Abstract
NHR-23, a conserved member of the nuclear receptor family of transcription factors, is required for normal development in C. elegans where it plays a critical role in growth and molting. In a search for NHR-23 dependent genes, we performed whole genome comparative expression microarrays on both control and nhr-23 inhibited synchronized larvae. Genes that decreased in response to nhr-23 RNAi included several collagen genes. Unexpectedly, several hedgehog-related genes were also down-regulated after nhr-23 RNAi. A homozygous nhr-23 deletion allele was used to confirm the RNAi knockdown phenotypes and the changes in gene expression. Our results indicate that NHR-23 is a critical co-regulator of functionally linked genes involved in growth and molting and reveal evolutionary parallels among the ecdysozoa.
Keywords: Nuclear hormone receptor, Caenorhabditis elegans, NHR-23, transcription, gene expression, development, hedgehog, molting, ROR
1. Introduction
Nuclear hormone receptors (NHRs) are transcription factors regulating metabolism and development in all metazoan species studied to date. NHR-dependent pathways include those involved in developmental patterning and timing. In insects, transitions through developmental stages are initiated by pulses of the steroid hormone 20-hydroxyecdysone (Ecdysone, 20E) that regulates ecdysis, the periodic exchange of exoskeleton that occurs during larval development [1–7]. Ecdysis, or molting, is a common mechanism used by insects, nematodes and other bilateral species that allows for growth and progression from larval stages to adult forms and is a defining feature of the clade ecdysozoa [8]. In Drosophila, pulses of ecdysone result in increased expression of early hormone response genes, including those encoding the ecdysone receptor (ECR) itself and two other nuclear receptors, DHR3 and E75B [2, 9]. ECR, a conserved member of the NHR family with greatest similarity to farnesyl X receptors (FXR, NR1H4), forms a heterodimer with Ultraspiracle (USP), the insect homologue of Retinoid X receptors (RXRs) [10, 11]. This pathway serves as a paradigm of developmental regulation by NHRs and their sterol ligands.
In the nematode C. elegans, molting and larval development are regulated by NHR-23, a close homologue of Drosophila DHR3 [12, 13]. Expression of nhr-23 oscillates, reaching peaks during intermolt periods and decreasing just prior to molting [13]. This timing roughly coincides with entry into lethargus, an approximately two-hour period during which movement and pharyngeal pumping decrease. The wave like pattern of nhr-23 expression during the third and fourth larval stages (L3/L4) is, in part, regulated by the micro RNAs (miRNAs) let-7 and miR-84 [14]. Moreover, the regulatory RNA let-7 is controlled by DAF-12, the nematode homologue of the vitamin D receptor [15] that in turn is regulated by the sterol dafachronic acid. Unlike Drosophila, C. elegans does not synthesize cholesterol, so DAF-12-mediated signaling and many other events regulating molting depend on exogenous sterols [16]. These nematode pathways illustrate that many upstream components and mechanisms for the regulation for ecdysis, including the critical involvement of NHRs and sterol ligands, are evolutionarily conserved among ecdysozoa.
To determine if other elements of ecdysis-related events were similarly conserved, we searched in C. elegans for genes acting downstream of NHR-23 using genome wide expression microarrays. Synchronized L2 larvae treated with nhr-23 RNAi by feeding from hatching were used as a source of mRNA for profiling. Among the potential NHR-23 target genes were those that were expected, including those encoding collagens, a principle component of the cuticle. However, we also identified several genes encoding hedgehog family-related proteins. Although, hedgehog-related proteins were previously identified as regulators of growth and molting [17–24], our results reveal the first clues about their co-regulation with collagens and their dependence on NHR-23. NHR-23 is likely a key positive regulator of molting and growth, orchestrating the expression of functionally linked epidermal genes in the nematode.
2. Materials and methods
2.1. C. elegans strains
Wild type (N2) Caenorhabditis elegans were obtained from the C. elegans Stock Center and maintained as described [25]. The mutant strain nhr-23(tm1323) was obtained from the National Bioresource Project, Tokyo, Japan. The mutant was backcrossed three times with N2 animals. The nhr-23(tm1323) deletion allele was confirmed by sequencing and the strain maintained as a heterozygote by scoring progeny mutant phenotypes.
