Abstract
V2/V3 conformational epitope antibodies that broadly neutralize HIV-1 (PG9 and PG16) have been recently described. Since an elicitation of previously known broadly neutralizing antibodies has proven elusive, the induction of antibodies with such specificity is an important goal for HIV-1 vaccine development. A critical question is which immunogens and vaccine formulations might be used to trigger and drive the development of memory B cell precursors with V2/V3 conformational epitope specificity. In this paper we identified a clonal lineage of four V2/V3 conformational epitope broadly neutralizing antibodies (CH01 to CH04) from an African HIV-1-infected broad neutralizer and inferred their common reverted unmutated ancestor (RUA) antibodies. While conformational epitope antibodies rarely bind recombinant Env monomers, a screen of 32 recombinant envelopes for binding to the CH01 to CH04 antibodies showed monoclonal antibody (MAb) binding to the E.A244 gp120 Env and to chronic Env AE.CM243; MAbs CH01 and CH02 also bound to transmitted/founder Env B.9021. CH01 to CH04 neutralized 38% to 49% of a panel of 91 HIV-1 tier 2 pseudoviruses, while the RUAs neutralized only 16% of HIV-1 isolates. Although the reverted unmutated ancestors showed restricted neutralizing activity, they retained the ability to bind to the E.A244 gp120 HIV-1 envelope with an affinity predicted to trigger B cell development. Thus, E.A244, B.9021, and AE.CM243 Envs are three potential immunogen candidates for studies aimed at defining strategies to induce V2/V3 conformational epitope-specific antibodies.
INTRODUCTION
The development of strategies to induce broadly neutralizing antibodies is a critical goal of HIV-1 vaccine development (22). Recent studies have demonstrated that up to 20% of chronically HIV-1-infected subjects make anti-envelope antibody responses that have some capacity for breadth of neutralization, and 1 to 3% of subjects mount high levels of broadly neutralizing antibody responses (11, 16, 17, 48). Individuals who make broadly neutralizing antibodies do not do so initially (37, 57); rather, it typically takes ∼2 to 3 years for broadly neutralizing antibodies to develop (16, 25, 40). The first well-studied HIV-1 broadly neutralizing antibodies were derived either from Epstein-Barr virus (EBV)-transformed B cells or from phage-displayed libraries, yielding several human monoclonal antibodies (MAbs) that bound to conserved targets of HIV-1 (3, 30, 52). Many of these antibodies have unusual characteristics, such as long heavy-chain complementarity-determining regions (HCDRs), polyreactivity, and high numbers of somatic mutations (11, 17, 18, 22, 29, 30, 38–40, 48). More recently, new broadly neutralizing antibodies have been derived from clonal or oligoclonal memory B cell cultures (56), from single-cell sorting of antigen-specific memory B cells (41, 42, 60), or from EBV-transformed memory B cell cultures (7, 35). These new broadly neutralizing MAbs also share many of the unusual characteristics of previously found broadly neutralizing antibodies and are not easily induced by immunizations with HIV-1 envelope glycoproteins (22). Thus, a possible strategy for rational immunogen design is to identify clonal lineages of broadly neutralizing antibodies and derive their common reverted unmutated ancestors (putative naïve B cell precursor B cell receptor genes) to determine potential immunogens that could bind to similar naïve B cells to elicit broadly neutralizing antibodies.
In this study, we combined memory B cell isolation, clonal or oligoclonal culture systems, single-cell flow cytometric sorting, EBV transformation, and recombinant antibody generation to isolate natural antibodies from immunoglobulin G-positive (IgG+) memory B cells of a broad neutralizer (46) African subject chronically infected with a clade A HIV-1 strain. By applying these technologies, we identified four members of a clonal lineage of broadly neutralizing antibodies. We have demonstrated that these antibodies recognize an epitope in the V2/V3 region of a single protomer that is usually, but not exclusively, conferred to the gp120 envelope glycoprotein by trimer formation and distinguished it from previously described V2/V3 conformational epitope-specific broadly neutralizing antibodies. We inferred the reverted unmutated ancestors of this clonal lineage and defined the need for somatic mutations for breadth of neutralization. We identified recombinant envelope glycoproteins that bind to putative reverted unmutated ancestor antibodies and that can potentially be used as candidate immunogens for the development of immunization regimens optimized to induce CH01- to CH04-like broadly neutralizing antibodies.
MATERIALS AND METHODS
Patient information.
Peripheral blood was collected from a female African subject (subject CH0219) during a screening of 308 chronically HIV-1-infected subjects in the CHAVI 008 and 001 protocols (G. D. Tomaras et al., unpublished data). The serum of subject CH0219, who was infected with a clade A HIV-1 strain, showed breadth of neutralization directed against gp120 Env epitopes (Tomaras et al., unpublished). No other coinfections were recorded for this individual throughout the study. All studies with human subjects were approved by the Kilimanjaro Christian Medical Centre Research Ethics Committee, the Tanzania National Institutes for Medical Research Ethics Coordinating Committee, and the Institutional Review Boards of the London School of Hygiene and Tropical Medicine and Duke University as well as by the NIH Human Subject Review Committee.
Cell cultures.
IgG+ memory B cells were isolated from frozen peripheral blood mononuclear cells (PBMCs) by two rounds of separations using magnetic beads. First, PBMCs were incubated for 30 min at 4°C with phycoerythrin (PE)-conjugated (BD Pharmingen, San Diego, CA) anti-CD2 (catalog number 555327), anti-CD14 (catalog number 555398), anti-CD16 (catalog number 555407), anti-CD235a (catalog number 555570), and anti-IgD (catalog number 555779) antibodies, followed by a 15-min incubation with anti-PE microbeads (Miltenyi Biotec, Auburn, CA) at 4°C. After washing, cells were run through magnetic columns using the Deplete_S program of the AutoMACs separator (Miltenyi Biotec). The negative fraction was then incubated for 15 min with anti-IgG microbeads (catalog number 130-047-501; Miltenyi Biotec). After washing, IgG+ memory B cells were positively selected by using the Possel_S program of the AutoMACs separator. All the steps of the separation procedure were carried out with phosphate-buffered saline (PBS)–1% bovine serum albumin (BSA) buffer. Cells were then resuspended in complete medium containing 2.5 μg/ml CpG ODN2006 (tlrl-2006; InvivoGen, San Diego, CA), 5 μM CHK2 kinase inhibitor (Calbiochem/EMD Chemicals, Gibbstown, NJ), and EBV (200-μl supernatant of B95-8 cells/104 memory B cells) and incubated in bulk overnight at 37°C in 5% CO2. After overnight incubation we seeded ∼30,000 viable IgG+ memory B cells at a concentration of 8 memory B cells/well in 96-well round-bottom tissue culture plates containing ODN2006 and the CHK2 kinase inhibitor in the presence of irradiated (7,500 cGy) CD40 ligand-expressing L cells (5,000 cells/well). Cells were refed with fresh medium containing 15% fetal calf serum, ODN2006, and the CHK2 kinase inhibitor at day 7 and harvested at day 14.
Culture conditions were optimized for high immunoglobulin yields from cells cultured at a near-clonal density, as predicted by the single-hit model of the Poisson distribution (19, 49) and as determined by heavy- and light-chain sequence analysis in preliminary experiments. The median IgG concentration of the cultures at the end of stimulation was 705 ng/ml (total IgG concentration range, 1 to >40,000 ng/ml) (see Fig. S1 in the supplemental material).
Selection and isolation of monoclonal antibodies CH01, CH02, CH03, and CH04.
After 14 days of culture, we selected 4 supernatants that most effectively neutralized a C.CAP45-pseudotyped lentivirus in vitro in TZM-bl cells (Fig. 1). To ensure the isolation of the neutralizing antibodies in each well, these 14-day-old cultures were processed as follows: (i) aliquots of cells from each well were transferred into RNAlater stabilization reagent (Qiagen, Valencia, CA) and cryopreserved at −80°C for the subsequent amplification and sequencing of the heavy and light chains from the bulk cultures, (ii) cells from positive cultures were single-cell sorted into 96-well plates containing 20 μl of reverse transcription (RT)-PCR Mastermix buffer (0.5 μl RNaseOut [Invitrogen], 5 μl 5× first-strand buffer [Invitrogen], 1.25 μl dithiothreitol [DTT] [Invitrogen], and 0.06 μl Igepal [Sigma-Aldrich, St. Louis, MO] in 13.25 μl distilled water [dH2O]) and cryopreserved at −80°C, and (iii) cells from selected cultures were used to establish hybridoma B cell lines.
