Abstract
This study reports for the first time qualitative and quantitative differences in carbonylated proteins shed into blood as a function of increasing levels of OS. Carbonylated proteins in freshly drawn blood from pairs of diabetic and lean rats were derivatized with biotin hydrazide, dialyzed, and enriched with avidin affinity chromatography. Proteins thus selected were used in several ways. Differences between control and diabetic subjects in relative concentration of proteins was achieved by differential labeling of tryptic digests with iTRAQ™ reagents followed by reversed phase chromatography (RPC) and tandem mass spectrometry (MS/MS). Identification and characterization of OS induced post-translational modification sites in contrast was achieved by fractionation of affinity selected proteins before proteolysis and RPC-MS/MS. Relative quantification of peptides bearing oxidative modifications was achieved for the first time by selective reaction monitoring (SRM). Approximately 1.7% of the proteins in Zucker diabetic rat plasma were selected by the avidin affinity column as compared to 0.98% in lean animal plasma. Among the thirty five proteins identified and quantified, Apo AII, clusterin, hemopexin precursor and potassium voltage-gated channel subfamily H member 7 showed the most dramatic changes in concentration. Seventeen carbonylation sites were identified and quantified, eleven of which changed more than 2 fold in oxidation state. Three types of carbonylation were identified at these sites; direct oxidative cleavage from reactive oxygen species, glycation and addition of advanced glycation end products, and addition of lipid peroxidation products. Direct oxidation was the dominant form of carbonylation observed while hemoglobin and murinoglobulin 1 homolog were the most heavily oxidized proteins.
INTRODUCTION
A plethora of diseases ranging from diabetes mellitus1 and neurodegenerative diseases (Alzheimer’s disease2, Parkinson’s disease3 and amyotrophic lateral sclerosis4) to inflammatory diseases (atherosclerosis5 and chronic lung disease6) and even cancer7 are associated with failure to regulate the redox potential of cells. The net result is over-production of reactive oxygen species (ROS) that damage DNA8, RNA9, unsaturated lipids10, and proteins11 to a degree that the signaling capacity of cells is reduced, the competence of proteasomes and lyzosomes to destroy oxidatively damaged proteins is diminished, and cellular viability is reduced, sometimes to the point of cell death12.
Proteins can be oxidized in at least 35 ways11. Among the many types of protein oxidation, carbonylation is one of the more prominent. Protein carbonylation is irreversible and often leads to loss of function and the need for degradation of damaged proteins. Carbonyl groups are introduced into proteins in vivo by i) direct oxidation of Pro, Arg, Lys, Thr, Glu, or Asp side chains or oxidative cleavage of the protein backbone, ii) introduction of 4-hydroxy-2-noneal (HNE), 2-propenal or malondialdehyde from lipid peroxidation to a Cys, His or Lys residue and iii) formation of advanced glycation end-products. Although there are some differences between individual subjects, both the number and level of oxidized proteins was found to be relatively reproducible in the blood plasma of 32-35 year old human male subjects.13 Moreover, the mechanism of carbonylation could be delineated by mass spectrometry as one of the three types noted above. Oxidation has even been traced to specific organs and tissues in some cases13, 14, suggesting that proteins are either being shed from cells or cell membranes are being breached in some way.
Interesting features of protein carbonylation are that it occurs without enzymatic catalysis and is more likely to occur in some proteins than others. How this occurs is unknown. Whether the probability and mechanism of carbonylation are constant or differ with increasing OS are still open questions. Cellular compartments in which oxidation occurs, protein abundance, and the propensity of proteins to undergo conformational change as they are oxidized could play an important role in protein oxidation. The objective of the work presented here was to address some of these questions in vivo; focusing on 1) the isolation of carbonylated proteins in blood, 2) their identification, 3) locating sites of oxidation, 4) determining types of modification, 5) quantifying the relative degree to which carbonylation changed between proteins and within several proteins during increasing OS, and 6) comparing protein carbonylation at normal and elevated levels of OS in vivo. Lean and diabetic rats were used as the model system for examining the impact of differentially increasing OS on protein carbonylation.
EXPERIMENTAL PROCEDURES
MATERIALS
Sodium cyanoborohydride, biotin hydrazide, ultralinked immobilized monomeric avidin, D-biotin, and Slide-A-Lyzer dialysis cassettes were purchased from Pierce Chemical Company (Rockford, IL, USA). Iodoacetamide, dithiothreitol (DTT), trypsin, dimethylglycine, α-cyano-4-hydroxy-cinnamic acid (CHCA), proteomics grade N-p-tosyl-phenylalanine chloromethyl ketone (TPCK)-treated trypsin, ammonium bicarbonate, guanidine, dithiothreitol, iodoacetamide acid (IAA), and L-cysteine were obtained from Sigma Chemical Co. (St. Louis, MO, USA). Protease inhibitor cocktail was purchased from Roche Diagnostics (Indianapolis, IN, USA). The ABI 4700 Proteomics Analyzer Calibration Mixture (4700) Cal Mix, bradykinin, angiotensin I, glu1-fibrinopeptide B, ACTH fragment 1-17, ACTH fragment 18-39, and ACTH fragment 7-38) and iTRAQ™ reagent multiplex kit were purchased from Applied Biosystems (ABI, Foster City, CA). Trifluoroacetic acid (TFA), and HPLC grade acetonitrile were purchased from Mallinckrodt Chemicals (Phillipsburg, NJ). Sodium phosphate, sodium chloride and formic acid (88%) were purchased from Mallinckrodt (St. Louis, MO, USA). Amicon Ultra-4 and Microcon Ultracel YM-3 centrifugal filter devices were purchased from Millipore (Billerica, MA).
METHODS
Animal Model
Zucker diabetic (ZDF) and lean rats were used in these studies as the animal model for investigating OS effects on protein carbonylation. Diabetic Zucker rats are homozygous for a leptin receptor defect, becoming obese and developing diabetes naturally at about 7 weeks of age. The Zucker lean rat is genetically matched and is heterozygous for the leptin receptor defect.15
Glucose assay
Glucose assays were done in a 96 well format utilizing a glucose oxidase reagent from Pointe Scientific (Canton, MI) as a means of validating OS. Both plasma and ultrafiltrate samples were analyzed using this method. Samples were stored at −80°C until the assay was performed.
Briefly, samples from diabetic animals and their lean controls were thawed on ice and used to prepare a standard curve using glucose solutions in benzoic acid (RICCA Chemical Company, Arlington, TX) diluted to 0, 100, 300 and 500 mg/dL. A reagent blank and positive controls were included on all assays. The positive control was Control Serum II™ obtained from Wako Chemicals USA, Inc. (Richmond, VA) and was diluted according to the manufacturer’s instructions. A microassay was utilized. Two μL of standard, positive control, and samples were loaded onto the plate in triplicate. Two hundred μL of pre-heated (37°C) glucose oxidase reagent were pipetted into all the wells. The plate was incubated at 37°C for 5 min and then read using a Powerwave X plate reader (Bio-Tek Instruments, Inc. Winooski, VT) at 500 nm. Baseline and lean rat samples were not diluted. All other samples were diluted 1:1 with deionized water to maintain the linearity specifications of the assay.
F2 isoprostane assay
Urinary isoprostanes were measured in duplicate at baseline, 3, and 6 weeks of the supplementation protocol using a commercial competitive ELISA kit from Oxford Biomedical Research (Oxford, MI) according to the manufacturer’s instructions. Briefly, two dilutions of the urine samples (1:4, 1:8) were prepared in the enhanced dilution buffer. Standards were prepared following the kit instructions, providing a range from 0 to 100 ng/mL. One hundred μL of each of the sample dilutions and standards were added to the pre-coated ELISA plate. In addition, 100 μL of the diluted 15-isoprostane F2t horse radish peroxidase (HRP) conjugate was added to all wells except the blank. The plate was covered and incubated at room temperature with gentle shaking for 2 hrs. After 2 hrs, the plate was washed 3 times with the kit wash buffer. Next, 200μL of the substrate was added to each well and allowed to incubate for 20 min. Fifty μL of 3M sulfuric acid was added to stop the reaction and absorbance was read at of 450nm using a Powerwave X (Bio-Tek Instruments, Inc. Winooski, VT). Concentrations of F2 isoprostanes at ng/24 hrs. urine levels were compared between diabetic and lean rats’ plasma samples using the Student t-test. A p-value of <0.05 on a 2-tailed test with 95% confidence intervals was considered statistically significant.
Biotinylation of the plasma samples
Diabetic and lean rats were scarified at the 6 months of age and blood collected in a venous blood Vacutainer™ collection tube coated with EDTA (Fisher Scientific, Hanover Park, IL, USA). Protease inhibitor cocktail was added to preclude proteolysis. Generally, each tablet of the protease inhibitor cocktail was dissolved in 1 mL of distilled water. Protease inhibitor solution was then mixed with plasma in a 1:10 (v/v) ratio, respectively. Because plasma samples were maintained at neutral pH during biotinylation and affinity selection there was no need to inhibit aspartate protease along with proteases that are only active at acidic pH. Samples were centrifuged at 1500 × g for 15 min. The supernatant was removed and centrifuged again at 2000 × g for 15 min after which 50 mM biotin hydrazide (BH) was added to a final concentration of 5 mM. Reaction of BH with carbonyl groups was allowed to proceed at room temperature for 2 hrs. after which sodium cyanoborohydride was added to a final concentration of 15 mM and incubated at 0°C for 1 hr. Samples were dialyzed three times against at least 200 volumes of PBS buffer to remove any unreacted BH.
