Abstract
The apicoplast of Plasmodium is an essential organelle with its own circular genome that must be faithfully replicated and segregated to its progeny during parasite sporogony and schizogony. DNA replication proteins are not encoded by its genome. Instead, the replication machinery must be imported from nuclear-encoded genes. A likely apicoplast DNA replication factor, PfPrex, bears a bipartite leader sequence for apicoplast trafficking and contains several DNA replication-related enzymatic domains. Here we analyze the domain structure of PfPrex and examine its trafficking and maturation within the parasite. A minimal primase domain of PfPrex is shown to contain functional zinc-binding and TOPRIM-fold domains, which in a recombinant form are sufficient to produce RNA primers from a single-stranded DNA template. PfPrex is shown to be extensively proteolytically matured within the parasite, which effectively separates its functional domains. Gene targeting attempts to knockout the P. yoelii ortholog of Prex were unsuccessful, indicating the apparent essentiality of this protein to the parasite. Finally, overexpression in P. falciparum of PfPrex’s trafficking and primase sequences yielded specific and dynamic localization to foci within the apicoplast. Taken together, these observations strongly suggest an essential role of PfPrex primase in the production of RNA primers for lagging strand DNA synthesis of the apicoplast genome.
Keywords: Plasmodium falciparum, malaria, apicoplast, DNA replication, primase, proteolytic processing
1. Introduction
Malaria is a leading cause of disease and death in the world. Over 300 million people are infected annually, resulting in approximately one million deaths per year [1, 2]. While five Plasmodium species infect humans and produce similar symptoms, P. falciparum is the most virulent and causes nearly all deaths associated with this disease [1]. Moreover, populations of P. falciparum that have developed resistance to commonly used anti-malarial drugs are spreading, thus compounding this medical problem and increasing the need for new chemotherapeutic drugs.
P. falciparum contains 14 nuclear, linear chromosomes and two circular genomes contained in specialized organelles, which collectively comprise the most A+T-rich genome sequenced to date [2]. One of these organelles, termed the apicoplast, is an essential, non-photosynthetic plastid that retains type II fatty acid and isoprenoid biosynthetic functions [2–6]. The apicoplast genome is comprised of a circular ~35kb dsDNA molecule that is replicated by both rolling circle and bidirectional D-loop mechanisms, reminiscent of the replication strategy of chloroplast genomes [7, 8]. The initiation of DNA synthesis in this plastid genome occurs at both segments of a discrete, inverted repeat region, although what marks this region as an origin of DNA synthesis is not known [9, 10]. Inhibitors of apicoplast DNA replication in the related species T. gondii result in the loss of the apicoplast, and the “delayed death” of the parasite in the next cell division cycle [11, 12]. Additionally, inhibitors of transcription from the apicoplast genome in P. falciparum result in the same death phenotype of the parasite [13]. These data, coupled with an evolutionarily distant origin of the organelle, make the apicoplast a promising target for the development of new anti-malarial drugs (recently reviewed by Dahl and Rosenthal [14]).
One attractive molecular target for drug development in the apicoplast is the putative DNA replicative machinery of its genome, termed Prex [15]. The P. falciparum orthologue of Prex (PfPrex, PF14_0112) is a large (~235kDa), multi-functional protein that is evolutionarily well-conserved among the Plasmodium genus and is present in other apicomplexans such as Theileria, Babesia, and Toxoplasma [16, 17]. A bioinformatics analysis of PfPrex identified potential primase, helicase, exonuclease and polymerase domains, which were indirectly or directly confirmed experimentally by in vitro assays [15]. Fusion of the bipartite leader sequence from PfPrex to GFP demonstrated its apicoplast targeting, likely via the secretory system [15]. PfPrex was also shown to undergo proteolytic maturation of a large portion of its C-terminus, but the subcellular location of this process was not determined (ibid). The fidelity of the polymerase activity (found in this processed C-terminal domain in vivo) was recently assessed biochemically using a recombinantly expressed protein, and was also found to rescue a temperature sensitive polymerase I growth defect in E. coli [18]. Taken together, this arrangement in which multiple domains with related, complementary functions are expressed as a poly-protein and are subsequently proteolytically processed is reminiscent of expression strategies employed by viruses such as poliovirus, hepatitis C virus, and HIV-1 (for a review, [19]). Inhibitors of the viral proteases responsible for the maturation of such poly-proteins have been shown to inhibit the subsequent enzymatic function of these proteins and are used in treating infections of these viruses [20, 21].
Here we examine the domain structure and cellular processing of PfPrex and test whether Prex activites are essential. PfPrex is proteolytically processed into several fragments comprising each of its enzymatic domains. Bioinformatic and biochemical approaches allowed re-annotation of the functional domains of PfPrex, which correlates with sequences that are well conserved across orthologs of the Plasmodium species. As with well-characterized bacterial primases, a recombinant form of the complete primase domain from PfPrex includes a zinc-binding element that contains evolutionarily conserved CXXC sequence motifs. Consistent with its proposed activity, this domain is competent to transcribe short RNA primers. The primase domain localizes in distinct foci within the apicoplast as the parasite progresses through the intraerythrocytic asexual stages. Interestingly, the gene encoding Prex in the P. yoelii rodent malaria model could not be deleted, which supports its probable essential nature. Taken together, this characterization of Prex permits a greater understanding of the mechanisms underlying plastid replication in Plasmodium spp., while also providing potential targets for development of specific chemotherapeutic agents.
2. Materials and Methods
2.1 Bacterial expression and purification of PfPrex primase derivatives
Sequences coding for PfPrex’s TOPRIM domain alone (residues 336–465) or complete primase domain (comprised of its Zinc-Binding and TOPRIM domains, residues 115–465; Figure 1) were PCR amplified from P. falciparum 3D7 genomic DNA, inserted between the NdeI and XhoI sites of pET28b+ (Novagen) and confirmed by sequencing. All primers used in this study are listed in Supplemental Table 1. Expression plasmids were transformed into the Rosetta2(DE3)pLysS strain of E. coli (Novagen). Significant quantities of soluble protein expression were only obtained by a multiple step induction process. In brief, bacteria were grown at 37 °C with shaking (220 rpm) until an OD600 of 0.7 was reached. The culture was then shifted to room temperature, shaking speed was reduced to 125 rpm, and chaperone expression was induced by the addition of benzyl alcohol (Sigma Aldrich) to 1mM for 15 minutes [22]. Expression of PfPrex derivatives was then induced for 20 hours by the addition of 0.5mM (final) isopropyl β-D-1-thiogalactopyranoside (IPTG).
