Summary
The recent development of next-generation sequencing technology has enabled significant progress in chromatin structure analysis. Here, we review the experimental and bioinformatic approaches to studying nucleosome positioning and histone modification profiles on a genome scale using this technology. These studies advanced our knowledge of the nucleosome positioning patterns of both epigenetically modified and bulk nucleosomes and elucidated the role of such patterns in regulation of gene expression. The identification and analysis of large sets of nucleosome-bound DNA sequences allowed better understanding of the rules that govern nucleosome positioning in organisms of various complexity. We also discuss the existing challenges and prospects of using next-generation sequencing for nucleosome positioning analysis and outline the importance of such studies for the entire chromatin structure field.
Keywords: chromatin, nucleosome positioning, histone modifications, next-generation sequencing
Introduction
Packaging of genomic DNA inside a eukaryotic cell is facilitated by the interaction between the DNA and a specific set of highly conserved architectural proteins, called histones [1]. Two copies of histones H2A, H2B, H3, and H4 are wrapped around by a 147-bp DNA fragment to form a nucleosome core particle, and histone H1/H5 binds to the linker DNA connecting the cores [2] (Figure 1). Placement of nucleosomes at specific locations can significantly affect transcriptional activity by hindering or facilitating binding of transcription factors to DNA [3-5]. Moreover, epigenetic marks such as covalent modifications of the histone tails or incorporation of specific histone variants are an essential part of transcriptional regulation. Therefore, mapping the dynamics of nucleosome positions and modifications of the histones on a genome-scale is crucial for understanding gene regulatory networks that underlie various biological processes both in normal development and disease [6].
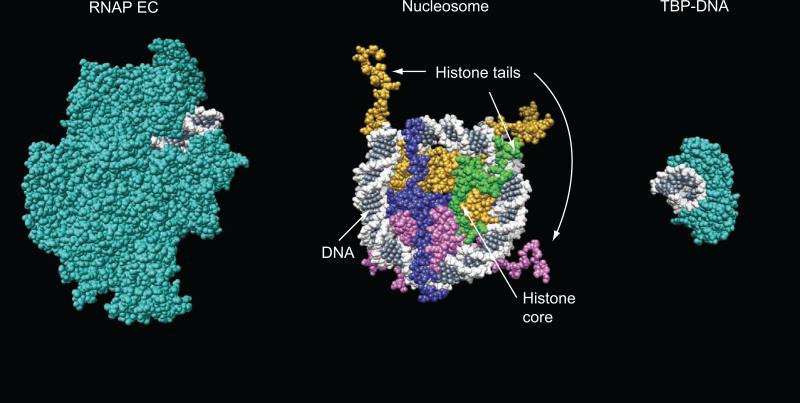
Figure 1.
Structural organization of the nucleosome core particle solved with x-ray crystallography to 1.9 A resolution [66] (shown in center). Histones H2A, H2B, H3, and H4 are shown in blue, magenta, yellow, and green respectively and DNA is shown in white and gray. The structures of RNA polymerase elongation complex [67] and TBP-DNA complex [68] are shown on the left and right of the nucleosome respectively. DNA is shown in white and gray and proteins are shown in cyan. The molecular images were generated using UCSF Chimera package [69].
Nucleosome mapping on a genome-scale can be performed by first fragmenting the chromatin extracted from a cell to preferentially release the DNA fragments associated with nucleosomes (Figure 2A). This is followed by identification of the exact locations of these fragments in the reference genome assembly, as described below. The two most common methods of chromatin fragmentation are enzymatic digestion with micrococcal nuclease (MNase) and sonication. Prior to chromatin fragmentation, histones can be fixed on DNA by reversible cross-linking. This procedure is required in the case of sonication, but a gentler action of MNase allows skipping of this step and ‘native’ chromatin is often used for nucleosome profiling. Chromatin immunoprecipitation (ChIP), a technique that enriches the DNA fragments associated with histones of specific type [7], enables profiling of the epigenetically modified nucleosomes, when the antibody specific to the modification of interest is available.
Figure 2.