Avidin purification of the biotinylated proteins
Ultralinked immobilized monomeric avidin (Pierce Chemical Company) was packed in a 4.6 × 100 mm PEEK column. The column was washed with PBS (0.15M pH 7.4) and then with 2 mM biotin to block any irreversible biotin binding sites. This was followed by washes with regenerating mobile phase (0.1M dimethylglycine, pH 2.5) and reequilibration buffer (PBS). The Bradford assay was used to estimate the protein concentration in rat plasma after which samples were adjusted to the same protein concentration and digested with trypsin before differential labeling samples according to their origin with iTRAQ™ reagents. The relative concentration of proteins thus isotopically coded between samples determined by isotope ratio measurements during mass spectral identification. Prior to characterization and quantification of OS induced post translational modifications, samples were enriched by avidin affinity chromatography. A total of 5 mg of plasma proteins was applied to the avidin column in PBS mobile phase (0.15 M phosphate buffered saline, pH 7.4) at a flow rate of 0.5 mL/min. Unbound and weakly bound proteins were removed by a 60 column volume wash. Strongly bound proteins were then eluted with 0.1M dimethylglycine (pH 2.5) during which detection was achieved by absorbance at 280 nm.
Quantitative comparison of protein abundance with iTRAQ™ and MALDI-TOF/TOF analysis
Tryptic digested proteins from avidin affinity fractions were labeled with iTRAQ™ reagent. The supplier’s guide lines (ABI) were followed for both trypsin digestion and labeling with iTRAQ™ reagent. Lean and diabetic rat plasma samples were labeled with the 114-dalton and 117-dalton iTRAQ™ labeling reagents respectively. The labeled peptides were then desalted and fractionated using an Agilent 1100 Series HPLC (Agilent Technologies). A Pepmap C18 trap column and a nano-column (Zorbax 300SB-C18, 3.5 μm, 100 μm i.d., 15 cm length column (Agilent Technologies, Santa Clara, CA) were used. Two solvents were used for the reversed phase separation, solvent A composed of 0.1% TFA in deionized water and solvent B composed of 0.1% TFA in acetonitrile. The RPC separation was achieved using a 40 min linear gradient from 98% solvent A: 2% solvent B to 60% solvent A: 40% solvent B at a flow rate of 800 nL/min. A post-column mixing tee was used to mix the peptides separated by RPC with MALDI matrix (α-cyano-4-hydroxycinnamic acid, 4 mg/mL in 60% ACN/0.1% TFA). A microfraction collector was used to spot eluent on MALDI plate. Peptides were analyzed in the positive ion mode using an ABI 4800 plus (4800 MALDI-TOF/TOF) Proteomics Analyzer equipped with a 200 Hz Nd:Yag laser. MS and MS/MS data was acquired using the 4000 Explorer software. Protein identification was achieved from MS/MS data utilizing the Pro Group™ algorithm (ABI) in the Protein Pilot software 2.0. The minimum acceptance criterion for peptide identification was based on a 95% confidence level. Again the Protein Pilot software 2.0 with the Pro Group™ algorithm was used, this time performing automated MS/MS data analysis for protein identification along with quantification of iTRAQ™ reporter ions. A decoy method was used to estimate the false discovery rate. The decoy for the samples was generally less than 5.0%.
Statistics
For each protein, a p-value was also generated via Student’s t test. The significantly changed proteins met two criteria (1) t test p-values ≤ 0.05, and (2) protein ratios change ≥1.5.
Reversed-phase separation of biotinylated proteins
Affinity selected proteins were desalted and separated with a Agilent Zorbax 300SB-C3 reversed-phase column. The reversed-phase column was equilibrated with 5 column volumes of buffer A (99.5% deionized H2O (dI H2O), 0.5 % acetonitrile (ACN) and 0.1% TFA). After a 5 column volume wash, a linear 50 min gradient was applied from 100% buffer A to 75% buffer B (5% dI H2O, 95 % ACN and 0.1% TFA) to elute proteins from the column. Collected fractions were vacuum dried and stored for digestion. A total of 12 fractions were collected for each of the pooled lean and diabetic samples.
Proteolysis
The samples were reconstituted in 6M guanidine HCl and 10 mM dithiothreitol. After 1 hr incubation at 70°C, iodoacetamide (at a final concentration of 10 mM) was added to the reaction and allowed to incubate for 30 min at 4 °C. Ammonium bicarbonate (0.1M, pH 8.0) was added to dilute samples 6 fold. Sequence grade trypsin (2%) was added and the reaction mixture incubated at 37°C for 18 hrs. Proteolysis was stopped by addition of tosyl lysine chloroketone (TLCK) (trypsin: TLCK, 1:1 (w/w)). The tryptic peptides were then used for characterization of oxidation sites using a nanoUPLC-QSTAR mass spectrometer and their relative quantification achieved using selective reaction monitoring (SRM).
LC/MS/MS of digested fractions and database searches
Tryptic peptides were separated on a nanoACQUITY UPLC BEH C18 column, (1.7 μm, 75 μm × 100mm) using a nano UPLC instrument (Waters instruments Inc., Milford, MA) at a flow rate of 0.2 μL/min. Solvent A was 0.01% TFA in deionized H2O (dI H2O) and solvent B was 95% ACN/0.01% TFA in dI H2O. The nanoUPLC instrument was coupled to a QSTAR workstation (Applied Biosystems, Framingham, MA) equipped with a nano ESI source. An 85 min linear gradient (from 0% B to 60% B) was used to separate the peptides. MS/MS spectra were obtained in the positive ion mode using an ionization voltage of 2000 V at a sampling rate of one spectrum per second. The top three peptides with charges ranging from 2 to 4 were monitored by MS/MS. Previously examined ions were excluded for 60 sec. The centroid value of MS/MS peaks was determined using the following parameters; the merge distance was set at 100 ppm with minimum and maximum widths of 10 ppm and 500 ppm, respectively. The percentage height was set at 50% and MASCOT was used for all searches.
Mascot database searching
The files were then sent to the Mascot Daemon software (version 2.2.2) where a merged file for each of the lean and the diabetic pooled fractions was produced and sent to an in-house MASCOT server (Version 2.2, Matrix Science16). The Rattus norvegicus taxonomy (24123 sequences) in the NCBI database (5532021 sequences and 1915541870 residues) was searched with trypsin being selected as the proteolytic enzyme. Mascot™ has the limitation of allowing only nine modifications per search. The database was searched three times. The “error tolerant” feature of Mascot was used in a firstpass search, finding oxidized methionine as the most likely biological modification on the proteins. The database was then searched for the oxidized methionine two more times in addition to carbonylation as the variable modifications. Carbamidomethyl cysteine was selected as a fixed modification in the first search with the variable modifications being biotinylated oxidized arginine, biotinylated oxidized lysine, biotinylated oxidized proline, biotinylated oxidized threonine and oxidized methionine. The second search included carbamidomethyl cysteine again as a fixed modification but with biotinylated 3-deoxyglucosone adduct, biotinylated HNE adduct, biotinylated glyoxal adduct, biotinylated methyl glyoxal adduct and oxidized methionine as variable modifications. The precursor mass tolerance was set at 100 ppm and the fragment mass tolerance was set to 0.6 Da. Biotin fragmentation produces noise (unassigned fragment ions that negatively impact the ion score) in the spectrum that increase the expectation values17-19. Consequently, biotin carrying carbonylated modifications were validated manually.
Relative quantification of carbonylation sites using selective reaction monitoring (SRM)
Tryptic peptides carrying carbonylation sites (as detected by the QSTAR workstation) in pooled lean and diabetic samples were assembled into a list. Skyline open source program (version 0.5.0.1245) was used to predict the most suitable transitions for these carbonylated peptides.20 The output was saved as an Excel file and imported into the MassHunter Optimizer™ (Agilent Technologies, Santa Clara, CA, USA, version B02.01). This software automatically optimized the Agilent 6410 Triple Quad LC/MS System parameters e.g. the collision energy and fragmentor to get maximal intensity of the SRM transitions. MassHunter Optimizer uses the equation
to predict the most suitable collision energy. Further optimization was achieved manually.
Tryptic peptides in fractions containing carbonylated peptides were separated with an HPLC-polymeric Chip (Agilent Technologies, Santa Clara, CA, USA). This Chip column has a volume of 40 nL, is of 75 m × 150 mm in dimensions, and is packed with 5 m particle diameter ZORBAX 300 SB-C18 particles. The column was gradient eluted with an Agilent series 1100 instrument (Agilent Technologies) at 0.4 μL/min. The mobile phase gradient started with 2% B, reached 35% B after 6 min, then reached 90% after another 2 min and was held constant for 2 min to rinse the column. Solvent A was water while solvent B was acetonitrile, both containing 0.1% (v/v) formic acid. Mobile phase composition was brought back to 2% B within 0.1 min. Finally the column was re-equilibrated with a post run time of 5 min. Prior to separation; analytes were enriched on the 40 nl Chip column using a capillary pump at 4 L/min, 0% B. The Chip was connected to a nanospray emitter tip which was connected to an Agilent 6410 triple quadrupole mass spectrometer equipped with an electrospray source.
Maximum sensitivity was achieved by adjusting the position of the Chip nanospray emitter tip and the capillary voltage to achieve good, direct spray. At least 1800 V was used on the capillary. MS/MS spectra were acquired in the positive ionization mode. Drying gas flow was set at 4L/min, 300 °C. The delta electron multiplier voltage was set to 200 V. The Agilent Mass Hunter ChemStation™ software (version B02.01) was used for data acquisition and processing. The ESI signal stability was tested by injecting 100 fmoles of a standard peptide (glu fibrinopeptide with a precursor ion of m/z 786) and monitoring the signal at both the total ion current level and the SRM signal of two fragment ions (m/z 1171 and m/z 684). The collision energy used was 30 and dwell time of 50 in both cases.
Two transitions were recorded for carbonylated peptides in each time window for acquisition. SRM transitions were acquired at unit resolution for both the first and third quadrupoles. The MassHunter workstation qualitative analysis software was used for the integration of the area under the curve with each of the transitions. The areas of the transitions for the carbonylated peptides in the diabetic rat plasma were divided by the corresponding areas in the lean rat plasma.
