Abstract
The pseudorabies virus (PRV) early protein UL54 is a homologue of herpes simplex virus 1 (HSV-1) immediate-early protein ICP27, which is a multifunctional protein that is essential for HSV-1 infection. In this study, the subcellular localization and nuclear import signals of PRV UL54 were characterized. UL54 was shown to predominantly localize to the nucleolus in transfected cells. By constructing a series of mutants, a functional nuclear localization signal (NLS) and a genuine nucleolar localization signal (NoLS) of UL54 were for the first time identified and mapped to amino acids 61RQRRR65 and 45RRRRGGRGGRAAR57, respectively. Additionally, three recombinant viruses with mutations of the NLS and/or the NoLS in UL54 were constructed based on PRV bacterial artificial chromosome (BAC) pBecker2 to test the effect of UL54 nuclear targeting on viral replication. In comparison with the wild-type virus, a recombinant virus harboring an NLS or NoLS mutation of UL54 reduced viral production to different extents. However, mutations of both the NLS and NoLS targeted UL54 to the cytoplasm in recombinant virus-infected cells and significantly impaired viral replication, comparable to the UL54-null virus. In addition, a virus lacking the NLS or the NoLS displayed modest defects in viral gene expression and DNA synthesis. However, deletion of both the NLS and the NoLS resulted in severe defects in viral gene expression and DNA synthesis, as well as production of infectious progeny. Thus, we have identified a classical NLS and a genuine NoLS in UL54 and demonstrate that the nuclear targeting of UL54 is required for efficient production of PRV.
INTRODUCTION
Pseudorabies virus (PRV), a typical alphaherpesvirus, is an economically important pathogen of swine. The neurotropic nature of PRV makes it a useful tracer of neuronal connections, and it is also a useful model for the study of herpesvirus pathogenesis (44). During the lytic cycle of PRV infection, the viral proteins are expressed in a cascade of three temporally distinct and functionally interdependent phases termed immediate-early (IE), early (E), and late (L) phase. It has been reported that UL54 is expressed with E kinetics (24), whereas its homologue from herpes simplex virus 1 (HSV-1), ICP27, belongs to the IE kinetic class. Furthermore, the amino acid sequence similarity between ICP27 and UL54 is only 41% (2). ICP27 is an essential protein and a multifunctional regulator of gene expression of HSV-1, given its different roles during infection (27, 37, 45). In contrast to ICP27-null HSV-1, UL54-null PRV is viable; however, it exhibits aberrant expression of several E and L gene products and is highly attenuated in a mouse model of PRV infection (47). Like its HSV-1 counterpart, UL54 contains a putative nuclear localization signal (NLS) and an RGG box RNA binding motif and shows RNA binding activity (25). All of these properties of UL54 make it of particular interest. Although ICP27 (27, 37, 45) and other herpesvirus homologues (4–6, 12, 19, 21, 28, 43, 52) have been extensively studied, the functions of UL54 are less known.
An important step toward understanding the detailed functions of a protein in vivo is to determine its precise subcellular localization. UL54 has been previously shown to target predominantly to the nuclei of infected and transfected cells (24, 25). In this study, we demonstrate that UL54 localizes predominantly to the nucleolus in transient transfected living cells but is distributed more widely throughout the nucleus in infected cells. By amino acid sequence analysis and construction of a series of UL54 variants, a functional NLS and nucleolar localization signal (NoLS) of UL54 were identified. In addition, by engineering NLS- and/or NoLS-mutated viruses based on a PRV bacterial artificial chromosome (BAC) (pBecker2), we found that inactivation of the NoLS is more detrimental to the replication of PRV at low multiplicities of infection than inactivation of the NLS. However, mutation of both the NLS and the NoLS significantly impairs viral replication, comparable to that of a UL54-null virus.
MATERIALS AND METHODS
Cells and viruses.
HEK293T, HeLa, PK-15, Vero, and COS-7 cells were grown in Dulbecco's modified minimal essential medium (DMEM; Gibco-BRL) supplemented with 10% fetal bovine serum (FBS; Gibco-BRL). PRV BAC (pBecker2) was a generous gift from Lynn W. Enquist (48).
Plasmid construction.
All enzymes used for cloning procedures were purchased from Takara (Dalian, China) except T4 DNA ligase (New England BioLabs, MA). The UL54 open reading frame (ORF) amplified from pBecker2 (48) was digested with EcoRI and BamHI and then inserted into the correspondingly digested green fluorescent protein variant mammalian expression vector pEYFP-N1 (Clontech) to create pUL54-EYFP (where EYFP is enhanced yellow fluorescent protein). Other plasmids used in this study were constructed similarly. Sequence information for all of the primers used is available upon request. pUL54(45–53)-EYFP was generated by ligating the annealed oligonucleotides into pEYFP-N1. pGFP-B23.1 was a generous gift from Julian A. Hiscox (14), and the ORF of B23.1 was subcloned into pECFP-N1 (Clontech) to yield B23.1-ECFP (where ECFP is enhanced cyan fluorescent protein). All constructs were verified by PCR, restriction analysis, and sequencing.
Western blot analysis.
Western blot analysis was performed as described in our previous studies (32, 53). Briefly, protein samples were separated by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes (Bio-Rad). The membranes were blocked with 5% skim milk in phosphate-buffered saline (PBS) overnight at 4°C; washed with PBST (PBS with 0.05% Tween 20) three times; incubated with anti-β-actin monoclonal antibody (MAb; Clontech), anti-YFP polyclonal antibody (pAb; Santa Cruz), anti-EP0 pAb (20), or anti-UL54 pAb (rabbit antiserum against the His-tagged N-terminal 66 amino acids [aa] of UL54) (unpublished data) at room temperature for 3 h; and then washed with PBST three times and incubated with secondary antibody conjugated with alkaline phosphatase (Kirkegaard and Perry Laboratories). Reactive bands were revealed with nitroblue tetrazolium bromochlorindodyl phosphate tablets (Sigma). Images were scanned and subsequently processed using Adobe Photoshop.
Transfection and fluorescence microscopy.
Transfection and fluorescence microscopy experiments were performed as described in our previous studies (31, 32). In the same experiment, each transfection was performed at least three times. Data shown are from one representative experiment. Samples were analyzed using a Zeiss Axiovert 200 M microscope (Germany). All the photomicrographs were taken under a magnification of ×400 unless otherwise specified. Each photomicrograph represents a vast majority of the cells with similar subcellular localizations. Light-translucent pictures show cellular morphology. Fluorescence images of EYFP and ECFP fusion proteins are presented in pseudocolor, green and red, respectively, and the merged images are shown to confirm the colocalization, indicated by yellow signals. Images were processed with Adobe Photoshop.
IFA.
Immunofluorescence assay (IFA) was applied to analyze the subcellular localization of PRV UL54 in transfected or infected cells. To investigate the subcellular localization of UL54 in transfected cells, two different fixation methods were performed, either methanol-acetic acid-based (25) or formaldehyde-based (19, 32). For the former, the transfected cells were fixed with 95% methanol and 5% glacial acetic acid, permeabilized with 0.25% Triton X-100 and 5% dimethyl sulfoxide (DMSO), and incubated with the anti-UL54 pAb and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (Zymed Laboratories). For the latter, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and incubated with the anti-UL54 or anti-EP0 pAb, respectively. To detect the subcellular localization of UL54 in infected cells, mock-infected PK-15 cells and cells infected with PRV at a multiplicity of infection (MOI) of 1 or 0.1 for several hours were subjected to formaldehyde-based (19), fixation-based IFA using the anti-UL54 or anti-EP0 pAb. After each incubation step, the cells were washed extensively with PBS. DAPI (4′,6-diamidino-2-phenylindole) stain was applied for the visualization of the cell nucleus, and cells were analyzed using a Zeiss Axiovert 200 M microscope. Each photomicrograph represents the majority of the cells. Images were processed with Adobe Photoshop.
Genetic manipulation of pBecker2 with recombineering technology.