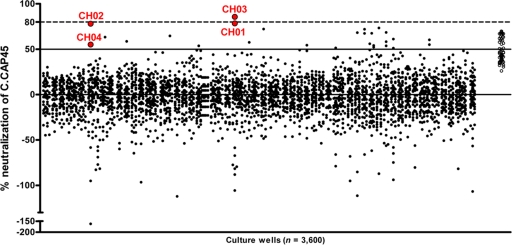
Fig. 1.
Neutralization screening of primary IgG+ memory B cell cultures. IgG+ memory B cells were isolated from the peripheral blood of a broad neutralizer African subject chronically infected with a clade A HIV-1 strain. IgG+ memory B cells were cultured at a density of 8 cells/well in 3,600 culture wells for 14 days as described in Materials and Methods. At the end of stimulation, crude supernatants were tested for neutralizing activity against reporter tier 2 HIV-1 clade C isolate CAP45, a difficult-to-neutralize virus that was effectively neutralized by this subject's serum (Tomaras et al., unpublished). Solid dots represent the percentage of neutralization of each of the 3,600 cultures. Monoclonal antibodies CH01 to CH04 were isolated from the cultures represented by circled red dots. Positive controls (HIV Ig) are shown as open circles on the far right.
Cultures that yielded MAbs CH01 and CH02 were monoclonal, whereas MAbs CH03 and CH04 were isolated from oligoclonal cultures through single-cell sorting. All the four antibodies shared the same V(D)J rearrangement (VH 3-20/Vκ 3-20), and when expressed in vitro as whole IgG1 antibodies, they all neutralized HIV-1 C.CAP45. Simultaneously, we generated a hybridoma cell line from the culture that contained MAb CH04 and reisolated the same antibody. The original isotype of MAbs CH01 to CH04 was IgG1 (Table 1).
Table 1.
Characteristics of the V heavy and V light chains of monoclonal antibodies CH01 to CH04
| MAb | V heavy chain |
V light chain |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| V | Db | J | HCDR3 lengtha | Mutation frequency | Isotype | V | J | κ/λ | LCDR3 lengtha | Mutation frequency | |
| CH01 | 3-20 | 3-10 | 2 | 24 | 0.133 | IgG1 | 3-20 | 1 | κ | 9 | 0.100 |
| CH02 | 3-20 | 3-10 | 2 | 24 | 0.143 | IgG1 | 3-20 | 1 | κ | 9 | 0.137 |
| CH03 | 3-20 | 3-10 | 2 | 24 | 0.120 | IgG1 | 3-20 | 1 | κ | 9 | 0.106 |
| CH04 | 3-20 | 3-10 | 2 | 24 | 0.115 | IgG1 | 3-20 | 1 | κ | 9 | 0.094 |
The HCDR3 and LCDR3 lengths are expressed as numbers of amino acids and reported according to the numbering system of Kabat et al. (20).
The D region may have resulted from the D-D fusion of the 3-10 gene segment with one of several other D gene segments.
Assurance that the amplified sequences accurately represented the natural repertoire in vivo was accomplished by the culturing of EBV-transformed B cells for ∼1 to 2 months. During the incubation period we sampled heavy- and light-chain pairs and consistently obtained sequences identical to those of cells sorted at the time of screening (day 14). In particular, the V(D)J gene sequences of MAb CH04 obtained after ∼2 months of culture were identical to those obtained from the original culture, demonstrating that MAb CH04 obtained from the hybridoma is true to the natural antibody. This observation also suggests that MAbs CH01 to CH03 did not mutate in vitro during the initial 14-day stimulation. To ensure that PCR and sequencing did not introduce mutations, we independently amplified and sequenced up to 240 single-cell-sorted cells from the 14-day-old cultures and obtained multiple, identical sequences. These data demonstrate that the strategy that we developed allowed the identification of neutralizing monoclonal antibodies after 14 days in culture.
Hybridoma cell line generation.
Hybridoma cell lines were generated by using a modification of a previously described electrofusion method (62). Briefly, K6H6/B5 myeloma partner cells and EBV-stimulated B cells were washed twice with an electrofusion medium (Cyto Pulse Sciences, Glen Burnie, MD) before fusion. A 1:2 B cell-to-myeloma cell ratio was used for fusion. Electrofusion was achieved by use of a PA-4000/PA-101 apparatus with an FE-20/800 electrode fusion chamber (Cyto Pulse Sciences). Prefusion dielectrophoresis was performed with an alternating current voltage of 75 V at 0.8 MHz for 15 s. Cells were fused with a single-square-wave direct-current voltage of 300 V for 0.04 ms. Postfusion dielectrophoresis was performed with an alternating current voltage of 20 V at 0.2 MHz for 30 s. After fusion, cells were harvested, distributed into flat-bottom 96-well plates at 4,000 B cells/well, and incubated in culture medium supplemented with 100 μM hypoxanthine, 0.4 μM aminopterin, 16 μM thymidine, and 0.5 μM ouabain. The resulting heterohybridomas were screened for the neutralization of tier 2 HIV-1 clade C isolate CAP45 and were cloned by using a standard limiting dilution method.
Isolation of V(D)J immunoglobulin regions.
RNA from positive cultures was extracted by using standard procedures (RNeasy minikit; Qiagen), and the genes encoding Ig V(D)J rearrangements were amplified by RT and nested PCR without cloning by use of a modification of a previously reported method (21, 47). Briefly, reverse transcription was performed at 55°C for 1 h after the addition of 50 units/reaction of Superscript III reverse transcriptase (InvitroGen) and 0.5 μM human IgG, IgM, IgD, IgA1, IgA2, Igκ, and Igλ constant-region primers. Separate reactions were used to amplify individual families of VH, Vκ, and Vλ genes from the cDNA template; this was performed by using two rounds of PCR (first round, 5 μl of RT reaction product, 5 units of HotStar Taq Plus [Qiagen], 0.2 mM deoxynucleoside triphosphates [dNTPs], 0.5 μM nested constant-region primers [IgH consisting of IgM, IgD, IgG, IgA1, IgA2, Igκ, or Igλ], and matched variable-region primers; second round, 2.5 μl of the first-round reaction product, 5 units of HotStar Taq Plus (Qiagen), 0.2 mM dNTPs, and 0.5 μM nested constant-region and nested variable-region primers), as previously described (21). First-round PCR was cycled as follows: 95°C for 5 min; 35 cycles at 95°C for 30 s, 55°C (VH and Vκ) or 50°C (Vλ) for 60 s, and 72°C for 90 s; and one cycle at 72°C for 7 min. Second-round PCR was similar, except that the extension step was performed at 58°C (VH), 60°C (Vκ), or 64°C (Vλ). Samples of VH, Vκ, and Vλ chain PCR products were analyzed on 2% agarose gels (Qiagen). Products were sequenced in the forward and reverse directions by using a BigDye sequencing kit on an ABI 3700 instrument (Applied Biosystems, Foster City, CA). Sequence base calling was performed by using Phred (13, 14). V, D, and J region genes and mutations were analyzed by using the SoDA information system (55). The HCDR3 length was expressed according to the numbering system described previously by Kabat et al. (20). When more than one heavy- and light-chain pair was obtained from a single culture, the natural pairs were identified from sorted single B cells of the respective cultures by using the same procedure described above. All primers used for the second round of PCR included tag sequences at the 5′ end of each primer to permit the assembly of the VH and VL genes into functional linear Ig gene expression cassettes by overlapping PCR as reported elsewhere (21).
Expression of recombinant antibodies.
Isolated Ig V(D)J gene pairs were assembled by PCR into linear full-length Ig heavy- and light-chain gene expression cassettes as previously described (21). The human embryonic kidney cell line 293T (ATCC, Manassas, VA) was grown to near confluence in 6-well tissue culture plates (Becton Dickinson, Franklin Lakes, NJ) and transfected with 2 μg per well of purified PCR-produced IgH and IgL linear Ig gene expression cassettes by using Effectene (Qiagen). The supernatants were harvested from the transfected 293T cells after 3 days of incubation at 37°C in 5% CO2 (21).
Reversion of CH01 to CH04 lineage antibodies to unmutated ancestors.
Reverted unmutated ancestor (RUA) antibody sequences were inferred by using a novel method that combines likelihood-based phylogenetic analysis and Bayesian inference of gene segments (T. B. Kepler et al., unpublished data). Briefly, we constructed a phylogenetic tree with the observed immunoglobulin sequences at the leaves (using both heavy- and light-chain genes) and the unknown common ancestor at the root. We use a standard likelihood-based approach for the phylogenetic component (2). Given the maximum-likelihood tree, we computed the posterior probability mass function over all nucleotide identities at each position in the ancestor by summing over all possible gene segment combinations. The RUAs used were those sequences constructed with the maximum posterior probability nucleotide identities at each position.