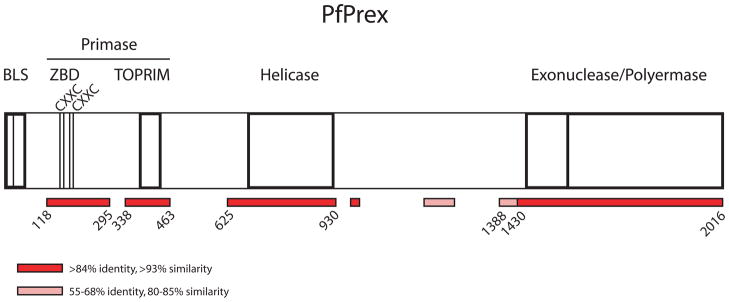
Figure 1. Schematic representation of PfPrex and its conserved enzymatic domains.
Prex traffics to the apicoplast via an N-terminal Bipartite Leader Sequence (BLS), and harbors evolutionarily conserved enzymatic domains with primase, helicase, exonuclease, and polymerase functions. The primase domain is here reannotated to include both the Zinc-Binding Domain (ZBD) and the Topoisomerase-Primase (TOPRIM) domain. Two zinc-binding CXXC motifs (AA155-158, 182–185) are noted within the ZBD. Those portions of PfPrex with high and moderate levels of sequence identity and/or similarity to P. berghei, P. yoelii, and P. chabaudi are noted in red and pink, respectively, with the amino acid positions of their boundaries noted.
Bacterial pellets were lysed by sonication in Nickel-NitriloTriAcetic acid (Ni-NTA) Loading Solution (25mM HEPES pH 7.5, 500mM NaCl, 150mM sodium formate, 10mM imidazole, 10% glycerol) with 1mM (final) phenylmethylsulfonyl fluoride (PMSF) and 1mM (final) benzamidine. The soluble fraction was separated from insoluble material by centrifugation (15,000 rpm, 4 °C, 20 minutes), and was then incubated in batch with Ni-NTA resin (Qiagen) on an orbital shaker for 1 hour at 4 °C. The resin was applied to a column and washed extensively (>20 column volumes) with Ni-NTA Loading Solution, and then five column volumes of Ni-NTA Washing Solution (25mM HEPES pH 7.5, 500mM NaCl, 150mM sodium formate, 250mM dextrose, 50mM imidazole, 10% glycerol). PfPrex derivatives were eluted by addition of five column volumes of Ni-NTA Elution Solution (25mM HEPES pH 7.5, 500mM NaCl, 150mM sodium formate, 300mM imidazole, 10% glycerol, 1mM DTT). PfPrex derivatives were then dialyzed against SPFF Buffer A (50mM MES pH 6.4, 500mM NaCl, 150mM sodium formate, 10% glycerol, 1mM DTT), diluted 1:5 in SPFF Buffer A, and purified via SPFF ion exchange chromatography using a linear gradient over 10 column volumes of 8–100% SPFF Buffer B (50mM MES pH 6.4, 1M NaCl, 150mM sodium formate, 10% glycerol, 1mM DTT). PfPrex derivatives eluted between 15–40% SPFF Buffer B and collected protein was concentrated to ~30μM. Purity of PfPrex derivatives was >90% as assessed by Coomassie Blue staining of a 12% SDS-polyacrylamide gel.
2.2 Generation of rabbit polyclonal antibodies to the TOPRIM domain of PfPrex
Bacterially expressed PfPrex TOPRIM was purified by Ni-NTA resin from inclusion bodies dissolved in 200mM HEPES pH 7.5, 500mM NaCl, 6M GuHCl. The resin was washed with an additional 20 column volumes of buffer, and then five column volumes of the same solution supplemented with 50mM imidazole. PfPrex TOPRIM was eluted with five column volumes of the same solution but containing 300mM imidazole, and was found to be >90% pure. Protein was precipitated by excessive dilution in distilled water and was washed in 5mM ammonium acetate prior to lyophilization. Pre-immune and post-immune sera were generated from two New Zealand white rabbits (Harlan BPS), and anti-TOPRIM antibodies were enriched by octanoic acid and ammonium sulfate precipitations and dialyzed into 1xPBS pH 7.2, 10% glycerol.
2.3 Colormetric detection of zinc ions by the PAR reagent
The amount of Zn2+ bound by PfPrex primase was determined spectrophotometrically by the use of 4-(2-pyridylazo)resorcinol (PAR), as previously described [23]. Briefly, the absorbance of PAR at 500nm increases linearly with the addition of 1–8μM zinc ions. Standard solutions of ZnCl2 were diluted in PAR Assay Solution (50mM HEPES pH7.5, 3M GuHCl) and were used to measure the amount of Zn2+ released from bacterially expressed and purified PfPrex primase (5μM) when unfolded. Measurements were conducted in triplicate.
2.4 Direct Primase Enzymatic Assay
The enzymatic activity of PfPrex primase was determined by direct measurement of [α 32P]ATP-incorporated RNA primers using a previously described method with modifications [24]. Primase reaction solutions (50mM HEPES pH 7.4, 5mM MgOAc, 1mM spermidine, 200mM NaCl, 10% glycerol, 0.5mM CTP, 0.5mM GTP, 0.5mM TTP, 0.3mM [α 32P]ATP) were incubated with 25ng of a single-stranded dT50 oligonucleotide template and 10μM (final) PfPrex primase or E. coli DnaG (positive control) at 37 °C for 1 hour. Reactions were phenol/chloroform treated, and then purified using Bio-Gel P-6 spin columns (GE Healthcare) to remove unincorporated [α 32P]ATP. Formamide loading solution was added to each reaction, which were then heated to 95 °C for 5–10 minutes and loaded on a 7M Urea/20% SDS-polyacrylamide gel with a 32P-labeled 4–20nt molecular weight marker (4,8,12,16,20 nucleotide long dT single-stranded DNA) run in parallel. Samples were electrophoresed at 4 °C, 250V for 6–8 hours. The gel was then dried onto Whatman paper and exposed to a storage phosphor screen. Signals were visualized by a Storm 860 PhosphorImager and ImageQuant 5.2 software (Molecular Dynamics).