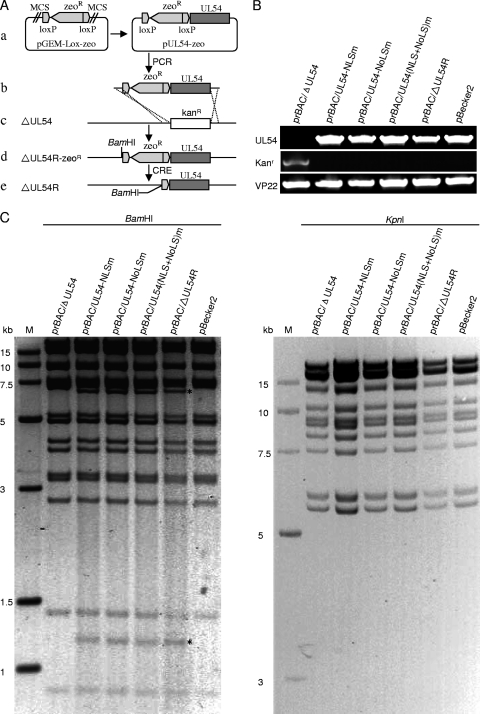
Wild-type (WT) and mutant PRV viruses were derived from the parental pBecker2 (strain Becker) BAC (48), and the strategy for introducing point mutations into pBecker2 by homologous recombination has been described previously (34, 54). Briefly, in the first step, a DNA fragment containing the Kanr expression cassette flanked by homology arms that target the UL54 coding sequence was amplified from the plasmid pGEM-oriV/Kan (54). The PCR product was digested with DpnI (New England Lab) and purified by a gel extraction kit (Qiagen). About 200 ng of the PCR product was transformed into Escherichia coli DY380 competent cells carrying pBecker2 via electroporation at 1.6 kV and 250 μF with a Bio-Rad Gene Pulser II. Recombinants were selected on agar plates containing kanamycin and chloramphenicol at 32°C. One positive clone, designated prBAC/ΔUL54 and identified by PCR, restriction analysis, and sequencing analysis, was employed for the second recombination step.
For the second step, to generate a UL54 deletion revertant (see Fig. 4A) and recombinants harboring desired mutations, the Kan cassette of prBAC/ΔUL54 was replaced by WT or mutant UL54 via homologous recombination (54). First, UL54, UL54-NLSm, UL54-NoLSm, and UL54-(NLS+NoLS)m sequences were amplified and subcloned into pGEM-Lox-Zeo to yield plasmids pUL54-Zeo, pUL54-NLSm-Zeo, pUL54-NoLSm-Zeo, and pUL54-(NLS+NoLS)m-Zeo. The resulting clones were verified by PCR, restriction analysis, and sequencing analysis. Next, the UL54-Zeor, UL54-NLSm-Zeor, UL54-NoLSm-Zeor, and UL54-(NLS+NoLS)m-Zeor cassettes were amplified from these plasmids. The PCR products were digested with DpnI to remove the plasmid template, purified, and electroporated into E. coli DY380/prBAC/ΔUL54 competent cells that had been induced at 42°C for 15 min (34, 54) for homologous recombination. The recombinants were selected on agar plates containing 50 μg/ml zeocin (Invitrogen) and 12.5 μg/ml chloramphenicol at 32°C. The PRV BAC prBAC/ΔUL54R, prBAC/UL54-NLSm, prBAC/UL54-NoLSm, and prBAC/UL54-(NLS+NoLS)m clones are resistant to chloramphenicol and zeocin and sensitive to kanamycin and ampicillin. The BAC DNAs from these clones were extracted and subjected to restriction analysis, PCR analysis, and sequencing analysis.
Fig. 4.
Construction of UL54-null BAC, UL54 revertant BAC, and BACs with mutations of NLS and/or NoLS in UL54. (A) Schematic diagram of the construction of a UL54 revertant clone, prBAC/ΔUL54R (ΔUL54R virus). (a) To generate the ΔUL54R clone, UL54 was amplified by PCR from the WT PRV BAC pBecker2 DNA. Then UL54 was cloned into the plasmid pGEM-Lox-Zeo to construct pUL54-Zeo. (b) Amplification of the UL54-Zeor cassette by PCR using a primer pair added 40 bp homology flanking UL54. (c) PCR product was transformed into E. coli DY380 competent cells carrying the prBAC/ΔUL54 via electroporation. (d) The Kanr gene was replaced with the UL54-Zeor cassette by homologous recombination to create the ΔUL54R clone prBAC/ΔUL54R. The introduced BamHI site within the UL54-Zeor cassette is also shown. (e) The Zeor gene was removed while generating virus from BAC DNA by cotransfecting a Cre recombinase-expressing plasmid. (B) PCR analysis of the BAC recombinants. The UL54, VP22, and Kanr genes were amplified. (C) Gel electrophoresis (0.8%) of BamHI- or KpnI-digested BAC recombinants. The restriction fragment length polymorphisms were observed at ∼1.3 kb and ∼6.5 kb in the BamHI-digested sample (marked with a black asterisk) because a BamHI site within the UL54-Zeor cassette was introduced.
Recombinant virus production.
To reconstitute recombinant viruses, confluent Vero cells were transfected with 2 μg of the corresponding BAC DNAs using a calcium phosphate transfection kit (Invitrogen) according to the manufacturer's manual. To remove the BAC vector and Zeor gene (flanked by two loxP sites) from the PRV genome, a Cre expression plasmid (pGS403) was cotransfected with the corresponding BAC DNAs as described previously (33). Following transfection, virus was harvested when the cytopathic effect (CPE) reached 90 to 95%. Lysates of transfected cells were then inoculated onto Vero cell monolayers in 10-cm-diameter dishes for 3 or 4 subsequent serial passages to increase virus titers.
Plaque assays and growth curve analysis.
Viruses were harvested and their titers were determined by plaque assay using crystal violet staining. PK-15 cells were infected with the vBecker2 reconstituted virus from parental pBecker2 and the vΔUL54, vΔUL54R, vNLSm, vNoLSm, and v(NLS+NoLS)m recombinant viruses from prBAC/ΔUL54, prBAC/ΔUL54R, prBAC/UL54-NLSm, prBAC/UL54-NoLSm, and prBAC/UL54-(NLS+NoLS)m, respectively. After 1 h of adsorption at 37°C, virus dilutions were washed off, and the plates were overlaid with 2× DMEM-2% FBS and white agar (1:1). After incubation at 37°C for 24 h (vBecker2, vΔUL54R, vNLSm, and vNoLSm viruses) or 72 h [vΔUL54 and v(NLS+NoLS)m viruses], the cell monolayers were fixed with methanol and stained with 0.1% crystal violet, and plaques were counted. IFA analysis was applied to detect the plaque phenotype with anti-EP0 pAb.
For the growth curve analyses, PK-15 cells were infected with different recombinant viruses at an MOI of 0.1 or 1. After 1 h of adsorption at 37°C, the cells were rinsed for 1 min with citrate buffer (40 mM Na citrate, 10 mM KCl, 135 mM NaCl, pH 3.0) to inactivate any unabsorbed virus. The cells were then incubated with fresh medium supplemented with 2% FBS for 4, 8, 12, and 24 h. Supernatant and cells were harvested at the indicated time points and were subjected to three freeze-thaw cycles to release infectious intracellular virus. Viral titers in all samples were determined in triplicate on a monolayer of PK-15 cells, and the average of each is shown.
Viral DNA isolation and analysis by PCR.
To examine viral DNA replication, PK-15 cells grown in 6-well plates were infected with either vΔUL54, vΔUL54R, vNLSm, vNoLSm, v(NLS+NoLS)m, or vBecker2 virus for 1 h at 37°C at an MOI of 0.1 or 1, and total cellular DNA was isolated at 16 h postinfection (hpi). To isolate total cellular DNA, infected cells were harvested, incubated with lysis buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 25 mM EDTA, 0.5% SDS, 0.1 g/ml proteinase K, 25 μg/ml RNase A) at 50°C overnight, and cell lysates were treated with equal volumes of phenol/chloroform/isoamyl alcohol (25:24:1) twice and chloroform/isoamyl alcohol (24:1) once. DNA was then precipitated by isopropyl alcohol at room temperature and dissolved in Tris-EDTA buffer (pH 8.0). Next, 25-μl PCR mixtures containing the extracted viral DNA and cellular DNA were analyzed using primers specific for gK and β-actin, respectively.
RNA isolation and semiquantitative RT-PCR.