Schematic illustration of the nucleosome mapping experiment. A. Chromatin is fragmented (indicated with double-headed orange arrows) so that nucleosome core particles are released and, if desired, the nucleosomes of a specific type are enriched with chromatin immunoprecipitation technique. The purified DNA fragments are processed according to sequencing protocol, which varies from platform to platform and involves such steps as library construction, sequencing itself, and base calling [58] . DNA strands of the nucleosomal fragments are separated in course of the library construction and either both or only one strand of a fragment can be sequenced. As a final step of this stage of the experiment, the sequenced tags are aligned to a reference genome. B, C. Tag distribution and nucleosome occupancy. B. The profile represents tag density around human gene TIMM17A from the sequencing data obtained for H3K4 tri-methylated nucleosomes [23] with an additional simulated peak representing experimental artifact emerging due to amplification bias (red peak on the right). The red profile corresponds to the density of 5'-ends of the tags aligned to the positive DNA strand and blue profile corresponds to the tags aligned to the negative strand. Transcription start of TIMM17A gene is marked with the vertical green line and the transcription direction is shown with the green arrow. Grey ovals represent stable positions of nucleosomes –1, +1,+2, which flank the nucleosome free region. C. The occupancy profile calculated according to the ‘tag extension’ approach: for each tag aligned to i-th position on the positive (negative) strand the occupancy was increased by one at positions [i:(i+147-1)] ([(i-147+1):i]). The stable nucleosome positions seen in (A) are correctly reflected in the resulting occupancy profile; at the same time the artificial peak is also reflected in the occupancy profile (indicated with red X) and needs to be filtered out before further analysis.
Until recently, hybridization on tiling microarrays was the most commonly used approach for large-scale identification of the DNA fragments obtained from the chromatin. Array studies allowed identification of the depletion in nucleosome density at active promoters in yeast [8;9] and human [10], followed by a complete map of nucleosome positioning for the yeast [11] and fly genomes [12]. More recently, high-throughput sequencing-based approaches have become available. Developed in the past three years, this massively parallel sequencing currently results in either hundreds of millions of short reads (35-50 bp, at the 5'-ends of DNA fragments) or about a million of longer reads (~100-500 bp, in most cases spanning the entire nucleosomal DNA) per run, at a much-reduced cost [13-15]. These techniques are collectively referred to as next-generation sequencing technology (NGS). Direct sequencing of the fragments rather than hybridization on a microarray platform results in a much improved spatial resolution of the nucleosome positioning maps. This has enabled significant advances in the field of chromatin structure analysis.
Available data and nucleosome positioning analysis
The nucleosomal datasets produced with the NGS platforms by August 2009 are summarized in Table 1. Both epigenetically modified nucleosomes and bulk nucleosomes that are not selected for any specific histone modification have been mapped in different organisms from yeast to human, and the list is growing rapidly due to the widespread availability of NGS. MNase digestion in combination with NGS (MNase-Seq) was used to profile bulk nucleosomes in the worm [16;17], yeast [18-20] and human genomes [21]. ChIP followed by sequencing (ChIP-seq) was used, either with sonication or MNase digestion, to profile nucleosomes that contain a histone variant or epigenetic modification. Nucleosomes with the histone variant H2A.Z were profiled in yeast [22], fly [12], and human [23]; a broad range of histone methylations and acetylations were profiled in human CD+ T cells [23;24] using the ChIP-seq approach. Most recently, the genomic locations enriched for nucleosomes containing the histone variant H3.3 and those additionally containing the histone variant H2A.Z were identified in the human HeLa cells [25].
Table 1.
Nucleosomal datasets produced with NGS by August 2009.
| Nucleosome type | Organism | Condition | Chromatin | Shearing method | Sequencing platform | Year | Ref | Comment |
|---|---|---|---|---|---|---|---|---|
| Bulk nucleosomes | C.elegans | in vivo | native | MNase | Roche 454 | 2006 | [16] | Mixed-stage population |
| 27 Histone methylations and H2A.Z variant | H.sapiens | in vivo | native | MNase | Solexa, single end | 2007 | [23] | CD4+ T cells |
| H2A.Z | S.cerevisiae | in vivo | cross-linked | MNase | Roche 454 | 2007 | [22] | mid-log phase cells |
| 5 Histone methylations and pan-H3 | M.musculus | in vivo | cross-linked | Sonication | Solexa, single end | 2007 | [65] | ES cells, neural progenitor cells, and embryonic fibroblasts |
| 18 Histone acetylations | H.sapiens | in vivo | native | MNase | Solexa, single end | 2008 | [24] | CD4+ T cells |
| Bulk nucleosomes | H.sapiens | in vivo | native | MNase | Solexa, single end | 2008 | [21] | CD4+T cells |
| Bulk nucleosomes | C.elegans | in vivo | native | MNase | SOLiD, single end | 2008 | [17] | Mixed-stage population, paired-end sequencing for genomic DNA as control |