2.2. RNA preparation
Synchronized populations of L1 larvae were plated with two sets of HT115 bacteria, one that had been transformed with the RNAi vector only (L4440 plasmid) and another that had been transformed with a vector targeting nhr-23 (clone 5174) [13]). Worms were kept on 2% agarose plates for 21 hr at 20°C, collected, and approximately 200µl of worms resuspended in PBS were used in each individual experiment. Total RNA was isolated from frozen pellets using a Mixer-Mill (Miller-Mill 300) following an RNeasy Mini Kit (Qiagen, Germantown, MD) according to manufacturer protocol. Aliquots of cultures used for RNA isolation were kept on nhr-23 RNAi plates to confirm the knockdown phenotypic changes occurred during subsequent molts.
2.3. Analysis of microarray results
C. elegans whole genome expression microarrays (Affymetrix, Santa Clara, CA) were used to profile gene expression from three independent, biological replicates for both experimental and control samples with all samples processed simultaneously. Microarray chip data was collected and analyzed by both Affymetrix MAS 5.0 suite software (≥1.6-fold change in mRNA expression) and Robust Multichip Average (RMA) (≥1.2-fold change in mRNA expression) as part of the Partek genomics suite software package, all with a p-value less than or equal to 0.05. Normalized data was further analyzed and visualized with Genespring software (Agilent Technologies, Santa Clara CA).
2.4. RT-qPCR
cDNA was prepared from 3µg of total RNA that was isolated as described above. Reverse transcription (RT) was performed as previously described [26] and quantitative PCR (qPCR) was performed using the Roche Universal Probe Library technique (Hoffmann-La Roche, Basel, Switzerland) [26]. Primers are given in Supplementary Table S1 and these primer sets interrogated the following transcripts: dpy-2, dpy-3, dpy-7, dpy-8, wrt-1, wrt-2, wrt-4. All samples were normalized against ama-1, the large subunit of RNA polymerase II as previously described [26].
3. Results
3.1. Identification of NHR-23 dependent genes
The microarray experiment evaluated 22,625 probe sets on the C. elegans whole genome expression arrays (Affymetrix) with triplicate RNA samples generated from wild type or an age-matched sample of nhr-23 RNAi treated animals. From these, 331 probes were decreased in at least two out of three RNAi experiments including 90 probes that were decreased in all three RNAi experiments. Probe sets that showed decreased values in two RNAi experiments but increased values in the third RNAi experiment were not evaluated further. The resulting 266 unique down regulated genes were identified based gene annotations in WS190 (Supplementary Table S2). Gene ontology analysis using the DAVID tool [27, 28] identified 10 clusters with enrichment scores greater than 2 with the main GO terms including molting cycle, collagen and cuticle genes, regulation of growth, and development. Genes annotated as constituents of hedgehog and sterol sensing signaling pathways were identified separately as significantly enriched GO clusters (Supplementary Table S3). A similar analysis of nhr-23 RNAi up-regulated transcripts yielded only 36 genes (not shown) that were grouped using DAVID software in only two GO clusters, both with enrichment scores less than 2.
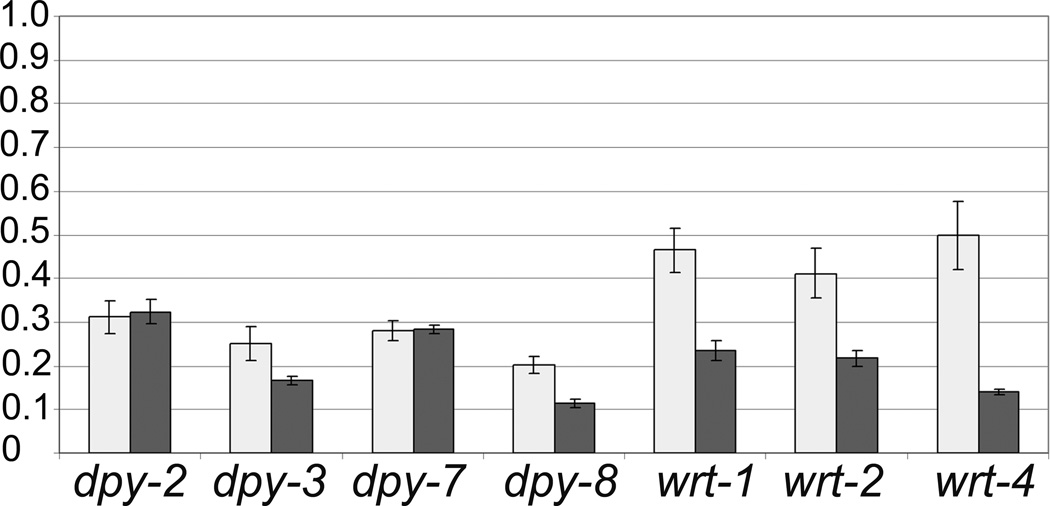
The list of nhr-23 RNAi down-regulated genes included four wrt genes, three grd genes, four grl genes, five ptr genes and the genes ptc-3 and qua-1. Additional genes known to be involved in molting regulation were also identified as NHR-23 dependent genes including mlt-8, mlt-9, mlt-10 and mlt-11. Interestingly, several collagen genes that are known to be co-expressed showed a dependence on NHR-23, including dpy-2, dpy-3, dpy-7, dpy-8, dpy-10, as did the dpy-5 gene. Finally, the hedgehog-related genes wrt-1, wrt-2 and wrt-4, which result in a Molt phenotype when mutated, were among those dependent on NHR-23. To validate the expression array data, selected collagen and hedgehog-related genes were analyzed by RT-qPCR in independent nhr-23 RNAi experiments. As shown in Fig. 1, these assays done at 21hr and 24hr of larval development confirmed that these genes were indeed down-regulated when NHR-23 activity was decreased by RNAi feeding. Thus, our expression array analysis of genes down-regulated by nhr-23 RNAi identified both known and unexpected genes involved in molting as downstream targets of NHR-23 regulation.