Total RNA was extracted from mock-infected or virus-infected PK-15 cells with TRIzol (Invitrogen) according to the manufacturer's instructions. Samples were digested with DNase I and subjected to reverse transcription (RT)-PCR. RNA was reverse transcribed using an oligo(dT) primer. Ten percent of the resulting cDNA was used as the template for PCR using specific primers for the EP0 and UL54 PRV E genes, the VP5 capsid gene, the VP22 tegument gene, and the gK envelope gene. β-actin was used as a housekeeping gene to establish a baseline against which target genes were compared between samples. PCR products were analyzed on a 2% agarose gel.
RESULTS
Subcellular localization of UL54 in the transfected cells.
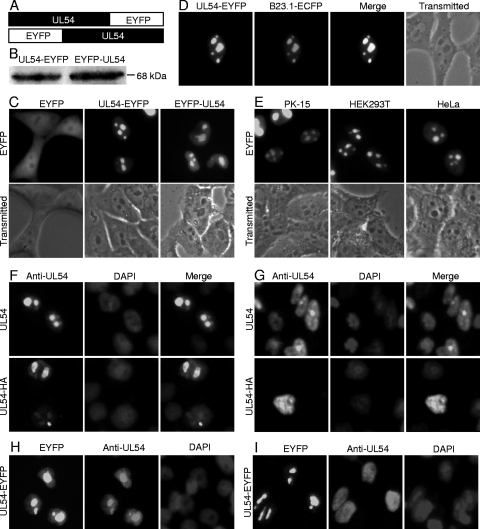
It is well known that the determination of subcellular localization is one way that potential roles of some proteins can be assessed. UL54 was shown to localize in the nucleus in transiently transfected cells (25). To further investigate the subcellular distribution of UL54 in transfected living cells, enhanced yellow fluorescent protein (EYFP)-tagged UL54 variants and fluorescence microscopy were applied. The plasmid encoding UL54 fused to the N terminus of EYFP was constructed (Fig. 1 A) and transfected into COS-7 cells to study the subcellular localization of UL54 in the absence of other viral proteins. UL54-EYFP exhibited mainly a speckle-like distribution in the nucleus that resembles the nucleolus (Fig. 1C). Western blot analysis with an anti-UL54 pAb showed that UL54-EYFP was expressed at the expected molecular size of about 68 kDa (Fig. 1B). Contrary to UL54-EYFP, control EYFP-N1 fluorescence was evenly distributed throughout the cytoplasm and the nucleus but not the nucleolus in cells transfected with pEYFP-N1 (Fig. 1C). In order to investigate whether the location of EYFP affects the localization of UL54 in cells, a DNA construct was also made to fuse EYFP to the N terminus of UL54 (EYFP-UL54) (Fig. 1A); Western blot analysis confirmed its expression (Fig. 1B). Fluorescence microscopy demonstrated identical subcellular distribution patterns of UL54-EYFP and EYFP-UL54 (Fig. 1C). Thus, subsequent experiments were performed using pUL54-EYFP. To investigate whether UL54 localizes to nucleolus, COS-7 cells were cotransfected with pUL54-EYFP and ECFP-tagged B23.1 (pB23.1-ECFP), which has been demonstrated to localize mainly to the nucleoli and is used as a nucleolar marker (1, 9, 14). Under these conditions, UL54 colocalized with B23.1 (Fig. 1D). Based on their size and numbers, these dense, dark-staining, irregular-shaped subnuclear organelles are nucleoli (Fig. 1D). To address whether the nucleolar accumulation of UL54 was an artifact of overexpression, a time course of expression showed that UL54 accumulated in the nucleolus as early as 4 h after transfection, when expression levels were low (data not shown).
Fig. 1.
Subcellular localization of UL54 in transfected cells. (A) Schematic diagram of UL54 fused with an EYFP monomer at its C terminus and N terminus. (B) Western blot analysis of the expressions of UL54-EYFP and EYFP-UL54 with an anti-UL54 pAb. The molecular mass is labeled to show the size of proteins. (C, D, and E) Subcellular localization of UL54 in transfected living cells. (C) Subcellular distribution of EYFP-UL54, UL54-EYFP, and EYFP. (D) Colocalization of UL54-EYFP and B23.1-ECFP. (E) Subcellular distribution of UL54-EYFP in PK-15, HEK293T, and HeLa cells. (F, G, H, and I) Subcellular localization of UL54 in transfected fixed cells. (F and H) COS-7 cells were transfected with pcDNA3.1-UL54, pCMV-HA-UL54, or pUL54-EYFP and fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and incubated with the anti-UL54 pAb. The cells were labeled with FITC (green)- or rhodamine (red)-conjugated goat anti-rabbit IgG and counterstained with DAPI to visualize the nuclei. (G and I) COS-7 cells were transfected with pcDNA3.1-UL54, pCMV-HA-UL54, or pUL54-EYFP and fixed with 95% methanol and 5% glacial acetic acid, permeabilized with 0.25% Triton X-100, and incubated with the anti-UL54 pAb. Each fluorescence image is representative of the vast majority of the cells observed.
To further investigate whether the subcellular localization of UL54 is cell type dependent in the absence of other viral proteins, different cell lines (PK-15, HEK293T, and HeLa) were transfected with pUL54-EYFP. In the three cell lines tested, UL54 was also found to localize almost exclusively to the nucleolus, indicating that there is a conserved mechanism for nucleolar localization in all four lines of cells (PK-15, HEK293T, COS-7, and HeLa) (Fig. 1E). As UL54 showed similar distribution in all cell types tested, the following transfection experiments were performed only in COS-7 cells, as they demonstrate the highest transfection efficiency.
Since EYFP is a relatively large tag, it might affect the nucleolar localization of UL54. To avoid this, plasmids encoding untagged UL54 or hemagglutinin (HA)-tagged UL54 (pUL54-HA) were constructed, and IFAs were performed to observe the subcellular localization of these UL54 proteins. In addition, it is well known that some fixation protocols may alter the subcellular localization of proteins, resulting in misleading conclusions in the analysis of the intracellular distribution of a specific protein (26). Therefore, two different IFA fixation methods were applied, namely, methanol-acetic acid based (25) or formaldehyde based (19, 32). Untagged UL54 or HA-tagged UL54 localized in the nucleolus (Fig. 1F) following formaldehyde-based fixation, as opposed to localization mainly in the nucleus and less in the nucleolus following methanol-acetic acid-based fixation (Fig. 1G). Therefore, the methanol-acetic acid-based fixation method may alter the subcellular localization of the UL54 protein. To corroborate this hypothesis, pUL54-EYFP was transfected into COS-7 cells, and the subcellular localization of UL54-EYFP was observed prior to fixation. Subsequently, IFA was performed using the two different fixation methods. The nucleolar localization of UL54-EYFP in living cells was changed by methanol-acetic acid-based fixation (Fig. 1I) but not by formaldehyde-based fixation (Fig. 1H), suggesting that the formaldehyde-based fixation method may be better to investigate the subcellular localization of UL54. Taken together, these results indicated that UL54 was a nucleolar-targeting protein.
Mapping and identification of a functional NLS and NoLS in the arginine-rich region of UL54.
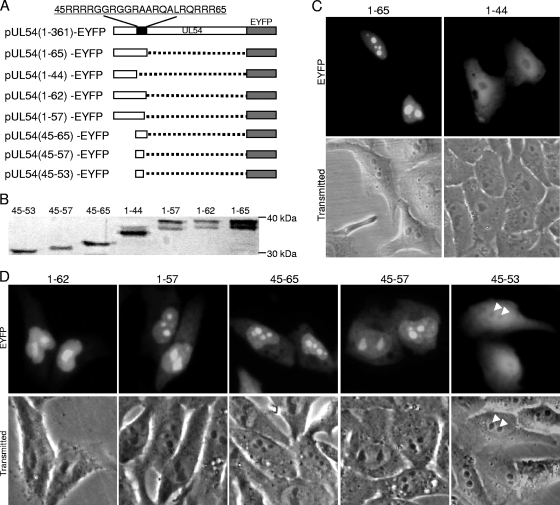
A previous study demonstrated that the N-terminal 83 aa of UL54 are responsible for its nuclear localization and contain an RGG box, which is critical for its RNA binding activity; the N-terminal 83 aa fused to green fluorescent protein (GFP) were also found to be enriched in the nucleolus (25). However, the precise NLS and NoLS of UL54 were still elusive. Sequence analysis using PSORT II (http://psort.nibb.ac.jp) (41) predicted that UL54 contained a classical NLS at aa 61 to 65 (RQRRR) and a short arginine- and glycine-rich sequence at aa 45 to 57, which resembles a functional NoLS as described for UL54's homologue ICP27 (35, 36). To verify these functional sequences, two recombinants encompassing aa 1 to 44 and 1 to 65 were constructed as in-frame fusions with EYFP (Fig. 2 A). The expression of these fusion proteins was confirmed by Western blot analysis using anti-YFP pAb (Fig. 2B). As predicted, aa1-44-EYFP showed a subcellular distribution similar to EYFP (Fig. 2C), whereas the subcellular distribution of aa1-65-EYFP was identical to that of UL54-EYFP (Fig. 1A), with the fluorescence enriched in the nucleoli (Fig. 2C), indicating that aa 1 to 65 are sufficient to target EYFP to the nucleolus.