Neutralization assay.
Neutralizing antibody assays with TZM-bl cells were performed as previously described (26). Antibodies were tested starting at a 50-μg/ml final concentration and titrated by using serial 3-fold dilutions. Pseudotyped lentiviruses were added to the antibody dilutions at a predetermined titer to produce measurable infection and incubated for 1 h. TZM-bl cells were added and incubated for 48 h before lysis, after which the supernatant was measured for firefly luciferase (Britelite Plus; Perkin-Elmer, Waltham, MA) activity by use of a luminometer. The data were calculated as a reduction in luminescence compared with control wells and are reported as the MAb 50% inhibitory concentration (IC50) in μg/ml (26). Further information on the viruses used for the neutralization screening is shown in Table S1 in the supplemental material. For the evaluation of effect of the kifunensine treatment on the neutralizing ability of the CH01 to CH04 MAbs, the assay was performed in the absence or presence of 50 μM kifunensine using two HIV-1 isolates, AE.CM244 and B.WITO, both produced in 293T cells.
Direct-binding ELISAs.
Three hundred eighty four-well plates (Corning Life Sciences, Lowell, MA) were coated overnight at 4°C with 15 μl of purified HIV-1 envelope proteins at optimized concentrations of either 1 or 2 μg/ml and blocked with assay diluent (PBS containing 4% [wt/vol] whey protein–15% normal goat serum–0.5% Tween 20–0.05% sodium azide) for 2 h at room temperature. The following HIV-1 envelope proteins were used for the enzyme-linked immunosorbent assay (ELISA) screening: gp140 00MSA, gp140 VRC A, and gp140 US-1 (subtype A); gp140 VRC B, gp140 JRFL, gp120 W61D, gp120 MN (generous gift of Carter Lee, Global Solutions for Infectious Diseases, South San Francisco, CA), and gp120 VBD2 (subtype B); gp120 A244 (generous gift of Carter Lee, Global Solutions for Infectious Diseases, South San Francisco, CA) and gp120 CM 243 (CRF01_AE); gp140 97CNGX2FΔCF, gp140 Du123, gp140CN54, and gp120 ZM651 (subtype C); gp140 DRCBL (subtype G); gp140 A1.con.env03ΔCF, gp140 AE.con.env03ΔCF, gp140 B.con.env03ΔCF, and gp140 C.con.env03ΔCF (subtype consensus); gp140 Con 6ΔCF and gp140 Con SΔCF (group M consensus based on 1999 and 2000 databases, respectively); and gp140 219, gp140 681-7, gp140 63521, gp140 6240, gp140 62357, gp140 9021, gp140 040, gp140 684-6, gp140 089C, gp140 1086C, and gp120 681-7 (transmitted/founders) (53; C. Y. Tsao, M. Bonsignori, and B. F. Haynes, unpublished data). Ten microliters per well of serial 5-fold dilutions of purified antibodies starting at 30 μg/ml was incubated for 2 h at room temperature on an orbital shaker, followed by washing with PBS–0.1% Tween 20. Fifteen microliters per well of horseradish peroxidase-conjugated goat anti-human IgG Fcγ (catalog number 109-035-098; Jackson ImmunoResearch Laboratories, West Grove, PA) at a 1:9,000 dilution in 5% goat serum–PBS was added for 1 h, washed, and detected with 30 μl/well of SureBlue Reserve TMB One Component microwell peroxidase substrate (catalog number 53-00-03; KPL, Gaithersburg, MD). Development was stopped with 15 μl/well of 0.1 M HCl (or 0.1 M H3PO4 in some assays), and plates were read at an optical density at 450 nm (OD450) with a VersaMax microplate reader (Molecular Devices, Sunnyvale, CA).
Surface plasmon resonance analysis of antibody reactivity.
Surface plasmon resonance (SPR) binding assays were performed with a BIAcore 3000 instrument (BIAcore Inc., Piscataway, NJ) maintained at 20°C. Anti-human IgG Fc protein was immobilized on a CM5 sensor chip to about 15,000 resonance units (RU), as previously described (1). Test antibodies were captured on each chip cell surface from 400 to 575 relative light units (RLUs) by flowing 5 μl of each antibody at a concentration of 100 μg/ml for 1 min. Envelope proteins were injected at 20 μl/min for 2 min over the captured antibodies. The specific binding responses of MAb binding were obtained following the subtraction of nonspecific binding on control surfaces. Bound envelope proteins were allowed to disassociate for 10 min before the chip surface was regenerated with glycin-HCl (pH 2.0). Rate constants were determined by repeating the SPR analysis for each antibody using serial dilutions of envelopes and global curve fitting analysis using the 1:1 Langmuir equation.
Binding antibody multiplex assay.
We used the Luminex platform to test for levels of polyreactivity of the purified antibodies as previously reported (51). Briefly, a total of 5 × 106 carboxylated fluorescent beads (Luminex Corporation, Austin, TX) were covalently coupled to 25 μg of each purified antigen tested (hepatitis C virus [HCV] E2, anaerobic and aerobic gut flora, and Wisconsin/influenza virus hemagglutinin) and incubated with purified antibodies at a concentration of 50 μg/ml. Antibody isotypes were detected with mouse anti-human IgG (Southern Biotech, Birmingham, AL), conjugated to phycoerythrin, at 4 μg/ml. Beads were then washed and acquired on a Bio-Plex instrument (Bio-Rad, Hercules, CA). Purified IgG proteins (Sigma-Aldrich) were utilized as positive controls in every assay. Background values (beads in the absence of detection antibody) and uninfected human plasma were utilized as negative controls.
Autoantibody Luminex assays.
Antibody binding to a panel of autoantigens was measured with a Luminex AtheNA Multi-Lyte ANA assay (Wampole Laboratories, Princeton, NJ) according to the kit manufacturer's protocol. Purified antibodies were tested in serial dilutions against the extractable nuclear antigens Sjogren's syndrome antigen A (SSA), SSB, Smith antigen (Sm), ribonucleoprotein (RNP), centromere B (Cent B), histone, scleroderma 70 (Scl 70), and Jo-1 and double-strand DNA (dsDNA).
HEp-2 cell staining.
Indirect immunofluorescence binding of MAbs to HEp-2 cells (Inverness Medical Professional Diagnostics, Princeton, NJ) was performed as previously described (18). Briefly, for each test antibody, 20 μl of antibody at a concentration of 50 μg/ml was aliquoted onto a predetermined spot on the surface of a slide (ANA HEp-2 kit). After incubation for 25 min at room temperature and washes, 20 μl of secondary antibody, goat anti-human Ig-fluorescein isothiocyanate (FITC) at 20 μg/ml (Southern Biotech), was added to each spot and incubated in a humid chamber for 25 min in the dark. After washing and drying, a drop of 33% glycerol was added to each spot, and the slide was covered with a 24- by 60-mm coverslip. Images were taken on an Olympus AX70 instrument with a SpotFlex FX1520 charge-coupled device (CCD) and with a UPlanFL 40× 0.75-numerical-aperture (NA) objective at 25°C in the FITC channel using SPOT software. All images were acquired for 12 s except for CH04 (10 s) and 2F5 (6 s). Image layout and scaling were performed with Adobe Photoshop without image manipulation.
Surface staining of cell surface-expressed gp160 envelope glycoproteins.
293T cells were transfected with gp160 proteins, and reactivity with MAbs was determined as previously described (58). Trimeric HIV-1 envelope glycoproteins were produced as previously described (27), by the transient transfection of 293T cells with pNL4-3.Luc.R−E− (6) and either a consensus C (conC) wild-type Env-expressing plasmid for homotrimeric Envs or a 1:2 mixture of conC wild-type and conC N160A-mutated Env-expressing plasmids for heterotrimeric Envs (56). Transfected 293T cells were used to test antibody binding to cell surface-expressed envelope trimers. We confirmed that both the homo- and heterotrimers were indeed expressed in a trimeric form by Western blot analysis of envelope glycoproteins, concentrated and processed from supernatants collected 2 days after transfection as previously described (27; data not shown).
Prediction of structures of monoclonal antibodies CH01 to CH04.
Structural models of the heavy and light chains of MAbs CH01 to CH04 were predicted by using a protocol previously described (31). Briefly, based on the sequence alignment, structural models were constructed by using Nest, a homology modeling program based on rigid-body optimization (34), and the model quality was evaluated by using a normalized DFIRE score (63).
Generation of CH01/CH02, CH01/CH03, CH02/CH03, and CH01 to CH03/PG16 chimeras.