2.5 Western blotting of P. falciparum lysates
Asynchronous P. falciparum (3D7 strain) parasites were grown in and isolated from human O+ erythrocytes by standard methods. Parasites were resuspended in 1xPBS pH 7.5 and lysed in an equal volume of 2x Sample Buffer (100mM Tris-HCl pH 6.8, 4% w/v SDS, 0.2% w/v bromophenol blue, 20% v/v glycerol) with sonication. Disulfide bonds were reduced by the addition of 5% (v/v, final) 2-mercaptoethanol. Lysates from a serial dilution of parasites were loaded on an 8% SDS-polyacrylamide gel with a molecular weight marker and 250ng bacterially expressed and purified PfPrex primase loaded in parallel. Samples were transferred to a PVDF membrane, probed with pre-immune or post-immune anti-TOPRIM rabbit sera, and visualized with a HRP-conjugated secondary antibody and SuperSignal West Pico ECL substrate (Thermo Scientific).
2.6 Indirect immunofluorescence assay (IFA)
P. falciparum (NF54 strain)-infected human RBCs (iRBCs) (4% parasitemia, 4% hematocrit) were processed for IFA using a previously described method [25]. Briefly, infected red blood cells (iRBCs) were pelleted initially (and between all steps) at 4000rpm in a microcentrifuge at room temperature for 1 minute. Cells were washed twice in 1xPBS, fixed in 1xPBS + 4% paraformaldehyde + 0.0075% glutaraldehyde for 30 minutes at room temperature, and permeabilized in 1xPBS + 0.1% Triton X-100 for 10 minutes at room temperature. A 1xPBS + 3% bovine serum albumin (BSA) blocking solution was applied at 4 °C overnight. A mouse anti-GFP monoclonal antibody (Roche, Cat# 11814460001) and a rabbit anti-P. falciparum acyl carrier protein (PfACP) polyclonal antibody (a kind gift of G. McFadden) were diluted in 1xPBS + 3% BSA solution at 1:200 and 1:100 respectively, and incubated with iRBCs for 1 hour with end-over-end rotation at room temperature. Following two washes with 1xPBS+ 3% BSA, Alexa Fluor-conjugated secondary antibodies (Invitrogen) specific to mouse IgG (Alexa Fluor 488, Green) and rabbit IgG (Alexa Fluor 594, Red) were diluted 1:500 in 1xPBS+ 3% BSA and applied to iRBCs for 30 minutes with end-over-end rotation at room temperature with shielding from light. Nucleic acid was then stained with 1μg/ml 4′, 6-diamidino-2-phenylindole (DAPI) in 1xPBS for 5–10 minutes at room temperature. Cells were washed three times with 1xPBS, and mixed 1:1 with VectaShield (Vector Laboratories) and applied to a glass slide and coverglass slip. Fluorescent and DIC images were acquired using a DeltaVision Spectris RT microscope (Applied Precision) using a 100x oil objective and were deconvolved using the softWoRx software package.
2.7 Experimental animals
Female Swiss Webster (SW) mice were purchased from Harlan Laboratories (Indianapolis, IN) and used for routine parasite maintenance and transfection experiments. Animal handling was conducted in accordance with protocols approved by the Seattle Biomedical Research Institute Institutional Animal Care and Use Committee.
2.8 Attempted generation of Prex knockout P. yoelii parasites
The P. yoelii 17XNL (nonlethal) strain was transfected using standard methods [26]. Regions of homology (800–900bp in length) located just 5′ and 3′ of the PyPrex open reading frame were PCR amplified, combined by SOE PCR [27], and inserted into the StuI site of pCR-Blunt (Invitrogen) for sequencing. The combined homology regions were released from pCR-Blunt by digestion with SacII and NotI and inserted into the same restriction sites of a B3d-derived plasmid. The B3d-PyPrexKO plasmid was linearized with SbfI, precipitated with ethanol, and 10μg was transfected in duplicate with an Amaxa electroporator into purified schizonts in three independent trials. One day post-transfection, parasites were selected by virtue of a T. gondii dihydrofolate reductase (TgDHFR) expression cassette with pyrimethamine cycling (3 days on, ≥3 days off), transferred to another mouse, and selected again with pyrimethamine cycling. After parasitemia increased above 1%, parasites were harvested by passage over CF11 resin and released from RBCs with saponin. Genomic DNA from parasites was isolated and purified (QiaAMP Blood DNA Mini Kit, Qiagen), precipitated with ethanol, and resuspended in ddH2O. The presence of transgenic parasites was assessed by genotyping PCR.
2.9 Transfection of the NF54 strain of P. falciparum with a protein overexpression vector
The pDC2_hsp86 vector was digested with BglII and AvrII to allow the in-frame fusion of PfPrexAA1-465 with GFP, with expression driven by the constitutively active Pf hsp86 5′untranslated region (UTR) [28]. This plasmid was transfected into synchronous ring-stage P. falciparum NF54 parasites as previously described [29]. Transgenic parasites were selected by virtue of a human DHFR expression cassette by the inclusion of the anti-folate drug WR99210 in the growth media, and passaged by standard methods [30].
3. Results and Discussion
3.1 Reannotation of the functional domains of PfPrex based upon conservation among Plasmodium species
PfPrex (PF14_0112) is a 2016-residue protein with ESTs present for all stages of the complex life cycle in many orthologs in Plasmodium species. Computational analyses predict that PfPrex is composed of several well-conserved domains encoding topoisomerase-primase (TOPRIM), helicase, exonuclease, and polymerase functions [31]. A discrete bipartite leader sequence (BLS) is predicted to be present at the N-terminus [32], which was previously shown to be sufficient to target GFP to the apicoplast [15]. Direct and indirect assays confirmed the presence of the predicted enzymatic activities in vitro using recombinant derivatives of the two halves of PfPrex[15]. As the primase domain of PfPrex bore similarity only to bacterial and phage proteins (Supplemental Figure 1), we focused our efforts upon it due to its potential as a drugable target.