Fig. 2.
The nucleolar localization signal locates in the arginine-rich region of UL54. (A) Schematic representation of the WT UL54 protein and its deletion mutants fused with EYFP; (B) Western blot analysis of the deletion derivatives of UL54 using anti-YFP pAb; (C) subcellular localization of deletion mutants aa1-65-EYFP and aa1-44-EYFP; (D) subcellular localization of the other UL54 mutants fused with EYFP (arrowheads, nucleoli of the cells). Each fluorescence image is representative of the vast majority of the cells observed.
To further confirm the motifs responsible for the nucleolar and nuclear localization of UL54, a series of fusion proteins, including aa1-62-EYFP, aa1-57-EYFP, aa45-65-EYFP, aa45-57-EYFP, and aa45-53-EYFP were constructed (Fig. 2A). The expression of these proteins was confirmed by Western blot analysis using anti-YFP Ab (Fig. 2B). As shown in Fig. 2D, aa 1 to 62, 1 to 57, 45 to 65, and 45 to 57 of UL54 can direct EYFP into the nucleolus; however, aa 45 to 53 exhibited a subcellular distribution pattern similar to that of EYFP, suggesting that aa 45 to 57 are a putative NoLS.
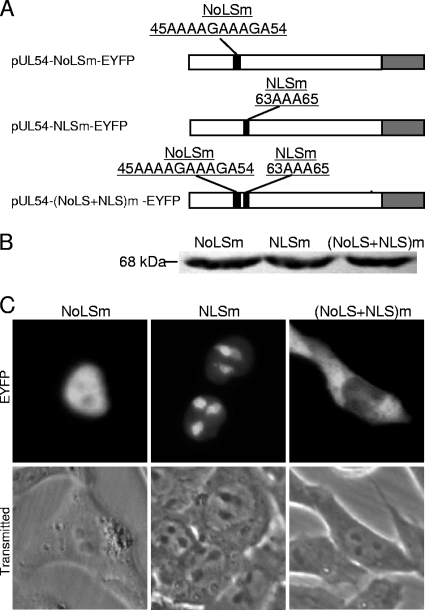
To confirm the results of the fusion proteins, three mutants of UL54 harboring mutations within the respective NLS and/or NoLS were made, namely, pUL54-NoLSm-EYFP, pUL54-NLSm-EYFP, and pUL54-(NoLS+NLS)m-EYFP (Fig. 3 A). The expression of these proteins was verified by Western blot analysis (Fig. 3B). Mutation of arginine to alanine in the NoLS significantly attenuated protein accumulation in the nucleoli (Fig. 3C). However, mutation of the NLS did not abrogate nuclear translocation of UL54-EYFP (Fig. 3C); consistent with the fusion protein data shown in Fig. 2D, the NoLS appears to possess a nuclear localization function in the absence of a functional NLS (19). When both the NoLS and NLS were mutated, UL54-(NoLS+NLS)m-EYFP was excluded from the nucleolus and nucleus (Fig. 3C). These results suggest that aa 61 to 65 (RQRRR) and 45 to 57 (RRRRGGRGGRAAR) are the functional NLS and NoLS of UL54, respectively.
Fig. 3.
Mutation of arginine residues in an arginine-rich motif abrogates the nuclear or nucleolar localization of UL54. (A) Schematic diagram of mutations of the arginine-rich domain in UL54; (B) Western blot analysis of the different mutants using the anti-UL54 pAb; (C) subcellular localization of these mutants. Each fluorescence image is representative of the vast majority of the cells observed.
Construction of a UL54 deletion BAC, a UL54 revertant BAC, and BACs with mutations of the NLS and/or NoLS in UL54.
To investigate the effect of subcellular localization of UL54 on PRV growth, the UL54 deletion BAC prBAC/ΔUL54 was constructed, and then UL54 revertant BAC prBAC/ΔUL54R (Fig. 4 A) and BACs [prBAC/UL54-NLSm, prBAC/UL54-NoLSm, and prBAC/UL54-(NLS+NoLS)m] with mutations of the NLS and/or NoLS in UL54 were constructed based on prBAC/ΔUL54. The expected recombinant clones were selected with zeocin and chloramphenicol double resistance. PCR analysis (Fig. 4B) and direct sequencing (data not shown) of each clone confirmed the expected mutations. Restriction fragment length polymorphism analysis of prBAC/ΔUL54, prBAC/ΔUL54R, prBAC/UL54-NLSm, prBAC/UL54-NoLSm, and prBAC/UL54-(NLS+NoLS)m showed similar patterns to that of the parental pBecker2 with KpnI digestion, whereas specific bands appeared with BamHI digestion, indicating that the proper recombination occurred at the expected locus (Fig. 4C). To rescue the viruses from the recombinant BACs, approximately 1 to 2 μg recombinant PRV BAC DNA and 0.5 μg Cre expression vector were cotransfected into Vero cells (Fig. 4A). The UL54 deletion mutant (prBAC/ΔUL54) produced the viable viral progeny vΔUL54 virus (data not shown) as previously described (47). All other recombinant viruses, vΔUL54R (Fig. 4A), vNLSm, vNoLSm, and v(NLS+NoLS)m, were also rescued from prBAC/ΔUL54R, prBAC/UL54-NLSm, prBAC/UL54-NoLSm, and prBAC/UL54-(NLS+NoLS)m clones, respectively (data not shown).
The subcellular localization of UL54-EYFP and EGFP-B23.1 in vBecker2-infected cells.