CH01/CH02/CH03 heavy-chain/light-chain chimeras and CH01 to CH03/PG16 heavy-chain/light-chain chimeras were generated as previously described (31). Briefly, 250 μg of light-chain plasmid and 250 μg of heavy-chain plasmid were mixed with 1 ml of 293fectin (Invitrogen) for 20 min before the DNA-293fectin complex was added to 0.9 liters of FreeStyle 293F cells. The transfected cells were returned to suspension and incubated for 2 days, and the culture was then fed with 50 ml of CellBoost-5 protein-expression-enriched medium (HyClone, Logan, UT) and the protein expression enhancer sodium butyrate at a final concentration of 2 nM (Sigma). After 6 days posttransfection, supernatants were harvested, centrifuged, filtered through a 0.22-μm membrane, and purified through a protein A column (Pierce protein A plus agarose; Thermo, Rockford, IL). Purified antibodies were dialyzed against PBS, analyzed, and stored at −80°C if not used immediately.
RESULTS
Antibodies CH01 to CH04 are members of the same clonal lineage.
Monoclonal antibodies (MAbs) CH01 to CH04 are members of the same clonal family, based on the following traits: (i) the same V(D)J rearrangement, (ii) the same length of HCDR3, and (iii) the high similarity of HCDR3 and light chain complementarity-determining region 3 (LCDR3). The characteristics of the heavy and light chains of MAbs CH01 to CH04 are summarized in Table 1. Heavy-chain analysis showed that the mutation frequency of the VDJ nucleotide sequences ranges from 11.5% to 14.3%. CH01 to CH04 are IgG1 antibodies and share the same VH 3-20*1/JH 2*01 rearrangements. Their HCDR3s are 24 amino acids (aa) long (numbering system of Kabat et al. [20]) with a similar nucleotide makeup (nucleotide and amino acid sequences are shown in Fig. S2A and S2B in the supplemental material). They also share a similar D region, which may have resulted from a D-D fusion of 3-10*1 with any of several other D gene segments. N-insertions into the heavy chains had a shared nucleotide makeup compatible with a clonal relationship (Fig. S2A).
We generated a phylogenetic tree of MAbs CH01 to CH04, rooted on the nucleotide sequence of the common reverted unmutated ancestor (RUA) antibody, as described in Materials and Methods (see Fig. S3 in the supplemental material). MAb CH02 accumulated the largest amount of somatic mutations, followed by MAbs CH03 and CH01. MAb CH04 diverged from the other members of the clonal lineage at an early stage of evolution and accumulated the smallest amount of somatic mutations (Table 1 and Fig. S3; see also nucleotide sequences in Fig. S2A in the supplemental material). Heavy chains were paired to light chains with the same Vκ3-20/Jκ1 rearrangement (Table 1) that had an LCDR3 of the same length (9 amino acids by the numbering of Kabat et al.) and similar N-insertions (Fig. S2C and S2D).
CH01 to CH04 are broadly neutralizing antibodies.
We tested the neutralization breadth of MAbs CH01 to CH04 against a panel of 91 HIV-1 primary isolates in a TZM-bl Env pseudovirus neutralization assay (Fig. 2). Tier 1A and 1B viruses are similar to laboratory-adapted strains and are very easy to neutralize (43). Tier 2 HIV-1 strains are derived from field isolates and are more difficult to neutralize (43). The virus panel was comprised of 4 tier 1A isolates, 3 tier 1B isolates (2 of clade B and 1 of CRF01_AE), and 84 tier 2 isolates, which included 10 clade A, 21 clade B, 25 clade C, 4 clade D, 5 clade G, 1 clade AD, 10 CRF01_AE, and 8 CRF02_AG viruses.
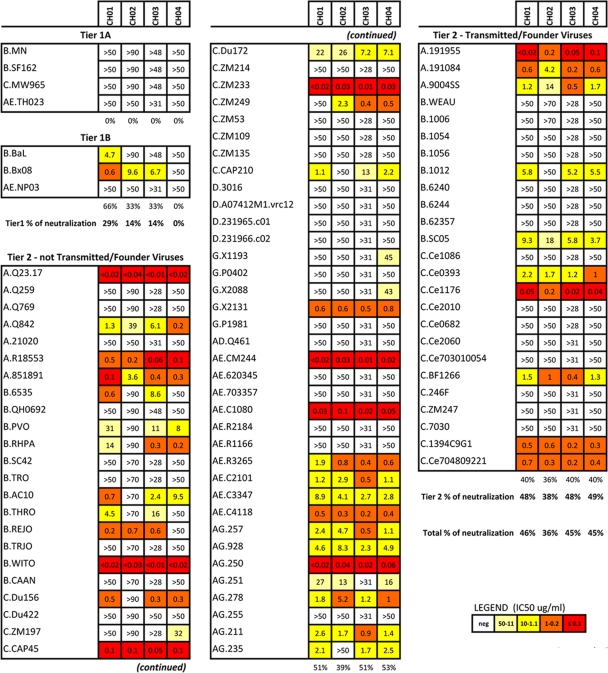
Fig. 2.
Neutralization profile of monoclonal antibodies CH01 to CH04. The neutralizing activity of MAbs CH01 to CH04 was tested on a panel of 91 pseudotyped lentiviruses, which comprised tier 1 and tier 2 isolates as well as transmitted founder viruses from multiple clades. Results are shown as IC50s (μg/ml) and color coded as shown in the key.
The four antibodies shared similar patterns of neutralization across multiple clades (Fig. 2), with breadth ranging from 46% (42/91 isolates) for MAb CH01 to 36% (33/91 isolates) for MAb CH02. Both MAbs CH03 and CH04 neutralized 45% (41/91) of the viruses. None of the antibodies neutralized tier 1A isolates. Tier 1B isolates were neutralized only by MAbs CH01 (2 out of 3), CH02, and CH03 (1 out of 3) but not by MAb CH04 (Fig. 2). Conversely, 49% of tier 2 viruses were neutralized. MAb CH01 preferentially neutralized CRF02_AG isolates (7/8; 88%), followed by clade A (7/10; 70%), CRF01_AE (6/10; 60%), clade B (9/21; 43%), clade C (10/25; 40%), and clade G (1/5; 20%) isolates. Clade D viruses were not neutralized by MAbs CH01 to CH04. It is important that MAbs CH01 to CH04 all strongly neutralized AE.CM244, the parental virus of the E.A244 gp120 envelope glycoprotein that, as described below, is strongly bound by MAbs CH01 to CH04 and their RUAs (Fig. 2). MAbs CH01, CH03, and CH04 also neutralized 10/25 (40%) transmitted/founder viruses: 3/3 (100%) clade A, 2/9 (22%) clade B, and 5/13 (39%) clade C viruses (Fig. 2). MAb CH02 neutralized the same transmitted/founder viruses, with the exception of B.1012 (Fig. 2). In terms of potency, MAbs CH01 to CH04 showed a mean IC50 ranging from 2.4 to 5.6 μg/ml, with MAb CH03 being the most potent (mean IC50 of 2.4 μg/ml and median IC50 of 0.46 μg/ml). These data indicate that the clonal family of antibodies CH01 to CH04 broadly neutralizes tier 2 viruses from multiple clades, including transmitted/founder viruses.
Since MAbs CH01 to CH04 had neutralization breadth for tier 2 primary isolates but poorly neutralized tier 1 HIV-1-pseudotyped lentiviruses, a characteristic of antibodies PG9 and PG16 (56), we compared them with MAbs PG9 and PG16 against a panel of 20 tier 2 pseudotyped lentiviruses. Figure S4 in the supplemental material shows that MAbs PG9 and PG16 have a broader spectrum of neutralization than MAbs CH01 to CH04 (20/20 versus 15/20 viruses neutralized). Viruses that were potently neutralized by MAbs CH01 to CH04 were also potently neutralized by MAbs PG9 and PG16 at IC50s of <0.02 μg/ml, but the IC50s of PG9 and PG16 were higher for viruses that were resistant to MAbs CH01 to CH04 (i.e., B.TRJO, G.P1981, and AD.Q461). These data suggest that MAbs CH01 to CH04 may recognize epitopes that are overlapping those of MAbs PG9 and PG16 and that MAbs CH01 to CH04 might be more sensitive than MAbs PG9 and PG16 to similar modifications in the conformation of the cognate epitope.
Breadth of binding of CH01 to CH04 to recombinant envelope glycoproteins.
