Summary
In mammalian meiosis, only a small fraction of programmed DNA double-strand breaks are repaired as inter-homolog crossovers (COs). To analyze another product of meiotic recombination, inter-homolog noncrossovers (NCOs), we performed high-resolution mapping of recombination events at an intensely active mouse hotspot in F1 hybrids derived from inbred mouse strains. We provide direct evidence that the vast majority of repair events are inter-homolog NCOs, consistent with models in which frequent inter-homolog interactions promote accurate chromosome pairing. NCOs peaked at the center of the hotspot, but were also broadly distributed throughout. In some hybrid strains, localized zones within the hotspot were highly refractory to COs and showed elevated frequency of co-conversion of adjacent polymorphisms in NCOs, raising the possibility of double-strand gap repair. Transmission distortion was observed in one hybrid, with NCOs providing a significant contribution. Thus, NCO recombination events play a substantial role in mammalian meiosis and genome evolution.
Introduction
Meiotic recombination occurs most often in specific genomic regions known as “hotspots”, which are thought to be preferred sites of DNA double-strand breaks (DSBs) generated by the Spo11 protein (Arnheim et al., 2007; Cole et al., 2010). Unlike in mitosis, meiotic recombination preferentially uses the homologous chromosome over the sister chromatid as the template for DSB repair and can result in reciprocal exchange to form a crossover (CO) (Hunter, 2006). COs promote accurate segregation of homologs in meiosis I, such that each chromosome requires at least one CO – the obligate CO – to avoid missegregation leading to aneuploid gametes. Human and mouse hotspots show substantial variation in CO recombination activity, ranging from 0.0004% to as high as 2% (Guillon and de Massy, 2002; Jeffreys et al., 2001).
COs represent only a fraction of meiotic recombination events. From cytological evidence in mammals, it is estimated that only 10% of DSBs are repaired as inter-homolog COs (Baudat and de Massy, 2007b). The remaining DSBs are inferred to be repaired largely by inter-homolog recombination without reciprocal exchange, resulting in noncrossovers (NCOs). However, where analyzed, the proportion of NCOs is lower than expected from the number of DSBs (Guillon et al., 2005; Jeffreys and May, 2004; Jeffreys and Neumann, 2005), raising the possibility that many DSBs are repaired instead by recombination involving only sister chromatids. COs and NCOs are thought to be generated by distinct pathways that diverge shortly after recombination initiation, with COs formed by canonical DSB repair (DSBR) through resolution of a double Holliday junction intermediate and NCOs formed by synthesis-dependent strand annealing (SDSA) resulting in unidirectional transfer of genetic information (Hunter, 2006).
NCOs can only be detected if they incorporate scoreable genetic markers, such that the low polymorphism density at many studied hotspots has limited the sensitivity and spatial resolution of NCO maps, especially given that NCO gene conversion tracts are presumed to be short (Guillon et al., 2005; Jeffreys and May, 2004; Jeffreys and Neumann, 2005). In contrast, COs alter the linkage of many markers in a single event, so they are more readily detected. Another barrier to NCO detection is that, whereas any CO within a hotspot can be specifically amplified from pools containing a large excess of nonrecombinants, NCOs cannot, unless selective methods are used to enrich for an NCO incorporating a particular polymorphism. As a consequence, prior studies focused on pre-selected subsets of all possible NCOs that could occur within a hotspot (Guillon et al., 2005; Jeffreys and May, 2003). Thus, even though NCOs may account for the majority of meiotic recombination events, they are largely unexplored.
To gain a more sophisticated understanding of meiotic recombination, we fine-scale mapped over 1500 inter-homolog recombination events at an intensely active and highly polymorphic hotspot in mouse using multiple inbred strain combinations. We observed a high NCO to CO ratio in all cases, demonstrating that most inter-homolog DSB repair at this hotspot occurs via NCO pathways. Significant disparity in transmission of genetic information (i.e., transmission distortion) was observed in one strain combination, much of which could be attributed to NCO gene conversion, implying that NCOs can contribute significantly to genome evolution.
Results
CO activity at a highly polymorphic meiotic recombination hotspot
The A3 hotspot on mouse Chr 1 was previously estimated by pedigree analysis to have CO activity several hundred-fold higher than the genome average of 0.55 centimorgans per megabasepair (cM/Mb) (Kelmenson et al., 2005), suggesting that it would be active enough to score both COs and NCOs in sperm DNA. Sequencing the A3 region in 9 inbred mouse strains identified two major haplotypes across the region (Figure S1, S2). The polymorphism density between the two haplotypes is high; for example, A/J and DBA/2J (hereafter A and D) have 32 polymorphisms within 2 kb, for a 1.6% polymorphism density (Figure S1, S2).
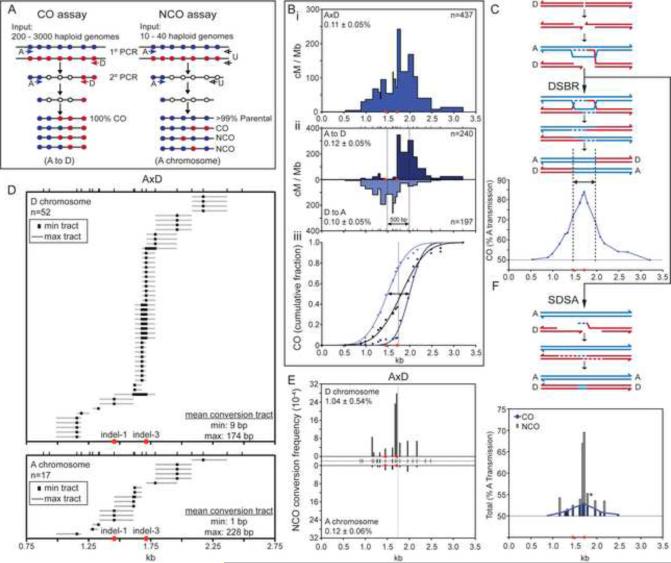
To position the hotspot, we used two rounds of nested, allele-specific PCR of pools equivalent to 200–3000 sperm genomes, followed by allele-specific hybridization to map CO exchange points (Figure 1A, left panel; Figure S3A). We identified 437 COs from 4 A×D F1 hybrid males with a total input equivalent to 376,000 sperm genomes, for a CO frequency of 0.11% per sperm genome (Figure 1B.i; Table S1). (Frequencies are Poisson-adjusted; see Experimental Procedures.) CO activity spanned 2.7 kb with 90% of exchange points within the central 1.5 kb. The activity across this central region averaged 78 cM/Mb, ~130-fold above genome average, and peaked at 242 cM/Mb. No COs were detected in somatic controls (spleen DNA; frequency <1 × 10−6), thus the COs isolated from sperm are bona fide meiotic recombinants and cannot be ascribed to PCR artifacts.
Figure 1. Recombination initiation bias for COs and NCOs in A×D results in transmission distortion.
