives (mean ± S.D.) are shown to the far right. C, first order decay plots of the data in B, from which half-lives were determined. Error bars, S.D.
An IDR from Chicken Fibrinogen-α Mimics the Degradation Function of the N-Terminal IDR of hTS
The primary sequence of the IDR at the N-terminal end of hTS varies extensively among mammalian species (26). Despite this high degree of variation, the region in several species is still capable of contributing to degron function, so long as it has a free N-terminal end and an Arg-Arg motif at or near residues 10–11 (26, 27). Thus, a disordered character rather than a specific primary sequence may be a central feature of the role of the IDR in degradation. This leads to the prediction that disordered domains from distinct proteins might be capable of replacing the hTS IDR with respect to its role in degradation.
To test this notion, we introduced a distinct disordered domain, specifically residues 20–38 from the chicken fibrinogen-α polypeptide (37), into hTS in place of amino acids 7–29 of the endogenous IDR. The resulting polypeptide, termed FIB-hTS, contains the first six amino acids of hTS (1MPVAGS6), the disordered region of fibrinogen-α, and residues 30–313 of hTS. The six hTS-derived residues were retained at the N terminus to protect against N-α-acetylation, a process that can lead to degradation resistance (26).
FIB-hTS exhibited a half-life of 3.7 h (Fig. 3, A and C), which is similar to that for wild-type hTS (26, 27). In addition, as is also shown in Fig. 3, A and C, the molecule was stabilized by a Pro-2 Ala replacement within the hTS moiety at the N terminus; this substitution is known to promote N-α-acetylation and to stabilize wild-type hTS (25, 26). Thus, the IDR of fibrinogen-α is capable of promoting degradation of full-length hTS and operates by a similar mechanism as the native IDR within hTS.
FIGURE 3.
An intrinsically disordered region from the chicken fibrinogen-α polypeptide contributes to degron function. Plasmids in which an intrinsically disordered region (residues 20–38) from the chicken fibrinogen-α polypeptide was inserted in place of the hTS IDR were stably transfected into cell line RJK88.13 and treated with CHX. Decay of exogenous expression over the indicated times was monitored by Western blotting (see “Experimental Procedures” for details). Schematic diagrams of the constructs are shown, along with representative blots. The half-lives (mean ± S.D.) are shown to the far right. A, constructs encoding FIB-hTS or a P2A derivative; B, constructs expressing eGFP fusion protein FIB-hA-eGFP or FIB-eGFP; C and D, first order decay plots of the data in A and B, respectively, from which half-lives were determined. Error bars, S.D.
To show that the fibrinogen-α segment contributes to degron activity, we inserted it into N30-hA-eGFP in place of amino acids 7–29 of the hTS IDR. The polypeptide, denoted FIB-hA-GFP, contains the first six amino acids of hTS (1MPVAGS6), followed by residues 20–38 of fibrinogen-α and the hA helix, all appended to the N-terminal end of the eGFP reporter. Again, the six residues of hTS were retained at the N terminus to protect against the potentially stabilizing impact of N-α-acetylation. The protein exhibited a half-life of ∼5 h (Fig. 3, B and D), which is similar to N30-hA-eGFP (see Fig. 1). As expected, degradation of FIB-hA-eGFP is proteasome-mediated and ubiquitin-independent (see supplemental Fig. 1). Thus, the fibrinogen-α segment can replace the IDR of hTS in promoting degradation of the eGFP reporter.
In order to test the dependence of FIB-hA-eGFP degradation on the hA helix, we generated a polypeptide, FIB-eGFP, which lacks the hA helical segment. This protein exhibited a half-life of >24 h (Fig. 3, B and D), indicating it to be at least 5-fold more stable than FIB-hA-eGFP. In all, the results indicate that the IDR of fibrinogen-α, like that of hTS, cooperates with a helical motif in forming a functional degron.