| H2A.Z | D.melanogaster | in vivo | cross-linked | MNase | Roche 454 | 2008 | [12] | 0-12h embryos |
| 2 Histone methylations | H.sapiens, M.musculus | in vivo | cross-linked | Sonication | Solexa, single end | 2008 | [26] | HeLa S3 IFNG-stimulated and untreated cells; mouse adult liver cells |
| Bulk nucleosomes | S.cerevisiae | in vivo | cross-linked | MNase | Solexa, single end | 2008 | [27] | Normal and heat-shocked cells grown in rich medium |
| Bulk nucleosomes | S.cerevisiae | in vivo | native | MNase | Roche 454 | 2008 | [18] | Log-phase cells |
| Bulk nucleosomes | S.cerevisiae, C.albicans | in vivo | native | MNase | Solexa, single end | 2009 | [19] | Log-phase cells |
| Bulk nucleosomes | S.cerevisiae | in vivo, in vitro | native | MNase | Solexa, single end | 2009 | [20] | cells grown on YPD medium and YP media with galactose or ethanol; additionally nucleosomes assembled on genomic DNA |
| Bulk nucleosomes | ovine gene, chicken histones | in vitro | native | MNase | Roche 454 | 2009 | [28] | nucleosomes mapped on linearised plasmid containing sheep beta-lactoglobulin gene |
| Bulk nucleosomes | A.fumigatus | in vivo | native | MNase | Solexa, paired ends | 2009 | [32] | cells grown on PD medium |
| H3.3, H2A.Z, H3.3/H2A.Z | H.sapiens | in vivo | native | MNase | SOLiD, singe end | 2009 | [25] | HeLa S3 |
| Bulk nucleosomes | S. cerevisiae, E.coli | in vitro | native | MNase | Solexa, single end | 2009 | [46] | nucleosomes assembled on yeast and bacterial genomic DNA |
Generally, characterization of nucleosome positioning comprises two aspects: (i) assessment of nucleosome occupancy or relative abundance of the nucleosomes of a specific epigenetic type to understand the overall impact of nucleosome positioning on the chromatin organization and function [17;19;23;25] and (ii) detection of stable positions of mono-nucleosomes in various genome regions to study the details of the regulatory pathways such as the interplay between nucleosome positioning and transcription factor binding [22;26;27] and to investigate the rules directing nucleosome positioning (discussed below). These aspects are not exclusive but rather complement each other, and NGS has proved to be useful for studying each of them.
The overall nucleosome occupancy can be assessed by directly counting the numbers of sequenced tags mapped to each position in the genome [23] or by using a more elaborate ‘tag extension’ approach [17]. In the latter approach, each sequenced tag represents an independent nucleosome fragment and is ‘extended’ by the expected mono-nucleosome size towards its 3'-end on the strand to which it maps. The number of overlapping nucleosome fragments at each position in the genome is assumed to correspond to the nucleosome occupancy averaged over the cell population used in the sequencing experiment. No ‘tag extension’ is required for constructing such occupancy profiles if the entire nucleosome fragments are sequenced, as in the studies that use Roche 454 pyrosequencing [12;16;28].
The profile of nucleosome occupancy could be used to determine stable nucleosome positions simply as peaks with sufficiently high tag counts [12;20;26]. However, the sequencing technology is not free of artifacts such as amplification bias, and this approach may result in a relatively large number of false positive calls of nucleosome positions (Figure 2B,C). A methodology for mapping of stable nucleosome positions in a more robust way has been developed based on the characteristic features in tag distribution on the DNA positive and negative strands [22;29;30]. As shown in Figure 2B, a specific pattern of tag density marks the sites that are protected against enzymatic digestion or other DNA shearing agents. This pattern comprises two peaks, one on each of the DNA strands, flanking the protected region. Scanning the tag distribution profile for such a signature pattern allows a reliable estimation of the stable nucleosome positions.
This approach can also help in identification of the length of the DNA sequence protected by the histone core against fragmentation [31]. This length represents the genome-wide mean for the nucleosome positions, averaged over the cell population. This information was used to show that the nucleosomes containing the H2A.Z variant are susceptible to asymmetric internal digestion [30], which is indicative of less tight DNA wrapping around the histone core. Concordantly, the shortened DNA protection was also reported for the nucleosomes at the transcription start sites of fungus genes based on the paired-end sequencing, which allows direct measurement of the DNA fragment lengths [32]. Other types of nucleosomes, e.g. those that carry a specific variant of histone H3 (CenH3) and are associated with centromoric regions of chromatin, or those that carry the histone variant H2A.Bbd and are associated with active chromatin, were previously reported to organize less DNA than canonical nucleosomes [33;34]. With more sequencing data available for various genomes, especially for the regions such as centromeres and heterochromatin in general, it should be possible to investigate local variability of nucleosomal DNA length. Accounting for such variability would be essential for correct reconstruction of the nucleosome occupancy profiles and predicting the exact locations of the stable nucleosomes.