Initial efforts to express the TOPRIM domain (AA336-465) of PfPrex yielded low amounts of soluble protein that had no primase activity (data not shown). This prompted a more thorough comparison of PfPrex with its orthologs across Plasmodium species, as well as with well-characterized bacterial primases. As shown in Figure 1, using cutoffs of >84% identity and >93% similarity (shaded red) and 55–68% identity and 80–85% similarity (shaded pink) allowed a precise prediction of the bounds of the conserved functional domains of PfPrex. Bacterial primases generally include a Zinc-Binding Domain (ZBD), TOPRIM domain, and a Helicase-Binding domain (HBD). Bacteriophage can vary from this domain arrangement; for example, gene 4 of bacteriophage T7 encodes both a primase and helicase and lacks such an HBD [33]. The conserved regions in PfPrex include both an apparent ZBD and TOPRIM but a well conserved apparent HBD was not observed. As the primase and helicase domains of PfPrex are expressed on the same polyprotein, it is possible that the two domains are part of a single protein like that in T7. The putative ZBD region of PfPrex (AA118-295) includes two well conserved CXXC motifs that are common zinc-binding motifs. Based upon this analysis, we have re-annotated the primase domain to be composed of both the ZBD and TOPRIM domain (AA118-463).
3.2 Recombinant derivatives of PfPrex primase can be expressed in soluble form and purified from E. coli Rosetta2(DE3)pLysS when cells are stressed by benzyl alcohol
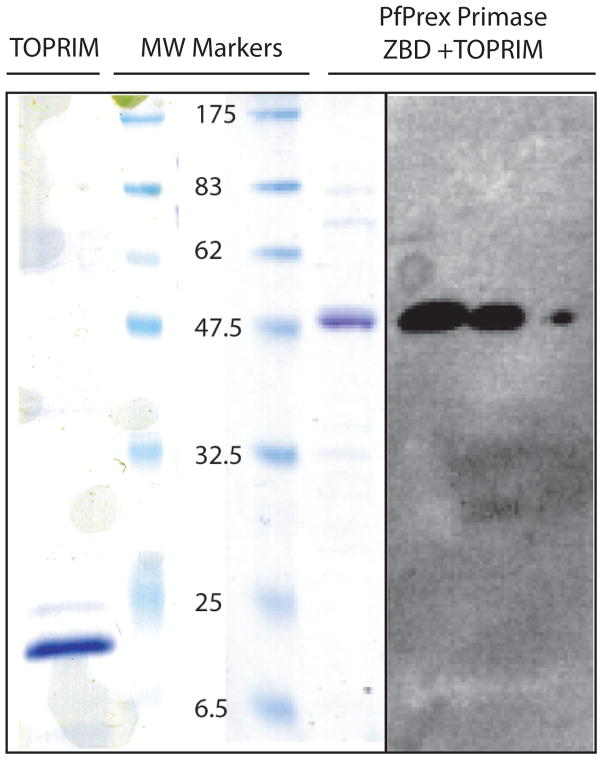
Using our re-annotated domain structure of PfPrex, an expression vector encoding for a protein with both the ZBD and TOPRIM domain (“PfPrex primase”, AA115-465) was created. Initial expression attempts using the Rosetta2(DE3)pLysS strain of E. coli yielded multiple smaller protein species that bound directly or indirectly to Ni-NTA resin. Tandem mass spectroscopic analysis revealed these to be premature translation termination products of PfPrex, with the major species identified as the bacterial chaperone protein GroEL (data not shown). To facilitate the production of full-length PfPrex primase derivative, protein expression was induced overnight at room temperature in the presence of benzyl alcohol, a potent membrane fluidizer and chaperone inducer [22]. Only in these conditions was PfPrex primase expressed in significant quantities in a soluble form. PfPrex primase was purified to >90% purity from the soluble fraction (Figure 2). A rabbit polyclonal antibody raised against the TOPRIM domain (AA336-465) reacted specifically with PfPrex primase in a serial dilution of bacterial lysates, as well as with purified protein (Figure 2).
Figure 2. Soluble recombinant TOPRIM domain and ZBD+TOPRIM domains of PfPrex can be purified from chaperone-induced bacteria.
Two derivatives of the primase domain of PfPrex were expressed in Rosetta2(DE3) pLysS bacteria during chaperone-induction by the addition of 1mM benzyl alcohol. Derivatives expressing either the TOPRIM domain alone or the ZBD+TOPRIM domains together were purified using standard methods to >90% purity as per Coomassie Blue staining of a 12% SDS-polyacrylamide gel (left). Rabbit polyclonal antibodies generated to the TOPRIM domain specifically react with a serial dilution of bacterial lysate harboring the complete primase domain (right).
3.3 PfPrex’s primase domain contains a functional ZBD
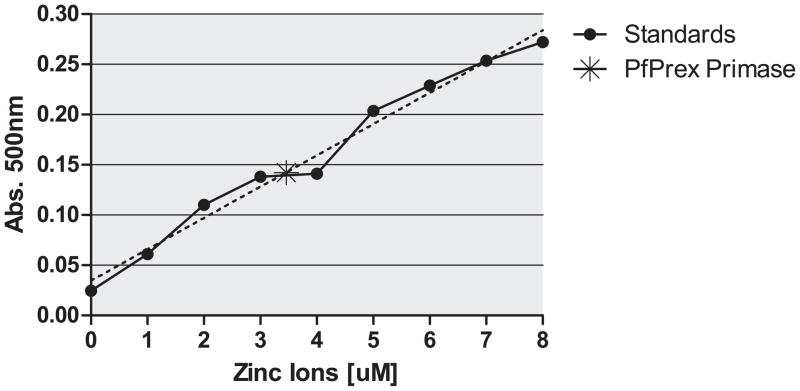
Purified PfPrex primase was tested to determine whether zinc was bound to the protein as predicted. Using an absorbance assay [23], PfPrex primase was found to have zinc bound at a 1:1 molar ratio (Figure 3). As the only source of this zinc was from the bacteria used to overexpress PfPrex primase, zinc is most likely incorporated into the structure of the protein as it folds. We predict that the ZBD participates in sequence-specific priming by PfPrex primase, as has been observed with the T7 phage primase [34].