The N-Terminal Domain of the E. coli lac Repressor Functions as a Degron Independently of the hA Helix
The highly mobile N-terminal “headpiece” domain of the E. coli lac repressor (residues 1–45) has been shown to provide a disordered region necessary for ubiquitin-directed proteasomal degradation (38). To test the ability of the domain to replace the IDR of hTS in driving degradation, we inserted it into hTS in place of amino acids 7–29 of the endogenous IDR. The encoded polypeptide, termed LAC-hTS, contains the first six amino acids of hTS (1MPVAGS6, to protect against N-α-acetylation) followed by residues 2–38 of the lac repressor and 30–313 of hTS. Degradation rate assays showed that LAC-hTS is a very unstable molecule, having a half-life of 0.56 h (Fig. 4, A and C). In addition, as is the case for wild-type hTS, it is stabilized by the acetylation-promoting Pro-2 Ala substitution within the small segment of hTS at its N-terminal end (Fig. 4, A and C). Thus, the lac repressor region effectively replaces the hTS IDR with regard to its ability to promote degradation. Data presented in supplemental Fig. 1 indicate that LAC-hA-eGFP degradation is proteasome-mediated and ubiquitin-independent.
FIGURE 4.
The N-terminal headpiece region of the lac repressor from E. coli functions as a degron. Plasmids containing the headpiece region (residues 2–38) of the E. coli lac repressor were stably transfected into cell line RJK88.13 and treated with CHX. Decay of protein expression was monitored over the indicated times by Western blotting (see “Experimental Procedures” for details). Schematic diagrams of the constructs are shown, along with representative blots. The half-lives (mean ± S.D.) are shown to the far right. A, constructs encoding LAC-hTS or a P2A mutant; B, constructs expressing eGFP fusion protein LAC-hA-eGFP or LAC-eGFP; C and D, first order decay plots of the data in A and B, respectively, from which half-lives were determined. Error bars, S.D.
To assess degron function, we inserted the repressor segment into N30-hA-eGFP in place of amino acids 7–29 of the hTS IDR. The encoded polypeptide, denoted LAC-hA-eGFP, contains the first six residues of hTS, followed by the repressor domain and the hA helix, all attached to the N-terminal end of the eGFP reporter. The molecule exhibited a very short half-life of 0.34 h (Fig. 4, B and D), indicating it to be significantly less stable than the native eGFP reporter. Thus, the lac repressor segment shows very strong degron activity, exceeding that of the hTS IDR.
To test whether or not degron activity exhibited by the repressor region depends upon the hA helix, we removed the hA segment. The resulting polypeptide, denoted LAC-eGFP, had a half-life of 1.0 h, which is slightly more stable than LAC-hA-eGFP (Fig. 4, B and D) but much less stable than eGFP (see Fig. 1). Thus, the lac repressor region exhibits strong degron activity even in the absence of the hA helix. The region acts on its own as a powerful degron independently of the hA helix.
The Helix-Loop-Helix Motif of the lac Repressor Contributes to Degron Function
Structural studies have shown that the headpiece region is not completely disordered but contains a helix-loop-helix motif (HLH) spanning residues 9–29 (39). To determine if this motif plays a role in degron function, we analyzed a mutant in which part of the region was deleted. We prepared a derivative of LAC-eGFP that is missing the first helix (residues 9–17) of its HLH domain. The encoded protein, denoted LACdel9–17-eGFP, exhibited a half-life of ∼10 h (Fig. 5, A and B), which is considerably longer than that for LAC-eGFP (see Fig. 4), indicating reduced degron activity. Introducing the hA helix into LACdel9–17-eGFP resulted in profound destabilization (LACdel9–17-hA-eGFP; Fig. 5, A and B). Thus, mutational alteration of the HLH motif within the lac repressor segment results in decreased degron function and dependence upon the hA helix. This suggests that the motif, or at least the first helix within it, provides a helical element necessary for degron function and explains the ability of the intact lac repressor region to promote degradation independently of the hA helix (see Fig. 4).
FIGURE 5.