(A) PCR strategy to amplify COs and NCOs at the A3 hotspot. Only one orientation is shown. Filled circles, polymorphisms on the A (A/J, blue) or D (DBA/2J, red) chromosome; filled arrows, allele-specific primers; open arrows, universal primers (U).
(B) CO breakpoints cluster in a 1.5 kb region, defining the A3 hotspot. (i) Total CO breakpoint map is shown. (ii) The CO breakpoint maps in the A to D (top) and D to A (bottom) orientations are shifted relative to each other, indicating reciprocal CO asymmetry. Vertical lines represent the midpoint for each orientation, as determined from the cumulative distributions (iii). The distance between the midpoints provides an estimate of the mean CO gene conversion tract length (500 bp, arrow). The vertical line in panel iii represents the average midpoint of CO gene conversion tracts, as determined from the midpoints for the two orientations. Numbers of COs examined and Poisson-adjusted CO frequencies (± S.D.) are indicated. Ticks represent positions of the 20 tested polymorphisms. Red circles, indels (see Figure S2D, E).
(C) Transmission distortion from COs in favor of the A chromosome arising from preferential DSB formation on the D chromosome (red) with the A chromosome (blue) serving as the donor of genetic information for DSBR. Only one chromatid from each homolog is shown for simplicity. The vertical lines represent the midpoints of the CO distributions for each orientation, as in (Bii).
(D) NCO gene conversion tracts on the D (top) and A (bottom) chromosomes.
(E) NCO frequencies for A×D are 10-fold higher than CO frequencies, with the D chromosome showing 9-fold more NCOs than the A chromosome. Total Poisson-adjusted NCO frequencies are indicated. NCO frequencies at each tested polymorphism are normalized for co-conversions. Ticks in the center represent the 19 polymorphisms tested. The vertical line represents the average midpoint of CO gene conversion tracts, as in (Biii).
(F) Transmission distortion in favor of the A chromosome derived from NCOs (gray bars) and COs (blue bars, derived from Figure 1C). Transmission distortion from NCOs arises from preferential DSB formation on the D chromosome (red) with the A chromosome (blue) serving as the donor of genetic information for SDSA.
Amplification of sperm DNA with an A-specific forward primer and a D-specific reverse primer (A to D orientation) gave rise to a shifted distribution of COs relative to D to A (Figure 1B.ii). Such asymmetric distribution in CO exchange points (known as reciprocal CO asymmetry) has been described for a few other mammalian hotspots (Arnheim et al., 2007) and is interpreted to reflect biased recombination initiation (i.e., DSB formation) in favor of one of the chromosomes (Jeffreys and Neumann, 2002). Thus, by the canonical DSBR model, the D chromosome undergoes the DSB and is the recipient of genetic information from the A chromosome (Figure 1C). As a result, D to A exchange points are located to the left of the conversion tract and A to D exchange points to the right. In cumulative CO distribution plots, the midpoints of the A to D and D to A orientations are shifted 500 bp (Figure 1B.iii), indicating that the average gene conversion tract length associated with crossing-over is ~500 bp. As a result of the biased direction of genetic information transfer, there is transmission distortion in favor of the A haplotype across this region of the hotspot (Figure 1C). At the peak of the distortion, ~80% of transmission favors the A polymorphism.
NCO analysis substantiates biased recombination initiation at the A3 hotspot
If biased DSB formation is indeed responsible for reciprocal CO asymmetry at A3, then inter-homolog NCOs should also show bias. Examining a single polymorphism, a previous study reported biased NCO formation on the chromosome expected to be favored for DSBs (Baudat and de Massy, 2007a). The high recombination activity and polymorphism density at A3 provide an ideal setting at which to fine map NCOs on both chromosomes. We performed two rounds of nested PCR on very small pools of sperm genomes using sequential sets of allele-specific forward primers and universal (U) reverse primers which recognize both chromosomes (Figure 1A, right panel) (Jeffreys and May, 2004). The universal primers allow amplification of both COs and NCOs, which are distinguished by genotyping polymorphisms by allele-specific hybridization (Figure S3B). We genotyped 19 polymorphisms, on average one per 100 bp. For A to U, we isolated 10 COs from 15,000 sperm genomes and for D to U, we isolated 7 COs from 6,000 genomes. This CO frequency (0.08%) was thus similar to that from the CO-specific assay.
The overall NCO frequency was 0.58% per sperm genome. Because four sperm are generated from each meiosis, but only one sperm inherits a recombinant DNA molecule when an NCO occurs, this value implies that a detectable NCO occurs in 2.3% of meioses. By contrast, the 0.11% frequency of CO molecules per sperm genome translates to a per-meiosis CO recombination frequency of 0.22%, because each CO recombination event generates two recombinant DNA molecules. Thus, NCO recombination occurs ≥10-fold more frequently than CO recombination. The NCO frequency from the D to U amplification was 1.04% per D chromosome, accounting for the majority of the NCOs. By contrast, the NCO frequency from the A to U amplification was significantly lower, at 0.12% per A chromosome (Figure 1E). These NCOs are bona fide meiotic recombination products because they were not observed in somatic controls (frequency <3.5 × 10−5). These results substantiate that biased recombination initiation in favor of one chromosome is responsible for the asymmetry in the CO distributions. A3 on the A chromosome is still a reasonably active hotspot: based on the 9:1 ratio of NCOs (D:A), it would contribute one tenth toward the total CO activity of 78 cM/Mb across the hotspot, which is ~13-fold above genome average.
NCOs map throughout the hotspot, but peak in the center only on the hotter chromosome
In 58 of 69 NCOs in A×D, only a single polymorphism was converted (Figure 1D), revealing that the gene conversion tracts were short. The remaining 11 were co-conversions, typically involving just 2 polymorphisms. The minimal conversion tracts (black boxes) are measured by considering just the converted polymorphisms themselves; for co-conversions, they also include the segments in between. The theoretical maximal conversion tracts (gray bars) additionally include the distance to the next polymorphism on each side that was not converted. Averaging the gene conversion tracts from both the A and D chromosomes, the mean minimal tract is 7 bp and the mean maximal tract is 187 bp.
We mapped the distribution of NCOs, normalizing for co-converted polymorphisms (Figure 1E). Intriguingly, the NCO distribution was wider than expected from previous reports (Baudat and de Massy, 2007b): NCO gene conversions mapped throughout a 1.5 kb region, corresponding to the area containing 90% of COs. For the D chromosome, 30 of 52 NCOs (58%) mapped to two polymorphisms located 34 bp apart, one of which, an insertion/deletion polymorphism (indel-3, Figure 1D), flanks the interval with highest CO activity.
The other 22 NCOs on the D chromosome were distributed to regions flanking the NCO peak (Figure 1D, E). If we assume that conversions occur preferentially close to the site where recombination initiated, then these results, coupled with the very short lengths of conversion tracts, imply that although DSBs may be highly favored at the center of a hotspot, they span a larger region than heretofore appreciated. By contrast, on the A chromosome, all 17 NCOs mapped to the flanking polymorphisms and were absent at the two polymorphisms at the center of the hotspot (Figure 1D, E). Thus, the reduction of NCOs on the A chromosome is disproportionately due to loss of the discrete peak in the hotspot center, indicating that a tight zone favored for DSB formation is lacking. This lack of DSB formation at the hotspot center would also account for the reciprocal CO asymmetry.