Knowledge Assembly Analysis
To determine whether the proteins that changed more than 1.5 fold could be correlated with specific diseases the GeneGo™ “disease biomarker networks” tool was used to reveal disease biomarkers as seed nodes for the network. The networks were set to the default value of 0.05 of significance level which indicates a false positive value of no more than 5% for the list of significant networks. The list of oxidized proteins identified was uploaded along with their calculated fold changes. All distributions were sorted by statistical significance using a P-val of 0.05 as the cutoff.
Annotating the Identified Proteins Based on Their Tissue of Origin
Differentially oxidized proteins in the diabetic Zucker rat plasma relative to their lean controls were searched for their human homologs. The human plasma atlas (version 7.1) was then used to annotate these proteins according to their expression in the different tissues inside the body. The degree of their expression can be strong, medium or weak. Supporting Figure 1 shows a distribution of these proteins in the different tissues.
RESULTS
Selection of animal model
Meeting the objectives of these studies required an in vivo model of increasing OS. Age, gender,21 environmental exposure,22 diet, exercise,23 rest,24 seasonal variability,25 surgery,26 smoking,27 and drugs (e.g. Adriamycin28 and Doxorubicin29) are all factors in OS. As a means of controlling these variables experiments were carried out with Zucker diabetic rats. This animal model of diabetes develops metabolic disturbances characteristic of diabetes in which the animals become hyperglycemic by seven weeks of age on a Purina 5008 chow diet. Hyperinsulinemia occurs during the development of diabetes in this animal and then decreases by nineteen weeks. As shown in Table 1, mean fasting glucose was significantly higher in the diabetic animal (449.3 mg/dl) than in a lean non-diabetic animal (93.4 mg/dl).
Table 1.
Mean fasting glucose level for five diabetic rats and their control lean rats at 16 weeks.
| Sample | Mean fasting glucose level at 16 weeks |
|---|---|
| Lean plasma | 93.4 mg/dL (SD=11.3) |
| Diabetic rat plasma | 449.3 mg/dL (SD=110.4 |
Sample preparation
Carbonyl groups on oxidized proteins are very reactive, easily forming a Schiff base nucleophilic amines in proteins samples. When sequestered in this manner carbonyl groups are not available for reaction with biotin hydrazide. Biotin hydrazide was thus added to fresh plasma to preclude in vitro loss of carbonyl groups. Additionally, it has been reported that intrinsic endoproteases digest plasma proteins during sample processing.30 Sample proteolysis by these cysteine and serine proteases was blocked by adding a mixture of inhibitors to plasma at collection. These protease inhibitors are removed before trypsin digestion during avidin affinity chromatography.
Abundant proteins are generally removed from samples during pretreatment in studies of the type described here. That was not done in this work for several reasons. First, high abundance proteins can complex with other proteins. It was recently shown that using immunoaffinity techniques to remove high abundance proteins from blood samples resulted in the capture of at least 129 other proteins not targeted by the immunosorbent.31 This “sponge effect” carried the risk of removing proteins that are oxidized and of interest in this work.32 Oxidized proteins complexed with or crosslinked to abundant proteins would be lost. Mathematical modeling has shown that washing an affinity column with at least 15 column volumes of the loading buffer will elute low affinity, nonspecifically bound proteins.33 This was deemed to be a better approach to abundant protein removal than use of immunosorbents that target abundant proteins. After biotinylated protein selection from plasma samples, avidin affinity columns were washed with approximately 60 column volumes of mobile phase. Elution of nonspecifically bound proteins was assessed by the degree to which absorbance had returned to zero after sample application.
Measurement of oxidative stress in rats via urinary isoprostanes
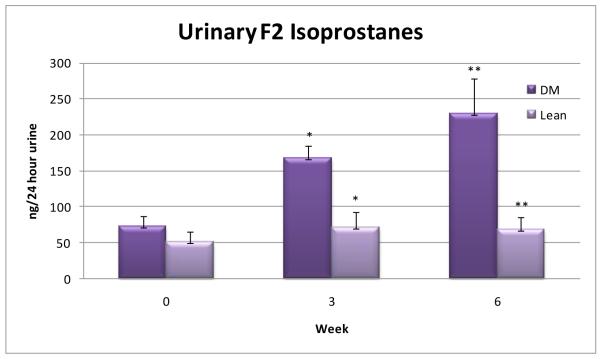
Oxidative stress was demonstrated by urinary isoprostanes using a competitive ELISA assay (Oxford Biomedical Research, Oxford, MI). Isoprostane is formed specifically as a consequence of free radical induced lipid peroxidation34 and is considered to be one of the most reliable indicators of in vivo OS.35 24-hr isoprostanes were significantly greater in the diabetic than controls rats at weeks 3 (p<0.05) and 6 (p<0.01) (Figure 1).
Figure 1.
Urinary isoprostanes (ng/24 hrs) measured at baseline, 3, and 6 weeks in diabetic (DM) and control (lean) rats. The difference in isoprostane concentration between diabetic and controls at 3 weeks was significant to the level of p < 0.05. At 6 weeks the difference was significant to the level of p < 0.01.
Analytical strategy
The blood proteome was chosen for analysis based on the rationale that i) proteins originating from multiple organs could be examined, ii) blood is readily available in the research setting, and iii) the techniques used will allow in vivo studies in a broad range of higher animals.
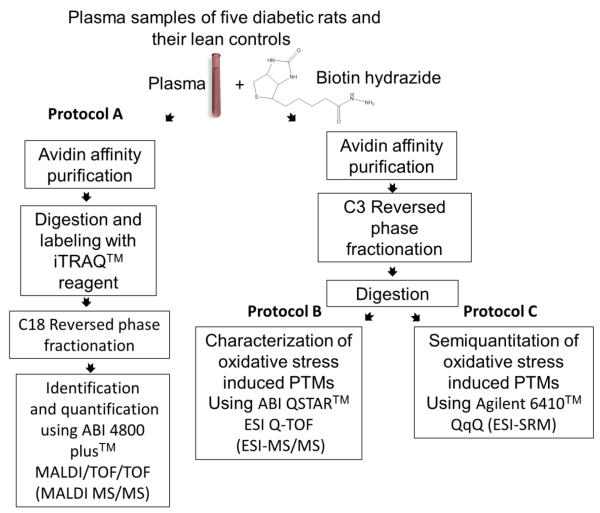
The analytical protocol used in these studies (Figure 2) is a modified version of a method first described for the analysis of oxidized proteins in the yeast proteome.36 Five diabetic Zucker rats were sacrificed and 6 mL of plasma withdrawn from each. The same procedure was used with non-diabetic control animals. After centrifugation, carbonylated proteins in plasma samples were biotinylated with biotin hydrazide (BH), Schiff base products of the reaction were reduced with sodium cyanoborohydride, and the samples were dialyzed to remove excess BH. Biotinylated proteins were subsequently selected from samples with avidin affinity chromatography (Figure 3).
Figure 2.
A schematic illustration of the strategy used for the identification, quantification, and characterization of carbonylated proteins and their oxidation sites in the plasma of diabetic and lean Zucker rats. For protein identification and quantifica tion, the samples were run individually, digested and then labeled with the iTRAQ™ reagent (protocol A). Characterization (protocol B) and semiquantification (protocol C) of oxidative post translational modifications were achieved using pooled samples.
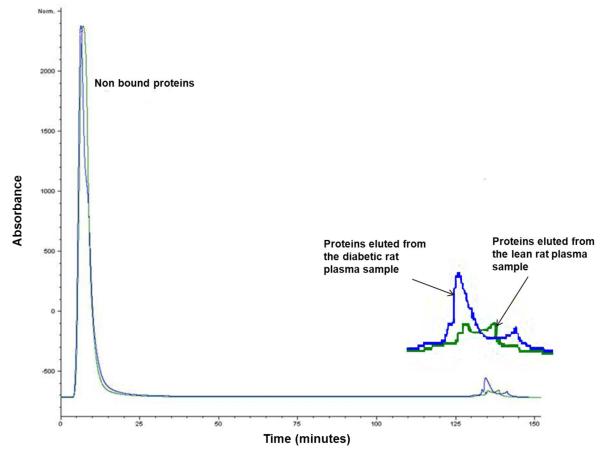
Figure 3.
Avidin affinity chromatogram of a Zucker diabetic rat plasma sample (blue line) overlayed on that of a lean rat plasma sample (green line). Plasma samples (each of 5mg total proteins content) were applied directly to a 4.6 × 100 mm column packed with UltraLink Biosupport™ to which avidin had been immobilized. The column was washed initially with 0.15 M phosphate buffered saline, (pH 7.4) at 0.5 mL/min for 120 min, then switched to a mobile phase containing 0.1M dimethylglycine / HCl(pH 2.5) for an additional 40 min at the same flow rate. Absorbance was monitored at 280 nm. Based on absorbance measurements, an average of 1% of the total protein from 5 lean rat plasma samples was captured by avidin affinity chromatography (SD= 0.46). The corresponding amount captured from 5 diabetic rat plasma samples was 1.7% (SD=0.0014).P-value= 0.0018
Proteins can be carbonylated at multiple sites, allowing for the existence of multiple isoforms. It is important to note that avidin affinity selection will isolate all the carbonylated isoforms of a protein together. No attempt was made to differentiate between the isoforms in the fractionation schemes used in this work.