Deletion of residues within the HLH motif of the lac repressor reduces degron function. A, plasmids expressing deletion mutants LACdel9–17-eGFP or LACdel9–17-hA-eGFP were stably transfected into cell line RJK88.13 and treated with CHX. Decay of eGFP concentrations over the indicated times was monitored by Western blotting (see “Experimental Procedures” for details). Schematic diagrams of the constructs are shown, along with representative blots. The half-lives (mean ± S.D.) are shown to the far right. B, first order decay plots of the data in A, from which half-lives were determined. Error bars S.D.
Degron Function Can Occur from either the N- or C-terminal End of the Target Polypeptide
The constructs examined in the preceding experiments contain degrons of interest at the N-terminal end of the eGFP reporter. It is pertinent to ask if these degrons function when appended to the C terminus. We therefore generated a polypeptide (eGFP-N30-hA) in which the hTS degron (i.e. residues 1–45) was attached to the C-terminal end of eGFP. The molecule exhibited a half-life of ∼7 h (Fig. 6, A and C), indicating that the degron is highly active at the C terminus. Furthermore, similar to what was observed when the degron was appended to the N terminus, proline substitutions at positions 40 and 41 within the hA segment caused loss of activity (eGFP-N30-hAP40/P41; Fig. 6, A and C), showing that a helical conformation is necessary. Finally, isoleucine substitutions at residues 39 and 42 within the hA helix resulted in profound destabilization of the protein (eGFP-N30-hAI39/I42; Fig. 6, A and C), again mimicking what is observed when the degron is at the N terminus.
FIGURE 6.
Degrons function at the C terminus of the substrate polypeptide. Constructs expressing eGFP fusion proteins containing degrons at their C termini were stably transfected into cell line RJK88.13 and treated with CHX. Decay of eGFP concentrations was monitored over the indicated times by Western blotting (see “Experimental Procedures” for details). Schematic diagrams of the constructs are shown, along with representative blots. The half-lives (mean ± S.D.) are shown to the far right. A, constructs encoding eGFP-N30-hA, eGFP-N30-hAP40/P41, or eGFP-N30-hAI39/I42; B, constructs expressing fusion protein eGFP-LAC; C and D, first order decay plots of the data in A and B, respectively, from which half-lives were determined. Error bars, S.D.
We tested whether or not the LAC degron is capable of functioning at the C-terminal end. A construct, denoted eGFP-LAC, was prepared containing the lac repressor domain appended to the C terminus of eGFP. The protein had a half-life of 2.1 h (Fig. 6, B and D), which is similar to that for LAC-eGFP, having the same degron at the N terminus (see Fig. 4).
Thus, both the hTS and LAC degrons are capable of exerting their degradation-promoting functions from either the N- or C-terminal ends of the target substrate. This indicates that the function of these domains is not terminus-dependent.
DISCUSSION
In the present work, we have extended earlier studies indicating that proteasomal degradation of hTS is mediated by an N-terminal degron composed of an IDR followed by an hA (24–27). Full degron activity requires both segments, indicating that they cooperate to promote degradation of the polypeptide to which they are attached. Importantly, we have shown that degron activity can be maintained by IDRs and helical domains derived from unrelated proteins, as long as both a disordered region and an α-helix are present. Thus, it is overall structure rather than a specific primary sequence that appears to predominate in degron function. Similar observations have recently been reported for the ubiquitin-independent degron of yeast ornithine decarboxylase (40).
The α-Helical Segment
The hA helix is an amphipathic segment that is highly conserved among a wide variety of prokaryotic and eukaryotic species (23, 26, 41). Results of the current study indicate that its degradative function is not mimicked by a second, tandem copy of the IDR (Fig. 1), indicating that it provides more than a simple extension of or scaffold for the IDR. Indeed, the observation that helix-destabilizing proline replacements result in complete loss of degradative function (Fig. 1) makes it apparent that an α-helical conformation is a prerequisite for degron activity.