We next assessed the binding reactivities of MAbs CH01 to CH04 to a panel of 32 recombinant chronic, consensus, and transmitted/founder gp120 or gp140 Envs by an ELISA (Table 2). PG9 and PG16 MAbs have been previously shown to bind to overlapping conformational epitopes (currently referred to as “quaternary”) preferentially, but not exclusively, expressed in the context of native trimers in the V2/V3 region of the gp120 envelope glycoprotein (9, 56). Only 3 out 32 Envs (9.4%) were bound by one or more of MAbs CH01 to CH04: MAbs CH01 to CH04 reacted well with E.A244 gp120 Env and weakly with another CRF01_AE gp120 Env, CM243. MAbs CH01 and CH02 also reacted weakly with a transmitted/founder gp140 Env, B.9021 (Table 2). The substantially narrower spectrum of binding reactivity to gp120 Envs despite a wide breadth of neutralization was another observation consistent with the hypothesis that MAbs CH01 to CH04 are PG9/PG16-like MAbs.
Table 2.
Binding of monoclonal antibodies CH01 to CH04 to gp120/gp140 monomeric envelopesa
| Strain | Source | Envelope glycoprotein | EC50 (μM) |
||||
|---|---|---|---|---|---|---|---|
| CH01 | CH02 | CH03 | CH04 | Synagis | |||
| A.00MSA | Chronic | gp140 | — | — | — | — | — |
| A.VRC | Chronic | gp140 | — | — | — | — | — |
| US-1b | Chronic | gp140 | — | — | — | — | — |
| B.VRC | Chronic | gp140 | — | — | — | — | — |
| B.JRFL | Chronic | gp140 | — | — | — | — | — |
| C.97CNGX2F | Chronic | gp140ΔCF | — | — | — | — | — |
| C.Du123 | Chronic | gp140 | — | — | — | — | — |
| C.CN54 | Chronic | gp140 | — | — | — | — | — |
| G.DRCBL | Chronic | gp140 | — | — | — | — | — |
| B.W61D | Chronic | gp120 | — | — | — | — | — |
| B.MN | Chronic | gp120 | — | — | — | — | — |
| B.VBD2 | Chronic | gp120 | — | — | — | — | — |
| E.A244 | Chronic | gp120 | 7.8 | 150 | 34.5 | 23.1 | — |
| C.ZM651 | Chronic | gp120 | — | — | — | — | — |
| AE.CM 243 | Chronic | gp120 | 12.7 | >666.7 | 97.3 | >666.7 | — |
| A1.con.env03 | Consensus | gp140ΔCF | — | — | — | — | — |
| AE.con.env03 | Consensus | gp140ΔCF | — | — | — | — | — |
| B.con.env03 | Consensus | gp140ΔCF | — | — | — | — | — |
| C.con.env03 | Consensus | gp140ΔCF | — | — | — | — | — |
| M.Con6 | Consensus | gp140ΔCF | — | — | — | — | — |
| M.ConS | Consensus | gp140ΔCFI | — | — | — | — | — |
| A.219 | T/F | gp140 | — | — | — | — | NA |
| B.681-7 | T/F | gp140 | — | — | — | — | — |
| B.63521 | T/F | gp140 | — | — | — | — | — |
| B.6240 | T/F | gp140 | — | — | — | — | — |
| B.62357 | T/F | gp140 | — | — | — | — | — |
| B.9021 | T/F | gp140 | 63.2 | 240 | — | — | — |
| B.040 | T/F | gp140 | — | — | — | — | — |
| B.684-6 | T/F | gp140 | — | — | — | — | — |
| C.089C | T/F | gp140 | — | — | — | — | — |
| C.1086C | T/F | gp140 | — | — | — | — | — |
| B.681-7 | T/F | gp120 | — | — | — | — | — |
The table shows 50% effective concentration (EC50) values determined from direct-binding ELISAs. “—” indicates negative results, as defined by OD450 readings below twice the background at the highest antibody concentration tested (see Materials and Methods). EC50 values reported as being >666.7 μM indicate that positive readouts were detected at the highest concentrations tested but that the predicted EC50 was higher than 666.7 μM and could not be accurately calculated. T/F, transmitted/founder envelope; NA, not applicable.
Simian immunodeficiency virus (SIV) gp140 envelope glycoprotein.
Antibodies CH01 to CH04 recognize a V2/V3 conformational epitope of the HIV-1 gp120 envelope.
We asked if MAbs CH01 to CH04 recognized a conformational epitope overlapping those of PG9 and PG16 (9, 10, 31, 33, 56). The criteria previously proposed by Walker et al. to define conformational PG9/PG16-like antibodies are (i) sensitivity to loss of N-linked glycans at amino acid 160 in the V2 envelope region, (ii) preferential MAb binding to native trimers versus monomers, and (iii) abrogation of the neutralization of viruses produced in cells treated with the mannosidase I inhibitor kifunensine, which prevents the processing of terminal mannose residues, resulting in the overexpression of high-mannose glycans on the envelope glycoproteins (10, 56). In addition, we threaded the CH01 amino acid sequence on the PG16 structure to determine if a similar antibody structure was predicted.
First, we asked if the point mutation of asparagine (N) to lysine in position 160 (N160K; HxB2 reference numbering) of gp120 V2 would abrogate the binding and neutralization of MAbs CH01 to CH04 (9, 10, 56, 59). Indeed, MAbs CH01 to CH04 did not neutralize (IC50 > 50 μg/ml) N160K mutant forms of otherwise neutralization-sensitive A.Q23.17 and B.JR-CSF isolates (see Fig. S5A in the supplemental material). In addition, neither MAb CH01 nor PG9/PG16 neutralized wild-type isolate B.JRFL. A single mutation at position 168 (E168K) reconstituted a properly conformed epitope and resulted in potent neutralization (IC50s of 0.044, 0.008, and 0.003 μg/ml for CH01, PG9, and PG16, respectively), but the subsequent introduction of the N160K mutation in B.JRFL.E168K reverted the effect of the E168K mutation, making isolate B.JRFL.E168K.N160K neutralization resistant (IC50 > 50 μg/ml) to MAbs CH01, PG9, and PG16 (Fig. S5A).
Next, we evaluated the effect of the gp120 N160Q mutation on the ability of these antibodies to bind cell surface-expressed HIV-1 envelope trimers (see Fig. S5B in the supplemental material). The binding of MAbs CH01 to CH04 to two gp145ΔCFI molecules, ZM233 and DU422 (which bind to both antibodies PG9 and PG16 [56]), was completely abrogated by the N160Q mutation.
Using flow cytometric staining, we asked whether MAbs CH01 to CH04 bound to a single protomer or cross-linked two gp120 subunits in the context of cell surface-expressed envelope trimers (32). We compared the levels of binding of MAbs CH01, CH02, and CH04 to wild-type consensus C (conC) trimeric envelope and conC heterotrimers containing both wild-type and N160A-mutated gp120 molecules cotransfected in 293T cells at a 1:2 ratio. IgG1b12 MAb, which is not sensitive to the N160A mutation in the conC gp120 envelope glycoprotein, bound to both the homo- and heterotrimers at comparable levels (see Fig. S5C in the supplemental material), indicating no differences in the overall levels of envelope cell surface expression. Conversely, MAb PG16 bound less efficiently to heterotrimers (Fig. S5C). The level of binding of MAbs CH01, CH02, and CH04 to the heterotrimers was reduced proportionally to the amount of input N160A envelope glycoprotein protomers in the mix (Fig. S5C). This result suggests binding to one gp120 subunit in the context of the trimeric spike rather than the cross-linking of two protomers, as previously shown by Walker et al. for MAbs PG9 and PG16 (56).
Next, we evaluated the role of HIV-1 glycosylation in the neutralizing activity of these antibodies by treating 293T cells producing pseudotyped lentiviruses with kifunensine, a mannosidase I inhibitor (10, 56). The production of AE.CM244- and B.WITO-pseudotyped lentiviruses in kifunensine-treated 293T cells abrogated the neutralization of these viruses by MAbs CH01 to CH04 (see Fig. S5D in the supplemental material).