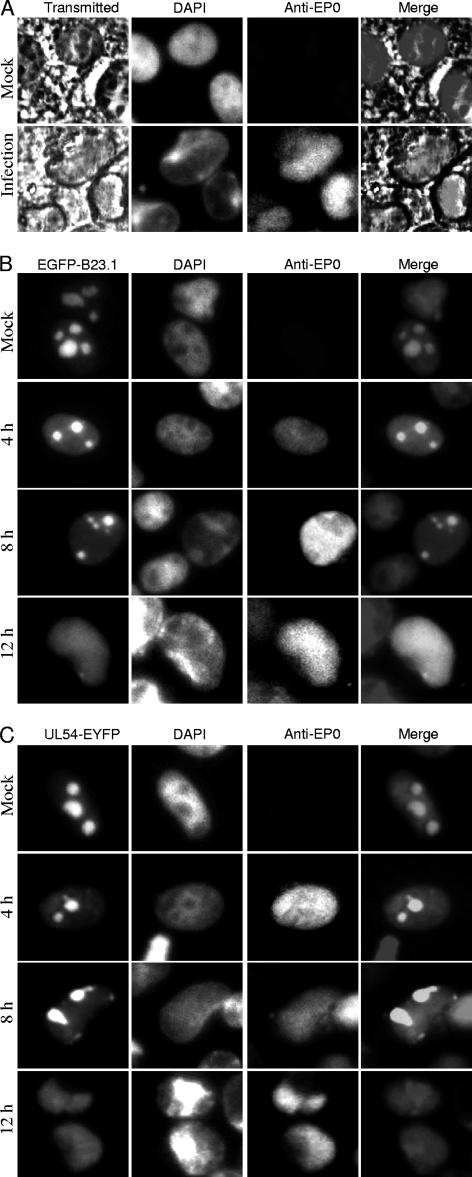
Previous studies have demonstrated that a number of morphological changes take place in the nucleus during HSV-1 infection, like the margination of host chromatin and profound modification of the nucleolus. These nuclear changes are usually accompanied by the formation of a viral replication compartment (VRC) that is essential for productive viral replication (3, 9, 14, 16, 22, 39, 46). The subcellular localization of the well-defined nucleolar protein EGFP-B23.1 was observed during the course of infection. DAPI staining (Fig. 5 A) revealed that the infected cell population contained a significant proportion of cells with well-developed chromatin-depleted regions, as described previously (9, 39). These appear to be much larger than nucleoli, the largest chromatin-depleted domains observed in mock-infected cells. As revealed by bright-field images (Fig. 5A), however, the nucleolar structure is obviously disrupted upon PRV infection. Expansion of the VRC is coincident with the presence of well-developed chromatin-depleted regions in infected cells (9, 39). Then the subcellular localization of UL54-EYFP was examined during the virus infection. COS-7 cells were first transfected with pEGFP-B23.1 and pUL54-EYFP, and 12 h after transfection, the cells were infected with WT vBecker2 virus at an MOI of 1. As a result of infection, EGFP-B23.1 and UL54-EYFP were redistributed from the nucleolus to the nucleus (Fig. 5B and C). EGFP-B23.1 and UL54-EYFP remained in structures that displayed a nucleolar morphology at an earlier time of infection (4 hpi). Then they localized in a structure that was devoid of the characteristic blue DAPI staining, especially at 12 hpi, indicating that this might be the VRC (9, 39). These results showed that UL54 localizes in the nucleolus in the absence of other viral proteins; however, it distributes to the nucleus in compartments that resemble the VRC during infection.
Fig. 5.
Dynamic localization of UL54-EYFP and EGFP-B23.1 during the course of vBecker2 infection. (A) DAPI staining of infected and uninfected PK-15 cell nuclei. The bright-field images show cellular morphology, and EP0 is used as a control for viral infection. (B and C) PK-15 cells were transfected with pEGFP-B23.1 and pUL54-EYFP for 12 h and then infected with vBecker2 at an MOI of 1. The cells were examined at different times postinfection (4, 8, and 12 h), and mock-infected PK-15 cells were used as a control.
The subcellular localization of UL54 recombinant viruses in PK-15 cells.
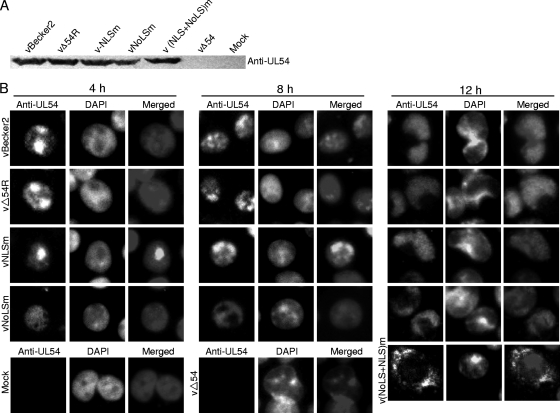
To determine whether UL54 mutants are expressed in virus-infected cells, Western blot analysis was employed. When the CPE of virus-infected cells reached 90 to 95%, cell lysates were produced and subjected to electrophoresis, transferred onto a nitrocellulose membrane, and reacted with the anti-UL54 pAb. As expected, the pAb could clearly detect an approximately 40-kDa protein (UL54) present in vBecker2, vΔUL54R, vNLSm, vNoLSm, and v(NLS+NoLS)m virus-infected cells, which did not exist in the mock-infected cells and vΔUL54 virus-infected cells (Fig. 6 A). In order to verify whether the NLS and/or NoLS are functional during infection, IFA was performed to investigate the exact subcellular localization of UL54 or its mutants in PK-15 cells infected with vBecker2 or other recombinant viruses. UL54 of vBecker2, vΔUL54R, and vNLSm viruses localized mainly in the nucleolus in the earlier times of infection, whereas it localized in the nucleus in the later times of infection (Fig. 6B). UL54 harboring a single NoLS mutation localized mainly to the nucleus during the course of vNoLSm infection (Fig. 6B). However, the mutant UL54-(NLS+NoLS)m protein encoding by v(NLS+NoLS)m localized mainly in the cytoplasm at 12 hpi, which is similar to the subcellular localization of UL54-(NLS+NoLS)m-EYFP in transfected living cells (Fig. 3C). In contrast, no specific staining was observed in mock-infected or UL54 deletion mutant-infected cells. These results indicate that the NoLS or both the NLS and NoLS function during virus infection.
Fig. 6.
Subcellular localization of UL54 in different recombinant virus-infected PK-15 cells during infection. (A) Western blot analysis of different recombinant virus-infected PK-15 cells. Monolayer PK-15 cells were infected with distinct recombinant viruses, including vBecker2, vΔUL54, vΔUL54R, vNLSm, vNoLSm, and v(NLS+NoLS)m viruses, and the cells were collected when CPE reached 90 to 95%. Then the cell lysates were subjected to Western blot analysis using an anti-UL54 pAb. (B) IFA was carried out to characterize the subcellular localization of UL54 in PRV-infected PK-15 cells. PK-15 cells infected with vBecker2, vΔUL54R, vNLSm, or vNoLSm virus were fixed at different time points postinfection (4, 8, and 12 h) with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and probed with anti-UL54 pAb. For v(NLS+NoLS)m virus, vΔUL54 virus, and mock infection, PK-15 cells were examined only at 12 hpi. Cells were treated with FITC-conjugated goat anti-mouse IgG and counterstained with DAPI to visualize the nuclei.
Nuclear targeting of UL54 is required for efficient production of PRV.
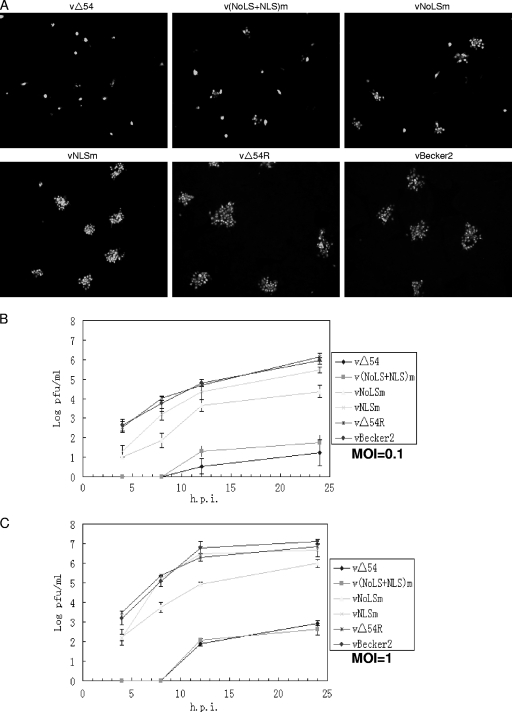
Next, we investigated whether the subcellular localization of UL54 affects viral replication of PRV. Stocks of WT virus vBecker2 and vΔUL54, vΔUL54R, vNLSm, vNoLSm, and v(NLS+NoLS)m recombinant viruses were prepared and their titers were determined. Plaque formation and viral proliferation characteristics were observed for each recombinant virus at an MOI of 0.1. Although the ΔUL54 virus was viable, plaques appeared later than with the WT virus. Plaque assays showed that the ΔUL54 virus could grow on PK-15 cells but resulted in an obvious reduction in plaque size as described previously (47). This phenotype was reverted to the WT when the UL54 allele was repaired (vΔUL54R virus) (Fig. 7 A), suggesting that the small plaque phenotype resulted from deletion of UL54. In addition, in comparison with the WT virus, all mutants exhibited a relative small-plaque phenotype; however, inactivation of the NoLS had more deleterious effects than that of the NLS on the production of infectious virus. Furthermore, mutation of both motifs was more deleterious than any single mutation (Fig. 7A). Thus, all of the mutations reduce the production of infectious virus, but the effect of each mutation differed at low MOI.
Fig. 7.