The NCO peak and the average midpoint of the CO gene conversion tracts are therefore at nearly identical positions (compare central line in Figures 1B.iii and 1E), despite the fact that the gene conversion tract lengths are substantially longer for COs (~500 bp). These results imply a preponderance of DSBs in the center of the hotspot leading to both COs and NCOs. As with COs, biased NCO formation leads to transmission distortion in favor of the A chromosome. The region of strongest distortion is narrower for the NCOs (Figure 1F), as expected from the relatively short gene conversion tracts, but NCOs nonetheless contribute a greater proportion to the total transmission distortion because of the high NCO to CO ratio. Considering the hotspot overall, NCOs contribute about 70% more to transmission distortion than COs (Figure 1F).
Regional deficiency of COs despite frequent NCOs defines a CO refractory zone
The question arises as to whether the recombination differences between the A and D chromosomes can be attributed to their distinct haplotypes at A3. C57BL/6J (hereafter, B) shares the same haplotype as A at A3 (Figure S1, S2). Thus, both have ~30 polymorphisms compared with D, but A and B differ by only 8 polymorphisms, notably a small indel (indel-2) located ~150 bp to the left of indel-3 (Figure S2).
To determine how recombination is affected by strain background, we examined recombination in B×D F1 hybrids. In marked contrast to A×D, the B×D hybrids did not show strong reciprocal CO asymmetry: the B to D CO distribution was very similar to the D to B distribution (Figure 2A), suggesting that both chromosomes initiate recombination at similar frequencies. Consistent with this interpretation, CO frequencies assayed in the B to D and D to B orientations were 0.33% and 0.22%, respectively, ~2-fold higher than observed for A×D, as expected from efficient DSB formation on both chromosomes. Also, cumulative distributions of the B to D and D to B COs were less shifted relative to each other compared to A×D (Figure 2B). (Because COs are likely initiating at similar frequencies on both chromosomes, this shift does not provide an accurate estimate of the average CO gene conversion tract.) The mild reciprocal CO asymmetry in B×D is in counterpoint to the patent asymmetry in A×D, and transmission distortion is thus much less pronounced (Figure S4C).
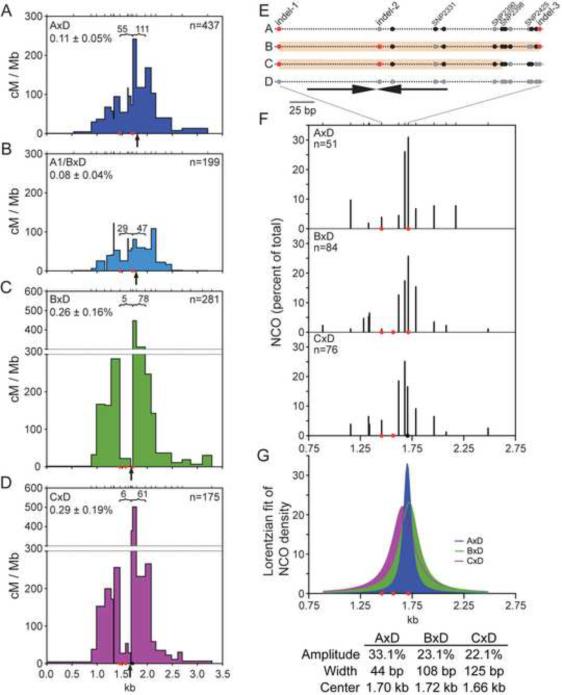
Figure 2. B×D shows CO refraction, but not biased recombination initiation.
(A) CO breakpoint maps from B×D in the B to D (top) and D to B (bottom) orientations. The CO refractory zone (salmon shading), is indicated. Ticks represent positions of the 16 tested polymorphisms.
(B) Cumulative CO distributions for all COs and for each orientation.
(C) NCO frequencies for the D (top) and B (bottom) chromosomes indicate similar frequencies of recombination initiation on both chromosomes. Ticks in the center represent the polymorphisms tested (20 and 16 for the D and B chromosome, respectively). The vertical line represents the midpoint of the total CO distribution, as in Figure 2B.
(D) NCO gene conversion tracts on the D chromosome, depicted as in Figure 1D. See Figure S4 for the B chromosome.
Strikingly, a dearth of COs was observed in the hotspot center. Similar observations have been made at other mammalian hotspots (Baudat and de Massy, 2007a; Jeffreys and Neumann, 2005; Paigen et al., 2008; Wu et al., 2010), indicating that holes in CO distributions may be a common characteristic of hotspots, although this phenomenon has not been systematically studied. The hole in A3 occurs just to the left of indel-3, which demarcates the interval of peak CO activity here as it did with A×D (compare Figures 3A,C): ~15-fold fewer COs were mapped to the 258 bp region encompassing the gap, as compared with the adjacent 249 bp region to the right of indel-3 (Figure 3C). By contrast, for A×D the difference between these two regions was only ~2-fold (Figure 3A).
Figure 3. CO refraction is linked to Chr 1 and is associated with a wider NCO distribution.
(A–D) Total CO breakpoint maps for the indicated hybrids. The number of CO breakpoints mapped to the regions indicated by brackets is given. The leftmost of these regions encompasses the CO refractory zone in B×D, except in panel D, where it encompasses the slightly shifted CO refractory zone in C×D. Breakpoints within the CO refractory zone are significantly reduced for B×D and C×D relative to either A×D or A1/B×D, p<0.0001 (Fisher's exact test). Arrows, midpoints of total CO breakpoints for each hybrid; red circles, indels; black circle, SNP2425.
(E) Schematic of polymorphisms for the A, B, or C versus D haplotypes in the area of the CO refractory zone. The three indels and the SNPs mentioned in the text are labeled, with the location of the ~140 bp inverted repeat indicated (arrows). Polymorphisms of the D genotype are shown in gray; SNPs differing from D are in black and insertions are in red. The CO refractory zones for B×D and C×D are shown with salmon shading on the B and C chromosomes, respectively.
(F) Comparison of NCO distributions for the D chromosome in the indicated hybrids. Only polymorphisms (ticks) shared by all three hybrids are plotted. NCOs are expressed as a percent of total and normalized for co-conversions.
(G) Lorentzian fit of the NCO distributions (F) showing an increased width for B×D and C×D compared with A×D. A slight shift to the left is observed for the C×D distribution. The amplitude (Amp), width, and the center of the fitted distributions are indicated.
To gain more insight into the B×D recombination patterns, we mapped NCOs on both chromosomes. The overall NCO frequency was 0.97% per sperm genome, implying that NCO recombination occurs 7.5-fold more frequently than CO recombination on a per-meiosis basis. Considering both NCOs and COs, the overall recombination frequency is >4.4% per meiosis, defining A3 as an intensely active hotspot. Most NCOs converted a single polymorphism (144 of 184 NCOs), with mean minimal and theoretical maximal conversion tracts of 9 and 168 bp, respectively, when averaging tracts from both chromosomes (Figure 2D, S4A).