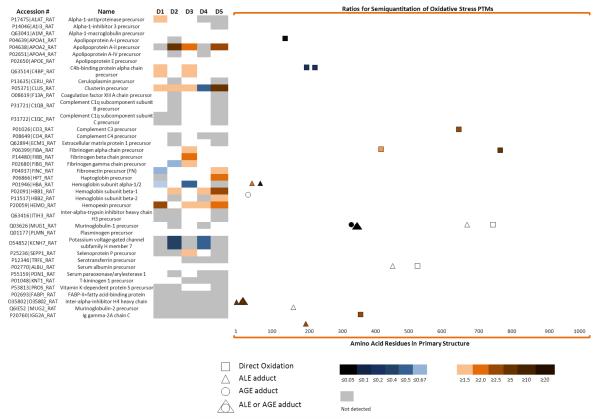
Three analytical protocols were used to identify oxidized proteins and the relative differences in oxidation at specific sites. Protocol A was used to examine samples from individual animals. Following selection of biotinylated proteins by avidin affinity chromatography in Protocol A, the selected proteins were digested with trypsin, labeled with iTRAQ™ coding agents, and further fractionated by C18 reversed phase chromatography (RPC). Tryptic peptides in fractions collected from the RPC column were identified and quantified based on relative differences in unoxidized peptides using an ABI 4800 plus™ (MALDI-TOF/TOF mass spectrometer). Protein Pilot™ was used for the analysis of the mass spectra as described in the METHODS. Because iTRAQ™ labeled, unoxidized peptides common to all isoforms of a protein were used for quantification, values being reported in Figure 4 and Table 2 below were the net change in all isoforms of a protein. Methods for measuring changes in oxidation at a specific site will be described below.
Figure 4.
A heat map of the quantification of the proteins detected and the semiquantitation of their oxidation sites. (Left) The first two columns show the proteins detected. D1-D5 is the fold change of these proteins in the diabetic versus lean controls as quantified by iTRAQ™ labeling. A list of proteins identified and quantified with iTRAQ™ and their corresponding p-values can be found in Table 2.(Right) Fold change at oxidation sites in diabetic rat plasma versus their lean controls. These oxidation sites can be either a product of: direct oxidation, Advanced Lipidperoxidation Endproducts (ALE) adducts or Advanced Glycation Endproducts (AGE) adducts. Methylglyoxal and glyoxal can be considered ALE or AGE. In all cases the color bar describes the intensity level of the proteins (left) or oxidation sites (right)
Table 2.
A list of proteins identified and quantified with iTRAQ™ detected with MALDI-TOF/TOF
| % Cov |
Accession # | Name | Number of unique peptides detected |
Species | Diabetic rat #1 versus its control | Diabetic rat #2 versus its control | Diabetic rat #3 versus its control | Diabetic rat #4 versus its control | Diabetic rat #5 versus its control | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ratio | P- value |
Ratio | P-value | Ratio | P- value |
Ratio | P-value | Ratio | P-value | |||||
| 15.8 | P17475|A1AT_RAT | Alpha-1-antiproteinase precursor |
3 | RAT | 1.6 | 1.5E-68 | 1.4 | 3.2E-01 | 1.2 | 7.0E-02 | ND | NA | ND | NA |
| 13.1 | P14046|A1I3_RAT | Alpha-1-inhibitor 3 precursor | 9 | RAT | 0.5 | 8.5E-01 | 1.0 | 1.5E-01 | 0.8 | 1.7E-01 | 1.2 | 3.8E-01 | ND | NA |
| 13.9 | Q63041|A1M_RAT | Alpha-1-macroglobulin precursor |
12 | RAT | 1.0 | 3.8E-01 | 0.9 | 1.6E-01 | 1.1 | 1.6E-06 | 1.0 | 3.1E-01 | 1.1 | 3.2E-01 |
| 45.2 | P04639|APOA1_RAT | Apolipoprotein A-I precursor | 8 | RAT | 1.1 | 1.8E-01 | ND | NA | 0.9 | 2.3E-01 | 0.8 | 4.8E-01 | 2.0 | 1.5E-01 |
| 21.6 | P04638|APOA2_RAT | Apolipoprotein A-II precursor | 2 | RAT | ND | NA | 9.0 | 4.0E-02 | 2.3 | 7.1E-03 | ND | NA | 4.9 | 4.0E-02 |
| 11.5 | P02651|APOA4_RAT | Apolipoprotein A-IV precursor |
2 | RAT | 0.9 | 7.3E-01 | ND | NA | 1.2 | 7.0E-01 | ND | NA | 2.1 | 4.4E-01 |
| 62.2 | P02650|APOE_RAT | Apolipoprotein E precursor | 9 | RAT | 1.0 | 2.0E-01 | 1.1 | 3.2E-01 | 0.8 | 1.5E-04 | 0.8 | 9.8E-01 | 1.2 | 1.2E-01 |
| 24.7 | Q63514|C4BP_RAT | C4b-binding protein alpha chain precursor |
10 | RAT | 1.9 | 1.2E-03 | 0.5 | 3.2E-01 | 1.6 | 2.1E-08 | 1.2 | 3.0E-02 | 0.8 | 1.7E-01 |
| 16.2 | P13635|CERU_RAT | Ceruloplasmin precursor | 4 | RAT | 1.5 | 5.3E-01 | ND | NA | 1.2 | 3.2E-01 | 1.0 | 3.3E-01 | ND | NA |
| 42.3 | P05371|CLUS_RAT | Clusterin precursor | 5 | RAT | 1.5 | 2.2E-03 | 1.5 | 8.0E-04 | 1.7 | 5.8E-06 | 0.6 | 2.1E-05 | 9.0 | 1.6E-04 |
| 19.8 | O08619 |F13A_RAT | Coagulation factor XIII A chain precursor |
2 | RAT | 1.3 | 1.5E-04 | ND | NA | 0.8 | 1.6E-02 | 0.9 | 8.8E-01 | ND | NA |
| 28.5 | P31721|C1QB_RAT | Complement C1q subcomponent subunit B precursor | 3 | RAT | 1.1 | 8.1E-02 | ND | NA | 1.3 | 5.0E-03 | 1.1 | 3.2E-01 | ND | NA |
| 41.2 | P31722|C1QC_RAT | Complement C1q subcomponent subunit C precursor | 6 | RAT | ND | NA | ND | NA | 1.4 | 2.1E-08 | 0.8 | 1.3E-01 | ND | NA |
| 37.6 | P01026|CO3_RAT | Complement C3 precursor | 12 | RAT | 0.9 | 3.6E-01 | 0.8 | 1.5E-01 | 0.8 | 2.9E-06 | 0.7 | 1.5E-01 | 0.9 | 1.3E-01 |
| 23.2 | P08649|CO4_RAT | Complement C4 precursor | 3 | RAT | 0.9 | 1.8E-01 | ND | NA | 0.9 | 7.5E-07 | ND | NA | ND | NA |
| 20.8 | Q62894|ECM1_RAT | Extracellular matrix protein 1 precursor |
3 | RAT | 1.0 | 4.1E-01 | ND | NA | 1.3 | 2.8E-03 | 1.3 | 7.0E-02 | ND | NA |
| 62.9 | P06399|FIBA_RAT | Fibrinogen alpha chain precursor |
218 | RAT | 0.7 | 1.6E-01 | 0.9 | 2.4E-02 | 1.9 | 4.3E-03 | 1.1 | 1.0E-02 | 1.2 | 3.2E-01 |
| 92.3 | P14480|FIBB_RAT | Fibrinogen beta chain precursor |
168 | RAT | 0.7 | 3.2E-01 | 0.7 | 7.1E-04 | 2.0 | 3.6E-79 | 1.2 | 7.0E-03 | 0.7 | 4.4E-02 |
| 84 | P02680|FIBG_RAT | Fibrinogen gamma chain precursor |
59 | RAT | 0.8 | 1.5E-08 | 0.6 | 1.4E-07 | 1.7 | 1.4E-33 | 1.1 | 3.1E-01 | 0.6 | 3.2E-01 |
| 70.8 | P04937|FINC_RAT | Fibronectin precursor (FN) | 229 | RAT | 0.6 | 1.5E-68 | 1.0 | 1.3E-03 | 1.2 | 2.5E-02 | 1.1 | 1.0E-03 | 1.5 | 1.3E-04 |
| 13.3 | P06866|HPT_RAT | Haptoglobin precursor | 2 | RAT | ND | NA | 1.1 | 5.9E-01 | 0.9 | 1.2E-01 | 0.7 | 3.2E-01 | 2.4 | 2.8E-01 |
| 47.9 | P01946|HBA_RAT | Hemoglobin subunit alpha- 1/2 |
16 | RAT | 0.8 | 1.2E-04 | 1.0 | 1.6E-01 | 0.5 | 1.2E-05 | 0.9 | 3.2E-01 | ND | NA |
| 69.4 | P02091|HBB1_RAT | Hemoglobin subunit beta-1 | 18 | RAT | 0.7 | 2.9E-05 | 1.6 | 3.2E-03 | 0.6 | 7.3E-01 | 1.5 | 1.9E-05 | 3.7 | 7.5E-02 |
| 50.3 | P11517|HBB2_RAT | Hemoglobin subunit beta-2 | 10 | RAT | 0.7 | 6.2E-02 | ND | NA | 0.7 | 1.3E-01 | ND | NA | 1.8 | 7.2E-02 |
| 58.3 | P20059|HEMO_RAT | Hemopexin precursor - Rattus norvegicus (Rat) |
37 | RAT | 3.9 | 1.7E-10 | 1.5 | 1.2E-02 | 1.6 | 1.8E-24 | 1.5 | 9.9E-06 | 2.4 | 2.5E-04 |
| 10.9 | Q63416|ITIH3_RAT | Inter-alpha-trypsin inhibitor heavy chain H3 precursor |
4 | RAT | ND | NA | ND | NA | 0.9 | 4.5E-03 | 1.1 | 8.0E-01 | ND | NA |
| 15.3 | Q03626|MUG1_RAT | Murinoglobulin-1 precursor | 7 | RAT | 1.2 | 8.4E-01 | ND | NA | 0.9 | 9.8E-03 | ND | NA | 1.5 | 3.2E-01 |
| 37.7 | Q01177|PLMN_RAT | Plasminogen precursor | 13 | RAT | 1.1 | 2.4E-01 | 1.1 | 1.6E-01 | 0.8 | 1.4E-01 | 1.1 | 3.0E-01 | 2.6 | 1.5E-01 |
| 10.5 | O54852 |KCNH7_RAT | Potassium voltage-gated channel subfamily H member 7 | 13 | RAT | ND | NA | 0.4 | 1.7E-04 | ND | NA | 0.6 | 4.1E-08 | ND | NA |
| 8.3 | P25236|SEPP1_RAT | Selenoprotein P precursor | 3 | RAT | ND | NA | ND | NA | 1.7 | 2.9E-04 | 1.3 | 1.7E-01 | ND | NA |
| 22.5 | P12346|TRFE_RAT | Serotransferrin precursor | 2 | RAT | 1.2 | 7.0E-02 | 1.6 | 3.3E-01 | ND | NA | ND | NA | ND | NA |
| 38.3 | P02770|ALBU_RAT | Serum albumin precursor | 32 | RAT | 1.1 | 9.5E-01 | 1.1 | 3.2E-01 | 0.8 | 3.9E-01 | 0.9 | 1.5E-01 | 1.9 | 1.6E-01 |
| 16.3 | P55159|PON1_RAT | Serum paraoxonase/arylesterase 1 |
3 | RAT | 0.9 | 1.3E-01 | ND | NA | 0.8 | 7.6E-02 | ND | NA | ND | NA |
| 18.8 | P01048|KNT1_RAT | T-kininogen 1 precursor | 2 | RAT | 1.6 | 1.1E-01 | 0.4 | 8.9E-01 | ND | NA | ND | NA | ND | NA |
| 7 | P53813|PROS_RAT | Vitamin K-dependent protein S precursor |
10 | RAT | ND | NA | ND | NA | 1.4 | 1.3E-01 | 1.1 | 8.1E-01 | ND | NA |
Samples from diabetic and lean rats respectively were pooled to facilitate oxidation site identification by using larger quantities of protein and examined by Protocol B. Five mg of pooled protein from either the diabetic or lean group of animals were applied to the avidin affinity column. Proteins thus selected were further fractionated on a C3 RPC column and collected for trypsin digestion. Following proteolysis, peptides from these fractions were examined by the LC-MS/MS using a Waters nano-UPLC coupled to a QSTAR Pulsar mass spectrometer. Identification of parent proteins was achieved with un-oxidized peptides whereas oxidatively modified peptides were used to identify types and sites of oxidation.