Nuclear targeting of UL54 is required for efficient production of PRV. (A) IFA analysis of the plaque phenotype of WT PRV vBecker2 and its derived recombinant viruses. Confluent PK-15 cells were infected with the indicated viruses at an MOI of 0.1. After 1 h of adsorption at 37°C, virus dilutions were washed off, and the plates were overlaid with 2× DMEM-2% FBS and white agar (1:1). After incubating at 37°C for 24 h, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, incubated with the anti-EP0 pAb, and then incubated with FITC-conjugated goat anti-rabbit IgG. All the samples were photographed with a Zeiss Axio 200 M microscope (Germany) under a magnification of ×100. (B and C) Growth curve analysis of WT PRV vBecker2 and its derived recombinant viruses. PK-15 cells were infected at an MOI of 0.1 (B) or 1 (C) with the WT vBecker2 or its recombinant PRV viruses, respectively. Supernatant and lysates of infected PK-15 cells were harvested at the indicated time points (4, 8, 12, and 24 h). The virus yields were enumerated by plaquing on PK-15 monolayers. The data plotted are the mean results from three independent experiments. The error bars represent the standard deviations of the means from three analyses.
To examine the molecular basis for the small-plaque phenotype of different mutant viruses, the growth kinetics of these viruses were determined at an MOI of 0.1. PK-15 cells were infected and harvested at the indicated time points (Fig. 7B). vΔUL54R exhibited comparable growth kinetics with vBecker2; however, vΔUL54, vNLSm, vNoLSm, and v(NLS+NoLS)m showed slower growth kinetics than the parental virus vBecker2. Twenty-four hours after infection, the titers of the vNLSm (∼3 × 105) and vNoLSm (∼2.6 × 104) mutants were approximately 3-fold and 35-fold lower than that of vBecker2 (∼9.3 × 105). Thus, both the NLS and NoLS mutations reduced production of infectious virus, with the effect of the NoLS mutation being more severe (Fig. 7B). In addition, viral replication of v(NLS+NoLS)m was severely impaired, which is comparable to that of the vΔUL54 virus (Fig. 7B).
Subsequently, we examined whether the growth defect of the UL54 mutant viruses is a multiplicity-dependent phenomenon. At an MOI of 1, the viral yield of vNoLSm was reduced by approximately 12-fold compared with that of its parental virus vBecker2, whereas the viral yield of vNLSm was almost equivalent to vBecker2 (Fig. 7C). However, compared to vBecker2, viral replication of v(NLS+NoLS)m and vΔUL54 was severely impaired (Fig. 7C). These results collectively show that the replication defect of vNLSm and vNoLSm could be overcome at a higher multiplicity of infection and that nuclear targeting of UL54 is important for efficient production of PRV.
Nuclear targeting of UL54 is required for efficient viral DNA replication and gene expression.
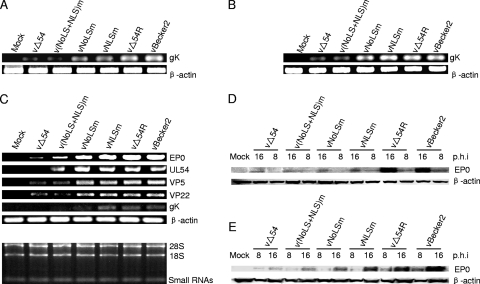
It is reported that ICP27 significantly enhances the levels of viral DNA synthesis (29). DNA replication is also reduced in cells infected with a UL54-null mutant (47). To investigate the effects of NLS and/or NoLS mutation of UL54 on DNA replication, total DNA of infected cells was extracted and the gK viral gene and β-actin host cell gene were amplified by PCR. Deletion of UL54 or mutation of either the NLS and/or the NoLS reduced viral DNA replication when the cells were infected with the corresponding viruses at an MOI of 0.1 (Fig. 8 A). However, compared with their parental virus vBecker2, the viral DNA levels of v(NLS+NoLS)m and vΔUL54 were severely impaired (Fig. 8B) at an MOI of 1, suggesting that expression and nuclear targeting of UL54 are required for efficient viral DNA replication.
Fig. 8.
Viral DNA replication and gene expression of WT PRV vBecker2 and its derived recombinant viruses. (A and B) DNA replication of WT PRV vBecker2 and its derived recombinant viruses. PK-15 cells were mock infected or infected with WT PRV vBecker2 and its derived recombinant viruses at an MOI of 0.1 (A) or 1 (B). At 16 hpi, total cellular DNA was purified, and PCR was performed with primers specific for gK to quantitate the levels of DNA. To ensure that equal amounts of DNA were used from each sample, the DNA from each sample was normalized with β-actin. (C) Expression of viral mRNAs of the WT PRV vBecker2 and its derived recombinant viruses. PK-15 cells were mock infected or infected with WT PRV vBecker2 and its derived recombinant viruses at an MOI of 1. At 16 hpi, total RNA was isolated, and equal amounts were analyzed by standard RT-PCR. The EP0, UL54, VP5, VP22, gK, and β-actin mRNA expression levels were assessed by RT-PCR. β-actin served as an internal control. (D and E) Protein synthesis of WT PRV vBecker2 and derived recombinant virus-infected PK-15 cells. PK-15 cells were mock infected or infected with WT PRV vBecker2 and its derived recombinant viruses at an MOI of 0.1 (D) or 1 (E). At 8 and 16 hpi, whole-cell extracts were prepared and subjected to analysis by SDS-PAGE. Protein accumulation was determined by Western blot analysis with antibodies specific for EP0 or β-actin. β-actin served as a loading control.
We next examined the effect of different UL54 mutant viruses on the expression of mRNAs of some E and L genes. For this end, we chose the true L gK gene, whose expression is dependent upon UL54 (47). In addition, the mRNAs of the EP0 gene (E gene), VP5 capsid gene (L gene), and VP22 tegument gene (L gene) were also analyzed. Total RNA from infected cells was isolated at 16 hpi, and UL54, EP0, VP5, VP22, and gK mRNA levels were assessed by RT-PCR. The mRNA expression of all selected genes was significantly decreased in v(NLS+NoLS)m- and vΔUL54-infected cells compared with that of its parental virus vBecker2 (Fig. 8C).
ICP27 and UL54 are important for accumulation of some viral proteins (29, 47). To investigate whether the nuclear targeting of UL54 is required for viral protein synthesis, the available EP0 pAb was used to analyze its expression following infection with different recombinant viruses. Infection with v(NLS+NoLS)m and vΔUL54 showed severe reduction in EP0 viral protein synthesis at both a low MOI of 0.1 (Fig. 8D) and a high MOI of 1 (Fig. 8E). Taken together, these pieces of evidence indicate that the nuclear targeting of UL54 is required for efficient viral DNA replication and gene expression of PRV.
DISCUSSION
UL54 homologues are important multifunctional proteins, which are conserved among alpha-, beta-, and gammaherpesviruses. In the present study, the exact subcellular localization of PRV UL54 was investigated, and we found that UL54 localized almost exclusively to the nucleolus in the absence of other viral proteins (Fig. 1D). In addition, different fixation methods, such as methanol-acetic acid and formaldehyde-based fixation, were applied, and we found that different fixation methods could result in different patterns of subcellular localization of untagged, small-tagged, and EYFP-tagged UL54. Specifically, the methanol-acetic acid-based fixation method (25) affects the subcellular localization of these proteins, i.e., the nucleolar localization of UL54-EYFP in living cells was not observed in fixed cells (Fig. 1I), whereas formaldehyde-based fixation methods yielded results consistent with localization observed within living cells (Fig. 1H). Therefore, UL54 is a genuine nucleolar-targeting protein, and the EYFP tag does not affect the subcellular localization of UL54.
Amino acid sequence analysis suggested that two clusters of arginine-rich sequences in the N terminus of UL54 might represent the potential NoLS/NLS. Mutation of the putative NoLS significantly attenuated accumulation of UL54 in the nucleolus. Mutation of both the NoLS and the NLS abrogated the nucleolar and nuclear localization of UL54, whereas mutation of the NLS alone did not alter the subcellular localization of UL54, suggesting that the NoLS also possesses nuclear localization function in the absence of a functional NLS, as we have demonstrated for its homologue BICP27 of bovine herpesvirus 1 (19). These results demonstrate that 45RRRRGGRGGR57 is a functional NoLS and 61RQRRR65 is a functional NLS. Surprisingly, doublets were detected, as shown in Fig. 2B, which might result from phosphorylation of Ser-rich residues or ubiquitination or sumoylation of Arg-rich residues in the N terminus of UL54.