Figure 3. PfPrex’s primase domain binds zinc.
A 5μM solution of PfPrex ZBD+TOPRIM was unfolded in 50mM HEPES pH 7.5, 3M GuHCl and the amount of zinc ion released was observed by the change in A500 in the presence of 100μM PAR. Values were normalized against a serial dilution of 1–8μM zinc ions, which fell within the linear range of detection (R2= 0.9817). The asterisk (A500 = 0.143+/−0.007, 3.46μM) represents the average signal observed over three replicates.
3.4 Recombinant PfPrex primase transcribes RNA primers from a ssDNA template
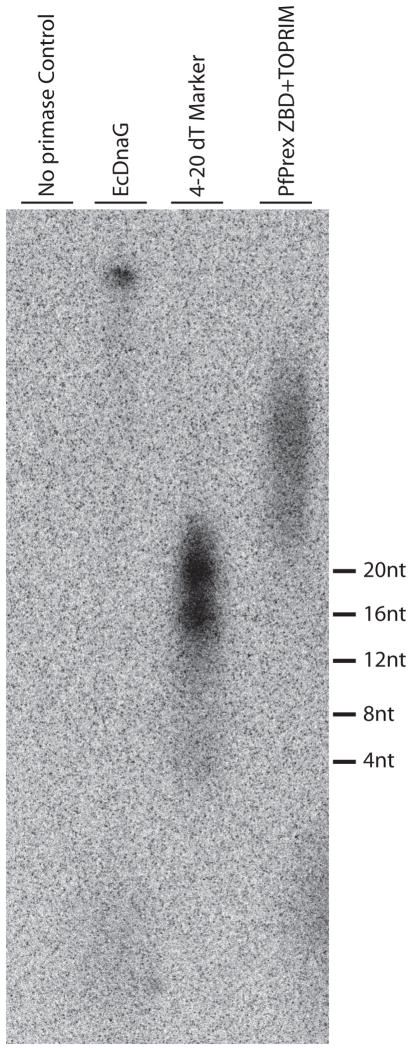
We next wanted to determine if recombinant PfPrex primase was able to catalyze DNA-dependent RNA synthesis. Previous work by Seow and colleagues had used a Klenow fragment extension assay to indirectly demonstrate that the N-terminal half of PfPrex contains a functional primase domain [15]. Here we measured primase activity directly by observing the incorporation of [α32P]ATP into RNA polymers produced using a dT50 single-stranded DNA template. As seen in Figure 4, RNA primers are produced by PfPrex and the positive control bacterial primase E. coli DnaG. In the absence of other components of replisome, it has been shown that EcDnaG can create RNA primers up to 60nt in length, which we observe as well [35]. PfPrex primase is also sufficient to transcribe RNA primers, which form in a variety of lengths > 20nts as evidenced by a smear of products. While a preferred initiation trinucleotide was not determined here due to the use of a dT50 single-stranded DNA template, it is not surprising that PfPrex primase produces a RNA polymer from this template due to the extremely A-T rich content of the P. falciparum genome. Therefore it is likely that the primase domain of PfPrex is capable of producing RNA primers to facilitate lagging-strand DNA synthesis within the parasite as well.
Figure 4. A PfPrex primase fragment that includes both the ZBD and TOPRIM domains is sufficient for primase activity.
Ten micromolar solutions of the purified recombinant PfPrex primase or E. coli DnaG were incubated at 37C with a dT(50) template and [α 32P]ATP/rNTP mix for 1 hour to produce radiolabeled RNA primers. Reaction products were phenol/chloroform treated, purified by size-exclusion chromatography (Bio-Gel P-6), and then electrophoresed through a 7M Urea/20% denaturing polyacrylamide gel. The dried gel was exposed to a storage phosphor screen and signals were visualized by a Storm 860 PhosphorImager and ImageQuant 5.2 software.
3.5 Testing the possible essential nature of Prex in P. yoelii
Having characterized PfPrex biochemically, we next sought to characterize it within the context of the parasite. The rodent malaria parasite P. yoelii is an excellent model system to utilize as it resembles P. falciparum in its infectivity, in the kinetics of the immune response against it, and in its overall genomic structure [26, 36, 37]. Moreover, a rapid and efficient transfection procedure is available and the complete genome sequence is known, making such assessments relatively quick and straightforward [26, 36]. To this end, the P. yoelii syntenic ortholog of Prex (PyPrex, PY00163) was targeted for gene deletion. Regions of homology to the 5′UTR and 3′UTR of PyPrex were inserted into the pB3d-DsRed plasmid [38], linearized to expose the regions of homology for double-strand DNA homologous recombination, and transfected into the non-lethal 17XNL strain of P. yoelii. Across three independent experiments each of which was conducted in duplicate, only wild-type parasites were recovered after pyrimethamine drug cycling as determined by genotyping PCR of the resulting parasites (Supplemental Figure 2). As a knockout parasite was never observed, this likely indicates that PyPrex is an essential protein, as would be predicted by its constitutive life-cycle expression pattern. While an indirect experiment, the repeated inability to disrupt the PyPrex gene is strongly indicative of its essentiality in the bloodstage.
