Abstract
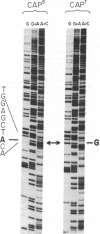
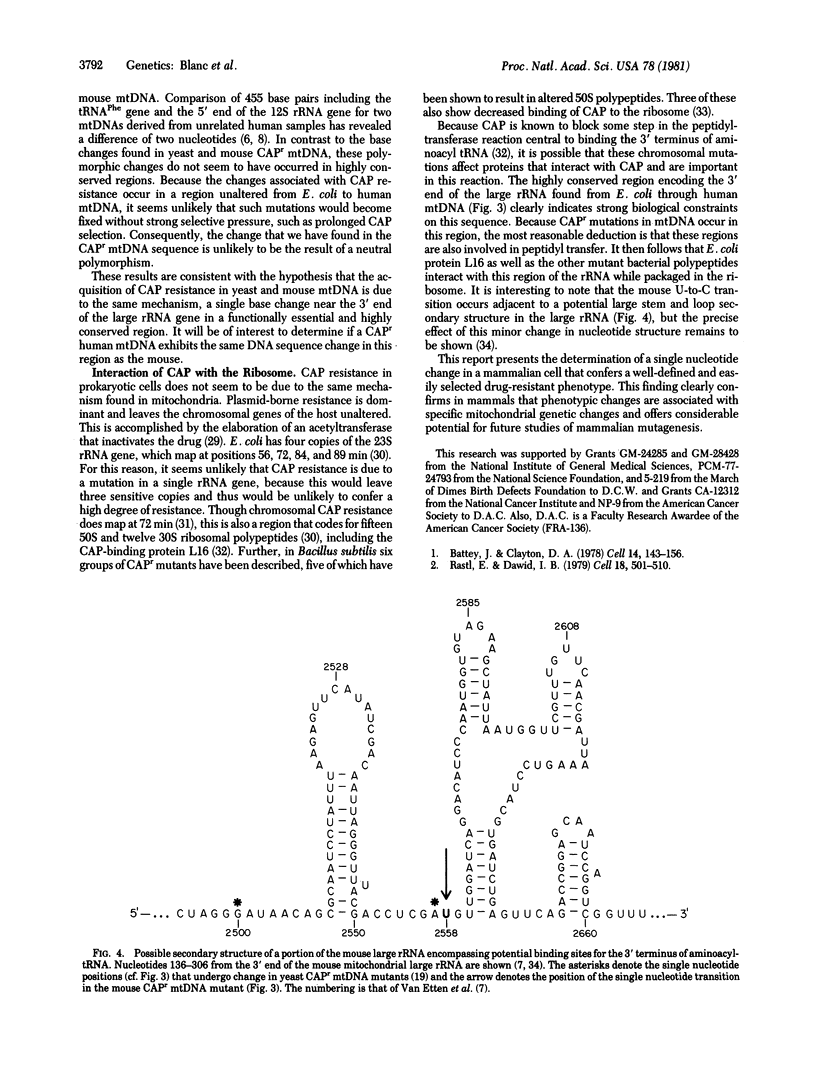
The complete DNA sequence of the rRNA genes of mouse L cell mtDNA provides a basis for the examination of the nucleotide sequence of this region in a mutant mouse cell line that is resistant to chloramphenicol, a known inhibitor of mitochondrial protein synthesis. Resistance to chloramphenicol (CAPr) is conferred by a cytoplasmic determinant that is linked to mtDNA restriction endonuclease site polymorphisms. We have determined the sequence of a 212-nucleotide region of mtDNA from a CAPr mouse cell line that encodes a portion of the 1582-nucleotide large rRNA. This sequence is located 107-318 nucleotides from the 5' end of the heavy strand coding sequence, which corresponds to the 3' end of the rRNA. There is a single nucleotide difference in the large rRNA gene from CAPr cells, an A-to-G transition 243 nucleotides from the 5' end of the coding sequence. This single transition is located within a region of 10 nucleotides tht is otherwise completely homologous to human and yeast mitochondrial large rRNAs and Escherichia coli 23S rRNA and is positioned immediately adjacent to a single nucleotide transversion known to occur in a yeast CAPr mutant. This characterization of a mammalian mitochondrial mutant at the nucleotide level directly demonstrates that a mutant phenotype may result from a single mtDNA nucleotide change in an animal cell.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R. J., Dubin D. T. Methylated regions of hamster mitochondrial ribosomal RNA: structural and functional correlates. Nucleic Acids Res. 1981 Jan 24;9(2):323–337. doi: 10.1093/nar/9.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell B. G., Bankier A. T., Drouin J. A different genetic code in human mitochondria. Nature. 1979 Nov 8;282(5735):189–194. doi: 10.1038/282189a0. [DOI] [PubMed] [Google Scholar]
- Battey J., Clayton D. A. The transcription map of human mitochondrial DNA implicates transfer RNA excision as a major processing event. J Biol Chem. 1980 Dec 10;255(23):11599–11606. [PubMed] [Google Scholar]
- Battey J., Clayton D. A. The transcription map of mouse mitochondrial DNA. Cell. 1978 May;14(1):143–156. doi: 10.1016/0092-8674(78)90309-4. [DOI] [PubMed] [Google Scholar]
- Baughman G. A., Fahnestock S. R. Chloramphenicol resistance mutation in Escherichia coli which maps in the major ribosomal protein gene cluster. J Bacteriol. 1979 Mar;137(3):1315–1323. doi: 10.1128/jb.137.3.1315-1323.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D., Clayton D. A. The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J Biol Chem. 1974 Dec 25;249(24):7991–7995. [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Noller H. F. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jan;77(1):201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunn C. L., Wallace D. C., Eisenstadt J. M. Cytoplasmic inheritance of chloramphenicol resistance in mouse tissue culture cells. Proc Natl Acad Sci U S A. 1974 May;71(5):1681–1685. doi: 10.1073/pnas.71.5.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunn C. L., Wallace D. C., Eisenstadt J. M. Mitotic segregation of cytoplasmic determinants for chloramphenicol resistance in mammalian cells. I: Fusion with mouse cell lines. Somatic Cell Genet. 1977 Jan;3(1):71–92. doi: 10.1007/BF01550988. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews S., Attardi G. The sequences of the small ribosomal RNA gene and the phenylalanine tRNA gene are joined end to end in human mitochondrial DNA. Cell. 1980 Mar;19(3):775–784. doi: 10.1016/s0092-8674(80)80053-5. [DOI] [PubMed] [Google Scholar]