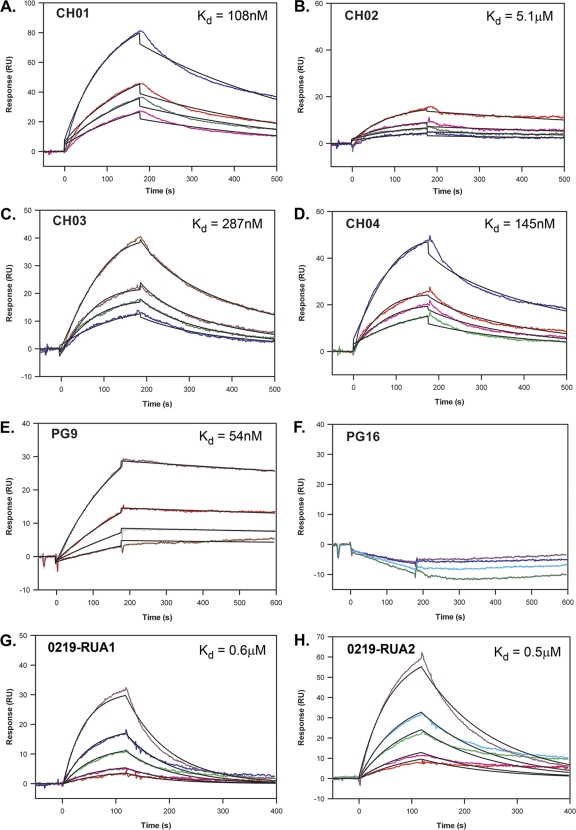
Finally, to confirm that MAbs CH01 to CH04 bind to an epitope that overlaps that of PG9, we determined the ability of MAbs CH01 to CH04 to compete with MAb PG9 for binding to the E.A244 gp120 envelope glycoprotein and vice versa in SPR competition assays. We first determined by SPR analysis that E.A244 gp120 Env bound to each of the mature antibodies (Fig. 3A to E) with dissociation constants (Kds) ranging from 108 to 287 nM for MAbs CH01, CH03, and CH04, while CH02 had a higher Kd of 5.1 μM. The dissociation constant for MAb PG9 was 54 nM, whereas MAb PG16 did not bind to E.A244 gp120 Env (Fig. 3F and G).
Fig. 3.
Determination of the dissociation constant (Kd) from the gp120 E.A244 envelope glycoprotein by surface plasmon resonance (SPR) analysis. Serial dilutions ranging from 100 to 10 μg/ml of MAbs CH01 to CH04 (A to D, respectively), PG9 (E), PG16 (F), and putative reverted unmutated ancestor antibodies of the clonal lineage of CH01 to CH04 (G and H) were tested by SPR to identify the dissociation constants from the gp120 E.A244 envelope glycoprotein. The global curve-fitting χ2 values were between 1.9 and 0.08. The data shown are representative of duplicate experiments.
Figure S5E in the supplemental material shows that MAbs CH01 to CH04 and PG9 blocked each other's binding to the E.A244 gp120 envelope glycoprotein. MAbs CH01 to CH04 blocked 39% (CH02) to 71% (CH04) of PG9 binding to E.A244 gp120 Env. The finding that CH02 was less efficient in blocking PG9 binding was not surprising, considering the fast off-rate of CH02 (Fig. 3B) and the 10-fold difference in the constants of dissociation between the two antibodies. Conversely, PG9 blocked the binding of MAbs CH01 to CH04 only partially (18 to 27%) (Fig. S5E). These data suggest that MAbs CH01 to CH04 and PG9 bind to overlapping epitopes.
Taken together, the data from the functional studies described above demonstrate that MAbs CH01 to CH04 recognize a conformational epitope on the HIV-1 envelope glycoprotein similar to those of MAbs PG9 and PG16.
To determine the level of similarity between MAbs CH01 to CH04 and PG9/PG16, we tested the neutralizing activities of chimeric antibodies with permutations of the heavy and light chains of MAbs from the CH and PG lineages (see Tables S2A to S2D in the supplemental material) and within the CH lineage (Tables S2E and S2F) to assess the individual contributions of the heavy and light chains of the CH MAbs to their activity, as previously described for MAbs 2909 and PG9/PG16 (4, 31). We found that all CH/PG chimeric antibodies lost their ability to neutralize the virus (Tables S2A to S2D), indicating a substantial lack of complementation between the heavy and light chains of the CH and PG MAbs (4). In contrast, chimeras obtained from the permutations of heavy and light chains of antibodies CH01, CH02, and CH03 did mediate neutralization at levels comparable to those of the natural antibodies (Tables S2E and S2F).
In addition, we evaluated the structural similarity of MAb CH01 and MAb PG16 compared with other anti-gp120 human MAbs by threading, a technique that can predict whether two sequences that do not have obvious sequence homology may share a similar structural conformation (34). The model was made by aligning each amino acid of the test sequence to the template structure and then evaluating how well the test sequence fits the template. Lower DFIRE scores indicated a better test sequence fit to the template structure (34). We aligned the heavy chain of MAb CH01 to that of MAb PG16 in two possible ways to optimally match either the “blade” (see alignment 1 in Fig. S6A in the supplemental material) or the “butt” (alignment 2 in Fig. S6A) of the PG16 HCDR3 structure (31, 33). The two alternative threaded models of the CH antibodies on the PG16 Fab structure produced lower DFIRE scores for the second alignment, indicating that the alignment of the CH antibodies matching the “butt” of the PG16 HCDR3 structure provided the best fit (Fig. S6B). Using the same modeling technique, we predicted structures of antibodies CH01 to CH04 threaded onto 7 anti-CD4i antibody structures (47e, 412d, 17b, 48d, E51, and X5) (Fig. S6C). We compared the DFIRE scores of the 7 models using score normalization as described in Materials and Methods. Briefly, a normalized DFIRE score of 1.0 indicated that the prediction matched a known structure, whereas higher normalized DFIRE scores indicated a lower probability that the prediction was accurate. The sequences of CH01 to CH04 fit the PG16 structure (normalized DFIRE scores of 1.1 and 1.2) better than any other structure (normalized DFIRE scores ranging from 1.4 to 4.7) (Fig. S6C).
Neutralization epitope mapping of CH01 to CH04.
The ability of MAbs CH01, CH02, and CH04 to neutralize consensus C (conC) viruses with point mutations in the V2 and V3 regions was assessed and compared to that of MAbs PG9 and PG16 (see Table S3 in the supplemental material). All antibodies neutralized the unmutated conC virus with IC50s ranging from <0.002 to 1.5 μg/ml (Table S3). CH lineage neutralization of the conC virus was completely abrogated by the V127A, F159A, N160A, K169E, K171A, and I181A mutations (Table S3). In addition, neutralization was decreased by the R166A and the K168A mutations, as measured by the >3-fold increase of the IC50 against these mutants (Table S3). CH MAbs were more sensitive to the mutations of V127, F159, K171, and I181 than were MAbs PG9 and PG16 (Table S3). In particular, it should be noted that, overall, MAb CH02 was more sensitive to mutations throughout the V2/V3 region; indeed, CH02 had the narrowest breadth of neutralization among MAbs CH01 to CH04 (Fig. 4). Analysis of amino acids at positions 127, 159, 160, 168, 169, 171, and 181 of the viruses used in the neutralization assay confirmed that the substitution of the asparagine in position 160 was always associated with the inability to neutralize virus but failed to indicate any other direct association between a specific amino acid in a given position and the ability of the CH MAbs to neutralize virus (data not shown). These results are in line with similar findings recently described by Euler and colleagues for MAbs PG9 and PG16 (12). Overall, these data indicated that despite the CH MAbs being in the same lineage and recognizing a conformational epitope overlapping those of PG9 and PG16, they have distinct fine specificities compared with MAbs PG9 and PG16. It is likely that these fine specificities reflect the differential potency and breadth of neutralization of each of MAbs CH01 to CH04 and of the CH lineage compared to MAbs PG9 and PG16.
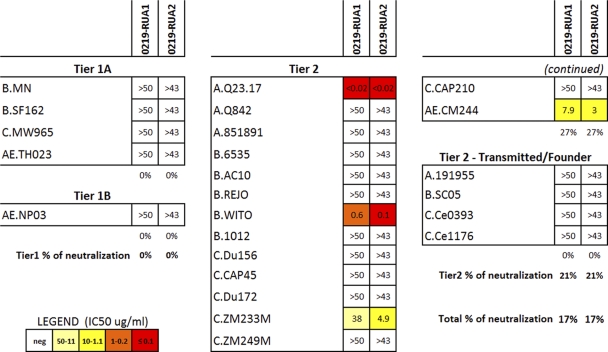
Fig. 4.
Neutralizing activity of the putative reverted unmutated ancestor antibody candidates of the CH01 to CH04 clonal family antibodies. Two putative reverted ancestor antibody sequences (0219-RUA1 and 0219-RUA2) common to MAbs CH01 to CH04 were inferred and expressed as described in Materials and Methods. The two RUAs differed by a single nonsynonymous nucleotide mutation. The breadth of neutralization of 0219-RUA1 and 0219-RUA2 was tested against a panel of 24 pseudotyped lentiviruses comprising tier 1 and tier 2 isolates. Results are shown as IC50s (μg/ml) and color coded as shown in the key.
Neutralizing and binding activities of the reverted unmutated ancestors of CH01 to CH04.