Peptides isolated in Protocol B were also used to evaluate relative changes in the concentration of specific carbonylated peptides through Protocol C. Quantification in Protocol C was achieved by selected reaction monitoring (SRM) using an Agilent Triple Quad 6410 LC/MS.
Avidin affinity selection
Biotinylated plasma samples were applied directly to the avidin affinity column without abundant protein removal. Up to 5 mg of protein was loaded on the column using PBS (0.15M, pH 7.4) loading buffer at a flow rate of 0.5 mL/min. Affinity selected proteins were then eluted by switching to 0.1 M dimethylglycine (pH 2.5). The affinity column was used in processing approximately 50 samples over six months without significant loss of capacity. Capacity was verified by the binding of standard biotinylated bovine serum albumin at periodic intervals.
Using absorbance at 280 nm and assuming that affinity selected and unbound proteins have the same extinction coefficient, approximately 1.7% (SD=0.0014) of the protein in Zucker diabetic rat plasma was bound to the avidin affinity column compared to 1% (SD=0.46) from lean rat plasma, p<0.01 (Figure 3). It should be noted that naturally biotinylated proteins, proteins naturally complexed with or crosslinked to the biotinylated proteins, and non-specifically bound proteins are included in this affinity selected fraction as well.36
Oxidized proteins in diabetic plasma
Using Protocol A an average of 2604 mass spectra were generated from plasma samples. Thirty five proteins were identified based on a minimum of two un-oxidized peptides found in two or more pairs of diabetic or lean animals with a confidence level higher than 95% (Figure 4 Table 2) and differences in oxidation quantified. Oxidized proteins of high abundance (e.g. fibrinogen alpha chain precursor), medium abundance (e.g. fibronectin) and low abundance (e.g. extracellular matrix protein 1 precursor) were identified.37 Fourteen of these proteins (alpha-1-macroglobulin precursor, apolipoprotein A-II precursor, apolipoprotein E precursor, C4b-binding protein alpha chain precursor, clusterin precursor, complement C3 precursor, fibrinogen alpha chain precursor, fibrinogen beta chain precursor, fibrinogen gamma chain precursor, fibronectin precursor, hemoglobin subunit alpha-1/2, hemoglobin subunit beta, plasminogen precursor and serum albumin precursor)were detected and quantified in five diabetic and lean pairs. Four proteins (alpha-1-inhibitor 3 precursor, apolipoprotein A-I precursor, haptoglobin precursor and hemopexin precursor) appeared in four diabetic and lean pairs. Eight proteins (alpha-1-antiproteinase precursor, apolipoprotein A-IV precursor, ceruloplasmin precursor, coagulation factor XIII A chain precursor, complement C1q subcomponent subunit B precursor, murinoglobulin-1 precursor, extracellular matrix protein 1 precursor and hemoglobin subunit beta-2) appeared in three diabetic and lean pairs and another nine proteins (complement C1q subcomponent subunit C precursor, complement C4 precursor, inter-alpha-trypsin inhibitor heavy chain H3 precursor, potassium voltage-gated channel subfamily H member 7, selenoprotein P precursor, serotransferrin precursor, serum paraoxonase/arylesterase 1, T-kininogen 1 precursor and vitamin K-dependent protein S precursor) appeared in two diabetic and lean pairs. Apolipoprotein AII (Apo AII) precursor, clusterin precursor, and hemopexin precursor were significantly elevated more than 1.5 fold in the diabetic animals relative to lean controls. Potassium voltage-gated channel subfamily H member 7 protein significantly decreased more than 1.5 fold. The highest sequence coverage for the proteins identified and quantified came from fribrinogen beta chain precursor (92.3% coverage), fibrinogen gamma chain precursor (84% coverage) and fibronectin precursor (70.8% coverage).
The fact that many oxidized proteins appeared at the same concentration in multiple animals (Figure 4 and Table 2) indicates a low level of biological variation between animals. This also means that in cases where concentration differences were observed between animals they were biologically relevant. The only proteins that were shown previously to be reproducibly oxidized in lean animal plasma were keratins14. One possible reason for the difference between these two studies may be the amount of plasma protein used in affinity selection. Five mg of protein were used here in contrast to a mg in the other study. Keratins were detected with an ESI-QSTAR (described later) but not MALDI-TOF/TOF mass spectrometry. Keratin identification was not reported because unambiguous identification of carbonylation sites was not achieved. Interestingly, twenty two of the proteins identified in this study were previously reported to be oxidized in normal human plasma.13 This suggests some similarity in protein oxidation between these two species.
Disease distribution
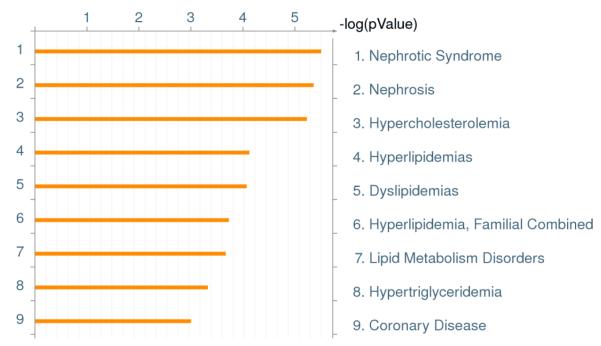
The GeneGo pathway analysis software was used to determine whether the proteins found to change significantly in concentration could be associated with any particular disease (Figure 5). GeneGo analysis of disease ontology revealed that the group of proteins oxidized 50% more than those in control samples were related to nephrotic syndrome, other kidney pathologies, and cardiovascular problems in the diabetic rats. This is as expected. As known from the literature, kidney disease and coronary heart disease are complications associated with the progression of diabetes.38
Figure 5.
Disease pathways identified by GeneGo™ (version 6.4) analyses using oxidized protein data from these studies. P value for the ontology was set to 0.05.
Tissues of origin
Tracing oxidized proteins back to their tissue of origin using the Protein Atlas database showed that kidney, liver and pancreas contributed more proteins than other organs (Supporting Figure 1). This confirms our GeneGo pathway analysis results that proteins related to the kidney complications in diabetes can be detected in plasma of diabetic rats.
Identification and semiquantitation of oxidation sites
Carbonylation sites were identified through Protocol B in which samples from multiple diabetic or lean animals were pooled and oxidized proteins enriched. After avidin affinity chromatography, the selected proteins were further resolved by RPC on a column with a C3 alkyl silane stationary phase. Fractions collected from the RPC column were trypsin digested and the resulting peptides identified by LC-MS/MS using a QSTAR ESI-MS/MS. Quantification was achieved by selected reaction monitoring (SRM) according to Protocol C. Unmodified peptides were used to identify the protein parent while biotin modified peptides were used to identify oxidation sites. In an early phase of this study, carbonylation sites were examined using both the ABI 4800 MALDI-TOF/TOF and ESI QSTAR. The later instrument was able to characterize a significantly larger number of oxidation sites than the MALDI-TOF/TOF. The reason for this may be that the large fragmentation energy of the hybrid TOF mass analyzer results in the loss of the fragments carrying biotin hydrazide labeled modifications.
An average of 10,625 spectra was obtained per analyses. As shown in Figure 4, Supporting Tables 1 and 2, seventeen carbonylation sites were detected using the QSTAR ESI-MS/MS. Label free quantification using the QSTAR ESI-MS/MS was not possible. iTRAQ™ based quantification of carbonylation sites through biotinylated peptides was equally ineffective. Fragmentation of the iTRAQ™ labeled peptides causes the generation of many unassigned MS/MS peaks.39 Fragmentation of biotin in the same peptides increased noise in the spectra even more, increasing expectation values (i.e. lowering scores).17-19 The presence of these two reagents in the same peptides prevented MASCOT™ from identifying any of the oxidation sites (data not shown). Carbonylated peptides were instead quantified with the Agilent 6410 QqQ ESI-MS/MS using a selected reaction monitoring (SRM) approach.