In stark contrast to A×D, NCOs were frequent on both chromosomes and similarly distributed (Figure 2C, 2D, S4A). For the D chromosome, 66 of 91 NCOs (72%) mapped to 6 polymorphisms spanning 166 bp at the center of CO activity. Many NCOs on the B chromosome also spanned this interval (46 of 93, 49%; 5 polymorphisms tested). The NCO frequency on the D chromosome was only marginally higher than that on B (1.02% versus 0.92%), consistent with the direction and magnitude of transmission distortion observed for COs (Figure S4C).
The hole in the CO distribution in B×D could result from different initiation of recombination (e.g., a lack of DSBs) or different outcomes of recombination relative to A×D. More than half of NCOs mapped within this hole (D chromosome: 55/91, 60%; B chromosome: 48/93, 52%; Figures 2D, S4A), despite the absence of polymorphisms between indel-1 and indel-2, indicating that recombination frequently initiated there. Moreover, a substantial fraction of these NCOs overlapped those in A×D, which did not exhibit a hole in the CO distribution. These results imply that recombination outcomes are different in B×D, with COs specifically excluded from this region. Therefore, we have defined this 258 bp interval as a CO refractory zone (Figure 3E).
CO refraction and recombination initiation bias are linked to Chr 1
Analysis of the A3 hotspot in A×D and B×D distinguished two distinct phenomena, recombination initiation bias and CO refraction. To gain more insight into the influences of strain background, we examined COs in other F1 hybrids, including a chromosome substitution strain that contains A/J Chr 1 in an otherwise C57BL/6J background (A1/B). Crossing this strain to DBA/2J allowed us to distinguish whether recombination differences between the A and B chromosomes are linked to Chr 1, which contains A3. A1/B×D showed a similar pattern of COs as A×D, i.e., strong reciprocal CO asymmetry (513 bp offset between midpoints of the CO distributions, Figure S4B), strong transmission distortion at the expense of D alleles (Figure S4C), and lack of a strong CO refractory zone (29 versus 47 COs in the two regions that flank indel-3; Figure 3B), linking these phenomena to Chr 1, possibly to A3 itself.
We examined a third strain, C3H/HeJ (C), which shares the same haplotype as A and B (Figure S1, S2). In C×D F1 hybrids, we found a similar total CO frequency (0.29% per sperm genome) and similar spatial pattern of COs as B×D (Figure 3D), i.e., mild reciprocal CO asymmetry (102 bp offset, Figure S5B), modest transmission distortion (Figure S4C), and a strong CO refractory zone, although the CO refractory zone is slightly contracted compared with B×D (Figure 3D, E). Similar to B×D, the majority of NCOs on the D chromosome in C×D occurred within the CO refractory zone (50 of 87, 57%; Figure S5C, D), indicating that recombination frequently initiated in this region despite the paucity of COs.
Only 2 polymorphisms are shared by B×D and C×D but not by A×D, indel-2 and a single nucleotide polymorphism (SNP2390), both in the CO refractory zone, raising the possibility that one or both of these polymorphisms cause CO refraction. Indel-2 is near the center of an ~140 bp imperfect inverted repeat, which has potential to form secondary structure (Figure 3E). The CO refractory zones in B×D and C×D are flanked by indel-1 on the left and contain indel-2 in the center (Figure 3E). The right edge of the CO refractory zone in B×D is indel-3; this indel is absent from C×D, but SNP2425, located 7 bp away, can be typed instead (Figure S2). In C×D, a significant number of COs mapped to the interval between SNP2398 and SNP2425 (379.3 cM/Mb), shifting the right edge of the CO refractory zone to the left relative to B×D (Figure 3E). These observations potentially link the presence of these indels to the presence and position of CO refraction.
CO refraction is associated with wider NCO distributions
Given that the D chromosome is common to all of the F1 hybrids, we expected to observe similar NCO patterns on this chromosome in all hybrids. However, the majority of A×D NCOs were tightly clustered at 2 polymorphisms 34 bp apart (Figure 1E), whereas in B×D and C×D (Figures 2C and S5C), NCOs were more broadly spread over 166 bp.
To better compare D chromosome NCO activity, NCO distributions were plotted to reflect only the polymorphisms shared by all 3 F1 hybrids, with NCOs expressed as percentages of total (Figure 3F). Because most polymorphisms are shared and those that differ are frequently incorporated within co-converted NCO tracts, over 90% of the NCOs were included in this analysis. A wider NCO distribution at the center of NCO activity is apparent for B×D and C×D as compared with A×D. Smoothing the distributions using a Lorentzian fit quantifies the differences in the distributions (Figure 3G). Whereas the width to amplitude ratio is 1.3 for A×D, the ratio is 4.7 and 5.6 for B×D and C×D, respectively. Also apparent is a small shift of the center of the NCO distribution for C×D relative to B×D (~60 bp to the left). Thus, even though the same chromosome was undergoing conversion, NCO distributions differed in the 3 hybrids. Wider NCO distributions were observed in both hybrids exhibiting CO refraction, raising the possibility that these two phenomena are mechanistically linked. The similar shifts in the NCO distribution and CO refractory zone in C×D provide further support for such a link.
NCOs with co-converted polymorphisms are enriched in the center of the A3 hotspot
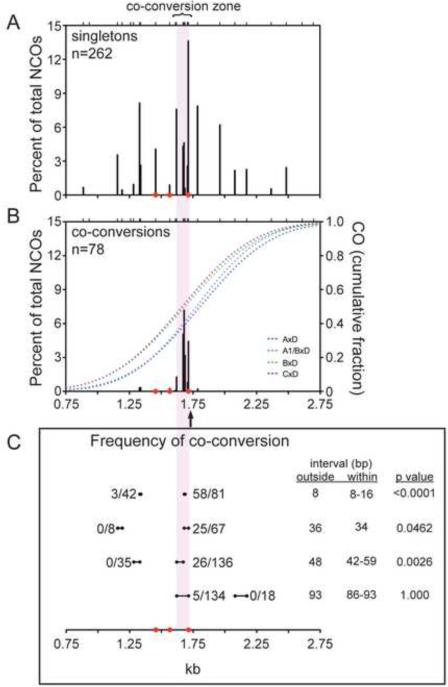
In all three hybrids, most NCOs converted a single polymorphism, but co-conversions formed a significant class, whether considering all chromosomes (78 of 340 NCOs, 23%) or just the D chromosome (64 of 230, 28%). Co-conversions were not equally distributed throughout A3, but were strongly enriched in the central ~100 bp, such that 92% of co-conversions were located within this “co-conversion zone” (compare Figure 4A, B). Consequently, 41% of the 184 NCOs in this zone were co-conversions. The D chromosome showed a co-conversion zone in all hybrids, but, interestingly, a greater fraction of NCOs in this zone were co-conversions in B×D and C×D. The co-conversion zone is the peak region of NCOs, with a density of 1.8 NCOs per bp. (Compare with 0.12 NCOs per bp in the flanking 1.3 kb or 0.53 NCOs per bp in the second highest region of NCOs). Although the mean minimal gene conversion tract in the co-conversion zone is longer than in flanking regions (16 bp versus 1 bp), conversion tracts within this zone are still very short, with a mean theoretical maximal gene conversion tract of 117 bp.