Two positively charged residues within the hA helix (His-39 and Arg-42), which face into the cleft at the interface of the two subunits of the hTS dimer, play important roles in degron function. Introducing hydrophobic amino acid substitutions at both of these positions results in shortened half-lives (Fig. 2), indicating that one or both wild-type residues attenuate degron activity. The fact that His-39 and Arg-42 are highly conserved among mammalian TS molecules (23, 26) may signal the operation of selective constraints that have maintained optimal stability for hTS (i.e. about 6 h) within the cell.
The Intrinsically Disordered Domain
The IDR at the N terminus of hTS is an extended domain that is specific to mammalian TS. It is hypervariable in primary sequence among mammalian species, although its disordered nature, high Pro content, and unblocked N terminus have been conserved (26). Its function as a determinant of degradation requires that it have a free, unblocked N-terminal end, along with an Arg-Arg motif at residues 10–11 (26, 27). It is likely that the Arg-Arg dipeptide is involved in one or more postdocking steps involved in substrate interaction with the proteasome (27). Recent studies (42) have indicated that following binding of ubiquitinylated substrates to the proteasome, a second step that results in these substrates progressing to a more tightly bound state, occurs; this step requires both ATP and a loosely structured conformation. Perhaps the Arg-Arg motif within the hTS IDR is involved in formation of a tight binding complex between the hTS polypeptide and the proteasome. Further studies will be necessary to confirm this point.
In the present study, we have shown that disordered segments from unrelated proteins can replace the degron function of the native IDR within hTS. A disordered region from the chicken fibrinogen-α polypeptide behaves similarly to that from hTS in forming a degron that is dependent upon the hA helix (Fig. 3), again showing that both an IDR and an α-helix are necessary. The headpiece domain of the lac repressor of E. coli also has the ability to functionally replace the IDR of hTS (Fig. 4). However, in this case, activity is not dependent upon the presence of the hA helix; rather, an HLH segment at residues 9–29 supplies the required helical motif, as deduced from the observation that a deletion within the motif caused dramatic loss of degron activity and rendered it dependent upon the presence of the hA helix (Fig. 4). That the HLH plays a critical role is further supported by the finding that introduction of Arg substitutions at residues 11 and 14 within the first helix causes loss of degradation activity.4
We have not determined the role, if any, of the second helix of the HLH. The fact that deletion of the first helix results in such profound stabilization of the substrate makes it clear that the second helix is incapable on its own of supporting degradation. However, whether or not it contributes to degron function in the presence of the first helix remains to be tested.
In all, it would appear that any disordered segment of adequate length can function as part of a degron, as long as an appropriate α-helix is present nearby. Although the degradative function of the hA helix can be mimicked by a heterologous helical segment (such as that from the lac repressor), not all α-helices are active. Indeed, as mentioned above, the second helix of the HLH within the lac repressor has little or no activity in promoting degradation. Furthermore, replacing the IDR of hTS with a helix from the SUMO-conjugating protein UBC9 results in slight, if any, destabilization of the reporter,4 suggesting that this particular helix is essentially inactive. Thus, there does appear to be some level of specificity with regard to the ability of an α-helix to function as a degron constituent.
Several studies have shown that IDRs play critical roles in both ubiquitin-dependent and -independent degradation by the proteasome (11, 38). For example, the half-life of ornithine decarboxylase, a ubiquitin-independent substrate, is conferred by a degron that is located at the C-terminal end of the polypeptide, is intrinsically disordered, and mediates both proteasome recognition and chamber entry (11–13). Ubiquitin-independent degradation of the TP53 suppressor protein is governed by a Pro-rich disordered domain located near its N-terminal end (43). Finally, the cyclin-dependent kinase inhibitor p21Waf1/Cip1 is a loosely structured protein whose degradation is initiated via binding of its C-terminal end to the α7 subunit of the 20 S core complex (44). There is, therefore, ample precedent for an important role of disordered protein segments in proteasome-mediated degradation.