Genome-wide maps of histone modifications and nucleosome positioning
The nucleosome profiling on a genome-scale has already revealed specific patterns in placement of both bulk and epigenetically modified nucleosomes at the regulatory regions such as transcription start and end sites (reviewed in Refs. [35;36;37]). These studies have highlighted the importance of the combinatorial patterns of various histone modifications and variants and their interplay with other factors such as DNA structural flexibility, methylation status, binding of small RNAs and other regulatory proteins.
For instance, the broad regions of compact, mostly inactive chromatin called heterochromatin are enriched in the nucleosomes that are di- and tri-methylated at lysine 9 of histone H3. The deposition of this methylation was shown to be mediated by RNA interference in a number of organisms [38] and the presence of these epigenetic marks is essential for binding of such proteins as HP1 that silence chromatin. Another methylation mark, tri-methylation of histone H3 at lysine 27, is associated with Polycomb-assisted silencing of chromatin [23]. The histone modification marks associated with active transcription are usually more localized and are focused at the regulatory and transcribed regions of the genome. The tri-methylation of histone H3 at lysine 36 is the signature of the actively transcribed regions which is the most pronounced at gene ends [23]. The mono-methylation of histone H3 at lysine 4 combined with binding of the protein p300 denotes the distant-acting enhancers, which modulate transcription activity in a tissue specific manner [39-41].
Perhaps the most characterized genomic regions in terms of nucleosome positioning and histone modifications patterns are around transcription start sites (TSS) [12;16;18;20-23]. Tri-methylation of histone H3 at lysine 4 as well as histone acetylations and enrichment in the histone variants H2A.Z and H3.3 mark the TSS of active genes [12;22-24]. Many studies have shown that nucleosomes are arranged in a specific pattern at gene starts. This pattern comprises the nucleosome-free region at the TSS, flanked by the stably positioned nucleosomes (an example of such a pattern is shown in Figure 2B,C for human gene TIMM17A). A recent study [25] argues, however, that the nucleosome free region can actually be occupied to a considerable extent by the nucleosomes simultaneously containing the two histone variants H3.3 and H2A.Z. These nucleosomes are very labile and could be easily displaced by transcription factors. Thus, presence of such nucleosomes would not suppress transcription while it helps to keep the region free from the stable canonical nucleosomes. Future studies will test this hypothesis and further reveal the role of epigenetically modified nucleosomes in the regulation of gene expression and other biological processes.
Rules of nucleosome positioning
Identification of large sets of the DNA sequences that wrap around the histone core in chromatin has led to an explosion in the number of studies focused on the principles that guide nucleosome positioning, in particular on the role of the DNA sequence itself in this process [16-18;20;22;30;42-46;47]. Previously known nucleosome positioning signals such as the 10-bp periodicity in the dinucleotide composition [48] was confirmed in the studies on yeast and worm genomes [16;20;22;42;45]. At the same time, such a periodicity was shown to be less pronounced in fly and even further diminished in human [12;30]. In addition, a positioning signal associated with the increased GC-content of the DNA incorporated in the core particle as compared to linker DNA was shown to be present in various genomes [11;30;43;49].
The rules of rotational positioning deduced from both in vitro and in vivo studies appear to be very similar, with AT-rich stretches tending to occupy the locations on the histone surface where DNA is bent into the minor groove and GC-rich stretches tending to occupy the locations where DNA is bent into the major groove [20;42;50]. On the other hand, the overall ability of DNA sequence to direct translational positioning of nucleosomes remains a subject of debate. In one study, nucleosome occupancy profile observed in yeast in vivo was compared to the profile obtained for reconstituted nucleosomes on bare DNA in vitro [20]. The two profiles showed a high level of correspondence, suggesting that DNA sequence plays an important role in determining chromatin structure at least in yeast. However, a related recent study argues that the DNA sequence contributes mainly to nucleosome rotational positioning and to formation of the nucleosome free regions in yeast promoters, e.g. through well-documented nucleosome exclusion by A-tracts (reviewed in Ref. [51]) and does not determine precise translational positioning of nucleosomes on genomic DNA [46]. Furthermore, a study on worm chromatin showed that exact nucleosome positioning is not preserved in cell population on a single-nucleotide scale [17].