Figure 4. Co-conversions in NCOs are enriched in the central 100 bp of the hotspot.
Compilation of NCOs with conversion of only a single polymorphism (singletons) (A) and co-conversions (B) for A3, plotted as a percent of total NCOs. Values are Poisson adjusted. The 100-bp zone enriched for co-conversions is indicated by the lilac shading. NCOs are derived from all chromosomes tested except the A chromosome.
(B) CO breakpoint cumulative distributions (right y-axis) for the indicated hybrids are also plotted; the arrow indicates the midpoint for all of the CO breakpoints.
(C) Comparison of the frequency of co-conversion within and outside of the co-conversion zone. Intervals of similar sizes are compared for the number of co-conversions relative to total conversions involving one or both polymorphisms. Interval sizes within the co-conversion zone may differ slightly between hybrids because of polymorphism variation. Co-conversions outside and within the co-conversion zone were compared using a Fisher's exact test.
To verify that these NCOs were bona fide co-conversions and not two separate NCOs in the same amplification pool, we cloned several NCOs and genotyped them by colony hybridization using allele-specific probes (data not shown). Six of 7 NCOs were verified to be co-conversions; the other, which appeared to be a discontinuous gene conversion, actually comprised two separate NCOs.
Because polymorphisms are not equally spaced, we quantified the co-conversion frequency for similarly spaced polymorphisms in the co-conversion zone and in flanking regions (Figure 4C). Strikingly, in the flanking region, 2 polymorphisms separated by 8 bp were co-converted in only 3 of 42 NCOs (7.1%), whereas in the co-conversion zone, 2 polymorphisms separated by the same or somewhat longer distance (8 to 16 bp) were co-converted in 58 of 81 NCOs (72%; p <0.0001, Fisher's exact test). Three progressively larger intervals showed the same trend, although the fraction of co-conversions decreased with distance. As the co-conversion zone is the peak region of NCOs, co-conversions may preferentially occur in regions of frequent DSB formation.
Discussion
A3 is an intensely active recombination hotspot with a high NCO to CO ratio
Here we define A3 as a very strong mouse meiotic recombination hotspot, with an overall recombination frequency as high as ≥4.4% per meiosis. Significantly, a nearly 10-fold excess of NCOs over COs was observed in high resolution recombination maps. Cytological estimates of DSB numbers previously suggested that inter-homolog NCOs could greatly exceed COs in both mouse and human (Baudat and de Massy, 2007b); however, studies examining this issue never approached this cytological estimate and in fact often reported fewer NCOs than COs (Baudat and de Massy, 2007a; Holloway et al., 2006; Jeffreys and May, 2004; Jeffreys and Neumann, 2005). Given these previous results, it seemed possible that inter-sister recombination could contribute more to meiotic recombination in mammals than thought from studies of budding yeast (Hunter, 2006). The large excess of NCOs we observe therefore is in agreement with that predicted from cytology and emphasizes that most DSBs are repaired by inter-homolog recombination that favors an NCO outcome.
Efficient NCO detection throughout the A3 hotspot is possible because polymorphisms are located on average every 30 bp in the hotspot center and are also frequent in the flanks. Only one prior report has documented more NCOs than COs for a mammalian hotspot, DNA3 in humans, where a 2.7:1 ratio was obtained, although the paucity of polymorphisms and short NCO conversion tracts led the authors to conclude that the actual ratio was likely higher (Jeffreys and May, 2004). While it is possible that A3 is unusual in having a high NCO to CO ratio, we consider it more likely that its highly polymorphic nature allows an unprecedented sensitivity for detecting NCOs that are representative of recombination across the genome. Thus, we predict that fine mapping of other suitably polymorphic mammalian hotspots will reveal similarly high NCO:CO ratios.
Implications of a high NCO to CO ratio
Because formation of just one CO per chromosome pair is sufficient for proper chromosome segregation, having only one DSB per chromosome pair (the “obligate” DSB) should suffice if every DSB could be efficiently converted to a CO. From this perspective, it seems that mouse meiocytes incur an enormous assault on genomic integrity. What is the purpose of these “extra” DSBs? One likely answer is that homology-dependent DNA interactions from multiple recombination events enforce proper homolog pairing. In most organisms, homologs form increasingly stable pairing interactions during meiotic prophase (Burgess, 2002). In mouse, similar to many fungi and plants but distinct from Drosophila or C. elegans, recombination is necessary for homologs to locate one another, to align along their lengths, and to become intimately synapsed (Baudat et al., 2000; Romanienko and Camerini-Otero, 2000). It is likely that having multiple recombination sites dispersed on each homolog pair suppresses interactions between non-allelic homologous sequences in mouse, as it does in yeast (Goldman and Lichten, 2000).
Another possible reason for a high NCO to CO ratio may be that it reflects elements of CO control. In every organism that has been examined, the number of DSBs is variable between cells, whereas CO numbers are tightly regulated (Martinez-Perez and Colaiacovo, 2009). In mice, the coefficient of variation in DSB markers is ~30%, compared with <10% for COs (F.C., unpublished results). It is plausible that a high set point for DSBs ensures that enough recombination events occur for pairing and CO formation even in meiocytes at the low end of the DSB spectrum, making meiosis robust. A related point is that having a relatively large number of DSBs per bivalent allows for regulated placement of COs. Thus, an excess of NCOs may be the natural consequence of having an excess of DSBs that provide a reservoir from which properly regulated crossing over can be executed.
CO and NCO gene conversion tracts
The fine mapping of recombination events at A3 allows us to gain insight into recombination mechanisms. Our results imply a preponderance of DSBs in the hotspot center leading to both COs and NCOs, but the patterns support models in which these recombination products arise by distinct pathways. This is particularly evident from the A×D hybrid, as it shows biased recombination initiation. The CO frequencies in the two orientations (A to D vs. D to A) are equivalent, consistent with reciprocal exchange as predicted by the DSBR model (Szostak et al., 1983). In contrast, the NCO frequency is much lower on the non-initiating chromosome, implying that most NCOs do not alter the donor chromosome, a central feature of the SDSA pathway (Paques and Haber, 1999).
Also consistent with distinct pathways, COs have longer gene conversion tracts than NCOs: for COs, conversion tracts are ~500 bp, while most NCOs convert just a single polymorphism even in the co-conversion zone, where the mean NCO tract length ranges from 16 to 117 bp. Short NCO conversion tracts are also seen in mitotic cells (Larocque and Jasin, 2010). At a human meiotic hotspot, NCO tract lengths were estimated to be longer, although this difference is likely a by-product of the lower polymorphism density (Jeffreys and May, 2004).