- Dujon B. Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell. 1980 May;20(1):185–197. doi: 10.1016/0092-8674(80)90246-9. [DOI] [PubMed] [Google Scholar]
- Eperon I. C., Anderson S., Nierlich D. P. Distinctive sequence of human mitochondrial ribosomal RNA genes. Nature. 1980 Jul 31;286(5772):460–467. doi: 10.1038/286460a0. [DOI] [PubMed] [Google Scholar]
- Giles R. E., Stroynowski I., Wallace D. C. Characterization of mitochondrial DNA in chloramphenicol-resistant interspecific hybrids and a cybrid. Somatic Cell Genet. 1980 Jul;6(4):543–554. doi: 10.1007/BF01539155. [DOI] [PubMed] [Google Scholar]
- Grivell L. A., Netter P., Borst P., Slonimski P. P. Mitochondrial antibiotic resistance in yeast: ribosomal mutants resistant to chloramphenicol, erythromycin and spiramycin. Biochim Biophys Acta. 1973 Jun 23;312(2):358–367. doi: 10.1016/0005-2787(73)90380-8. [DOI] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nierhaus D., Nierhaus K. H. Identification of the chloramphenicol-binding protein in Escherichia coli ribosomes by partial reconstitution. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2224–2228. doi: 10.1073/pnas.70.8.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D. K., Montoya J., Attardi G. The putative mRNA for subunit II of human cytochrome c oxidase starts directly at the translation initiator codon. Nature. 1980 Sep 4;287(5777):79–82. doi: 10.1038/287079a0. [DOI] [PubMed] [Google Scholar]
- Ojala D., Merkel C., Gelfand R., Attardi G. The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell. 1980 Nov;22(2 Pt 2):393–403. doi: 10.1016/0092-8674(80)90350-5. [DOI] [PubMed] [Google Scholar]
- Osawa S., Takata R., Tanaka K., Tamaki M. Chloramphenicol resistant mutants of Bacillus subtilis. Mol Gen Genet. 1973 Dec 20;127(2):163–173. doi: 10.1007/BF00333664. [DOI] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M. Restriction endonuclease cleavage maps of rat and mouse mitochondrial DNAs. Nucleic Acids Res. 1977;4(5):1291–1299. doi: 10.1093/nar/4.5.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastl E., Dawid I. B. Expression of the mitochondrial genome in Xenopus laevis: a map of transcripts. Cell. 1979 Oct;18(2):501–510. doi: 10.1016/0092-8674(79)90067-9. [DOI] [PubMed] [Google Scholar]
- Robberson D. L., Clayton D. A. Replication of mitochondrial DNA in mouse L cells and their thymidine kinase - derivatives: displacement replication on a covalently-closed circular template. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3810–3814. doi: 10.1073/pnas.69.12.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V. The enzymatic acetylation of chloramphenicol by extracts of R factor-resistant Escherichia coli. J Biol Chem. 1967 Feb 25;242(4):687–693. [PubMed] [Google Scholar]
- Spolsky C. M., Eisenstadt J. M. Chloramphenicol-resistant mutants of human HeLa cells. FEBS Lett. 1972 Sep 15;25(2):319–324. doi: 10.1016/0014-5793(72)80514-3. [DOI] [PubMed] [Google Scholar]
- Van Etten R. A., Walberg M. W., Clayton D. A. Precise localization and nucleotide sequence of the two mouse mitochondrial rRNA genes and three immediately adjacent novel tRNA genes. Cell. 1980 Nov;22(1 Pt 1):157–170. doi: 10.1016/0092-8674(80)90164-6. [DOI] [PubMed] [Google Scholar]
- Wallace D. C., Bunn C. L., Eisenstadt J. M. Cytoplasmic transfer of chloramphenicol resistance in human tissue culture cells. J Cell Biol. 1975 Oct;67(1):174–188. doi: 10.1083/jcb.67.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]