Many factors other than DNA sequence influence nucleosome positioning. The arrays of positioned nucleosome that are consistently observed at the transcription starts [12;22;23] can be a result of ‘statistical’ positioning [52] caused by the presence of a ‘barrier’ in the form of a single stably positioned nucleosome or a bound transcription-related protein [44;46;53]. ATP-dependent chromatin remodeling complexes can be recruited to displace nucleosomes and form nucleosome free regions at gene starts [54]. Other proteins can contribute to nucleosome positioning as well. For example, binding of insulator protein CTCF was demonstrated to facilitate nucleosome positioning in human genome [31]. At the same time, the DNA sequence could evolve to ‘imprint’ the active chromatin structure at the starts of the most actively transcribed genes [19]. It remains to be determined which fraction of stable nucleosomes is positioned by the signals encoded in genomic DNA, as those discussed above, in the genomes of organisms of various complexity.
Interestingly, the epigenetically modified nucleosomes of different types from the human genome were shown to be preferentially associated with the DNA sequences characterized by distinct dinucleotide patterns [55]. Clearly, the DNA sequence cannot be the sole determinant of the epigenetic state of a DNA locus. However, the sequence may reflect an increased probability for a DNA region to have a certain epigenetic state. For instance, the GC-rich sequences of CpG islands tend to be enriched for histone modifications associated with open chromatin. Furthermore, biophysical properties of the sequence may favor incorporation of particular nucleosome types. It was shown, for example, that the sequence organization of the DNA from human H2A.Z nucleosomes differs from the organization of the DNA associated with bulk nucleosomes [30]. Such a sequence specificity of the H2A.Z-containing nucleosomes was suggested to facilitate nucleosome repositioning upon the histone variant deposition which would constitute an important mechanism for fine-tuning of the chromatin structure at regulatory regions.
Limitations and challenges of nucleosome mapping studies
The biggest obstacle in these studies so far has been the large amount of sequencing that is needed to profile bulk nucleosomes in a large genome. To profile human bulk nucleosomes [21], more than 140 million Illumina/Solexa tags were generated, but it was only sufficient to analyze the aggregate tag distributions averaged over large groups of genes. To understand nucleosome dynamics in development and differentiation, nucleosomes have to be mapped in a myriad of epigenomes. It may be possible to carry out a focused profiling of selected regions of the genome using tiling arrays as a first step to capture the appropriate fragments [56;57]; however, additional studies are required before such an approach become available to a broad community. The amount of sequencing necessary for epigenetically modified nucleosomes is generally smaller, but it still requires a substantial number of tags and high-quality antibodies. For most experimental laboratories, storing, handling and analyzing such large data sets pose perhaps the most difficult challenge [58].
The nucleosome profiling experiment has a number of internal biases, which need to be addressed (Figure 3A). An important issue is the salt concentration used for chromatin extraction. It was shown [59] that the experiments performed under low and high salt concentrations resulted in profiling of chromatin regions that have different physical properties, with the inactive chromatin regions being less soluble on average than the active regions (we note, however, that some fraction of active regions was reported to be present in the least soluble chromatin fraction). Using different salt conditions can be a tool for capturing epigenetic dynamics of chromatin as was demonstrated by studies of the distributions of histone variants H3.3 and H2A.Z in the fly and human genomes [25;59].
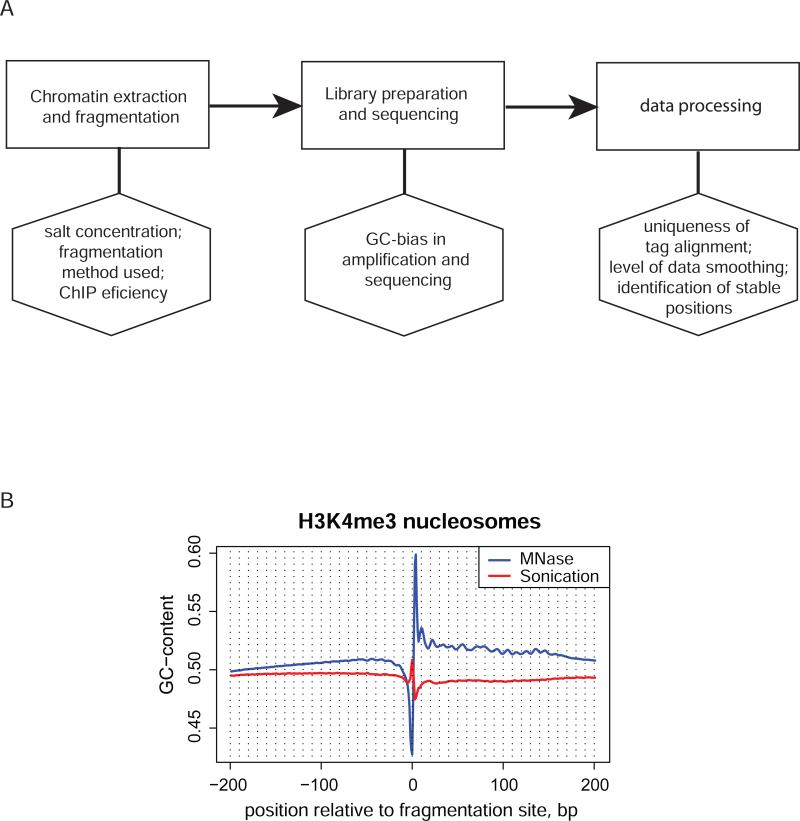
Figure 3.