Figure 1.
Analysis of the expression of selected collagen and hedgehog-related genes by RT-qPCR at 21 hours (grey columns) and 24 hours (dark columns) of larval development. Mean values of quadruplicates (21 hours) and triplicates (24 hours) are expressed as the ratio of the normalized expression values relative to the ama-1 gene in RNAi inhibited cultures compared to controls. SD are indicated by vertical bars.
Previously we have shown that NHR-23 binds the core DNA sequence AGGTCA and we provided evidence that NHR-23 likely functions as a transcriptional activator [12]. We analyzed the putative promoter regions, defined as 2000 bp upstream of the start codon, of the putative NHR-23 target genes encoding collagens and hedgehog-related factors that were down-regulated in response to nhr-23 RNAi. Specifically, we looked for the NHR-23 binding site sequence, which is identical the ROR alpha site from mammals, that consists of the half-site PuGGTCA preceded by the AT-rich consensus sequence (A/G/T)(T/A)(A/T)(T/A)C(A/T) [29]. The program TFsearch [30] that is based on the Transfac database [31] identified monomeric NHR-23/ROR alpha binding sites in dpy-2, dpy-3, dpy-7, dpy-8, dpy-10, wrt-1, wrt-2 and wrt-4. To determine if these binding sites were evolutionarily conserved between multiple nematode species, we used the tool EvoPrinterHD [32]. The NHR-23/ROR alpha sites for all of these genes were conserved in five nematode species (C. elegans, C. briggsae, C. brenneri, C. remanei and C. japonica) with the exception of dpy-10, which is conserved only in four species (not in C. japonica) and wrt-4, which contains the site only in C. elegans (Fig. 2). Analysis of all promoter regions from genes down-regulated in nhr-23 RNAi experiment identified potential NHR-23/ROR alpha binding sites in 150 of the 266 genes (56 %). A similar analysis of the promoters for the up-regulated genes following nhr-23 RNAi identified 12 of 36 total genes (33%) with the NHR-23/ROR alpha site. Of these, two genes had conserved binding sites in all five nematode species (F53B3.5 and ZK1290.11) and one (alh-9) is partially conserved (one base is not conserved). As a control for this analysis, we chose two sets of genes. One set consisted of 100 genes unrelated to molting that showed no change in expression in our analysis. The other was a set of 151 collagen genes that also showed no significant change in expression in our microarray experiment. The control set of 100 genes unrelated to molting had the NHR-23/ROR site in 33% of the promoters compared to 38% among the 151 unchanged collagen genes. We concluded that the promoters associated with genes down-regulated after nhr-23 RNAi had a significantly (p=0.0003) higher frequency of potential NHR-23/ROR alpha binding sites than control sets and that these sites were most often evolutionarily conserved.
Figure 2.
Analysis of predicted promoters of selected genes. The program TFSearch [30] identified monomeric ROR alpha response elements (grey shading) in promoters of the genes indicated to the left. Analysis with the program EvoprinterHD [32] was used to determine the conservation of the NHR-23/ROR alpha sites and flanking regions across five nematode species (see text). Conserved bases in five out of five nematode species, indicated by capital letters, reveal extensive sequence conservation within and around the putative NHR-23/ROR alpha binding sites.