Mechanism of Degron Function
The currently accepted “two-component” model of proteasomal degradation posits two phases in degron function (45, 46): (i) proteasome recognition and binding and (ii) initiation of insertion into the proteolytic chamber. For ubiquitin-dependent degrons, the recognition/binding phase is accomplished by covalently attached polyubiquitin moieties that recognize one or more subunits of the proteasomal complex (46, 47). For ubiquitin-independent degradation, mechanisms that directly involve the target substrate's polypeptide chain are utilized (12, 18, 44). The initiation phase involves unfolding of the proteasome-bound substrate, “threading” of the substrate through the pore that leads to the proteolytic chamber, and entry into the chamber itself, where proteolysis ensues (11, 38, 46). These steps are typically mediated by disordered segments that are often (although not always) located at one or the other end of the protein (11, 38). Our observation that degradation requires cooperation between an α-helical motif and an intrinsically disordered region is consistent with this model. The α-helical component may govern proteasome recognition and binding, whereas the disordered region directs substrate unfolding and chamber entry. In this scheme, certain amino acids within each segment (e.g. Arg-10/Arg-11 within the IDR, His-39/Arg-42 within the hA helix) represent critical residues that mediate the functions of the respective segments.
A precedent for this scheme may be the prokaryotic ubiquitin-like protein (PUP) of actinobacteria. PUP is an intrinsically disordered protein with a length of 64 residues that, similar to ubiquitin, undergoes covalent ligation to lysine residues on target proteins, rendering them susceptible to degradation by a proteasome-like complex (48–50). Recent elegant structural analyses have shown that, upon ligation to a target substrate, PUP forms what is essentially a “two-component” degron; its C-terminal region binds regulatory subunits of the proteasome via formation of an helix, whereas the N terminus remains disordered and governs initiation of chamber entry (51, 52). Sequence alignments indicate that the helix-forming region of PUP is more conserved among bacterial species than is the disordered region (51).
Both the hTS and PUP degrons share the property that their C-terminal regions are α-helical and well conserved among species, whereas their N-terminal regions are disordered and hypervariable. The major distinction between the two degrons is that the α-helical element of hTS preexists, whereas that in PUP forms upon proteasomal docking. Further work will be necessary to determine if, in fact, the degrons of PUP and hTS operate in a similar fashion.
Finally, an interesting finding having implications for the mechanism by which the hTS degron operates is that it, as well as the LAC degron, functions at either end of the substrate polypeptide (Fig. 6). Thus, there appears to be little, if any, specificity for one or the other terminus. For hTS, this raises an interesting issue regarding the requirement that the IDR have a free N-terminal end (26). When the hTS degron is appended to the reporter's C terminus, the IDR becomes positioned between the hA helix and the body of the reporter and therefore lacks a free N-terminal end. The fact that the degron is functional although its N terminus is “blocked” implies a mechanism that is distinct from that operating from the substrate's other end. Although proteasomal degradation is commonly initiated from one or the other end of a target molecule, initiation via an endoproteolytic mechanism has been described in certain circumstances and can result in bidirectional proteolysis that progresses toward each terminus (38, 53). Whether or not this is the case for the hTS degron when located at the C-terminal end of a protein remains to be elucidated.
Supplementary Material
Acknowledgments
We thank Dr. Marj Peña for numerous discussions and insightful suggestions during the course of this work. We also thank Yang Yang Xing for technical help in conducting the experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant CA44013.
This paper is dedicated to Gabriela Garzon-Melo.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
S. Melo, unpublished results.
- TS
- thymidylate synthase
- hTS
- human thymidylate synthase
- CHX
- cycloheximide
- eGFP
- enhanced green fluorescent protein
- hA
- helix A
- IDR
- intrinsically disordered region
- HLH
- helix-loop-helix
- PUP
- prokaryotic ubiquitin-like protein.