Xiao et al. postulated previously that one reason why broadly neutralizing antibodies are so difficult to induce is that their putative ancestral naïve B cell receptors do not bind to HIV-1 envelope glycoproteins (61). The inference of reverted unmutated ancestors (RUAs) for individual Ig genes may have a high uncertainty in V(D)J due to both somatic mutations and N-nucleotide additions; this uncertainty may be reduced by utilizing information available within clonal lineage families with multiple members, as in the case of MAbs CH01 to CH04 (Kepler et al., unpublished). The inferred RUAs of the family of CH MAbs were uncertain at one nonsynonymous position (G/T at position 401 in HCDR3) (see Fig. S2A in the supplemental material); therefore, we synthesized two candidate RUAs (0219-RUA1 and 0219-RUA2) (nucleotide and amino acid sequences of the heavy and light chains are shown in Fig. S2A to S2D in the supplemental material). In contrast to the mature antibodies, the inferred RUAs did not show breadth of neutralization but retained the ability to potently neutralize a few HIV-1 strains: AE.CM244 (IC50 < 10 μg/ml), A.Q23.17 (IC50 < 0.02 μg/ml), and B.WITO (IC50 < 1 μg/ml) (Fig. 4). Interestingly, C.ZM233M, which is neutralized by the V-gene-reverted PG9 antibody (31), was also neutralized, with IC50s of 38 μg/ml for 0219-RUA1 and 4.88 μg/ml for 0219-RUA2.
To evaluate the ability of RUAs to bind to monomeric envelope glycoproteins, we measured the dissociation constants of the two inferred RUAs by SPR, employing the higher sensitivity of this technique due to the predicted low affinity of the RUAs (8, 15, 44, 50). We found that both 0219-RUA1 and 0219-RUA2 bound to the E.A244 gp120 molecule with affinities sufficient to trigger the affinity maturation of unmutated B cells in vivo (Kd = 0.6 μM and 0.5 μM, respectively) (Fig. 3H and I) (8, 15, 44, 50). It should be noted that AE.CM244 is the parental virus of the E.A244 gp120 envelope glycoprotein.
Polyreactivity profile of broadly neutralizing antibodies CH01 to CH04.
Finally, we evaluated the auto- and polyreactivities of MAbs CH01 to CH04 (see Fig. S7A to S7C in the supplemental material). MAb CH03 reacted with histones, ribonucleoprotein (RNP), centromere B autoantigens (Fig. S7A), and, weakly, human epithelial HEp-2 cells (Fig. S7B). MAbs CH01, CH02, and CH03 bound to the hepatitis C virus E2 envelope glycoprotein and to whole-cell extracts of anaerobic and aerobic gut flora (Fig. S7C). These data demonstrate fluctuation in the degree of autoreactivity and polyreactivity within the same clonal lineage family. Neither MAb PG9 nor PG16 was autoreactive or polyreactive in the above-described assays.
DISCUSSION
In this study, we identified a set (CH01 to CH04) of broadly neutralizing HIV-1 antibodies specific for V2/V3 conformational epitopes that were recovered from clonally related memory B cells of a broad neutralizer African subject chronically infected with a clade A HIV-1 strain.
MAbs CH01 to CH04 recognize a PG9/PG16-like conformational epitope and neutralized 36% (CH02) to 47% (CH04) of 89 tier 2 pseudotyped viruses. Despite notable similarities, differences in breadths of neutralization and sensitivities to amino acids at positions 127, 159, 171, and 181 indicate either that MAbs CH01 to CH04 bind to a discretely different epitope or that they approach the same epitopes of PG9 and PG16 but in a different orientation. Similarly to MAbs PG9 and PG16, MAbs CH01 to CH04 recognize an epitope on a single protomer that is usually, but not exclusively, conferred to the gp120 envelope glycoprotein by trimer formation (9, 56). Interestingly, the two recombinant Env proteins that reacted with all members of the clone, the E.A244 and AE.CM243 gp120 Env proteins, were both AE_01 recombinant envelope glycoproteins. The E.A244 gp120 envelope glycoprotein was a component of the RV144 Thai trial that showed 31% efficacy (36). It will be of interest to determine if conformational V2/V3 epitopes expressed on the E.A244 gp120 envelope glycoprotein were immunogenic in the RV144 trial and if it gave rise to any component of the neutralizing antibody response.
Unlike MAbs PG9 and PG16, MAb CH03 was autoreactive for ribonucleoprotein, centromere B, and histone antigens, and MAbs CH01 to CH03 were polyreactive with the hepatitis C virus E2 protein or gut flora antigens, raising the possibility that MAbs CH01 to CH04, like broadly reactive gp41-neutralizing antibodies (54), may be subjected to tolerance mechanisms. Whether the degree of autoreactivity of MAb CH03 is sufficient to engage peripheral tolerance mechanisms such as anergy or deletion is not yet known (8, 15, 44, 50). The high degree of somatic mutations of the VDJ nucleotide sequences of CH01 to CH04 (11.5 to 14.3%) is similar to those of MAbs PG9 (11.9%) and PG16 (13.2%), and, as has been noted elsewhere, this level of mutation is considerable and may have implications for eliciting these kinds of responses (31). Antibodies with this degree of mutation may be less common for two reasons: (i) the need to have survived multiple rounds of tolerance elimination to delete autoreactive cells or (ii) because individuals capable of making broadly neutralizing antibodies due to a decreased capacity to eliminate such cells may be less common. The finding that MAbs CH01 to CH04 are not autoreactive, with the exception of CH03, which shows some degree of autoreactivity, implies the former, i.e., that these are uncommon events. However, more studies will be needed to address these points.
While the precise etiology of long HCDR3 in immune responses is still unknown, several contributing factors have been proposed, such as prolonged antigen stimulation (41), D-D fusions (5, 23), or a lack of selection against long HCDR3 (24, 45). That MAbs CH01 to CH04 have a long HCDR3 (24 aa), like PG9 and PG16 (28 aa), suggests a common mechanism of generation or selection of antibodies with a long HCDR3. Interestingly, the long HCDR3 of CH01 to CH04 appears to have resulted from a D-D fusion of 3-10*1 with one of several other D gene segments.
Of great importance is the quest to find immunogens that can be used to attempt to induce conformational-epitope-specific antibodies such as PG9, PG16, and CH01 to CH04 (9, 12, 28, 31). This class of antibodies is characterized by high levels of somatic mutations, which suggests that the profiles of binding of the naïve B cell precursor and the mature antibodies to monomeric envelope glycoproteins used as immunogens might differ. Therefore, one can envision a vaccine strategy comprising immunogen components that both trigger the naïve B cell and drive development toward the desired mature antibody. This four-member clonal lineage of broadly neutralizing antibodies allowed us to more accurately infer the common reverted unmutated ancestor, to test it against a panel of recombinant envelope glycoprotein monomers and oligomers, as well as to discern its abilities to neutralize HIV-1.
The RUAs of MAbs CH01 to CH04 bound to E.A244 gp120 Env at an affinity anticipated to be biologically significant for naïve B cell triggering (8, 44). In addition, isolate C.ZM233 was neutralized by the RUAs of CH01 to CH04 and was the only isolate also neutralized by the PG9 RUA (31), suggesting that this isolate expresses epitopes recognized by RUAs of both CH01 to CH04 and PG9. Envelope glycoprotein E.A244 gp120 was also bound by all the mature MAbs CH01 to CH04 as well as AE.CM243 gp120 Env. In addition, a transmitted/founder virus Env gp140, B.9021, weakly bound to MAbs CH01 and CH02. In a different study, we also found two additional HIV-1 gp140 envelope glycoproteins from transmitted/founder viruses that bind to MAbs PG9 and PG16 (B.63521 and B.6240) (53; Tsao et al., unpublished). Thus, an immunization strategy where one or more of the E.A244, AE.CM243, B.9021, B.63521, and B.6240 Envs are used to prime and boost alone, together or in a sequence, to trigger and drive the development of naïve B cells may be capable of eliciting broadly neutralizing antibodies reacting with V2/V3 conformational epitopes. Certainly, the ability of the RUAs to neutralize 4 of the HIV-1 isolates tested suggests that the putative RUAs of MAbs CH01 to CH04 are excellent targets for vaccine design, but, as with MAbs PG9 and PG16 (31), affinity maturation is required to develop breadth of neutralization.