SRM analysis offered two advantages. One was that it allowed precise, reliable, and highly sensitive quantification of peptides below the detection limit of conventional LC-MS/MS methods. A second was that carbonylated peptide fragments could be detected which were not seen in the initial product ion scan by conventional LC/MS/MS. This appears to be the first use of SRM-MS to quantify carbonylated peptides (Figure 4, Supporting Tables 1 and 2).An example of the transitions used to quantify carbonylated peptides is shown in Figure 6.
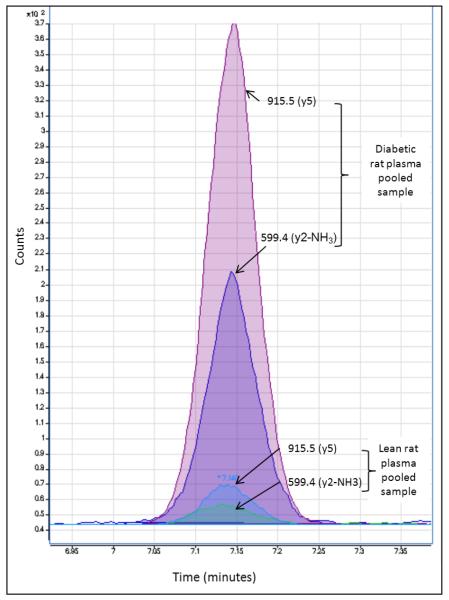
Figure 6.
Relative quantification of the carbonylated peptide KVADALAK using selective reaction monitoring (SRM). The peptide was modified in vivo with HNE. During the analytical work flow, the parent protein bearing this HNE modified peptide was biotinylated with biotin hydrazide and after avidin affinity selection biotinylated peptide were released from their parent proteins by trypsin digestion. Quantification was based on two CID transitions, the fragment ion at m/z = 915.5 (y5) and m/z = 599.4 (y2-NH3).
In all cases, QSTAR-MS/MS experiments were used to predict the major charge state of the precursor ion. As shown in Figure 4, seventeen peptides were quantified using this strategy. Eight peptides carrying nine carbonylation sites increased at least 2 fold in the diabetic pooled plasma compared to the lean pooled plasma (K69 of hemoglobin alpha 2 chain with a ratio of 20, K12 of hemoglobin alpha 2 chain with a ratio of 2, P356 of murinoglobulin 2 with a ratio of 2.1,R770 of fibrinogen alpha polypeptide isoform 1 with a ratio of 5.9, R419 of fibrinogen alpha polypeptide isoform 1 with a ratio of 2, K8 and K17 of fatty acid binding protein with a ratio of 9.4, K195 of Ig gamma-2A chain C region with a ratio of 4.2, and R688 of complement component 3 with a ratio of 2.6). Three peptides carrying five carbonylation sites decreased at least 2 fold in the diabetic pooled plasma compared to the lean pooled plasma (K224 and R228 of C4b-binding protein alpha chain precursor with a ratio of 0.1, K347 and K352 of murinoglobulin1 homolog with a ratio of 0.01 and T147 of preapolipoprotein A-I with a ratio of 0.01). Finally, a total of six peptides carrying a total of six carbonylation sites didn’t show any significant change (K49 of hemoglobin beta chain which remained unchanged, P543 of albumin with a ratio of 1.1, K161 of inter-alpha-inhibitor H4 heavy chain with a ratio of 1, R731 of murinoglobulin 1 homolog with a ratio of 1.2, K682 of murinoglobulin 1 homolog with a ratio of 0.9and C461 of albumin with a ratio of 1.3. Again, the presence of a number of carbonylated peptides that showed no significant change suggests that alterations in carbonylation are not random.
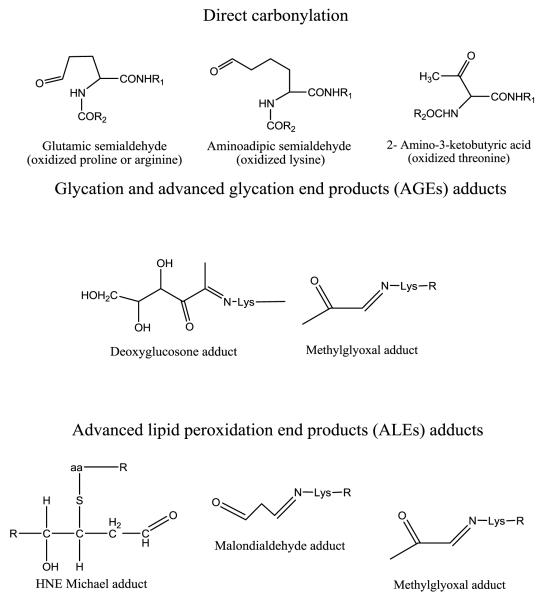
All three mechanisms by which protein carbonylation occurs were noted to contribute to increased protein oxidation in diabetic rat plasma. Among the changed modifications (i.e. either increased or decreased), seven carbonylation sites (P356 of murinoglobulin 2, R419 and R770 of fibrinogen alpha polypeptide isoform 1, K224 and R228 of C4b-binding protein alpha chain precursor, R688 of complement component 3 and T147 of preapolipoprotein A-I) originated from direct oxidation of an amino acid side chain. Four carbonylation sites (K69 of hemoglobin alpha 2 chain, K12 of hemoglobin alpha 2 chain, K8 of fatty acid binding protein, and K195 of Ig gamma-2A chain C region) involved the formation of advanced lipid peroxidation product adducts (e.g. malondialdehyde and HNE adducts). One carbonylation site (K347 of murinoglobulin 1 homolog) was formed by glycation and oxidation of an advanced glycation end product (e.g. deoxyglucosone adduct). Two sites (K17 of fatty acid binding protein and K352 of murinoglobulin 1 homolog) arose as a result of methylglyoxal adducts. This modification arises from either advanced glycation or advanced lipid peroxidation end products (Figure 7). Figure 8 shows an example MS/MS spectrum for one carbonylated peptide where we were able detect the oxidation of P543 in albumin to glutamic semialdehyde.
Figure 7.
Structures of carbonylation products detected in this study. R refers to the sequence of polypeptides, aa referes to lysine, histidine or cysteine that can form Michael adducts with 4-HNE.
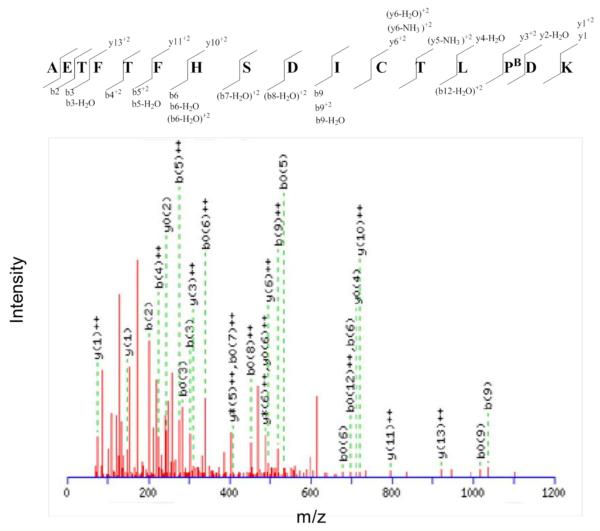
Figure 8.
MS/MS spectrum of the biotinylated peptide AETFTFHSDICTLPBDK from albumin. PB indicates a biotinylated oxidized proline residue. The difference between the masses of the y3+2 and y2-H2O ions equals the mass of biotinylated oxidized proline residue.
Among the proteins identified, hemoglobin and murinoglobulin 1 homolog had the largest number of carbonylation sites, possessing sites derived from all three types of carbonylation; e.g. direct carbonylation (e.g. oxidized arginine), glycoxidation of advanced glycation endproducts (AGE (e.g. 3-deoxyglucosone adducts) and advanced lipid peroxidation endproduct (ALE) adducts (e.g. malondialdehyde). The diabetic to lean ratio at carbonylation sites in hemoglobin was surprisingly diverse. Whereas the deoxyglucosone adduct ratio was unchanged at 1.0, HNE adduct, and malondialdehyde ratios of 20, and 2.0, respectively were seen in diabetic animals compared to lean controls. Similar results were observed with murinoglobulin 1 homolog, while, deoxyglucosone adduct and methylglyoxal adduct had a ratio of nearly 0.01, the malondialdehyde adduct and oxidized arginine remained unchanged. Fibrinogen alpha polypeptide isoform 1 provided a similar example. The ratio of arginine oxidation between diabetic and lean animals at sites 419 and 770 was 2.0 and 5.9, respectively.
There is always the question with proteins that change in association with disease progression whether such changes are tightly connected to the disease. The GeneGo pathway analysis software recognized oxidized proteins whose concentration changed more than 50% in diabetics relative to leans controls as being associated with nephrotic syndrome, other kidney pathologies, and cardiovascular problems. As known from the literature, kidney disease and coronary heart disease are complications associated with the progression of diabetes.38 There is potential that with large scale validation some set of these oxidized proteins could potentially be biomarkers.
DISCUSSION
The objective of the work described here was to examine and compare both i) protein carbonylation in general and ii) carbonylation at specific amino acid residues in a protein as a function of increasing OS using diabetic and lean animals.