Possible biases in the sequencing experiment. A. Flow chart showing major stages of the nucleosome profiling experiment (upper row, rectangles) and internal biases that may be associated with them (lower row, hexagons). B. GC-content profile of the sequences around fragmentation sites determined in two independent ChIP-seq experiments profiling H3K4 tri-methylated nucleosomes in human. Blue line represents the data obtained for MNase digested chromatin from CD4+ T-cells [23] and red line represents the data obtained for sonicated chromatin from HeLa cells [26]. Only the tags mapped to unique regions of human genome, as defined in UCSC genome browser [70;71], were taken for analysis to avoid possible bias when repetitive sequences are included. The DNA fragmentation was performed with MNase digestion of native chromatin in the case of CD4+ T cells and with sonication of cross-linked chromatin in the case of HeLa cells. Note the difference in the characteristic patterns at the fragmentation site and the overall GC-content of the two sequence sets.
Estimated nucleosome positions can also be affected to a high degree by the procedure used for DNA fragmentation. In the most studies, the MNase digestion is used for these purposes (Table 1) because it produces nucleosome maps with near base-pair resolution and does not require cross-linking. However, this enzyme has pronounced sequence preferences [60;61], which may lead to bias in the nucleosome occupancy profile. Although several computational approaches have been suggested to compensate for such a bias, they do not address the issue completely [22;30]. Sonication also appears to have sequence-specific bias, which has not been comprehensively studied yet. As a result, different sequence organization may be revealed for the DNA fragments associated with the same type of nucleosomes depending on whether MNase digestion or sonication is used for chromatin fragmentation. The GC-content profiles for the sequences around the 5’-ends of nucleosome fragments obtained by MNase digestion or sonication of human chromatin are shown in Figure 3B. Not only the two fragmentation techniques produce distinct sequence signatures at the site of the DNA breakage but also the overall nucleotide composition is different in the adjacent DNA regions. We note that these experiments using the two fragmentation methods were carried out on the cells of different types, CD4+ T-cells and HeLa cells, but biases shown are likely to be present regardless. The issue clearly requires further investigation.
Another important issue is the GC-bias. The GC-rich sequences tend to be over-represented in the final set of sequenced tags, while the extremely AT-rich genomic regions may be poorly covered [62]. Such a bias is explained, at least partly, by the DNA amplification which is used during sample preparation by most of the sequencing platforms (see Figures 2B,C and the related text for a discussion of the effect of the amplification bias on nucleosome detection). A recently developed Helicos platform does not require amplification and seems to be a promising technique for addressing the GC-bias. However, to date there was no published study which would use this platform in a nucleosome positioning experiment.
Future perspective
In the next several years, a large amount of epigenetic data will be generated. Nucleosomes of various types are being currently profiled in several human cell lines in the ENCODE (Encyclopedia of DNA elements) consortium project [63]. A related effort, Model Organism ENCODE, is underway for fruit fly and worm [64]. A new NIH initiative, Roadmap Epigenomics Program, will promote the discovery of novel epigenetic marks in mammalian cells and is expected to advance our understanding of epigenomics of human health and disease. Analysis and integration of the data generated in the ongoing and future epigenetic projects will require further development of the experimental and bioinformatic approaches some of which are discussed above. The new data will facilitate direct comparison of the nucleosome positioning and histone modification profiles obtained for different cell types and developmental stages of the same organism and the profiles of both bulk and epigenetically modified nucleosomes in organisms of various complexity. It is anticipated that these projects will provide novel mechanistic insights into the subject of nucleosome positioning and bring our understanding of the pathways of epigenetic regulation to a new level.
Executive Summary.
Introduction
Next-generation sequencing technologies have enabled significant progress in chromatin structure field, allowing precise genome-wide mapping of nucleosomes and histone modification.
Profiling nucleosome density and stable nucleosome positions is important for understanding the overall impact of primary chromatin structure on biological processes and for elucidating epigenetic pathways.