A high polymorphism density is required for fine mapping recombinants, but it is possible that sequence divergence affects recombination frequencies or outcomes (Borts and Haber, 1987; Dooner, 2002). For A3, however, the high polymorphism density does not obviously affect CO frequencies. Previous CO estimates by pedigree analysis in B×C hybrids (Kelmenson et al., 2005) were similar to what we measured in B×D and C×D, despite a lower polymorphism density (Figure S1). Conversely, B × Mus musculus spretus hybrids, which have a high polymorphism density (2.4%), gave a higher CO frequency. Hence, there is not a simple relationship between polymorphism density and CO frequency. Polymorphism density could also potentially affect gene conversion tract lengths, although there is no evidence for this in mitotic cells (Larocque and Jasin, 2010).
An unexpected finding was that while NCOs were concentrated in the center of A3, nearly half occurred in the flanking 1.5 kb. Given short conversion tracts, NCOs likely approximate the position of DSBs, suggesting that Spo11 has preferred sites for DSB formation within A3 but is not restricted to just one or a few nearby cleavage sites. In yeast, DSBs also occur at multiple sites within hotspots, although they cluster over smaller regions of 75–250 bp (Liu et al., 1995). Intriguingly, NCOs were largely absent from the hotspot center for the A chromosome in the A×D hybrid, but were still present in the flanks. This pattern suggests that sites of preferred cleavage within the B, C, and D haplotypes do not undergo frequent cleavage in the A haplotype, but that less frequent cleavage at the subordinate sites is maintained. The alternative possibility, that NCOs on the A chromosome are derived from DSBR events initiated on the D chromosome, seems unlikely because the conversion tracts are short and do not extend from the center of the hotspot (Paques and Haber, 1999).
It is not clear what leads to lower Spo11 cleavage on the A chromosome. Only one polymorphism, SNP2331, is specific for the A haplotype, so this sequence variant may be the cause of lower DSB formation. Alternatively, the A haplotype may have sequence variants elsewhere on Chr 1 that act in cis at a distance to promote DSB formation within A3. In human hotspots DNA2 and NID1, single SNPs appear to directly affect recombination initiation (Jeffreys and Neumann, 2002, 2005). Both of these SNPs have been proposed to disrupt DNA binding by PRDM9 (Baudat et al., 2010), an H3K4 trimethyltransferase implicated in hotspot determination in both humans and mice (Neale, 2010). It is unclear whether A3 contains a PRDM9 binding site, given that the mouse consensus is degenerate (Baudat et al., 2010).
Transmission distortion and the contribution of NCOs and COs
Because of biased DSB formation at A3 in A×D hybrids, the A chromosome is more frequently the donor of genetic information, leading to its transmission at a higher rate. Several factors govern the magnitude of such transmission distortion. The overall frequency of recombination and the degree of biased DSB formation between haplotypes are the most important factors. However, given that conversion tract lengths differ so dramatically for COs and NCOs, the relative frequencies of COs and NCOs are also important. Transmission distortion presumed to be from biased DSB formation has previously been observed for COs at other mouse and human hotspots (Baudat and de Massy, 2007a; Jeffreys and Neumann, 2002; Webb et al., 2008), as we observed at A3. What has not been rigorously evaluated, however, is the extent to which NCOs also contribute. At A3, we find that NCOs have a 1.9-fold greater contribution than COs. Interestingly, an ~2-fold difference is what might be expected genome-wide, i.e., an ~10-fold NCO to CO ratio is offset by ~5-fold longer CO gene conversion tracts.
Transmission distortion can lead to fixation of alleles and hotspot extinction (Coop and Myers, 2007). Given the high recombination frequency at A3, the probability of fixation of A/J alleles is 100% for many of the polymorphisms, if one assumes that the alleles subject to transmission distortion are themselves the cause of biased DSB formation. For example, if we consider a polymorphism in which NCOs and COs have a similar contribution to transmission distortion (asterisk, Figure 1F), simulations suggest that fixation would be reached after ~3100 generations (see Supplementary Experimental Procedures). The neighboring polymorphism, indel-3, which owes a much greater fraction of its transmission distortion to NCOs, would reach fixation in a mere 1200 generations. Thus, even though NCOs have been more difficult to identify, they have the potential to substantially influence mammalian genome evolution.
CO refraction: suppressing COs in repetitive regions
We defined a strong CO refractory zone in the middle of A3 in two F1 hybrids. In principle, the dearth of COs could arise from a local lack of DSBs, but the abundance of NCOs indicates that recombination initiation is frequent within and near this zone and that instead, the outcome of recombination is different. Holes in CO distributions have been seen within many mammalian hotspots (Baudat and de Massy, 2007a; Bois, 2007; Jeffreys and Neumann, 2005; Paigen et al., 2008), and, although NCOs have not been tested in these other cases, it may be that CO refractory zones are common.
It is important to note that recombination initiated on either chromosome shows strong CO refraction in the B×D and C×D hybrids (>10-fold), but only weak refraction in the A×D hybrids (≤2-fold). The differential behavior of the D chromosome depending on what sequence(s) is present on its homolog suggests that CO refraction is best explained as a phenomenon involving the direct interaction of DNA sequences from both haplotypes, most likely within heteroduplex DNA-containing recombination intermediates. A notable feature of the A3 CO refractory zone is the presence of an imperfect inverted repeat of ~140 bp in all of the hybrids. However, only two polymorphisms in A3 are associated with strong CO refraction, including indel-2, which is close to the center of the inverted repeat (Figure 3E) and has the potential to alter the structural characteristics of this region.
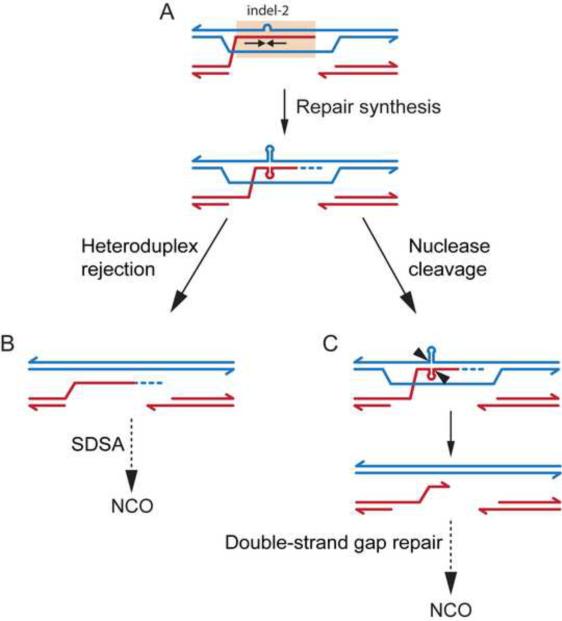
We favor a model in which strand invasion creates heteroduplex DNA which encompasses the inverted repeat, with indel-2 making the heteroduplex susceptible to DNA unwinding caused by torsional stress from repair synthesis to form a cruciform structure (Figure 5A) (Kouzine et al., 2008). We envision two possible scenarios for resolving the heteroduplex intermediate: heteroduplex rejection, promoting SDSA and thereby favoring NCOs at the expense of COs (Figure 5B); or cleavage at the base of the cruciform by a structure-specific endonuclease, leading to a double-strand gap (Figure 5C). The second scenario might promote NCOs at the expense of COs, as shown, or may alter CO placement by constraining CO breakpoints to fall outside the CO refractory zone.