Thus, in summary, we report a combination strategy of memory B cell near-clonal cultures, single-cell sorting of memory B cell cultures, and EBV transformation to isolate multiple members of a clonal lineage of V2/V3 gp120 Env conformational epitope-specific broadly neutralizing antibodies. From the analysis of the reactivities of mature and RUA antibodies, we identified recombinant envelope glycoproteins that react, providing candidate immunogens for the development of immunization regimens optimized to induce V2/V3 conformational epitope-specific broadly neutralizing antibodies.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Ryan Meyerhoff, Miroslawa Bilska, Robert J. Parks, Andrew Foulger, Michele Donathan, Thaddeus Gurley, Josh A. Eudailey, and Daniel M. Kozink for expert technical assistance; Kara Anasti and Frederick Jaeger for SPR measurements; and Feng Gao for Env sequence alignments. We thank Carter Lee of Global Solutions for Infectious Diseases (GSID) for kindly providing the A244 and MN gp120 envelope glycoproteins. We thank James Robinson, Micah Luftig, and Jerome Kim for expert advice and discussions.
Single-cell sorting was performed in the Research Flow Cytometry Core Facility at the Duke Human Vaccine Institute, partly supported by the NIH-funded Duke Center for AIDS Research (CFAR) (grant AI64518), and directed by J.F.W. This work was supported by the NIH, NIAID, the Division of AIDS with the Center for HIV/AIDS Vaccine Immunology (CHAVI) (grant U19 AI067854); by a Collaboration for AIDS Vaccine Discovery grant to B.F.H. from the Bill and Melinda Gates Foundation; in part by a grant from the intramural research program of the Vaccine Research Center, NIAID, NIH, to J.R.M., P.D.K., and G.J.N.; and by a grant from the International AIDS Vaccine Initiative to J.R.M., P.D.K., and S.P.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 27 July 2011.
REFERENCES
- 1. Alam S. M., et al. 2007. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 178:4424–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bryant D., Galtier N., Poursat M. A. 2007. Likelihood calculation in molecular phylogenetics, p. 33–62 In Gascual O. (ed.), Mathematics of evolution and phylogeny. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 3. Burton D. R., et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027 [DOI] [PubMed] [Google Scholar]
- 4. Changela A., et al. 2011. Crystal structure of human antibody 2909 reveals conserved features of quaternary structure-specific antibodies that potently neutralize HIV-1. J. Virol. 85:2524–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collins A. M., et al. 2004. Partitioning of rearranged Ig genes by mutation analysis demonstrates D-D fusion and V gene replacement in the expressed human repertoire. J. Immunol. 172:340–348 [DOI] [PubMed] [Google Scholar]
- 6. Connor R. I., Chen B. K., Choe S., Landau N. R. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944 [DOI] [PubMed] [Google Scholar]
- 7. Corti D., et al. 2010. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One 5:e8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dal Porto J. M., Haberman A. M., Kelsoe G., Shlomchik M. J. 2002. Very low affinity B cells form germinal centers, become memory B cells, and participate in secondary immune responses when higher affinity competition is reduced. J. Exp. Med. 195:1215–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davenport T. M., et al. 2011. Binding interactions between soluble HIV envelope glycoproteins and quaternary-structure-specific MAbs PG9 and PG16. J. Virol. 85:7095–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doores K. J., Burton D. R. 2010. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J. Virol. 84:10510–10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doria-Rose N. A., et al. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84:1631–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Euler Z., et al. 2011. Activity of broadly neutralizing antibodies, including PG9, PG16, and VRC01, against recently transmitted subtype B HIV-1 variants from early and late in the epidemic. J. Virol. 85:7236–7245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ewing B., Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 14. Ewing B., Hillier L., Wendl M. C., Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 15. Gay D., Saunders T., Camper S., Weigert M. 1993. Receptor editing: an approach by autoreactive B cells to escape tolerance. J. Exp. Med. 177:999–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gray E. S., et al. 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 85:4828–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gray E. S., et al. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J. Virol. 83:8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haynes B. F., et al. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308:1906–1908 [DOI] [PubMed] [Google Scholar]
- 19. Henderson E., Miller G., Robinson J., Heston L. 1977. Efficiency of transformation of lymphocytes by Epstein-Barr virus. Virology 76:152–163 [DOI] [PubMed] [Google Scholar]
- 20. Kabat E. A., Wu T. T., Perry H. M., Gottesman K. S., Foeller C. 1991. Sequences of proteins of immunological interest, 5th ed. National Institutes of Health, U.S. Department of Health and Human Services, Bethesda, MD [Google Scholar]
- 21. Liao H. X., et al. 2009. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J. Virol. Methods 158:171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McElrath M. J., Haynes B. F. 2010. Induction of immunity to human immunodeficiency virus type-1 by vaccination. Immunity 33:542–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meek K., Hasemann C., Capra J. D. 1989. Novel rearrangements at the immunoglobulin D locus. J. Exp. Med. 170:39–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meffre E., et al. 2001. Immunoglobulin heavy chain expression shapes the B cell receptor repertoire in human B cell development. J. Clin. Invest. 108:879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mikell I., et al. 2011. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 7:e1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Montefiori D. C. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. 12:12.11. [DOI] [PubMed] [Google Scholar]
- 27. Moore P. L., et al. 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80:2515–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moore P. L., et al. 2011. Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop. J. Virol. 85:3128–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mouquet H., et al. 2010. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature 467:591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muster T., et al. 1994. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J. Virol. 68:4031–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pancera M., et al. 2010. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J. Virol. 84:8098–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pancera M., Wyatt R. 2005. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology 332:145–156 [DOI] [PubMed] [Google Scholar]
- 33. Pejchal R., et al. 2010. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl. Acad. Sci. U. S. A. 107:11483–11488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Petrey D., et al. 2003. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins 53(Suppl. 6):430–435 [DOI] [PubMed] [Google Scholar]
- 35. Pietzsch J., et al. 2010. Human anti-HIV-neutralizing antibodies frequently target a conserved epitope essential for viral fitness. J. Exp. Med. 207:1995–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rerks-Ngarm S., et al. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 37. Richman D. D., Wrin T., Little S. J., Petropoulos C. J. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sanders R. W., et al. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saphire E. O., et al. 2001. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293:1155–1159 [DOI] [PubMed] [Google Scholar]
- 40. Sather D. N., et al. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scheid J. F., et al. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636–640 [DOI] [PubMed] [Google Scholar]
- 42. Scheid J. F., et al. 2009. A method for identification of HIV gp140 binding memory B cells in human blood. J. Immunol. Methods 343:65–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seaman M. S., et al. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shih T. A., Meffre E., Roederer M., Nussenzweig M. C. 2002. Role of BCR affinity in T cell dependent antibody responses in vivo. Nat. Immunol. 3:570–575 [DOI] [PubMed] [Google Scholar]
- 45. Shiokawa S., et al. 1999. IgM heavy chain complementarity-determining region 3 diversity is constrained by genetic and somatic mechanisms until two months after birth. J. Immunol. 162:6060–6070 [PubMed] [Google Scholar]
- 46. Simek M. D., et al. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smith K., et al. 2009. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 4:372–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stamatatos L., Morris L., Burton D. R., Mascola J. R. 2009. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat. Med. 15:866–870 [DOI] [PubMed] [Google Scholar]
- 49. Sugden B., Mark W. 1977. Clonal transformation of adult human leukocytes by Epstein-Barr virus. J. Virol. 23:503–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tiegs S. L., Russell D. M., Nemazee D. 1993. Receptor editing in self-reactive bone marrow B cells. J. Exp. Med. 177:1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tomaras G. D., et al. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82:12449–12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Trkola A., et al. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tsao C., et al. 2010. Antigenicity and immunogenicity of transmitted/founder HIV envelope oligomers compared to chronic HIV envelopes. AIDS Res. Hum. Retroviruses 10.01:A27 [Google Scholar]
- 54. Verkoczy L., et al. 2009. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc. Natl. Acad. Sci. U. S. A. 107:181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Volpe J. M., Cowell L. G., Kepler T. B. 2006. SoDA: implementation of a 3D alignment algorithm for inference of antigen receptor recombinations. Bioinformatics 22:438–444 [DOI] [PubMed] [Google Scholar]
- 56. Walker L. M., et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wei X., et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 58. Wu L., et al. 2009. Enhanced exposure of the CD4-binding site to neutralizing antibodies by structural design of a membrane-anchored human immunodeficiency virus type 1 gp120 domain. J. Virol. 83:5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu X., et al. 2011. Immunotypes of a quaternary site of HIV-1 vulnerability and their recognition by antibodies. J. Virol. 85:4578–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu X., et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xiao X., et al. 2009. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem. Biophys. Res. Commun. 390:404–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yu X., McGraw P. A., House F. S., Crowe J. E., Jr 2008. An optimized electrofusion-based protocol for generating virus-specific human monoclonal antibodies. J. Immunol. Methods 336:142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhou H., Zhou Y. 2002. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 11:2714–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.