Biological role and rules of nucleosome positioning
Nucleosome positioning studies have revealed specific patterns in placement of both bulk and epigenetically modified nucleosomes at the regulatory regions such as transcription start and end sites and allowed identification of broad regions of enrichment for certain histone modification marks associated with silent chromatin.
These studies highlight the importance of the combinatorial patterns of various histone modifications and variants and their interplay with other factors such as DNA methylation, binding of small RNAs and regulatory proteins.
Analysis of large sets of the DNA sequences that preferentially wrap around the histone core has shed light on the rules that govern nucleosome positioning. The extent to which DNA sequence directs nucleosome positioning is still debated and it remains to be determined which fraction of nucleosomes is positioned by the signals encoded in DNA in organisms of varying complexity.
Challenges and prospects of chromatin structure analysis with next-generation sequencing
The large amount of sequencing required to profile nucleosomes in a large genome, and consequently its high cost, is a major obstacle.
Details of chromatin preparation procedure such as the salt conditions for chromatin extraction or the method for chromatin fragmentation can influence the relative representation of different chromatin regions in the final data.
Data sets from a sequencing experiment are large, and managing and analyzing them pose a substantial bioinformatic problem.
Several large-scale consortium projects were launched to study epigenomes of various organisms. These projects will lead to development of new experimental and bioinformatic approaches for analysis of chromatin structure within a single cell type as well as comparison of different epigenomes. The data generated in these projects are expected to bring new mechanistic insights into epigenetic pathways and to significantly advance our knowledge about how the genome functions.
References
- 1.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98(3):285–94. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Beato M, Eisfeld K. Transcription factor access to chromatin. Nucleic Acids Res. 1997;25(18):3559–63. doi: 10.1093/nar/25.18.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kornberg RD, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Dev. 1999;9(2):148–51. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 7.Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53(6):937–47. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 8.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36(8):900–5. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 9*.Yuan GC, Liu YJ, Dion MF, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309(5734):626–30. doi: 10.1126/science.1112178. [The first genome-wide study of in vivo nucleosome positioning.] [DOI] [PubMed] [Google Scholar]
- 10.Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol. 2007;25(2):244–8. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 11.Lee W, Tillo D, Bray N, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39(10):1235–44. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 12.Mavrich TN, Jiang C, Ioshikhes IP, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453(7193):358–62. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16(6):545–52. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 15.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 16.Johnson SM, Tan FJ, McCullough HL, Riordan DP, Fire AZ. Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin. Genome Res. 2006;16(12):1505–16. doi: 10.1101/gr.5560806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Valouev A, Ichikawa J, Tonthat T, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18(7):1051–63. doi: 10.1101/gr.076463.108. [High-resolution nucleosome profiling with deep sequencing to access the fraction of stably positioned nucleosomes in the worm genome.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Field Y, Kaplan N, Fondufe-Mittendorf Y, et al. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4(11):e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Field Y, Fondufe-Mittendorf Y, Moore IK, et al. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat Genet. 2009;41(4):438–45. doi: 10.1038/ng.324. [Provides evidence of the connection between gene expression divergence and changes in nucleosome positioning during yeast evolution.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20*.Kaplan N, Moore IK, Fondufe-Mittendorf Y, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458(7236):362–6. doi: 10.1038/nature07667. [Together with Ref. 46 provides genome-wide data on positioning of nucleosomes assembled in vitro on purified DNA.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schones DE, Cui K, Cuddapah S, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132(5):887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albert I, Mavrich TN, Tomsho LP, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446(7135):572–6. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 23*.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. doi: 10.1016/j.cell.2007.05.009. [Large scale profiling of histone modifications and H2A.Z histone variant in the human genome.] [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin C, Zang C, Wei G, et al. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41(8):941–5. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson AG, Bilenky M, Tam A, et al. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 2008;18(12):1906–17. doi: 10.1101/gr.078519.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27*.Shivaswamy S, Bhinge A, Zhao Y, et al. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6(3):e65. doi: 10.1371/journal.pbio.0060065. [Reports changes in nucleosome poisoning on genome scale as a result of a physiological perturbation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraser RM, Keszenman-Pereyra D, Simmen MW, Allan J. High-resolution mapping of sequence-directed nucleosome positioning on genomic DNA. J Mol Biol. 2009;390(2):292–305. doi: 10.1016/j.jmb.2009.04.079. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Shin H, Song JS, Lei Y, Liu XS. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics. 2008;9:537. doi: 10.1186/1471-2164-9-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.Tolstorukov MY, Kharchenko PV, Goldman JA, Kingston RE, Park PJ. Comparative analysis of H2A.Z nucleosome organization in the human and yeast genomes. Genome Res. 2009;19(6):967–77. doi: 10.1101/gr.084830.108. [Direct comparison of nucleosome organization in organisms of different complexity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4(7):e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishida H, Motoyama T, Yamamoto S, Aburatani H. Osada H: Genome-wide maps of mono- and di-nucleosomes of Aspergillus fumigatus. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp413. [DOI] [PubMed] [Google Scholar]