Figure 5. CO refraction models.
(A) Spo11-generated DSB formation is inferred to occur most often to the right of the CO refractory zone (salmon shading). Strand invasion generates heteroduplex DNA encompassing the imperfect inverted repeat (arrows), with indel-2 near the center. DNA repair synthesis causes torsional stress resulting in cruciform extrusion.
(B) Heteroduplex rejection favors NCOs at the expense of COs. Recognition of the aberrant secondary structure by a helicase promotes dissociation of the invading strand and leads to SDSA, generating an NCO.
(C) Nuclease cleavage leading to double-strand gap formation. Cleavage by a structure-specific nuclease, followed by dissociation, results in a double-strand gap which extends from the base of the inverted repeat to the Spo11 DSB site. Gap repair without reciprocal exchange generates an NCO with a longer gene conversion tract than normal, incorporating multiple polymorphisms (i.e., co-conversions).
CO refraction affects only a subset of COs, hence these scenarios cannot be distinguished by frequency comparisons. However, consistent with an increase in NCOs at the expense of COs, a wider NCO distribution is observed in hybrids that exhibit CO refraction, including NCOs within the CO refractory zone. Further support for an effect on the CO/NCO decision comes from the shift in the location of both the CO refractory zone and NCO distribution in C×D compared with B×D, as well as an increase in NCOs outside of the CO refractory zone. These findings suggest that DNA secondary structure within recombination intermediates could influence recombination throughout the hotspot. Notably, the NID1 hotspot in humans also contains a dearth in COs within a repetitive palindromic minisatellite (Jeffreys and Neumann, 2005). We speculate that CO refraction at A3 might reflect mechanisms that help suppress meiotic genomic rearrangements between secondary structure-forming repetitive regions.
Co-conversions may reflect double-strand gap repair
Although most NCOs at A3 convert only a single polymorphism, a significant class convert ≥2 polymorphisms and these are enriched in the central ~100 bp of the hotspot, delineating a co-conversion zone. This zone is also the peak region of NCOs, suggesting that co-conversions may preferentially occur in regions of frequent DSB formation. One attractive possibility, especially for hotspots with potential to form secondary structure, is that cleavage of heteroduplex DNA in recombination intermediates by structure-specific nucleases or mismatch repair factors may produce double-strand gaps (Figure 5C). Indeed, co-conversions are more frequent in hybrids with strong CO refraction. An alternative is that two or more adjacent DSBs could be formed on the same DNA molecule in a fraction of cells, creating a gap. Such events would be expected to occur more often with higher-frequency cleavage sites, as observed at A3. In both cases, formation of double-strand gaps would necessitate more extensive resynthesis templated by the intact recombination partner, thereby lengthening the gene conversion tract.
Conclusions
In summary, our analysis describes a mammalian hotspot that approaches the ratio of NCOs to COs predicted from cytological estimates of DSB numbers. Examination of other highly polymorphic, intensely active mammalian hotspots will provide further insight into CO/NCO ratios and homologous recombination mechanisms.
Experimental Procedures
Polymorphism identification and animal husbandry
In addition to SNPs from the dbSNP database (NCBI) and (Kelmenson et al., 2005), SNPs and indels were also identified by sequencing genomic DNA from mouse strains from The Jackson Laboratory (primers in Table S1). Hybrid mice were either directly purchased (B×D, C×D) or bred from parental strains (A, A1/B, B, D) from The Jackson Laboratory. F1 hybrids in this study are identified as haplotype 1 × haplotype 2 (e.g., A×D) regardless of the parent of origin (Table S4). All experiments were performed in accordance with relevant regulatory standards and were approved by the MSKCC Institutional Animal Care and Use Committee.
DNA isolation and determination of amplifiable DNA concentration
Allele-specific (AS) PCR primers were designed and optimized as described (Kauppi et al., 2009). Cauda epididymides were dissected from adult mice, and sperm DNA was extracted. Somatic DNA was extracted from spleen. DNA was handled and purified as described (Kauppi et al., 2009). The number of amplifiable DNA molecules/pg was determined by performing 48 PCR reactions per sample seeded with 12 pg per reaction with AS primers against universal primers (Cole and Jasin, 2010). Spleen DNA from the same mouse was used at total DNA inputs equivalent to sperm DNA throughout, as a negative control.
CO assays
For CO assays, primary PCRs were performed with input pools of 200–3000 amplifiable sperm DNA molecules, using AS primers (Table S2) targeted to SNPs flanking A3 (arrowheads, Figure S2B). Primary PCR products were digested with S1 nuclease, diluted, and reamplified in a nested secondary AS PCR. CO-positive PCRs were identified by gel electrophoresis, confirmed in many cases by Southern blotting (Figure S3A). CO rates were calculated using Poisson correction to account for multiple COs. The standard deviation was estimated using the normal approximation of the Poisson distribution (Baudat and de Massy, 2009; Cole and Jasin, 2010). All positive secondary sperm PCRs, along with all somatic and no-DNA controls, were subjected to a third round of PCR with nested U primers. PCRs were transferred onto membranes, and CO breakpoints were mapped by hybridization with AS oligonucleotides (Table S3). Methods for detecting and mapping COs are detailed in (Kauppi et al., 2009). For a breakdown of CO calculations from each individual DNA sample, see Table S4. All CO breakpoint maps are plotted with COs from the CO assay and do not include those identified in the NCO assay. For every strain combination described, DNA was extracted and assayed from at least 2 mice (A×D, 4 mice; A1/B×D, 2 mice; B×D, 4 mice; C×D, 2 mice).
NCO assays
For NCO assays, primary PCRs were performed with input pools of 10–40 amplifiable sperm DNA molecules, using AS primers targeted to SNPs to the left side (centromere proximal) of A3 (Table S2) against U primers (Table S1). Primary PCRs were diluted and reamplified in a secondary PCR with nested AS and U primers. All secondary PCRs along with positive controls were transferred to membranes and probed with AS oligonucleotides directed toward the donor genotype (Figure S3B). Methods for detecting and mapping NCO molecules are detailed in (Kauppi et al., 2009), and NCO and CO rates were calculated using Poisson correction to account for multiple events (Cole and Jasin, 2010). NCOs with co-conversions were normalized by dividing by the number of converted polymorphisms to avoid overrepresentation of these NCOs. For every strain combination described, DNA was extracted and assayed from at least 2 mice (A×D, 3 mice; B×D, 5 mice; C×D, 2 mice). Multiple co-converted NCO recombinants were confirmed by cloning and genotyping as described in (Cole and Jasin, 2010).