REFERENCES
- 1. Ciechanover A. (1998) EMBO J. 17, 7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Glickman M. H., Ciechanover A. (2002) Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 3. Rechsteiner M., Hill C. P. (2005) Trends Cell Biol. 15, 27–33 [DOI] [PubMed] [Google Scholar]
- 4. Cheng Y. (2009) Curr. Opin. Struct. Biol. 19, 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Finley D. (2009) Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pickart C. M., Cohen R. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 177–187 [DOI] [PubMed] [Google Scholar]
- 7. Braun B. C., Glickman M., Kraft R., Dahlmann B., Kloetzel P. M., Finley D., Schmidt M. (1999) Nat. Cell Biol. 1, 221–226 [DOI] [PubMed] [Google Scholar]
- 8. Strickland E., Hakala K., Thomas P. J., DeMartino G. N. (2000) J. Biol. Chem. 275, 5565–5572 [DOI] [PubMed] [Google Scholar]
- 9. Jariel-Encontre I., Bossis G., Piechaczyk M. (2008) Biochim. Biophys. Acta 1786, 153–177 [DOI] [PubMed] [Google Scholar]
- 10. Orlowski M., Wilk S. (2003) Arch. Biochem. Biophys. 415, 1–5 [DOI] [PubMed] [Google Scholar]
- 11. Takeuchi J., Chen H., Coffino P. (2007) EMBO J. 26, 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takeuchi J., Chen H., Hoyt M. A., Coffino P. (2008) Biochem. J. 410, 401–407 [DOI] [PubMed] [Google Scholar]
- 13. Zhang M., Pickart C. M., Coffino P. (2003) EMBO J. 22, 1488–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Basbous J., Jariel-Encontre I., Gomard T., Bossis G., Piechaczyk M. (2008) Biochimie 90, 296–305 [DOI] [PubMed] [Google Scholar]
- 15. Bossis G., Ferrara P., Acquaviva C., Jariel-Encontre I., Piechaczyk M. (2003) Mol. Cell. Biol. 23, 7425–7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen X., Barton L. F., Chi Y., Clurman B. E., Roberts J. M. (2007) Mol. Cell 26, 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X., Amazit L., Long W., Lonard D. M., Monaco J. J., O'Malley B. W. (2007) Mol. Cell 26, 831–842 [DOI] [PubMed] [Google Scholar]
- 18. Yuksek K., Chen W. L., Chien D., Ou J. H. (2009) J. Virol. 83, 612–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mathes E., O'Dea E. L., Hoffmann A., Ghosh G. (2008) EMBO J. 27, 1357–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Truhlar S. M., Mathes E., Cervantes C. F., Ghosh G., Komives E. A. (2008) J. Mol. Biol. 380, 67–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baugh J. M., Viktorova E. G., Pilipenko E. V. (2009) J. Mol. Biol. 386, 814–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berger F. G., Berger S. H. (2006) Cancer Biol. Ther. 5, 1238–1241 [DOI] [PubMed] [Google Scholar]
- 23. Carreras C. W., Santi D. V. (1995) Annu. Rev. Biochem. 64, 721–762 [DOI] [PubMed] [Google Scholar]
- 24. Forsthoefel A. M., Peña M. M., Xing Y. Y., Rafique Z., Berger F. G. (2004) Biochemistry 43, 1972–1979 [DOI] [PubMed] [Google Scholar]
- 25. Peña M. M., Xing Y. Y., Koli S., Berger F. G. (2006) Biochem. J. 394, 355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peña M. M., Melo S. P., Xing Y. Y., White K., Barbour K. W., Berger F. G. (2009) J. Biol. Chem. 284, 31597–31607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Melo S. P., Yoshida A., Berger F. G. (2010) Biochem. J. 432, 217–226 [DOI] [PubMed] [Google Scholar]
- 28. Nussbaum R. L., Walmsley R. M., Lesko J. G., Airhart S. D., Ledbetter D. H. (1985) Am. J. Hum. Genet. 37, 1192–1205 [PMC free article] [PubMed] [Google Scholar]