3.2. Validation of expression data in the nhr-23(tm1323) deletion mutant
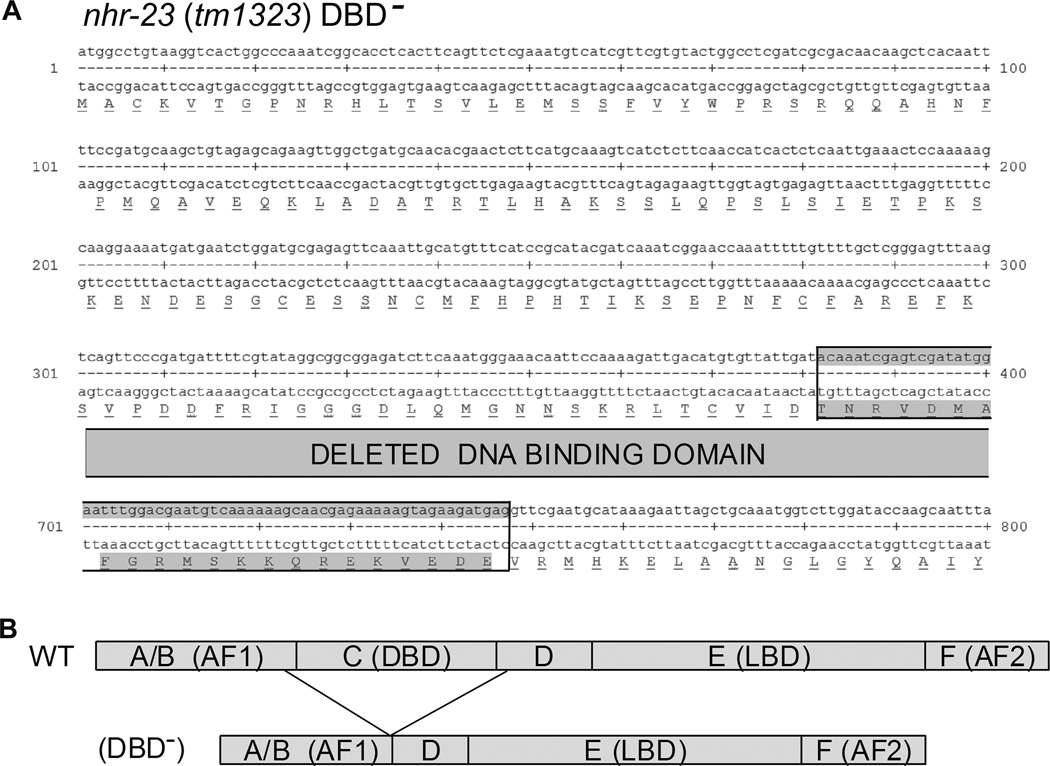
To further confirm that changes in gene expression were dependent on NHR-23, we turned to a mutant strain harboring the nhr-23 deletion allele tm1323 kindly provided by the National Bioresource Project, Tokyo, Japan. As reported, and confirmed by our independent sequencing, this mutant allele has an in-frame deletion that eliminates the coding region for the central part of the NHR-23 resulting in protein product lacking the entire DNA binding domain and a small portion of the adjacent parts of the A/B and hinge regions (Fig. 3). This allele was out-crossed three times with wild type animals prior to further characterization and Western blot analysis confirmed production of the predicted mutant protein product (data not shown).
Figure 3.
Schematic representation of the nhr-23(tm1323) allele. A – The deletion extending from exons 3 to 5 creates a novel exon and results in an encoded protein that lacks the complete DNA binding domain and small adjacent portions of domains A/B and D. B – Schematic representation of the wild type (WT) and mutant NHR-23 (tm1323) DBD−.