Previous studies have shown that UL54 targeted mainly to the nucleus in infected cells (24). Our results show that transfected UL54 and the nucleolar protein B23.1 are delocalized from the nucleolus to the nucleus in a structure that might resemble the VRC during infection (Fig. 5 and 6) (9, 39). It is reported that several nucleolar proteins are redistributed out of the nucleoli as a consequence of HSV-1 infection (9, 39). Moreover, it is well known that different types of viruses can induce important alterations of nucleolar and nuclear architecture and that these alterations may participate directly in specific processes that are crucial for viral production (9, 22, 23). We speculate that UL54 is a nucleolar protein in infected cells, but its localization might be changed during infection to assist with efficient PRV production.
A previous study has shown that the accumulation of gC is decreased in cells infected with UL54-null PRV compared with that of cells infected with the WT virus and that UL54 also possibly regulates the expression of UL53 and UL52 at the transcriptional level (47), suggesting that UL54 may be a potential viral gene regulator. In the present study, the nuclear targeting of UL54 was demonstrated to be required for efficient viral DNA replication, viral mRNA accumulation, and protein expression. Interestingly, increasing numbers of key proteins from both DNA and RNA viruses have been reported to localize to the nucleolus. Nucleolar targeting of herpesvirus saimiri (HVS) (6) and KSHV (5) ORF57 are essential for the nuclear export of intronless herpesvirus mRNA. In addition, the HIV regulatory proteins Rev and Tat employ the nucleolus for trafficking of HIV-1 RNA (38). Furthermore, the nucleolar localization of HIV Rev and Rex proteins is involved in posttranscriptional regulation of viral mRNA (13). The nucleolar targeting of the HIV Tat protein is implicated in proviral DNA transcription (18). Indeed, localization to the nucleolus has been proved to be part of the strategy for virus to regulate both virus and host subgenomic RNA translation (22). Many viruses divert nucleolar functions by redirecting specific host nucleolar proteins from the nucleolus to different cell compartments, where they play essential roles in the virus life cycle (18). Therefore, the redistribution of UL54 from the nucleolus to the nucleus could be necessary to implement its function. Overexpression of the cellular mRNA export receptor TAP/NXF1 can promote the nuclear export of UL54 (32), suggesting that UL54 may be involved in trafficking of viral mRNA and act as a posttranscriptional regulation factor.
It is reported that nucleolar-resident proteins often contain Arg-, Lys-, and/or Gly-rich motifs, and several proteins' RGG domains are responsible for the interaction with distinct ribosomal proteins or RNA and/or involved in its nucleolar targeting (30, 51). Thus, the UL54 RGG motif is reminiscent of a nucleolar localization signal. Our results show that the RGG motif of UL54 directs EYFP to the nucleolus (Fig. 2D), demonstrating that it acts as an NoLS. Internal mutation of the RGG motif confirmed this verdict, as mutated protein was excluded from the nucleolus (Fig. 3C). Moreover, viral infection results also show that the NoLS is important for the nucleolar localization of UL54 (Fig. 6B). Furthermore, inactivation of the NoLS resulted in slower growth kinetics than inactivation of the NLS. This difference might result from the more disruptive intramolecular effect of the NoLS mutation on the overall protein fold. It also could result from the stronger effect of the NoLS mutation on virus or host protein translocation or expression, acting in combination to interfere with virion formation and giving a comparatively more pronounced phenotype than that of the NLS mutation. In addition, the NoLS mutation may result in inactivating the RNA binding and trafficking functions of UL54 (11, 29, 49, 50), and that may be the root cause of the viral replication defects. Further detailed investigations will be required to clarify these points. For HSV-1 ICP27, several roles have been attributed to the RGG motif, such as nucleolar localization, RNA binding, efficient viral RNA export, and regulation of HSV-1 replication (11, 29, 49, 50). In addition to its RNA binding activity (25), the RGG motif of UL54 is also critical for its nucleolar localization and efficient PRV replication.
In our studies, the kinetics of UL54-null virus growth was slower than that of a previous study, which showed no obvious difference between UL54-null virus and WT virus and suggested that UL54 was not essential for viral growth in cell culture (47). Although we also show that UL54 is not essential for PRV growth, the deletion mutants of UL54 exhibited severe decreased DNA replication, virus yields, and plaque sizes and reduced accumulation of viral mRNA and proteins (47). Therefore, we believe that our findings are at least in partial agreement with previous studies, because this previous study utilized partial deletion of the UL54 protein, whereas we completely removed UL54 from the PRV genome. In addition, this discrepancy can probably be explained by subtle differences in experimental design, such as the employ of a different recombinant system and cell lines.
A multiplicity-dependent growth phenotype has been observed for several viruses, including HSV-1 (8, 10, 15), human cytomegalovirus (HCMV) (7), African swine fever virus (40), and adenoviruses (17, 42). The defect of UL82 severely restricted viral replication of the UL82-deficient mutant virus of HCMV at low input MOI but can be rescued by higher input MOI (7). Similarly, the severe replication defect of an ICP0-null HSV-1 mutant in BHK or Vero cells at low MOI also can be overcome at high MOI (8, 10, 15).
Taken together, this study suggests that UL54's function as a viral gene regulator may not be limited to the transcriptional level, it can also act at the posttranscriptional level. The comparatively more disruptive effects of mutating the NoLS suggests that the NoLS has a major function other than, or in addition to, its role as an NoLS. UL54 association with the nucleolus may provide new leads to uncovering other novel activities.
ACKNOWLEDGMENTS
This work was supported by grants from the Start-up Fund of the Hundred Talents Program of the Chinese Academy of Sciences (grant no. 20071010141) and National Natural Science Foundation of China (grant no. 30900059 and 81000736). We gratefully acknowledge the support of K. C. Wang Education Foundation, Hong Kong, China.
We thank Lynn W. Enquist, Zhu Hua, Julian A. Hiscox, and Angus I. Lamond for the generous gifts of pBecker2, pGEM-Lox-Zeo, and pGFP-B23.1. We thank Zhou Rui for the generous gift of anti-EP0 pAb. We also thank Zhang Cun and Zhu Hua for their helpful suggestions on BAC construction. We thank Rozanne M. Sandri-Goldin and Karen L. Mossman for critical reviews of the manuscript. Anonymous reviewers helped improve the manuscript.
Footnotes
Published ahead of print on 27 July 2011.