3.6 PfPrex is proteolytically matured into separate domains in vivo
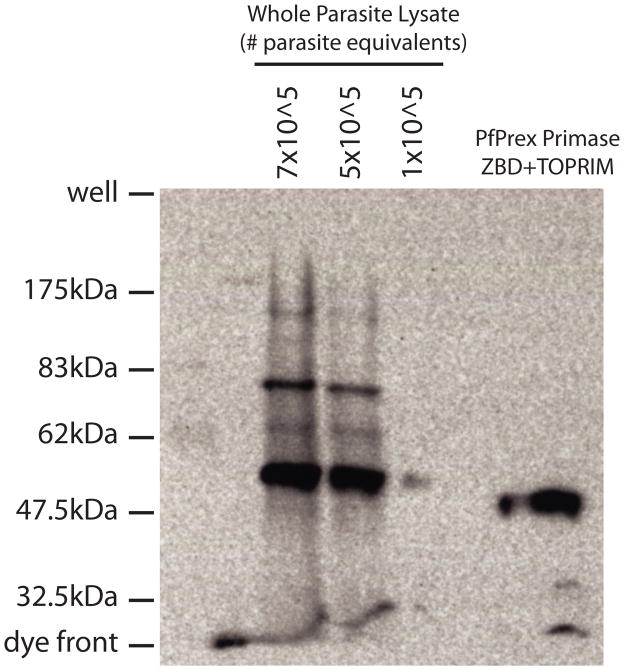
PfPrex is encoded by a large open reading frame composed of several related functional domains, which is reminiscent of the polyprotein strategy employed by several viruses [19]. It was previously shown that the C-terminal Exonuclease/Polymerase (“Exo/Pol”) domain of PfPrex is proteolytically separated in vivo by Western Blotting [15]. In order to assess if any further proteolytic maturation of PfPrex occurs in the parasite, we used a rabbit polyclonal antibody specific to the TOPRIM domain of PfPrex to probe lysates from RBCs infected with an asynchronous population of P. falciparum (Figure 5). When compared with the pre-immune sera (which yielded no reactive bands, Supplemental Figure 3), anti-TOPRIM antibody reacted specifically with recombinantly expressed PfPrex primase, as well as with several protein species from the parasite lysate. The major species migrated slightly more slowly than did recombinant PfPrex primase, which could reflect a protein cleaved downstream of its bipartite leader sequence and/or between the primase and helicase domains. Other larger protein species observed also carry the epitopes from the TOPRIM domain, and may correspond to intermediate forms of PfPrex. For instance, it is possible that the band observed at ~80kDa could represent the primase-helicase portion of PfPrex prior to proteolysis. Thus, the extensive proteolytic maturation observed with PfPrex separates it minimally into its Primase and Exo/Pol functional domains, and by extension also separates its helicase domain as well. Such proteolysis of PfPrex may have many possible functions, including allowing regulation of enzymatic activities and/or providing a means to obtain the necessary/optimal stoichiometries of each component in subsequent complexes. Further work is warranted to appropriately explore the role of these proteolytic processes. As with viral polyproteins, if proteolytic processing of Prex is prevented or inhibited its enzymatic functions might be compromised, and thus the putative maturase of Prex might constitute a good therapeutic target.
Figure 5. PfPrex is proteolytically processed in multiple locations in vivo.
Whole parasite lysates or purified, bacterially-expressed PfPrex Primase were electrophoresed through an 8% SDS-polyacrylamide gel, transferred to a PVDF membrane, and probed with rabbit polyclonal anti-PfPrex TOPRIM and a HRP-conjugated secondary antibody. Reactive species corresponding to proteolytically processed forms of PfPrex were visualized by standard ECL reagents. No specific signals were recognized by preimmune sera from parasite lysate nor from purified PfPrex Primase (Supplemental Figure 3). Pre-stained size markers were run in parallel, and are denoted to the left of the blot.
3.7 PfPrex primase traffics to the apicoplast and forms multiple foci in vivo
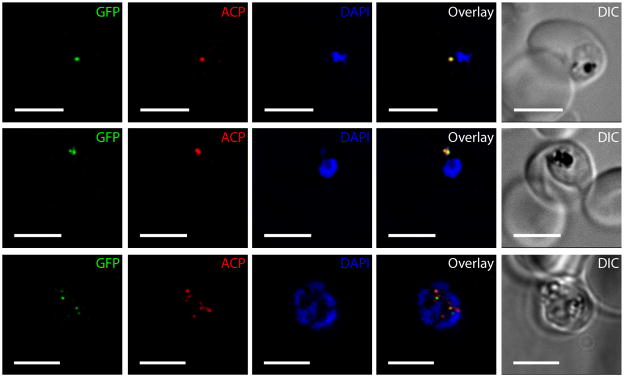
Because PfPrex is proteolytically matured, we sought to determine the localization of PfPrex primase within the parasite. To this end, we used the hsp86 promoter to overexpress PfPrex primase with its bipartite leader sequence intact (AA1-465), and with GFP fused to its C-terminus. Indirect immunofluorescence microscopy of asynchronous parasite cultures revealed a strong co-localization of PfPrex primase with the apicoplast resident acyl carrier protein (ACP) in early ring stages, as well as with DAPI signal derived from staining of the apicoplast genome (Figure 6, upper row). However, as parasites progressed through the asexual blood stage schizogony, two discrete foci of PfPrex primase formed in trophozoites, which flanked and partially overlapped the anti-ACP signal (middle row). These foci became less intense as the apicoplast underwent branching, and subsequently they reformed in late-stage schizonts when foci paired with daughter merozoites (lower row).
Figure 6. PfPrex primase traffics to the apicoplast and forms two foci during the intraerythrocytic cycle.
The complete primase domain (AA1-465) of PfPrex was genetically fused to GFPmut2 and expressed from a nuclear plasmid in P. falciparum NF54 parasites. Indirect immunofluorescence microscopy with DAPI and antibodies against GFP and PfACP demonstrates clear apicoplast localization of PfPrex throughout the intraerythrocytic cycle. Differential Interference Contrast (DIC) images show typical appearances of both the parasite and the host RBC. Two foci appear within the apicoplast during the early trophozoite stage and then segregate to daughter merozoites during schizogony. Scale bar is 5μm.