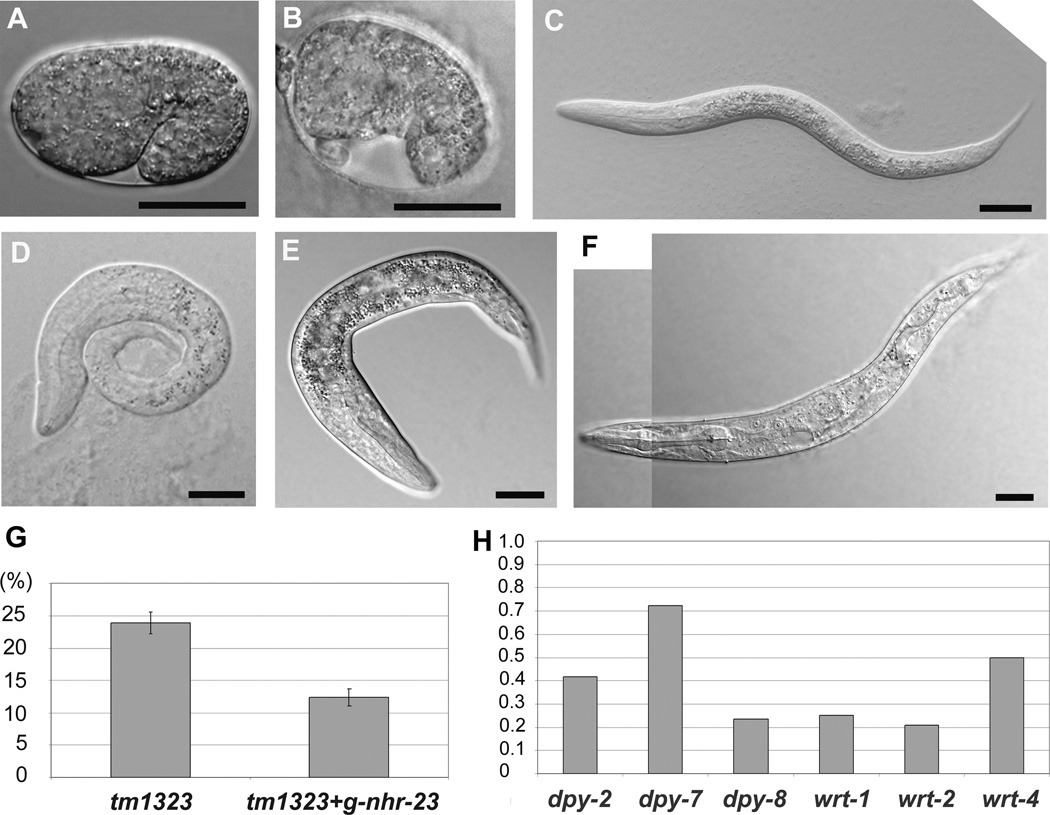
We further characterized the nhr-23(tm1323) mutant because little was previously known other than it was homozygous inviable with larval arrest. We found that almost all homozygous mutants die during the embryonic or L1 larval stages, with a very small proportion of homozygous mutant larvae developing into the L2 stage prior to arrest (n= 2678, affected progeny 23.74% SD=2.07, embryonic lethality 11.37% SD=3.15, L1 arrest 8.52% SD=3, L2 arrest 3.54% SD=1.96). As we showed previously by nhr-23 RNAi [12, 13], homozygous nhr-23(tm1323) embryos had a range of phenotypes with arrest and death from the comma stage to 3-fold stage with severe morphological defects common. Arrested homozygous L1 larvae also had variable phenotypes, but often included a Dpy phenotype and/or variable bulges and constrictions along the length of the body (Fig. 4). The small number of homozygous larvae that reached the L2 stage also had a severe Dpy phenotype (Fig. 4). To confirm that these phenotypes were indeed due to the loss of NHR-23 activity, heterozygote hermaphrodites were injected with an amplified genomic sequence containing the wild type nhr-23 gene (6,253 bp) in a rescue assay commonly used [33–36]. We observed rescue of the embryonic and early larval lethality in approximately 50% of offspring (Fig. 4 G), demonstrating that the mutant phenotypes described were due to loss of NHR-23 activity.
Figure 4.
Phenotypic characterization of homozygous nhr-23(tm1323) mutants. A – A wild type embryo at the 1.5 fold stage of development. B – An embryo arrested at the comma stage shows the abnormal positions for cells on the ventral side of the embryo and other morphological defects. C – Wild type L1 larva. D to F – nhr-23(tm1323) mutant larvae. Panel D shows the mutant L1 Dpy phenotype and irregular body shape with constrictions and bulges. Panel E shows an L2 larva with a Dpy phenotype, vacuoles in the head and irregular bulges. Panel F shows an L2 larva at the arrest point with numerous vacuoles throughout the body. Bars represent 20 µm. G – Rescue of the mutant lethality with a wild type transgene. Transgenic animals had a significantly decreased penetrance of nhr-23 loss-of-function phenotypes scored as the percentage of L1/L2 arrest (n = 2307) compared to non-rescued controls (n = 6,125). Mean value of the percent affected progeny and SD are indicated, P < 0.0001. H – RT-qPCR of dpy-2, dpy-7, dpy-8, wrt-1, wrt-2 and wrt-4 expression in 200 manually selected mutated or control larvae. Results are shown as the ratio of the normalized expression values relative to the ama-1 gene in nhr-23 (tm1323) mutant larvae compared to controls.