The blood proteome was chosen as a sample source because of the ease with which it could be obtained and still provide oxidized proteins originating from multiple organs. At the general level involving the assay of all carbonylated isoforms of proteins together by Protocol A, the data shows clearly that there is no relationship between protein abundance in plasma and protein carbonylation. Although oxidized serum albumin was found, this highly abundant protein was not a major contributor to the carbonylated plasma proteome. Abundant proteins in plasma are either not as easily oxidized or carbonylation occurs at local sites and oxidized proteins are exported into plasma. The later seems the most likely based on the fact that so many proteins in plasma are of tissue origin. This makes localized OS a major source of carbonylated proteins in plasma. Compartmentalization seems to be important in protein carbonylation. It also explains why the level of some oxidized proteins in plasma is elevated more than others. They experience differing levels of localized OS in the compartments where they reside.
Actually the importance of the location of proteins at the time of oxidation and the source of oxidants has been reported in the case of intestinal fatty acid binding protein (I-FABP). This protein plays an important role in the transport of fatty acids to mitochondria and their subsequent β-oxidation.40 The close proximity of I-FABP to mitochondria and fatty acids provides a potential explanation for how malondialdehyde and methylglyoxal adducts are formed in diabetic rats. A previous study has shown that carbonylation of A-FABP (adipocyte FABP) with 4-HNE is elevated in vivo in adipose tissue of insulin resistant mice.41
Hemopexin, clusterin and Apo AII were observed to have experienced the greatest increases in carbonylation among plasma proteins. Presumably this is due to elevated ROS levels having oxidized a greater proportion of these proteins in diabetic rats versus lean controls but increased expression might have been a factor as well. Hemopexin42 and clusterin43 expression has been found to increase in the plasma of type II diabetes. Another study showed that increased expression of Apo AII was a factor in insulin resistance.44 A 1.5 fold increase in oxidized protein isoforms has been shown to be strongly correlated with kidney and coronary heart diseases. Whether oxidized proteins cause or are the result of these phenomena remains to be determined. A study quantifying both changes in protein expression and carbonylation is needed to clarify the mechanism of how the elevation of these proteins occurred in diabetics.
With specific proteins data from these studies shows that i) that oxidation is specific and reproducible, and ii) that the extent of oxidation at any one site as a function of increasing OS is quantitatively independent of that at other sites. Some sites are dramatically more labile than others as seen in the cases of hemoglobin, murinoglobulin and fibrinogen. There is also no relationship between the mole fraction of carbonylation across all sites in a protein and changes at individual sites. It’s currently impossible to detect all the carbonylation sites of a protein therefore; the net change of all oxidized isoforms of a protein can’t be correlated to the oxidation levels at different sites.
One of the most distinguishing features of protein carbonylation was that oxidation patterns were protein specific in terms of i) the dominant oxidation mechanism, ii) the amino acids being modified, and iii) the oxidation mechanisms involved.
With hemoglobin for example, OS induced post-translational modifications by the ALE mechanism were elevated 20 fold in diabetic subjects relative to lean controls at residue K69 (by HNE addition) and 2 fold at residue K12 (with malondialdehyde addition). Deoxyglucosone modification at reside K49 was the same in lean and diabetic subjects. It is surprising that in a diabetic subject with elevated glucose levels and accelerated ROS production that carbonylation by glucose induced AGE mechanisms was the same as in lean controls. The dramatic increase in carbonylation from lipid peroxidation in contrast is expected based on the elevation of OS in diabetics. The presence of iron in hemoglobin could also increases the potential for oxidation, as would be the case with other metal containing proteins such as serotransferrin, ceruplasmin, hemopexin, and fibrinogen.45
Changes in AGE based carbonylation were equally dramatic with murinoglobulin 1 homolog. Although there was no significant change in total carbonylation at all sites, carbonylation by AGE mechanisms at residues K347 and K352 decreased 100 fold in diabetic subjects while ALE based malondialdehyde adduct formation and direct carbonylation at residues 682 and 731 respectively were unchanged. Again, a decline in carbonylation by the AGE mechanism when ROS and glycation are increasing with diabetic progression is not expected in a diabetic. Why ALE based carbonylation at residues K69 and K12 in hemoglobin was substantially elevated in diabetics while there was a significant reduction in AGE base adduction formation at residue K347 and K352 in murinoglobin 1 is a mystery.
Enhanced degradation of oxidized proteins could explain the selective decrease of some oxidized proteins. Previous reports indicated that there is an increase in proteolysis of some oxidized and glycated proteins in the plasma of diabetic patients, particularly in the case of methyl glyoxal adducts.49 Moreover, the activity of oxidized protein hydrolase (OPH) in the serum of diabetic rats increased nearly in parallel to the increase of blood glucose levels.50 In addition, oxidized proteins are ubiquitinated and form aggregates in the cytoplasm of pancreatic β cells that degrade through autophagy.51 Diabetes induced OS in the retinal endothelial cells and up-regulated the ubiquitin proteasomal system in yet another study.52
Fibrinogen alpha polypeptide isoform 1 showed still another pattern, there was no significant change in total carbonylation at all sites while arginine oxidation in diabetic subjects increased 2 fold at residue 419 and 6 fold at residue 770, respectively.
Protein conformation along with changes in tertiary structure seems to be another factor in protein oxidation. A recent study showed that glyceraldehyde-3-phosphate dehydrogenase changes its conformation in vivo according to the degree of OS.46 With this protein, oxidatively induced changes in conformation increase the probability of oxidation at other sites.
CONCLUSIONS
It is concluded from this work that proteins in rat plasma are carbonylated at a number of sites that vary widely in oxidative labiality. Moreover, site specific liability is due more to i) the protein itself, ii) the site of an amino acid in the protein, and iii) localized oxidation potential than the amino acid itself. The extent of oxidation at any one site as a function of increasing OS is quantitatively independent of that at other sites. There is also no relationship between the mole fraction of carbonylation across all sites in a protein and changes at individual sites. Moreover, there is no relationship between the concentration of any oxidatively modified form of a protein and the relative abundance of the parent protein in plasma. Carbonylation at any particular site in a protein can occur by any of three OS induced post-translational modification modes. The ratio of these modifications varies between proteins and changes within a single protein with increasing OS. It is further concluded that most carbonylated proteins in plasma are locally oxidized and exported into the circulatory system. Finally it is concluded that it is not possible at present to predict oxidation sites in proteins.
Supplementary Material
ACKNOWLEDGEMENT
The authors are thankful for Drs Mike Sturek and Mouhamad Alloosh (Indiana University School of Medicine) for supplying them with swine plasma samples used in the initial phases of this study. Special thanks to Dr H. Dorota Inerowic for managing the Purdue Proteomics Facility and Catherine P Riley for useful discussions. The authors gratefully acknowledge the support of this work by the National Cancer Institute (grant number 1U24CA126480-01), the national institute of aging (grant number 5R01AG025362-02), the Purdue University-University of Alabama at Birmingham Botanical Center for Age Related Diseases funded by the Office of Dietary Supplements and NCCAM grant P50 AT 00477 and the Indiana Clinical and Translational Sciences Institute funded in part by grant # RR 02576 from the National Institutes of Health, National Center for Research Resources.
Footnotes
SUPPORTING INFORMATION
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Wei W, Liu QJ, Tan Y, Liu LC, Li XK, Cai L. Oxidative Stress, Diabetes, and Diabetic Complications. Hemoglobin. 2009;33(5):370–377. doi: 10.3109/03630260903212175. [DOI] [PubMed] [Google Scholar]
- 2.Bonda DJ, Wang XL, Perry G, Nunomura A, Tabaton M, Zhu XW, Smith MA. Oxidative stress in Alzheimer disease: A possibility for prevention. Neuropharmacology. 2010;59(4-5):290–294. doi: 10.1016/j.neuropharm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Jenner P. Oxidative stress in Parkinson’s disease. Ann Neurol. 2003;53(Suppl 3):S26–36. doi: 10.1002/ana.10483. discussion S36-8. [DOI] [PubMed] [Google Scholar]
- 4.Baillet A, Chanteperdrix V, Trocme C, Casez P, Garrel C, Besson G. The role of oxidative stress in amyotrophic lateral sclerosis and Parkinson’s disease. Neurochemical Research. 2010;35(10):1530–7. doi: 10.1007/s11064-010-0212-5. [DOI] [PubMed] [Google Scholar]
- 5.Harrison D, Griendling KK, Landmesser U, Hornig B, Drexler H. Role of oxidative stress in atherosclerosis. Am J Cardiol. 2003;91(3A):7A–11A. doi: 10.1016/s0002-9149(02)03144-2. [DOI] [PubMed] [Google Scholar]