As was observed with the bipartite leader sequence alone, PfPrex primase also conclusively traffics to the apicoplast and changes its localization therein over the course of the asexual intraerythrocytic life cycle. It is also noteworthy that overexpression of PfPrex AA1-465 had no negative impact upon the parasite, permitting such an analysis. We postulate that PfPrex associates with other components of the apicoplast replisome, and contributes to the replication of its 35kb circular genome by provision of RNA primers for DNA synthesis. The two discrete foci observed in trophozoites may therefore be stationary replication loci used to duplicate the plastid genome into the several copies eventually needed for each daughter merozoite.
3.8 Conclusions
In this study, we have characterized several important aspects of PfPrex in vitro, as well as in its native context. Specifically, we have demonstrated that the re-annotated primase domain of PfPrex (AA115-465) is composed of both a functional ZBD and a TOPRIM fold domain. This recombinant form of PfPrex is sufficient to transcribe RNA primers from a single-stranded DNA template. When coupled with its apparent essentiality, apicoplast localization and proteolytic separation from the other enzymatic domains of PfPrex in vivo, these observations implicate a role of PfPrex primase in the production of RNA primers for the replication of the apicoplast genome. Further inspection of the other enzymatic domains of PfPrex, along with the role of its extensive proteolytic maturation, will continue to build upon our understanding of the replicative processes of Plasmodium’s apicoplast and its genome. Moreover, inhibition of PfPrex’s enzymatic functions or its proteolytic processing may prove to be good therapeutic targets.
Supplementary Material
PfPrex primase (residues 115–465) was aligned by ClustalW (v. 1.83) via the T-coffee multiple sequence alignment package (www.tcoffee.org) with related proteins from bacteria and bacteriophage. Consensus residues are denoted by * (asterisk, identity), : (double dot, conserved within physiochemical groups), . (single dot, semi-conserved within physiochemical groups).
A. Diagram of the knockout strategy by replacement. Primer sets used below in Panel B are represented by arrows, with the resulting PCR product sizes listed above the joining line. Regions of homology on the 5′ and 3′ side of the PfPrex ORF are noted by crossed lines. B. A representative gel image from genotyping PCR of a PyPrex knockout attempt. Genomic DNA was isolated and purified from transfected parasite populations and used as a template for genotyping PCR. Primer sets are noted in panel A that specifically amplify sequences from wild-type (WT) or transgenic (TG) genomes. Control PCRs using the targeting plasmid as a template with primers used to create the 5′ and 3′ regions of homology for recombination are included to show primer competence. Products are compared to Molecular Weight (MW) standards (New England Biolabs, 1kb+ Marker).
Whole parasite lysate (7×10^5 parasite equivalents) or purified, bacterially-expressed PfPrex Primase (50ng) were electrophoresed through an 8% SDS-polyacrylamide gel, transferred to a PVDF membrane, and probed with rabbit preimmune sera and a HRP-conjugated secondary antibody.
Acknowledgments
We thank G. McFadden for providing the anti-PfACP antibody, and J. Berger for purified recombinant EcDnaG. We also thank the Keck and Kappe labs for critical discussion of this work. Special thanks to Drew MacKellar for microscopy assistance, and to Nelly Camargo for training in the transfection of P. falciparum. This investigation was supported by the NIH under a Ruth L. Kirschstein National Research Service Award (F32GM083438) to SEL and by an R01 award (GM068061) to JLK. SEL performed research and wrote the report; SEL, JLK, ML, and SK designed research, and analyzed and interpreted data.
Abbreviations
- ZBD
Zinc-Binding Domain
- TOPRIM
topoisomerase-primase
- PAR
4-(2-pyridylazo)resorcinol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.The world malaria report. Geneva, Geneva: World Health Organization; 2005. in (unpublished document WHO/HTM/MAL/2005.1102) [Google Scholar]
- 2.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419(6906):498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardner MJ, et al. Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum. Science. 1998;282(5391):1126–32. doi: 10.1126/science.282.5391.1126. [DOI] [PubMed] [Google Scholar]
- 4.Jomaa H, et al. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science. 1999;285(5433):1573–6. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- 5.Smith S. The animal fatty acid synthase: one gene, one polypeptide, seven enzymes. Faseb J. 1994;8(15):1248–59. [PubMed] [Google Scholar]
- 6.Waller RF, et al. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. Embo J. 2000;19(8):1794–802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolodner RD, Tewari KK. Chloroplast DNA from higher plants replicates by both the Cairns and the rolling circle mechanism. Nature. 1975;256(5520):708–11. doi: 10.1038/256708a0. [DOI] [PubMed] [Google Scholar]
- 8.Williamson DH, et al. The plastid DNA of the malaria parasite Plasmodium falciparum is replicated by two mechanisms. Mol Microbiol. 2002;45(2):533–42. doi: 10.1046/j.1365-2958.2002.03033.x. [DOI] [PubMed] [Google Scholar]
- 9.Singh D, Chaubey S, Habib S. Replication of the Plasmodium falciparum apicoplast DNA initiates within the inverted repeat region. Mol Biochem Parasitol. 2003;126(1):9–14. doi: 10.1016/s0166-6851(02)00251-7. [DOI] [PubMed] [Google Scholar]
- 10.Singh D, et al. Multiple replication origins within the inverted repeat region of the Plasmodium falciparum apicoplast genome are differentially activated. Mol Biochem Parasitol. 2005;139(1):99–106. doi: 10.1016/j.molbiopara.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Fichera ME, Roos DS. A plastid organelle as a drug target in apicomplexan parasites. Nature. 1997;390(6658):407–9. doi: 10.1038/37132. [DOI] [PubMed] [Google Scholar]
- 12.He CY, et al. A plastid segregation defect in the protozoan parasite Toxoplasma gondii. Embo J. 2001;20(3):330–9. doi: 10.1093/emboj/20.3.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McConkey GA, Rogers MJ, McCutchan TF. Inhibition of Plasmodium falciparum protein synthesis. Targeting the plastid-like organelle with thiostrepton. J Biol Chem. 1997;272(4):2046–9. doi: 10.1074/jbc.272.4.2046. [DOI] [PubMed] [Google Scholar]