- 29. Cole C., Barber J. D., Barton G. J. (2008) Nucleic Acids Res. 36, W197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bryson K., McGuffin L. J., Marsden R. L., Ward J. J., Sodhi J. S., Jones D. T. (2005) Nucleic Acids Res. 33, W36–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moreland J. L., Gramada A., Buzko O. V., Zhang Q., Bourne P. E. (2005) BMC Bioinformatics 6, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chou P. Y., Fasman G. D. (1978) Adv. Enzymol. Relat. Areas Mol. Biol. 47, 45–148 [DOI] [PubMed] [Google Scholar]
- 33. Kumar S., Bansal M. (1998) Proteins 31, 460–476 [DOI] [PubMed] [Google Scholar]
- 34. Mahalingam S., Khan S. A., Murali R., Jabbar M. A., Monken C. E., Collman R. G., Srinivasan A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 3794–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Phan J., Steadman D. J., Koli S., Ding W. C., Minor W., Dunlap R. B., Berger S. H., Lebioda L. (2001) J. Biol. Chem. 276, 14170–14177 [DOI] [PubMed] [Google Scholar]
- 36. Schiffer C. A., Clifton I. J., Davisson V. J., Santi D. V., Stroud R. M. (1995) Biochemistry 34, 16279–16287 [DOI] [PubMed] [Google Scholar]
- 37. Yang Z., Kollman J. M., Pandi L., Doolittle R. F. (2001) Biochemistry 40, 12515–12523 [DOI] [PubMed] [Google Scholar]
- 38. Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., Matouschek A. (2004) Nat. Struct. Mol. Biol. 11, 830–837 [DOI] [PubMed] [Google Scholar]
- 39. Slijper M., Bonvin A. M., Boelens R., Kaptein R. (1996) J. Mol. Biol. 259, 761–773 [DOI] [PubMed] [Google Scholar]
- 40. Gödderz D., Schäfer E., Palanimurugan R., Dohmen R. J. (2011) J. Mol. Biol. 407, 354–367 [DOI] [PubMed] [Google Scholar]
- 41. Hardy L. W., Finer-Moore J. S., Montfort W. R., Jones M. O., Santi D. V., Stroud R. M. (1987) Science 235, 448–455 [DOI] [PubMed] [Google Scholar]
- 42. Peth A., Uchiki T., Goldberg A. L. (2010) Mol. Cell 40, 671–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tsvetkov P., Reuven N., Prives C., Shaul Y. (2009) J. Biol. Chem. 284, 26234–26242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Touitou R., Richardson J., Bose S., Nakanishi M., Rivett J., Allday M. J. (2001) EMBO J. 20, 2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Inobe T., Fishbain S., Prakash S., Matouschek A. (2011) Nat. Chem. Biol. 7, 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schrader E. K., Harstad K. G., Matouschek A. (2009) Nat. Chem. Biol. 5, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Elsasser S., Finley D. (2005) Nat. Cell Biol. 7, 742–749 [DOI] [PubMed] [Google Scholar]
- 48. Burns K. E., Darwin K. H. (2010) Subcell. Biochem. 54, 149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen X., Solomon W. C., Kang Y., Cerda-Maira F., Darwin K. H., Walters K. J. (2009) J. Mol. Biol. 392, 208–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liao S., Shang Q., Zhang X., Zhang J., Xu C., Tu X. (2009) Biochem. J. 422, 207–215 [DOI] [PubMed] [Google Scholar]
- 51. Sutter M., Striebel F., Damberger F. F., Allain F. H., Weber-Ban E. (2009) FEBS Lett. 583, 3151–3157 [DOI] [PubMed] [Google Scholar]
- 52. Wang T., Darwin K. H., Li H. (2010) Nat. Struct. Mol. Biol. 17, 1352–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Piwko W., Jentsch S. (2006) Nat. Struct. Mol. Biol. 13, 691–697 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.