- 6.Ballard PL, Truog WE, Merrill JD, Gow A, Posencheg M, Golombek SG, Parton LA, Luan X, Cnaan A, Ballard RA. Plasma biomarkers of oxidative stress: relationship to lung disease and inhaled nitric oxide therapy in premature infants. Pediatrics. 2008;121(3):555–61. doi: 10.1542/peds.2007-2479. [DOI] [PubMed] [Google Scholar]
- 7.Lawless MW, O’Byrne KJ, Gray SG. Targeting oxidative stress in cancer. Expert Opin Ther Targets. 2010;14(11):1225–45. doi: 10.1517/14728222.2010.526933. [DOI] [PubMed] [Google Scholar]
- 8.Danielsen PH, Loft S, Jacobsen NR, Jensen KA, Autrup H, Ravanat JL, Wallin H, Moller P. Oxidative stress, inflammation and DNA damage in rats after intratracheal instillation or oral exposure to ambient air and wood smoke particulate matter. Toxicol Sci. 2010 doi: 10.1093/toxsci/kfq290. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, Wu J, Deleo CJ. RNA damage and surveillance under oxidative stress. IUBMB Life. 2006;58(10):581–8. doi: 10.1080/15216540600946456. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Wang X, Shigenaga MK, Yeo HC, Mori A, Ames BN. Immobilization stress causes oxidative damage to lipid, protein, and DNA in the brain of rats. FASEB J. 1996;10(13):1532–8. [PubMed] [Google Scholar]
- 11.Madian A, Regnier F. Proteomic Identification of Carbonylated Proteins and Their Oxidation Sites. Journal of Proteome Research. 2010;9(8):3766–3780. doi: 10.1021/pr1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullineaux PM, Baker NR. Oxidative stress: antagonistic signaling for acclimation or cell death? Plant Physiol. 2010;154(2):521–5. doi: 10.1104/pp.110.161406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madian AG, Regnier FE. Profiling Carbonylated Proteins in Human Plasma. Journal of Proteome Research. 2010;9(3):1330–1343. doi: 10.1021/pr900890k. [DOI] [PubMed] [Google Scholar]
- 14.Mirzaei H, Baena B, Barbas C, Regnier F. Identification of oxidized proteins in rat plasma using avidin chromatography and tandem mass spectrometry. Proteomics. 2008;8(7):1516–1527. doi: 10.1002/pmic.200700363. [DOI] [PubMed] [Google Scholar]
- 15.Peterson RG, Shaw WN, Neel MA, Little LA, Eichberg J. Zucker diabetic fatty as a model for non-insulindependent diabetes mellitus. ILAR News. 1990;32:16–19. doi: 10.1093/ilar.32.3.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 17.Borisov Oleg V, Goshe Michael B, Conrads Thomas P, Rakov VS, Veenstra Timothy D, Smith Richard D. Low-energy collision-induced dissociation fragmentation analysis of cysteinyl-modified peptides. Analytical Chemistry. 2002;74(10):2284–92. doi: 10.1021/ac010974p. [DOI] [PubMed] [Google Scholar]
- 18.Han B, Stevens Jan F, Maier Claudia S. Design, synthesis, and application of a hydrazide-functionalized isotope-coded affinity tag for the quantification of oxylipid-protein conjugates. Analytical Chemistry. 2007;79(9):3342–54. doi: 10.1021/ac062262a. [DOI] [PubMed] [Google Scholar]
- 19.Mirzaei H, Regnier F. Protein:protein aggregation induced by protein oxidation. Journal of Chromatography, B: Analytical Technologiesin the Biomedical and Life Sciences. 2008;873(1):8–14. doi: 10.1016/j.jchromb.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 20.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guevara R, Santandreu FM, Valle A, Gianotti M, Oliver J, Roca P. Sex-dependent differences in aged rat brain mitochondrial function and oxidative stress. Free Radic Biol Med. 2009;46(2):169–75. doi: 10.1016/j.freeradbiomed.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 22.Migliore L, Coppede F. Environmental-induced oxidative stress in neurodegenerative disorders and aging. Mutat Res. 2009;674(1-2):73–84. doi: 10.1016/j.mrgentox.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 23.Bloomer RJ, Fisher-Wellman K. The role of exercise in minimizing postprandial oxidative stress in cigarette smokers. Nicotine Tob Res. 2009;11(1):3–11. doi: 10.1093/ntr/ntn005. [DOI] [PubMed] [Google Scholar]
- 24.Siu PM, Pistilli EE, Alway SE. Age-dependent increase in oxidative stress in gastrocnemius muscle with unloading. J Appl Physiol. 2008;105(6):1695–705. doi: 10.1152/japplphysiol.90800.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossner P, Jr., Svecova V, Milcova A, Lnenickova Z, Solansky I, Sram RJ. Seasonal variability of oxidative stress markers in city bus drivers. Part I. Oxidative damage to DNA. Mutat Res. 2008;642(1-2):14–20. doi: 10.1016/j.mrfmmm.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Kucukakin B, Gogenur I, Reiter RJ, Rosenberg J. Oxidative stress in relation to surgery: is there a role for the antioxidant melatonin? J Surg Res. 2009;152(2):338–47. doi: 10.1016/j.jss.2007.12.753. [DOI] [PubMed] [Google Scholar]
- 27.Ozguner F, Koyu A, Cesur G. Active smoking causes oxidative stress and decreases blood melatonin levels. Toxicol Ind Health. 2005;21(1-2):21–6. doi: 10.1191/0748233705th211oa. [DOI] [PubMed] [Google Scholar]
- 28.Julka D, Sandhir R, Gill KD. Adriamycin-induced oxidative stress in rat central nervous system. Biochem Mol Biol Int. 1993;29(5):807–20. [PubMed] [Google Scholar]
- 29.Wolf MB, Baynes JW. The anti-cancer drug, doxorubicin, causes oxidant stress-induced endothelial dysfunction. Biochim Biophys Acta. 2006;1760(2):267–71. doi: 10.1016/j.bbagen.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Yi J, Kim C, Gelfand Craig A. Inhibition of intrinsic proteolytic activities moderates preanalytical variability and instability of human plasma. Journal of Proteome Research. 2007;6(5):1768–81. doi: 10.1021/pr060550h. [DOI] [PubMed] [Google Scholar]
- 31.Gong Y, Li X, Yang B, Ying W, Li D, Zhang Y, Dai S, Cai Y, Wang J, He F, Qian X. Different Immunoaffinity Fractionation Strategies to Characterize the Human Plasma Proteome. Journal of Proteome Research. 2006;5(6):1379–1387. doi: 10.1021/pr0600024. [DOI] [PubMed] [Google Scholar]
- 32.Issaq Haleem J, Xiao Z, Veenstra Timothy D. Serum and plasma proteomics. Chemical reviews. 2007;107(8):3601–20. doi: 10.1021/cr068287r. [DOI] [PubMed] [Google Scholar]
- 33.Cho W, Jung K, Regnier FE. Use of Glycan Targeting Antibodies To Identify Cancer-Associated Glycoproteins in Plasma of Breast Cancer Patients. Analytical Chemistry (Washington, DC, United States) 2008;80(14):5286–5292. doi: 10.1021/ac8008675. [DOI] [PubMed] [Google Scholar]
- 34.Khosrowbeygi A, Zarghami N. Seminal plasma levels of free 8-isoprostane and its relationship with sperm quality parameters. Indian J. Clin. Biochem. 2008;23(1):49–52. doi: 10.1007/s12291-008-0012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vassalle C, Botto N, Andreassi Maria G, Berti S, Biagini A. Evidence for enhanced 8-isoprostane plasma levels, as index of oxidative stress in vivo, in patients with coronary artery disease. Coron Artery Dis. 2003;14(3):213–8. doi: 10.1097/01.mca.0000063504.13456.c3. [DOI] [PubMed] [Google Scholar]
- 36.Mirzaei H, Regnier F. Affinity chromatographic selection of carbonylated proteins followed by identification of oxidation sites using tandem mass spectrometry. Analytical Chemistry. 2005;77(8):2386–2392. doi: 10.1021/ac0484373. [DOI] [PubMed] [Google Scholar]
- 37.Polanski M, Anderson NL. A list of candidate cancer biomarkers for targeted proteomics. Biomark Insights. 2007;1:1–48. [PMC free article] [PubMed] [Google Scholar]
- 38.Gotch PM. Atherosclerosis as a complication of diabetes. Implications for cardiovascular nursing. Focus Crit Care. 1986;13(4):9–15. [PubMed] [Google Scholar]
- 39.Aggarwal K, Choe LH, Lee KH. Shotgun proteomics using the iTRAQ isobaric tags. Briefings in Functional Genomics & Proteomics. 2006;5(2):112–120. doi: 10.1093/bfgp/ell018. [DOI] [PubMed] [Google Scholar]
- 40.Montoudis A, Seidman E, Boudreau F, Beaulieu J-F, Menard D, Elchebly M, Mailhot G, Sane A-T, Lambert M, Delvin E, Levy E. Intestinal fatty acid binding protein regulates mitochondrion Î2-oxidation and cholesterol uptake. Journal of Lipid Research. 2008;49(5):961–972. doi: 10.1194/jlr.M700363-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Grimsrud PA, Picklo MJ, Sr., Griffin TJ, Bernlohr DA. Carbonylation of adipose proteins in obesity and insulin resistance: identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Mol Cell Proteomics. 2007;6(4):624–37. doi: 10.1074/mcp.M600120-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Van Campenhout A, Van Campenhout C, Lagrou Albert R, Abrams P, Moorkens G, Van Gaal L, Manuel-y-Keenoy B. Impact of diabetes mellitus on the relationships between iron-, inflammatory- and oxidative stress status. Diabetes/Metabolism Research and Reviews. 2006;22(6):444–54. doi: 10.1002/dmrr.635. [DOI] [PubMed] [Google Scholar]
- 43.Trougakos IP, Poulakou M, Stathatos M, Chalikia A, Melidonis A, Gonos ES. Serum levels of the senescence biomarker clusterin/apolipoprotein J increase significantly in diabetes type II and during development of coronary heart disease or at myocardial infarction. Experimental Gerontology. 2002;37(10-11):1175–1187. doi: 10.1016/s0531-5565(02)00139-0. [DOI] [PubMed] [Google Scholar]
- 44.Castellani LW, Goto AM, Lusis AJ. Studies with apolipoprotein A-II transgenic mice indicate a role for HDLs in adiposity and insulin resistance. Diabetes. 2001;50(3):643–651. doi: 10.2337/diabetes.50.3.643. [DOI] [PubMed] [Google Scholar]
- 45.Shacter E, Williams JA, Lim M, Levine RL. Differential Susceptibility of Plasma-Proteins to Oxidative Modification - Examination by Western-Blot immunoassay. Free Radical Biology and Medicine. 1994;17(5):429–437. doi: 10.1016/0891-5849(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 46.Pierce A, Mirzaei H, Muller F, De Waal E, Taylor AB, Leonard S, Van Remmen H, Regnier F, Richardson A, Chaudhuri A. GAPDH Is Conformationally and Functionally Altered in Association with Oxidative Stress in Mouse Models of Amyotrophic Lateral Sclerosis. Journal of Molecular Biology. 2008;382(5):1195–1210. doi: 10.1016/j.jmb.2008.07.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.