- 14.Dahl EL, Rosenthal PJ. Apicoplast translation, transcription and genome replication: targets for antimalarial antibiotics. Trends Parasitol. 2008;24(6):279–84. doi: 10.1016/j.pt.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Seow F, et al. The plastidic DNA replication enzyme complex of Plasmodium falciparum. Mol Biochem Parasitol. 2005;141(2):145–153. doi: 10.1016/j.molbiopara.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukhopadhyay A, et al. The Toxoplasma gondii plastid replication and repair enzyme complex, PREX. Parasitology. 2009;136(7):747–55. doi: 10.1017/S0031182009006027. [DOI] [PubMed] [Google Scholar]
- 18.Kennedy SR, et al. The Biochemistry and Fidelity of Synthesis by the Apicoplast Genome Replication DNA Polymerase Pfprex from the Malaria Parasite Plasmodium falciparum. J Mol Biol. 2011;410(1):27–38. doi: 10.1016/j.jmb.2011.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patick AK, Potts KE. Protease inhibitors as antiviral agents. Clin Microbiol Rev. 1998;11(4):614–27. doi: 10.1128/cmr.11.4.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fear G, Komarnytsky S, Raskin I. Protease inhibitors and their peptidomimetic derivatives as potential drugs. Pharmacol Ther. 2007;113(2):354–68. doi: 10.1016/j.pharmthera.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamarre D, et al. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature. 2003;426(6963):186–9. doi: 10.1038/nature02099. [DOI] [PubMed] [Google Scholar]
- 22.de Marco A, et al. Native folding of aggregation-prone recombinant proteins in Escherichia coli by osmolytes, plasmid- or benzyl alcohol-overexpressed molecular chaperones. Cell Stress Chaperones. 2005;10(4):329–39. doi: 10.1379/CSC-139R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCall KA, Fierke CA. Colorimetric and fluorimetric assays to quantitate micromolar concentrations of transition metals. Anal Biochem. 2000;284(2):307–15. doi: 10.1006/abio.2000.4706. [DOI] [PubMed] [Google Scholar]
- 24.Corn JE, et al. Crosstalk between primase subunits can act to regulate primer synthesis in trans. Mol Cell. 2005;20(3):391–401. doi: 10.1016/j.molcel.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Tonkin CJ, et al. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol Biochem Parasitol. 2004;137(1):13–21. doi: 10.1016/j.molbiopara.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Jongco AM, et al. Improved transfection and new selectable markers for the rodent malaria parasite Plasmodium yoelii. Mol Biochem Parasitol. 2006;146(2):242–50. doi: 10.1016/j.molbiopara.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Mikolajczak SA, et al. An efficient strategy for gene targeting and phenotypic assessment in the Plasmodium yoelii rodent malaria model. Mol Biochem Parasitol. 2008;158(2):213–6. doi: 10.1016/j.molbiopara.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Lee MC, et al. Plasmodium falciparum Sec24 marks transitional ER that exports a model cargo via a diacidic motif. Mol Microbiol. 2008;68(6):1535–46. doi: 10.1111/j.1365-2958.2008.06250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.VanBuskirk KM, et al. Preerythrocytic, live-attenuated Plasmodium falciparum vaccine candidates by design. Proc Natl Acad Sci U S A. 2009;106(31):13004–9. doi: 10.1073/pnas.0906387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193(4254):673–5. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 31.Aurrecoechea C, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37(Database issue):D539–43. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuegge J, et al. Deciphering apicoplast targeting signals--feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene. 2001;280(1–2):19–26. doi: 10.1016/s0378-1119(01)00776-4. [DOI] [PubMed] [Google Scholar]
- 33.Zhu B, Lee SJ, Richardson CC. An in trans interaction at the interface of the helicase and primase domains of the hexameric gene 4 protein of bacteriophage T7 modulates their activities. J Biol Chem. 2009;284(35):23842–51. doi: 10.1074/jbc.M109.026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kusakabe T, Richardson CC. The role of the zinc motif in sequence recognition by DNA primases. J Biol Chem. 1996;271(32):19563–70. doi: 10.1074/jbc.271.32.19563. [DOI] [PubMed] [Google Scholar]
- 35.Zechner EL, Wu CA, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. III. A polymerase-primase interaction governs primer size. J Biol Chem. 1992;267(6):4054–63. [PubMed] [Google Scholar]
- 36.Carlton JM, et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419(6906):512–9. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- 37.Khan ZM, Vanderberg JP. Role of host cellular response in differential susceptibility of nonimmunized BALB/c mice to Plasmodium berghei and Plasmodium yoelii sporozoites. Infect Immun. 1991;59(8):2529–34. doi: 10.1128/iai.59.8.2529-2534.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobs-Lorena VY, et al. A dispensable Plasmodium locus for stable transgene expression. Mol Biochem Parasitol. 2010;171(1):40–4. doi: 10.1016/j.molbiopara.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PfPrex primase (residues 115–465) was aligned by ClustalW (v. 1.83) via the T-coffee multiple sequence alignment package (www.tcoffee.org) with related proteins from bacteria and bacteriophage. Consensus residues are denoted by * (asterisk, identity), : (double dot, conserved within physiochemical groups), . (single dot, semi-conserved within physiochemical groups).
A. Diagram of the knockout strategy by replacement. Primer sets used below in Panel B are represented by arrows, with the resulting PCR product sizes listed above the joining line. Regions of homology on the 5′ and 3′ side of the PfPrex ORF are noted by crossed lines. B. A representative gel image from genotyping PCR of a PyPrex knockout attempt. Genomic DNA was isolated and purified from transfected parasite populations and used as a template for genotyping PCR. Primer sets are noted in panel A that specifically amplify sequences from wild-type (WT) or transgenic (TG) genomes. Control PCRs using the targeting plasmid as a template with primers used to create the 5′ and 3′ regions of homology for recombination are included to show primer competence. Products are compared to Molecular Weight (MW) standards (New England Biolabs, 1kb+ Marker).
Whole parasite lysate (7×10^5 parasite equivalents) or purified, bacterially-expressed PfPrex Primase (50ng) were electrophoresed through an 8% SDS-polyacrylamide gel, transferred to a PVDF membrane, and probed with rabbit preimmune sera and a HRP-conjugated secondary antibody.