Transmission distortion
For COs, transmission distortion was determined by adding one-half the cumulative distribution fraction (CDF) in one orientation to one-half of (1−CDF) in the other orientation, on a per polymorphism basis. Thus, transmission of A alleles in an A×D hybrid is given by:
For NCOs, transmission of A alleles in the A×D hybrid was calculated for each polymorphism from the following formula:
where fNCOtot is the co-conversion-normalized frequency of NCOs for the entire hotspot; and fD and fA are the frequencies of NCO conversions detected at a given polymorphism on the D and A chromosomes, respectively, not normalized for co-conversion. This formula expresses transmission on the basis of a pair of recombining chromatids, which is directly comparable to the values for transmission distortion from COs.
Supplementary Material
Acknowledgments
We thank members of the Jasin and Keeney laboratories, especially Liisa Kauppi and Erika Brunet, for discussions, and Aaron Gabow and Alex Lash of the MSKCC Bioinformatics Core for calculations of fixation. This work was supported by a Ruth L. Kirschstein National Research Service Award F32HD51392 (F.C.), National Institutes of Health Grants R01HD40916 (M.J., S.K.) and R01HD53855 (S.K., M.J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnheim N, Calabrese P, Tiemann-Boege I. Mammalian meiotic recombination hot spots. Annu Rev Genet. 2007;41:369–399. doi: 10.1146/annurev.genet.41.110306.130301. [DOI] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, de Massy B. Cis- and trans-acting elements regulate the mouse Psmb9 meiotic recombination hotspot. PLoS Genet. 2007a;3:e100. doi: 10.1371/journal.pgen.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2007b;15:565–577. doi: 10.1007/s10577-007-1140-3. [DOI] [PubMed] [Google Scholar]
- Baudat F, de Massy B. Parallel detection of crossovers and noncrossovers in mouse germ cells. Methods Mol Biol. 2009;557:305–322. doi: 10.1007/978-1-59745-527-5_19. [DOI] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- Bois PR. A highly polymorphic meiotic recombination mouse hot spot exhibits incomplete repair. Mol Cell Biol. 2007;27:7053–7062. doi: 10.1128/MCB.00874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borts RH, Haber JE. Meiotic recombination in yeast: alteration by multiple heterozygosities. Science. 1987;237:1459–1465. doi: 10.1126/science.2820060. [DOI] [PubMed] [Google Scholar]
- Burgess SM. Homologous chromosome associations and nuclear order in meiotic and mitotically dividing cells of budding yeast. Adv Genet. 2002;46:49–90. doi: 10.1016/s0065-2660(02)46004-x. [DOI] [PubMed] [Google Scholar]
- Cole F, Jasin M. Isolation of meiotic recombinants from mouse sperm. Methods Mol Biol. 2010 doi: 10.1007/978-1-61779-129-1_15. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F, Keeney S, Jasin M. Evolutionary conservation of meiotic DSB proteins: more than just Spo11. Genes Dev. 2010;24:1201–1207. doi: 10.1101/gad.1944710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Myers SR. Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 2007;3:e35. doi: 10.1371/journal.pgen.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner HK. Extensive interallelic polymorphisms drive meiotic recombination into a crossover pathway. Plant Cell. 2002;14:1173–1183. doi: 10.1105/tpc.001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman AS, Lichten M. Restriction of ectopic recombination by interhomolog interactions during Saccharomyces cerevisiae meiosis. Proc Natl Acad Sci U S A. 2000;97:9537–9542. doi: 10.1073/pnas.97.17.9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon H, Baudat F, Grey C, Liskay RM, de Massy B. Crossover and noncrossover pathways in mouse meiosis. Mol Cell. 2005;20:563–573. doi: 10.1016/j.molcel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Guillon H, de Massy B. An initiation site for meiotic crossing-over and gene conversion in the mouse. Nat Genet. 2002;32:296–299. doi: 10.1038/ng990. [DOI] [PubMed] [Google Scholar]
- Holloway K, Lawson VE, Jeffreys AJ. Allelic recombination and de novo deletions in sperm in the human beta-globin gene region. Hum Mol Genet. 2006;15:1099–1111. doi: 10.1093/hmg/ddl025. [DOI] [PubMed] [Google Scholar]
- Hunter N. Meiotic Recombination. In: Aguilera A, Rothstein R, editors. Topics in Current Genetics, Molecular Genetics of Recombination. Springer-Verlag; Heidelberg: 2006. pp. 381–442. [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, May CA. DNA enrichment by allele-specific hybridization (DEASH): a novel method for haplotyping and for detecting low-frequency base substitutional variants and recombinant DNA molecules. Genome Res. 2003;13:2316–2324. doi: 10.1101/gr.1214603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36:151–156. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Neumann R. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat Genet. 2002;31:267–271. doi: 10.1038/ng910. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Neumann R. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum Mol Genet. 2005;14:2277–2287. doi: 10.1093/hmg/ddi232. [DOI] [PubMed] [Google Scholar]
- Kauppi L, May CA, Jeffreys AJ. Analysis of meiotic recombination products from human sperm. Methods Mol Biol. 2009;557:323–355. doi: 10.1007/978-1-59745-527-5_20. [DOI] [PubMed] [Google Scholar]
- Kelmenson PM, Petkov P, Wang X, Higgins DC, Paigen BJ, Paigen K. A torrid zone on mouse chromosome 1 containing a cluster of recombinational hotspots. Genetics. 2005;169:833–841. doi: 10.1534/genetics.104.035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- Larocque JR, Jasin M. Mechanisms of recombination between diverged sequences in wild-type and BLM-deficient mouse and human cells. Mol Cell Biol. 2010;30:1887–1897. doi: 10.1128/MCB.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wu TC, Lichten M. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. Embo J. 1995;14:4599–4608. doi: 10.1002/j.1460-2075.1995.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E, Colaiacovo MP. Distribution of meiotic recombination events: talking to your neighbors. Curr Opin Genet Dev. 2009;19:105–112. doi: 10.1016/j.gde.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ. PRDM9 points the zinc finger at meiotic recombination hotspots. Genome Biol. 2010;11:104. doi: 10.1186/gb-2010-11-2-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Szatkiewicz JP, Sawyer K, Leahy N, Parvanov ED, Ng SH, Graber JH, Broman KW, Petkov PM. The recombinational anatomy of a mouse chromosome. PLoS Genet. 2008;4:e1000119. doi: 10.1371/journal.pgen.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Webb AJ, Berg IL, Jeffreys A. Sperm cross-over activity in regions of the human genome showing extreme breakdown of marker association. Proc Natl Acad Sci U S A. 2008;105:10471–10476. doi: 10.1073/pnas.0804933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZK, Getun IV, Bois PR. Anatomy of mouse recombination hot spots. Nucleic Acids Res. 2010;38:2346–2354. doi: 10.1093/nar/gkp1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.