To validate changes in gene expression resulting from nhr-23 RNAi, we analyzed transcript abundance in homozygous nhr-23(tm1323) animals compared to wild type controls. This analysis was limited to only a few genes because of the difficulty of isolating large populations of homozygous progeny. Therefore, we decided to focus on a few of the hedgehog-related genes as their deregulation as assayed by microarrays was unanticipated. From nhr-23(tm1323) heterozygote parental animals, we picked separately 200 L1 animals displaying either a wild type or Dpy phenotype, isolated total RNA, and assayed gene expression by RT-qPCR. Whereas mRNA corresponding to genes wrt-1, wrt-2 and wrt-4 was easily detectable in animals with a wild type phenotype, these genes were decreased or undetectable in the Dpy animals (Fig. 4 H) relative to the ama-1 positive control. Thus, although very limited, our analysis of gene expression in homozygous nhr-23 mutants was consistent with whole genome expression array results following nhr-23 RNAi.
4. Discussion
C. elegans NHRs form a very large family of transcription factors encoded by approximately 300 genes. This gene family includes a small set (< 20) that is conserved between various animal phyla and a large set (>250) that are likely products of intensive multiplication of an ancestor gene related to hepatocyte nuclear factor 4 (HNF4). NHR-23 is a member of the small set of conserved NHRs and shares many functional similarities with its Drosophila homolog (DHR3), including the regulation of molting and ecdysis in the nematode and fly, respectively. Our current work identifies several genes dependent on NHR-23 for proper regulation that extends this evolutionarily conserved pathway to the hedgehog-related genes, further underscoring the ancient nature of this growth and developmental regulatory module.
NHR-23 appears to be a powerful regulator of genes required for embryonic and larval development. RNA interference applied at various stages of C. elegans development reveals the critical requirement of NHR-23 for late embryogenesis, growth and molting during all four larval stages [12, 13]. The morphological characterization of a putative null allele of nhr-23 in the present study agrees well with the previously reported effects of nhr-23 RNAi in the embryonic and L1 larval stages and together demonstrate the essential role for this factor. Thus, the identification of NHR-23 target genes is important for providing mechanistic insight into the function of this conserved regulatory pathway. One such insight comes from the co-regulation of functionally linked collagen genes that has been studied previously in some detail [37]. Our current work strongly suggests that NHR-23 may be a critical master regulator that orchestrates the expression of such specific gene groups throughout development. Presumably NHR-23 is acting in concert with other transcription factors that dictate which subset of NHR-23 targets are activated during specific molting cycles.
Another insight from our work is the connection between NHR-23, molting, and the hedgehog-related genes. Hedgehog signaling is an ancient metazoan pathway that employs sterols as structural molecules and sterol transport for signaling mechanisms in development [23]. Nematodes have orthologues of many hedgehog signaling proteins (encoded by wrt, grd, grl, and qua genes), and sterol-sensing receptors homologous to Patched proteins (the ptr genes) [21] while some proteins of the hedgehog signaling pathway seem to be absent. Decreased activity of many of the C. elegans hedgehog-related genes result in growth and molting phenotypes as does sterol restriction [18–20, 24]. Our current work provides a strong connection between sterol signaling, the hedgehog-related pathways and NHR-23, thereby linking the signaling and structural components of molting with a common and evolutionarily conserved NHR regulator.
Regulation of molting by NHR-23 in C. elegans shares several features with its vertebrate homologues, the RORs. In addition to their conserved DNA response element sequence and predominant function as transcriptional activators [12], there appears to be conservation of downstream target genes. For example, hedgehog signaling was found in the ROR alpha regulatory pathway [38] and our current works links hedgehog-related pathways to NHR-23. Interestingly, hedgehog signaling is also involved in the execution of tissue specific developmental regulation in amphibian metamorphosis, perhaps reflecting a distant evolutionary link to ecdysis and molting in flies and nematodes, respectively. Once again we see that there is a limited repertoire of molecular mechanisms regulating signaling and morphogenesis that have evolved to control a variety of growth-related processes in animals. Understanding the details of these mechanisms in each model system will, therefore, shed light on all.
Highlights.
> NHR-23 is a critical regulator of nematode development and molting. > The manuscript characterizes the loss-of-function phenotype of an nhr-23 mutant. > Whole genome expression analysis identifies new potential targets of NHR-23. > Hedgehog-related genes are identified as NHR-23 dependent genes. > New link between sterol mediated signaling and regulation by NHR-23 is found.