- 33.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5(8):e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bao Y, Konesky K, Park YJ, et al. Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA. Embo J. 2004;23(16):3314–24. doi: 10.1038/sj.emboj.7600316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10(3):161–72. doi: 10.1038/nrg2522. [Reviews impact of nucleosome positioning on gene regulation and evaluates different models of organization of nucleosome at genes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9(3):179–91. doi: 10.1038/nrg2270. [Reviews chromatin organization features such as distribution and function of chromatin modifications in the genomes of different organisms, from yeast to human.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segal E, Widom J. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat Rev Genet. 2009;10(7):443–56. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8(4):272–85. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 39.Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 40.Hon G, Ren B, Wang W. ChromaSig: a probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput Biol. 2008;4(10):e1000201. doi: 10.1371/journal.pcbi.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visel A, Blow MJ, Li Z, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457(7231):854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Segal E, Fondufe-Mittendorf Y, Chen L, et al. A genomic code for nucleosome positioning. Nature. 2006;442(7104):772–8. doi: 10.1038/nature04979. [First genome scale analysis of the sequence-directed nucleosome positioning in vivo (also see ref. 16).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peckham HE, Thurman RE, Fu Y, et al. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17(8):1170–7. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mavrich TN, Ioshikhes IP, Venters BJ, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18(7):1073–83. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gabdank I, Barash D, Trifonov EN. Nucleosome DNA bendability matrix (C. elegans). J Biomol Struct Dyn. 2009;26(4):403–11. doi: 10.1080/07391102.2009.10507255. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y, Moqtaderi Z, Rattner BP, et al. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol. 2009;16(8):847–52. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chung HR, Vingron M. Sequence-dependent nucleosome positioning. J Mol Biol. 2009;386(5):1411–22. doi: 10.1016/j.jmb.2008.11.049. [DOI] [PubMed] [Google Scholar]
- 48**.Trifonov EN, Sussman JL. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980;77(7):3816–20. doi: 10.1073/pnas.77.7.3816. [Pioneering study on sequence-directed nucleosome positioning.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kharchenko PV, Woo CJ, Tolstorukov MY, Kingston RE, Park PJ. Nucleosome positioning in human HOX gene clusters. Genome Res. 2008;18(10):1554–61. doi: 10.1101/gr.075952.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986;191(4):659–75. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 51.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19(1):65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fedor MJ, Lue NF, Kornberg RD. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988;204(1):109–27. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- 53.Rando OJ, Ahmad K. Rules and regulation in the primary structure of chromatin. Curr Opin Cell Biol. 2007;19(3):250–6. doi: 10.1016/j.ceb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 54.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–73. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 55.Yuan GC, Liu JS. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput Biol. 2008;4(1):e13. doi: 10.1371/journal.pcbi.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27(2):182–9. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herman DS, Hovingh GK, Iartchouk O, et al. Filter-based hybridization capture of subgenomes enables resequencing and copy-number detection. Nat Methods. 2009;6(7):507–10. doi: 10.1038/nmeth.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58*.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009 doi: 10.1038/nrg2641. in press. [A complete and up-to-date review of ChIP-Seq technology and its applications in chromatin analysis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59*.Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19(3):460–9. doi: 10.1101/gr.087619.108. [Highlights an important issue of experimental design, which should be accounted for in nucleosome positioning studies.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wingert L, Von Hippel PH. The conformation dependent hydrolysis of DNA by micrococcal nuclease. Biochim Biophys Acta. 1968;157(1):114–26. doi: 10.1016/0005-2787(68)90270-0. [DOI] [PubMed] [Google Scholar]
- 61.Dingwall C, Lomonossoff GP, Laskey RA. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981;9(12):2659–73. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hillier LW, Marth GT, Quinlan AR, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5(2):183–8. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 63.Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Celniker SE, Dillon LA, Gerstein MB, et al. Unlocking the secrets of the genome. Nature. 2009;459(7249):927–30. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319(5):1097–113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 67.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448(7150):157–62. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 68.Nikolov DB, Chen H, Halay ED, et al. Crystal structure of a human TATA box-binding protein/TATA element complex. Proc Natl Acad Sci U S A. 1996;93(10):4862–7. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 70.Karolchik D, Kuhn RM, Baertsch R, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36(Database issue):D773–9. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]