REFERENCES
- 1. Andersen J. S., et al. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1–11 [DOI] [PubMed] [Google Scholar]
- 2. Baumeister J., Klupp B. G., Mettenleiter T. C. 1995. Pseudorabies virus and equine herpesvirus 1 share a nonessential gene which is absent in other herpesviruses and located adjacent to a highly conserved gene cluster. J. Virol. 69:5560–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boisvert F. M., van Koningsbruggen S., Navascues J., Lamond A. I. 2007. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8:574–585 [DOI] [PubMed] [Google Scholar]
- 4. Boyne J. R., Colgan K. J., Whitehouse A. 2008. Herpesvirus saimiri ORF57: a posttranscriptional regulatory protein. Front. Biosci. 13:2928–2938 [DOI] [PubMed] [Google Scholar]
- 5. Boyne J. R., Whitehouse A. 2009. Nucleolar disruption impairs Kaposi's sarcoma-associated herpesvirus ORF57-mediated nuclear export of intronless viral mRNAs. FEBS Lett. 583:3549–3556 [DOI] [PubMed] [Google Scholar]
- 6. Boyne J. R., Whitehouse A. 2006. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. U. S. A. 103:15190–15195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bresnahan W. A., Shenk T. E. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. U. S. A. 97:14506–14511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cai W., Schaffer P. A. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66:2904–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Calle A., et al. 2008. Nucleolin is required for an efficient herpes simplex virus type 1 infection. J. Virol. 82:4762–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen J., Silverstein S. 1992. Herpes simplex viruses with mutations in the gene encoding ICP0 are defective in gene expression. J. Virol. 66:2916–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Corbin-Lickfett K. A., Souki S. K., Cocco M. J., Sandri-Goldin R. M. 2010. Three arginine residues within the RGG box are crucial for ICP27 binding to herpes simplex virus 1 GC-rich sequences and for efficient viral RNA export. J. Virol. 84:6367–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ding Q., et al. 2010. Characterization of the nuclear import and export mechanisms of bovine herpesvirus-1 infected cell protein 27. Virus Res. 149:95–103 [DOI] [PubMed] [Google Scholar]
- 13. Dundr M., et al. 1995. The roles of nucleolar structure and function in the subcellular location of the HIV-1 Rev protein. J. Cell Sci. 108:2811–2823 [DOI] [PubMed] [Google Scholar]
- 14. Emmott E., et al. 2008. Viral nucleolar localisation signals determine dynamic trafficking within the nucleolus. Virology 380:191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Everett R. D., Boutell C., Orr A. 2004. Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J. Virol. 78:1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Everett R. D., Maul G. G. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fang L., Stevens J. L., Berk A. J., Spindler K. R. 2004. Requirement of Sur2 for efficient replication of mouse adenovirus type 1. J. Virol. 78:12888–12900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greco A. 2009. Involvement of the nucleolus in replication of human viruses. Rev. Med. Virol. 19:201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo H., et al. 2009. Characterization of the nuclear and nucleolar localization signals of bovine herpesvirus-1 infected cell protein 27. Virus Res. 145:312–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo H., Zhou R., Xi Y., Xiao S., Chen H. 2009. Transcriptional suppression of IE180 and TK promoters by the EP0 of pseudorabies virus strains Ea and Fa. Virus Genes 38:269–275 [DOI] [PubMed] [Google Scholar]
- 21. Hamel F., Simard C. 2003. Mapping of the bovine herpesvirus 1 glycoprotein C promoter region and its specific transactivation by the viral BICP27 gene product. Arch. Virol. 148:137–152 [DOI] [PubMed] [Google Scholar]
- 22. Hiscox J. A. 2002. The nucleolus—a gateway to viral infection? Arch. Virol. 147:1077–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hiscox J. A. 2007. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5:119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang C., Wu C. Y. 2004. Characterization and expression of the pseudorabies virus early gene UL54. J. Virol. Methods 119:129–136 [DOI] [PubMed] [Google Scholar]
- 25. Huang Y. J., Chien M. S., Wu C. Y., Huang C. 2005. Mapping of functional regions conferring nuclear localization and RNA-binding activity of pseudorabies virus early protein UL54. J. Virol. Methods 130:102–107 [DOI] [PubMed] [Google Scholar]
- 26. Izumi M., Yatagai F., Hanaoka F. 2004. Localization of human Mcm10 is spatially and temporally regulated during the S phase. J. Biol. Chem. 279:32569–32577 [DOI] [PubMed] [Google Scholar]
- 27. Johnson K. E., Song B., Knipe D. M. 2008. Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology 374:487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Juillard F., et al. 2009. Epstein-Barr virus protein EB2 contains an N-terminal transferable nuclear export signal that promotes nucleocytoplasmic export by directly binding TAP/NXF1. J. Virol. 83:12759–12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lengyel J., Guy C., Leong V., Borge S., Rice S. A. 2002. Mapping of functional regions in the amino-terminal portion of the herpes simplex virus ICP27 regulatory protein: importance of the leucine-rich nuclear export signal and RGG box RNA-binding domain. J. Virol. 76:11866–11879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leung A. K., Andersen J. S., Mann M., Lamond A. I. 2003. Bioinformatic analysis of the nucleolus. Biochem. J. 376:553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li M., Wang L., Ren X., Zheng C. 2011. Host cell targets of tegument protein VP22 of herpes simplex virus 1. Arch. Virol. 156:1079–1084 [DOI] [PubMed] [Google Scholar]
- 32. Li M. L., Wang S., Cai M. S., Guo H., Zheng C. F. 2011. Characterization of molecular determinants for nucleocytoplasmic shuttling of PRV UL54. Virology. [Epub ahead of print.] doi:10.1016/j.virol.2011.06.004 [DOI] [PubMed] [Google Scholar]
- 33. Marchini A., Liu H., Zhu H. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. May K. F., Jr., et al. 2007. B7-deficient autoreactive T cells are highly susceptible to suppression by CD4(+)CD25(+) regulatory T cells. J. Immunol. 178:1542–1552 [DOI] [PubMed] [Google Scholar]
- 35. Mears W. E., Lam V., Rice S. A. 1995. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J. Virol. 69:935–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mears W. E., Rice S. A. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 70:7445–7453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Melchjorsen J., Siren J., Julkunen I., Paludan S. R., Matikainen S. 2006. Induction of cytokine expression by herpes simplex virus in human monocyte-derived macrophages and dendritic cells is dependent on virus replication and is counteracted by ICP27 targeting NF-kappaB and IRF-3. J. Gen. Virol. 87:1099–1108 [DOI] [PubMed] [Google Scholar]
- 38. Michienzi A., Cagnon L., Bahner I., Rossi J. J. 2000. Ribozyme-mediated inhibition of HIV-1 suggests nucleolar trafficking of HIV-1 RNA. Proc. Natl. Acad. Sci. U. S. A. 97:8955–8960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Monier K., Armas J. C., Etteldorf S., Ghazal P., Sullivan K. F. 2000. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2:661–665 [DOI] [PubMed] [Google Scholar]
- 40. Moore D. M., Zsak L., Neilan J. G., Lu Z., Rock D. L. 1998. The African swine fever virus thymidine kinase gene is required for efficient replication in swine macrophages and for virulence in swine. J. Virol. 72:10310–10315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakai K., Horton P. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34–36 [DOI] [PubMed] [Google Scholar]
- 42. Nevins J. R. 1981. Mechanism of activation of early viral transcription by the adenovirus E1A gene product. Cell 26:213–220 [DOI] [PubMed] [Google Scholar]
- 43. Ote I., Piette J., Sadzot-Delvaux C. 2010. The varicella-zoster virus IE4 protein: a conserved member of the herpesviral mRNA export factors family and a potential alternative target in antiherpetic therapies. Biochem. Pharmacol. 80:1973–1980 [DOI] [PubMed] [Google Scholar]
- 44. Pomeranz L. E., Reynolds A. E., Hengartner C. J. 2005. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 69:462–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sandri-Goldin R. M. 2008. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 13:5241–5256 [DOI] [PubMed] [Google Scholar]
- 46. Schwartz J., Roizman B. 1969. Concerning the egress of herpes simplex virus from infected cells: electron and light microscope observations. Virology 38:42–49 [DOI] [PubMed] [Google Scholar]
- 47. Schwartz J. A., Brittle E. E., Reynolds A. E., Enquist L. W., Silverstein S. J. 2006. UL54-null pseudorabies virus is attenuated in mice but productively infects cells in culture. J. Virol. 80:769–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smith G. A., Enquist L. W. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 97:4873–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sokolowski M., Scott J. E., Heaney R. P., Patel A. H., Clements J. B. 2003. Identification of herpes simplex virus RNAs that interact specifically with regulatory protein ICP27 in vivo. J. Biol. Chem. 278:33540–33549 [DOI] [PubMed] [Google Scholar]
- 50. Souki S. K., Gershon P. D., Sandri-Goldin R. M. 2009. Arginine methylation of the ICP27 RGG box regulates ICP27 export and is required for efficient herpes simplex virus 1 replication. J. Virol. 83:5309–5320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tillemans V., Leponce I., Rausin G., Dispa L., Motte P. 2006. Insights into nuclear organization in plants as revealed by the dynamic distribution of Arabidopsis SR splicing factors. Plant Cell 18:3218–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Toth Z., Stamminger T. 2008. The human cytomegalovirus regulatory protein UL69 and its effect on mRNA export. Front. Biosci. 13:2939–2949 [DOI] [PubMed] [Google Scholar]
- 53. Xing J., et al. 2011. Comprehensive characterization of interaction complexes of herpes simplex virus type 1 ICP22, UL3, UL4, and UL20.5. J. Virol. 85:1881–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang Z., Huang Y., Zhu H. 2008. A highly efficient protocol of generating and analyzing VZV ORF deletion mutants based on a newly developed luciferase VZV BAC system. J. Virol. Methods 148:197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]