Supplementary Material
Acknowledgements
We thank Dr. A. Fire for vector L4440 and HT115 cells host. This work was supported by Grant 0021620806 from the Ministry of Education, Youth and Sports of the Czech Republic, and Grant 304/08/0970, from the Czech Science Foundation. NAK and JN were supported by the grant SVV262502 from the Charles University in Prague. MWK is supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the National Institutes of Health, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huet F, Ruiz C, Richards G. Sequential gene activation by ecdysone in Drosophila melanogaster: the hierarchical equivalence of early and early late genes. Development. 1995;121:1195–1204. doi: 10.1242/dev.121.4.1195. [DOI] [PubMed] [Google Scholar]
- 2.Lam G, Hall BL, Bender M, Thummel CS. DHR3 is required for the prepupalpupal transition and differentiation of adult structures during Drosophila metamorphosis. Dev Biol. 1999;212:204–216. doi: 10.1006/dbio.1999.9343. [DOI] [PubMed] [Google Scholar]
- 3.Schubiger M, Carre C, Antoniewski C, Truman JW. Ligand-dependent derepression via EcR/USP acts as a gate to coordinate the differentiation of sensory neurons in the Drosophila wing. Development. 2005;132:5239–5248. doi: 10.1242/dev.02093. [DOI] [PubMed] [Google Scholar]
- 4.Schubiger M, Tomita S, Sung C, Robinow S, Truman JW. Isoform specific control of gene activity in vivo by the Drosophila ecdysone receptor. Mech Dev. 2003;120:909–918. doi: 10.1016/s0925-4773(03)00134-5. [DOI] [PubMed] [Google Scholar]
- 5.Ghbeish N, Tsai CC, Schubiger M, Zhou JY, Evans RM, McKeown M. The dual role of ultraspiracle, the Drosophila retinoid X receptor, in the ecdysone response. Proc Natl Acad Sci U S A. 2001;98:3867–3872. doi: 10.1073/pnas.061437798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schubiger M, Truman JW. The RXR ortholog USP suppresses early metamorphic processes in Drosophila in the absence of ecdysteroids. Development. 2000;127:1151–1159. doi: 10.1242/dev.127.6.1151. [DOI] [PubMed] [Google Scholar]
- 7.Schubiger M, Wade AA, Carney GE, Truman JW, Bender M. Drosophila EcRB ecdysone receptor isoforms are required for larval molting and for neuron remodeling during metamorphosis. Development. 1998;125:2053–2062. doi: 10.1242/dev.125.11.2053. [DOI] [PubMed] [Google Scholar]
- 8.Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, Sorensen MV, Haddock SH, Schmidt-Rhaesa A, Okusu A, Kristensen RM, Wheeler WC, Martindale MQ, Giribet G. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 9.King-Jones K, Thummel CS. Nuclear receptors--a perspective from Drosophila. Nat Rev Genet. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- 10.Carney GE, Wade AA, Sapra R, Goldstein ES, Bender M. DHR3, an ecdysone-inducible early-late gene encoding a Drosophila nuclear receptor, is required for embryogenesis. Proc Natl Acad Sci U S A. 1997;94:12024–12029. doi: 10.1073/pnas.94.22.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam GT, Jiang C, Thummel CS. Coordination of larval and prepupal gene expression by the DHR3 orphan receptor during Drosophila metamorphosis. Development. 1997;124:1757–1769. doi: 10.1242/dev.124.9.1757. [DOI] [PubMed] [Google Scholar]
- 12.Kostrouchova M, Krause M, Kostrouch Z, Rall JE. CHR3: a Caenorhabditis elegans orphan nuclear hormone receptor required for proper epidermal development and molting. Development. 1998;125:1617–1626. doi: 10.1242/dev.125.9.1617. [DOI] [PubMed] [Google Scholar]
- 13.Kostrouchova M, Krause M, Kostrouch Z, Rall JE. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2001;98:7360–7365. doi: 10.1073/pnas.131171898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayes GD, Frand AR, Ruvkun G. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development. 2006;133:4631–4641. doi: 10.1242/dev.02655. [DOI] [PubMed] [Google Scholar]
- 15.Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yochem J, Tuck S, Greenwald I, Han M. A gp330/megalin-related protein is required in the major epidermis of Caenorhabditis elegans for completion of molting. Development. 1999;126:597–606. doi: 10.1242/dev.126.3.597. [DOI] [PubMed] [Google Scholar]
- 17.Aspock G, Kagoshima H, Niklaus G, Burglin TR. Caenorhabditis elegans has scores of hedgehog-related genes: sequence and expression analysis. Genome Res. 1999;9:909–923. doi: 10.1101/gr.9.10.909. [DOI] [PubMed] [Google Scholar]
- 18.Hao L, Aspock G, Burglin TR. The hedgehog-related gene wrt-5 is essential for hypodermal development in Caenorhabditis elegans. Dev Biol. 2006;290:323–336. doi: 10.1016/j.ydbio.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 19.Hao L, Johnsen R, Lauter G, Baillie D, Burglin TR. Comprehensive analysis of gene expression patterns of hedgehog-related genes. BMC Genomics. 2006;7:280. doi: 10.1186/1471-2164-7-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao L, Mukherjee K, Liegeois S, Baillie D, Labouesse M, Burglin TR. The hedgehog-related gene qua-1 is required for molting in Caenorhabditis elegans. Dev Dyn. 2006;235:1469–1481. doi: 10.1002/dvdy.20721. [DOI] [PubMed] [Google Scholar]
- 21.Burglin TR, Kuwabara PE. Homologs of the Hh signalling network in C. elegans. WormBook. 2006:1–14. doi: 10.1895/wormbook.1.76.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burglin TR. The Hedgehog protein family. Genome Biol. 2008;9:241. doi: 10.1186/gb-2008-9-11-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burglin TR. Evolution of hedgehog and hedgehog-related genes, their origin from Hog proteins in ancestral eukaryotes and discovery of a novel Hint motif. BMC Genomics. 2008;9:127. doi: 10.1186/1471-2164-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zugasti O, Rajan J, Kuwabara PE. The function and expansion of the Patched-and Hedgehog-related homologs in C. elegans. Genome Res. 2005;15:1402–1410. doi: 10.1101/gr.3935405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vohanka J, Simeckova K, Machalova E, Behensky F, Krause MW, Kostrouch Z, Kostrouchova M. Diversification of fasting regulated transcription in a cluster of duplicated nuclear hormone receptors in C. elegans. Gene Expr Patterns. 2010;10:227–236. doi: 10.1016/j.gep.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang D, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giguere V, Tini M, Flock G, Ong E, Evans RM, Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8:538–553. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- 30.Akiyama Y. TFSEARCH: Searching Transcription Factor Binding Sites, based on the TRANSFAC databases (ref. 33) [August 5, 2011];1998 Date of the last accession http://www.cbrc.jp/research/db/TFSEARCH.html.
- 31.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odenwald WF, Rasband W, Kuzin A, Brody T. EVOPRINTER, a multigenomic comparative tool for rapid identification of functionally important DNA. Proc Natl Acad Sci U S A. 2005;102:14700–14705. doi: 10.1073/pnas.0506915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maryon EB, Coronado R, Anderson P. unc-68 encodes a ryanodine receptor involved in regulating C. elegans body-wall muscle contraction. J Cell Biol. 1996;134:885–893. doi: 10.1083/jcb.134.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maryon EB, Saari B, Anderson P. Muscle-specific functions of ryanodine receptor channels in Caenorhabditis elegans. J Cell Sci. 1998;111(Pt 19):2885–2895. doi: 10.1242/jcs.111.19.2885. [DOI] [PubMed] [Google Scholar]
- 35.Mercer KB, Flaherty DB, Miller RK, Qadota H, Tinley TL, Moerman DG, Benian GM. Caenorhabditis elegans UNC-98, a C2H2 Zn finger protein, is a novel partner of UNC-97/PINCH in muscle adhesion complexes. Mol Biol Cell. 2003;14:2492–2507. doi: 10.1091/mbc.E02-10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe N, Nagamatsu Y, Gengyo-Ando K, Mitani S, Ohshima Y. Control of body size by SMA-5, a homolog of MAP kinase BMK1/ERK5, in C. elegans. Development. 2005;132:3175–3184. doi: 10.1242/dev.01895. [DOI] [PubMed] [Google Scholar]
- 37.McMahon L, Muriel JM, Roberts B, Quinn M, Johnstone IL. Two sets of interacting collagens form functionally distinct substructures within a Caenorhabditis elegans extracellular matrix. Mol Biol Cell. 2003;14:1366–1378. doi: 10.1091/mbc.E02-08-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gold DA, Baek SH, Schork NJ, Rose DW, Larsen DD, Sachs BD, Rosenfeld MG, Hamilton BA. RORalpha coordinates reciprocal signaling in cerebellar development through sonic hedgehog and calcium-dependent pathways. Neuron. 2003;40:1119–1131. doi: 10.1016/s0896-6273(03)00769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.